Andersen et al. identify a novel genetic etiology of herpes encephalitis in an adult patient carrying a heterozygous loss-of-function mutation in the IRF3 gene. This mutation results in impaired INF production in response to viral infection
Abstract
Herpes simplex encephalitis (HSE) in children has previously been linked to defects in type I interferon (IFN) production downstream of Toll-like receptor 3. Here, we describe a novel genetic etiology of HSE by identifying a heterozygous loss-of-function mutation in the IFN regulatory factor 3 (IRF3) gene, leading to autosomal dominant (AD) IRF3 deficiency by haploinsufficiency, in an adolescent female patient with HSE. IRF3 is activated by most pattern recognition receptors recognizing viral infections and plays an essential role in induction of type I IFN. The identified IRF3 R285Q amino acid substitution results in impaired IFN responses to HSV-1 infection and particularly impairs signaling through the TLR3–TRIF pathway. In addition, the R285Q mutant of IRF3 fails to become phosphorylated at S386 and undergo dimerization, and thus has impaired ability to activate transcription. Finally, transduction with WT IRF3 rescues the ability of patient fibroblasts to express IFN in response to HSV-1 infection. The identification of IRF3 deficiency in HSE provides the first description of a defect in an IFN-regulating transcription factor conferring increased susceptibility to a viral infection in the CNS in humans.
Herpes simplex encephalitis (HSE) is the most common form of sporadic viral encephalitis in the Western world, with an estimated incidence of 1/250,000 individuals per year (Whitley and Lakeman, 1995). HSE is caused by HSV-1 and is a devastating disease with a mortality reaching 70% when untreated, and remaining at 25% even in the presence of antiviral therapy with acyclovir (Whitley and Lakeman, 1995; Griffin, 2005). Although 80% of adults are seropositive for HSV-1, neuroinvasion and establishment of CNS infection during either primary or secondary infection is a rare event, and knowledge of determinants of disease has remained sparse. Both innate and adaptive immune responses are essential for immune control of herpesviruses, including type I IFNs, NK cells, and cytotoxic T cells (Paludan et al., 2011).
The innate immune system utilizes pattern recognition receptors (PRRs) to detect pathogen–associated molecular patterns (PAMPs) to mount protective immune responses, including production of cytokines and IFN (Mogensen, 2009). Different classes of PRRs are involved in recognition of virus infections, including membrane-associated TLRs, cytosolic RNA-sensing RIG-like receptors (RLRs), and DNA sensors (Mogensen, 2009). Each of these classes of PRRs activate IFN regulatory factor 3 (IRF3) through unique adaptor molecules, known as TRIF, MAVS, and STING, respectively, to which IRF3 binds to become phosphorylated (Liu et al., 2015). Phosphorylation of IRF3 at specific serine residues in the C-terminal domain leads to dimerization, nuclear localization, and transcription of genes, including IFNs and IFN-stimulated genes (ISGs), with antiviral activity. The IRF family of transcription factors is essential for induction of type I (IFN-α/β) and type III (IFN-λ) IFN expression, with IRF3 and IRF7 being ascribed particularly important roles (Honda and Taniguchi, 2006). Importantly, IRF3-deficient mice are susceptible to HSV-1 infection in the brain (Menachery et al., 2010).
In recent years, it has emerged that single-gene inborn errors of innate immunity are associated with enhanced susceptibility to specific infections (Sancho-Shimizu et al., 2011a). Seminal studies have demonstrated that mutations in genes encoding proteins in the TLR3 pathway confer susceptibility to HSE in childhood (Casanova and Abel, 2007; Casrouge et al., 2006; Zhang et al., 2007; Pérez de Diego et al., 2010; Guo et al., 2011; Sancho-Shimizu et al., 2011b; Audry et al., 2011; Herman et al., 2012). Most of the identified mutations are inherited by an autosomal dominant (AD) mechanism, but cases of autosomal recessive (AR) mutations in the TLR3 pathway have also been identified (Casrouge et al., 2006; Guo et al., 2011; Sancho-Shimizu et al., 2011b; Lim et al., 2014). These findings may help explain why HSV becomes neuroinvasive to cause encephalitis in a small minority of individuals. At present, mutations in UNC93B1, TLR3, TRAF3, TRIF, TBK1, STAT1, and IKBKG (NEMO) genes have been identified in children with HSE. Notably, these genetic defects often appear to display incomplete penetrance (Abel et al., 2010; Zhang et al., 2013). Common to the identified genetic defects is that they lead to reduced IFN responses in cell culture after HSV-1 infection or stimulation through the TLR3 pathway. Moreover, a nonredundant role for TLR3 immunity has been demonstrated in induced pluripotent stem cell–derived CNS cells (Lafaille et al., 2012). Here, we identify a novel genetic etiology of HSE in an adolescent with HSE by demonstrating a heterozygous loss-of-function mutation in the IRF3 gene and impaired IFN production in response to HSV-1 and TLR3 stimulation in patient cells.
RESULTS AND DISCUSSION
Identification of a heterozygous mutation in IRF3
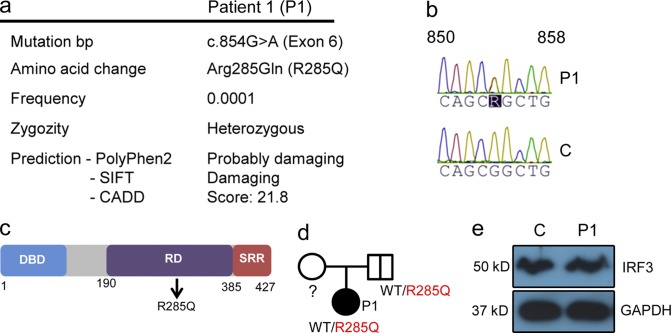
The patient (P1) was a 15-yr-old adolescent, who was part of a larger study involving whole-exome sequencing (WES) of a total of 16 adults with previous HSE. For a detailed medical history of P1, see Table S1 and supplemental text. WES was performed on patient DNA followed by bioinformatical analysis of the sequence data. Because HSE occurs with low frequency, disease-associated mutations were assumed to be novel or rare (<0.001 of the population). We identified a heterozygous mutation in IRF3 (frequency < 0.0001 in dbSNP) at base pair position 854, causing a G-to-A substitution in exon 6 of the molecule (NM_001571.5 IRF3 c.854G>A), resulting in an amino acid change at the highly conserved position 285 from arginine to glutamine (R285Q; Fig. 1 a). The identified mutation was confirmed by Sanger sequencing (Fig. 1 b). The R285Q mutation is located in the regulatory domain of the IRF3 protein (Fig. 1 c) and predicted to be damaging by PolyPhen-2 and SIFT software. Additionally, CADD software predicts a score of 21.8, indicating that the mutation belongs to the 1% of most deleterious mutations. The father was found to be a healthy carrier of the R285Q IRF3 mutation (Fig. 1 d), thus demonstrating incomplete penetrance. We did not have access to material from the patient’s mother, and there were no siblings. No mutations were found by analysis of WES data in the coding exons of TLR3, UNC-93B1, TRAF3, TRIF, TBK1, STAT1, or IKBKG (NEMO) known to be involved in the IFN-inducing and -responsive pathways and previously associated with childhood HSE. Importantly, despite careful analysis of WES data, we did not find any homozygous or compound heterozygous mutations. The expression of IRF3 protein in PBMCs from P1 was similar to a healthy age- and gender-matched control (Fig. 1 e).
Figure 1.
Identification of a heterozygous mutation in IRF3 by WES. (a) Summary of information on the IRF3 mutation in P1. (b) Sanger sequencing of the IRF3 gene in P1 and healthy control. (c) Schematic diagram of the IRF3 protein consisting of an N-terminal DNA-binding domain (DBD), a central regulatory domain (RD), and C-terminal serine-rich region (SRR). The identified R285Q mutation is localized in the RD. (d) Family pedigree with allele segregation. Family members heterozygous for the mutation are indicated by a bold vertical line (e) Whole-cell lysates from PBMCs from P1 and a healthy control were subjected to Western blotting and probed with anti-IRF3 and anti-GAPDH.
Impaired IFN responses through nucleic acid–activated pathways in patient PBMCs and fibroblasts
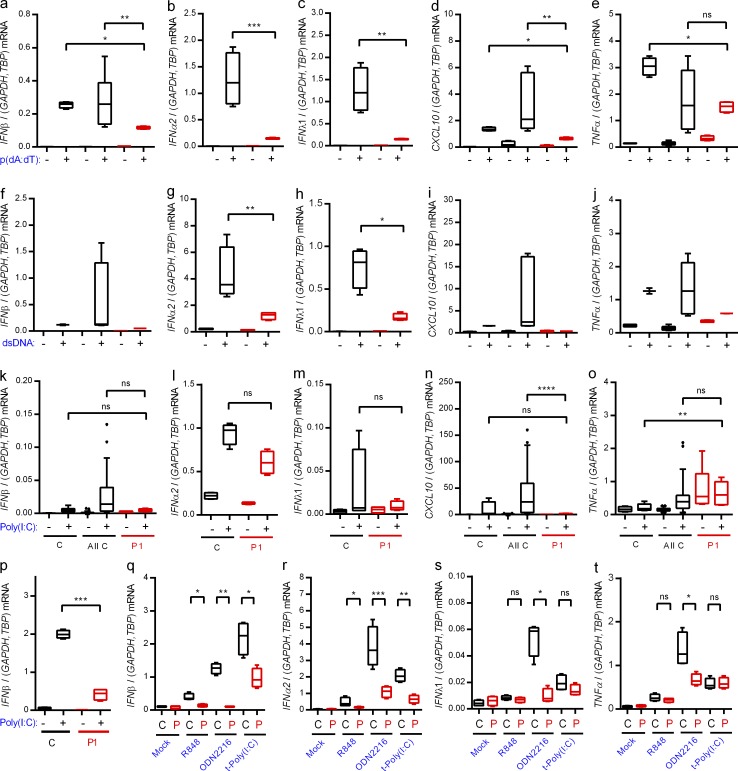
To examine the functional consequences of the identified heterozygous IRF3 mutation, PBMCs from P1 and controls were examined for expression of IFN and inflammatory cytokines. PBMCs express a wide panel of PRRs, and it has often not been possible to demonstrate impaired responsiveness through relevant pathways in patient PBMCs, most likely due to redundancy (Casanova and Abel, 2007; Casrouge et al., 2006; Zhang et al., 2007; Pérez de Diego et al., 2010; Audry et al., 2011; Guo et al., 2011; Sancho-Shimizu et al., 2011b; Herman et al., 2012; Lafaille et al., 2012). PBMCs were stimulated with nucleic acid PAMPs sensed by PRRs known to be activated by herpesvirus infection (Paludan et al., 2011). Induction of type I and III IFNs and CXCL10 in cells from P1 was significantly impaired in response to poly(dA:dT), an agonist of DNA sensors, particularly Pol III, as well as in response to dsDNA, a ligand of DNA sensors, primarily cGAS (Fig. 2, a–d and f–i). Although the TLR3 agonist extracellular poly(I:C) was generally a weak inducer in the PBMCs, we did observe highly significant reduction in CXCL10 induction in cells from P1 when comparing with the set of pooled controls (Fig. 2, k–n). The induction of TNF expression by DNA in PBMCs from P1 was partially reduced (Fig. 2, e and j). The ability of the IFN pathway to prime expression of inflammatory cytokines is well established (Osterlund et al., 2005). Extracellular poly(I:C) did not stimulate TNF expression (Fig. 2 o). Importantly, fibroblasts from healthy controls responded to stimulation with extracellular poly(I:C) with strong induction of IFN-β expression, whereas this response was severely impaired in fibroblasts from P1 (Fig. 2 p).
Figure 2.
Impaired IFN induction through nucleic acid–sensing pathways in patient cells. (a–o and q–t) PBMCs from the patient and controls were stimulated with 4 µg/ml poly(dA:dT) (a–e), 4 µg/ml HSV-1–derived dsDNA (f–j), 50 µg/ml extracellular poly(I:C) (k–o), 1 µg/ml R848, 5 µM ODN2216, or 4 µg/ml transfected poly(I:C) (q–t). Total RNA was harvested 6 h later and subjected to RT-qPCR for measurement of IFN-β (a, f, k, and q), IFNα2 (b, g, l, and r), IFNλ1 (c, h, m, and s), CXCL10, (d, i, and n), or TNF (e, j, o, and t). Cytokine mRNA levels were normalized and compared with an age- and gender-matched control or the pooled results of a total of 12 controls. (p) Fibroblasts from the patient and controls were stimulated with extracellular poly(I:C). Total RNA was harvested 6 h later and subjected to RT-qPCR for measurement of IFN-β. Data are shown as box plots with median, first, and third quartiles. Error bars represent minimum and maximum values. The pooled controls are illustrated as the 5–95% population, with outliers shown as independent dots. For all data, similar results were obtained in at least two independent experiments. Nonparametric Mann-Whitney ranked sum test was used for statistical analysis. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. C, control. P, patient. t-, transfected.
Finally, we tested a series of other nucleic acid–based PAMPs for induction of IFN-β and IFNα2 in PBMCs. Patient PBMCs responded with reduced expression after stimulation with the TLR7/8 agonist R848, the TLR9 agonist ODN2216, or the RLR agonist transfected poly(I:C) compared with control (Fig. 2, q and r). These treatments did not stimulate strong expression of IFNλ1 and TNF, and only for ODN2216 was the response reduced in cells from the patient (Fig. 2, s and t).
Collectively, PBMCs and fibroblasts from P1 exhibited impaired ability to evoke IFN responses after stimulation with synthetic PAMPs.
HSV-1–induced IFN expression is impaired in patient cells
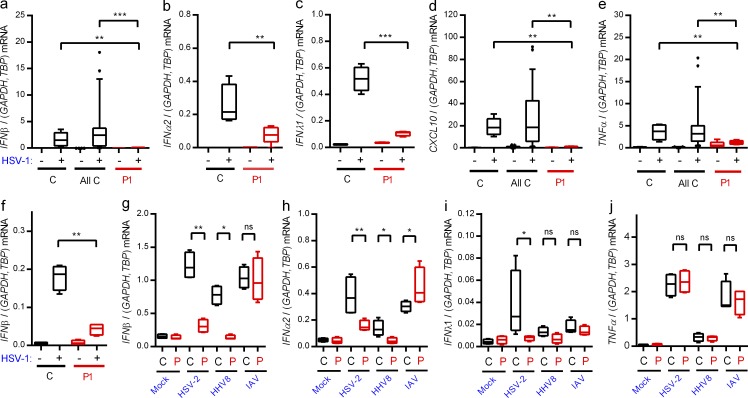
Infection of PBMCs from controls with HSV-1 induced expression of IFN-β, -α2, -λ1, and CXCL10, and this response was largely abrogated in cells from P1 (Fig. 3, a–e). When examining the response in fibroblasts, which—like the CNS—are ectodermally derived, we found that HSV-1 induced IFN-β expression in control but not patient fibroblasts (Fig. 3 f).
Figure 3.
Abolished IFN responses to HSV-1 in patient cells. (a–e and g–j) PBMCs from the patient and controls were infected with HSV-1 (MOI 9), HSV-2 (MOI 9), HHV8 (30 genomes per cell), or IAV (MOI 3) as indicated for 6 h, and RNA was isolated for measurement of IFN-β (a and g), IFNα2 (b and h), IFNλ1 (c and i), CXCL10 (d), and TNF expression (e and j). Cytokine mRNA levels were normalized and compared with an age- and gender-matched control or the pooled results of a total of 12 healthy controls. (f) Fibroblasts were infected with HSV-1 (MOI 9). Total RNA was harvested 6 h later and subjected to RT-qPCR for measurement of IFN-β. Data are shown as box plots with median, first, and third quartiles. Error bars represent minimum and maximum values. The pooled controls are illustrated as the 5–95% population, with outliers shown as independent dots. For all data, similar results were obtained in at least two independent experiments. Non-parametric Mann-Whitney ranked sum test was used for statistical analysis. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (g–j) C, control; P, patient.
Next, we infected PBMCs with the human pathogenic viruses HSV-2, human herpes virus (HHV) 8, and influenza A virus (IAV). All these viruses induced expression of IFN-β and -α2, and for HSV-2 and HHV8 we observed significantly lower responses in cells from P1 (Fig. 3, g and h). Interestingly, type I IFN expression in response to IAV infection was not reduced in cells from P1 (Fig. 3, g and h). IFNλ1 was only weakly induced by the three viruses, and only for HSV-2 was this response reduced in cells from P1 (Fig. 3 i). Finally, the levels of virus-induced TNF were not affected in patient cells (Fig. 3 j). Thus, expression of IFNs and ISGs in response to HSV-1 infection was largely abolished in PBMCs and fibroblasts from P1. This may be because IRF3 is localized at a position downstream of several PRR signaling pathways, which may cause a more extensive inhibition of IFN production than an isolated defect of the TLR3 pathway. In addition, cells of the CNS may be particularly sensitive to reduced IFN responses, thus leading to more pronounced susceptibility to CNS infections as compared with systemic viral infections. The differential responses, including almost entirely abolished responses to HSV-1, partially reduced responses to other human pathogenic herpes viruses, and normal responses to IAV, are in agreement with the narrow infectious phenotype and medical history.
The R285Q IRF3 mutant is not phosphorylated, dimerized, or transcriptionally activated upon stimulation
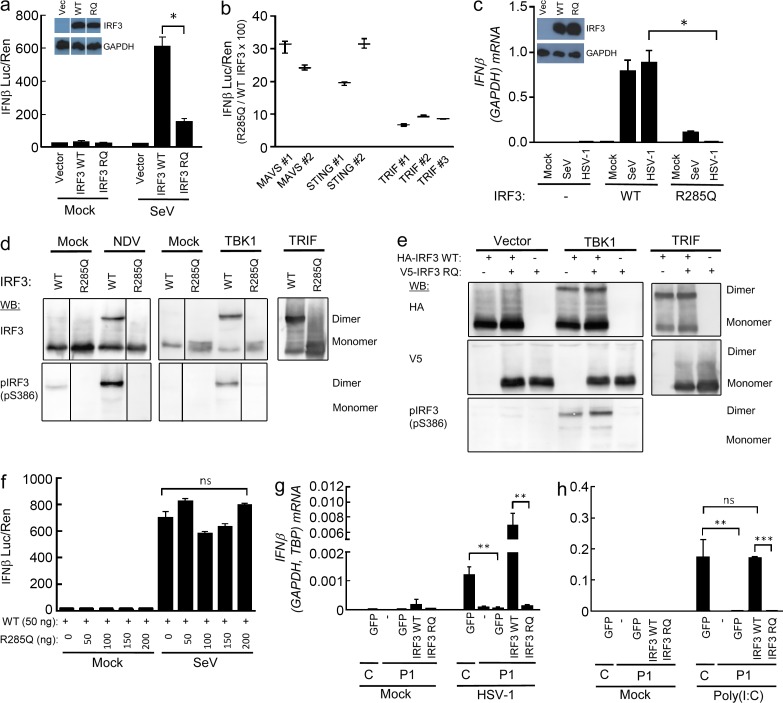
With the aim to identify the molecular mechanism underlying impaired IFN production in cells harboring the R285Q IRF3 mutant, WT IRF3 and R285Q IRF3 were transiently expressed in IRF3-deficient HEK293T cells, which were infected with Sendai virus (SeV). Whereas expression of R285Q IRF3 only marginally enabled infection to increase IFN-β promoter activity, expression of WT IRF3 allowed robust IFN-β promoter activation (Fig. 4 a). Likewise, expression of each of the three key IFN-stimulating PRR adaptor proteins TRIF, MAVS, or STING lead to markedly reduced IFN-β promoter activity in cells transfected with R285Q IRF3 as compared with IRF3 WT, with ∼30% of the activity of WT IRF3 in response to MAVS and STING overexpression and <10% of WT IRF3 in response to TRIF overexpression, thus strongly suggesting that the TLR3–TRIF pathway is affected the most by the R285Q mutation (Fig. 4 b). Finally, in IRF3-deficient THP1-derived monocytes, reconstitution with WT IRF3 but not R285Q IRF3 restored the ability of HSV-1 to induce IFN-β expression, whereas some degree of IFN-β expression was observed in R285Q IRF3–expressing cells infected with SeV (Fig. 4 c).
Figure 4.
The R285Q IRF3 mutant is not phosphorylated, dimerized, or transcriptionally activated upon stimulation. (a, b, and f) HEK293T-derived, IRF3-deficient cells were transiently transfected with IFN-β promoter luciferase reporter, β-actin promoter, Renilla luciferase reporter, and plasmids encoding WT IRF3, R285Q IRF3, MAVS, STING, or TRIF as indicated. (a and f) Cells were infected with SeV (20 HAU/well) and luciferase activities were measured 16 h later (a, b, and f). Firefly luciferase activity was normalized to Renilla luciferase activity and presented as either means from triplicate cultures ± SD or percentage stimulation in cells expressing R285Q IRF3 relative to cells expressing WT IRF3 ± SD. Data from 2–3 independent experiments are shown. (c) IRF3-deficient THP1-derived monocytes transduced with WT and R285Q IRF3 were infected for 6 h with HSV-1 (MOI 3) or SeV (5 HAU/well). (d and e) L929 cells were transfected with empty vector, IRF3 WT or IRF3 R285Q as indicated. The cells were stimulated by infection with Newcastle disease virus (MOI 100) or cotransfection with human TBK1 or TRIF. Cells were lysed and subjected to native-PAGE followed by immunoblotting with antibodies against IRF3 and IRF3 S386-P. (g and h) Fibroblasts from P1 and a control were transduced with lentiviral vectors encoding eGFP, WT IRF3, or R285Q IRF3. The cells were infected with HSV-1 (g; MOI 9) or stimulated with extracellular poly(I:C) (h; 50 µg/ml). Total RNA was harvested 6 h later and subjected to RT-qPCR for measurement of IFN-β. Data are shown as means of four measurements ± SD. Data are from at least two independent experiments. Nonparametric Mann-Whitney ranked sum test was used for statistical analysis. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. RQ, R285Q.
To evaluate the ability of R285Q IRF3 to become phosphorylated and form homodimers, cells were transfected with WT IRF3 or R285Q IRF3, and the pathway was stimulated with virus infection or by co-transfection with the IRF3 kinase TBK1 or TRIF. Extracts were analyzed by native-PAGE and Western blotting. Interestingly, unlike WT IRF3, the R285Q IRF3 mutant did not form homodimers upon infection or pathway-specific stimulation (Fig. 4 d). Furthermore, the R285Q IRF3 mutant failed to become phosphorylated at S386, which is involved in activation and dimerization of IRF3 (Mori et al., 2004; Fig. 4 d). These data demonstrate that the mutant R285Q IRF3 fails to become phosphorylated and also does not undergo dimerization in response to stimulation by TRIF, TBK1, or virus infection.
To examine whether the R285Q IRF3 mutant may interfere with the function of endogenous IRF3 and exert a dominant-negative effect, we coexpressed WT and mutant IRF3 in IRF3-deficient cells. The stimulation-induced IRF3 dimers were exclusively constituted of WT IRF3 and the R285Q IRF3 mutant did not impact on the efficiency of IRF3 dimer formation or IFN-β promoter activation (Fig. 4, e and f). Thus, in the model systems used, the R285Q IRF3 mutant did not exhibit a dominant-negative effect upon WT IRF3.
Finally, we wanted to examine the effect of WT IRF3 expression in fibroblasts from the patient. The cells were transduced with lentiviral vectors encoding GFP, WT, or R285Q IRF3 and stimulated with extracellular poly(I:C) or infected with HSV-1. Importantly, expression of WT IRF3, but neither GFP nor the R285Q IRF3 mutant, reconstituted the ability of patient fibroblasts to express IFN-β in response to the TLR3 agonist poly(I:C) or HSV-1 infection (Fig. 4, g and h). These data strongly suggest that the impaired IFN responses in patient cells are caused by the identified R285Q IRF3 mutation, thus proposing a causal relationship.
The HSE patient in the present study harbored a single base-pair mutation c854G>A in IRF3, resulting in an arginine-to-glutamine substitution, and thus a change from a positively charged to an uncharged amino acid, at position 285 in the regulatory domain of the IRF3 protein. Our data demonstrated abolished S386 phosphorylation and no detectable IRF3 dimerization in cells expressing the R285Q mutant. This is in agreement with previous work demonstrating a critical role for IRF3 S386 as a target for TBK1-mediated phosphorylation required for IRF3 dimerization. (Mori et al., 2004; Takahasi et al., 2010). A recent study showed that IRF3 is recruited to MAVS, STING, and TRIF through electrostatic interactions between a phosphorylated surface on the adaptors and a positively charged surface on IRF3, which includes R285 (Liu et al., 2015). This positions IRF3 for phosphorylation by TBK1. We compared the efficiency with which R285Q IRF3 induced IFN-β promoter activity in response to overexpression of each of MAVS, STING, and TRIF and found the TLR3–TRIF pathway to be most extensively inhibited. This may explain why we I patient cells observed significantly impaired responses to HSV-1, where the TLR3 pathway is essential, in the setting of an almost normal response to the RNA virus IAV, signaling mainly through RLRs, and agrees with the infectious phenotype of the patient. Overall, differential impairment in the recruitment of mutant IRF3 to the adaptor molecules may explain different effects on these signaling pathways and the IFN-inducing ability of different pathogens.
The finding that the R285Q IRF3 mutation is of functional significance despite being heterozygous indicates a mechanism involving either dominant-negative activity of the mutant protein or haploinsufficiency, with the former previously being suggested in the case of AD TRAF3 deficiency and AD partial TLR3 deficiency in HSE (Pérez de Diego et al., 2010; Herman et al., 2012). Although cells from the patient exhibited largely abrogated responsiveness to HSV-1 infection and several PAMPs, we did not observe a dominant-negative effect of the mutant R285Q IRF3 in the HEK293T cell model system. However, we were able to fully reconstitute the IFN response of patient fibroblasts with expression of WT IRF3. RNA sequencing revealed equal transcription from the WT and mutant IRF3 alleles (unpublished data). These data suggest that haploinsufficiency rather than a dominant-negative effect explains the infectious phenotype of the patient. This is in agreement with the father of the patient being a healthy carrier, because incomplete penetrance has been described in the majority of AD immunological conditions involving haploinsufficiency, including defects in the TLR3 pathway in HSE patients (Abel et al., 2010; Rieux-Laucat and Casanova, 2014).
A panel of criteria has recently been proposed for deciding if clinical and experimental data suffice to establish a causal relationship based on only one case (Casanova et al., 2014). Central among these is to rescue a normal phenotype from the disease-relevant phenotype by reintroducing the WT molecule in question. In the present work, we demonstrated that expression of WT IRF3 enabled P1 fibroblasts to produce IFN in response to HSV-1 infection. These data strongly argue for a causal relationship between the cellular and clinical phenotype of the patient and the identified R285Q IRF3 mutation. Other criteria fulfilled in the present case in favor of a causal relationship are that the identified mutation is very rare, that R285 is an evolutionarily highly conserved residue within IRF3, and that IRF3 is part of a known pathway well established in conferring increased susceptibility to HSE. In conclusion, the present study provides the first identification of a mutation in a member of the IRF family of IFN-inducing transcription factors in viral CNS infections in humans and adds a novel genetic etiology to HSE.
MATERIALS AND METHODS
Primary cells.
The 15-yr-old female patient of Danish (Caucasian) origin described in this study was identified within a study including a total of 16 adult individuals with previously diagnosed HSE by abnormal cerebrospinal fluid (pleocytosis and detection of HSV-1 by PCR), as well as a clinical history in agreement with HSE. PBMCs from the patient, as well as age- and gender-matched controls (obtained from the Danish blood bank) were purified from heparin-stabilized blood by a Ficoll density gradient. PBMCs were thawed at 37°C and transferred into a falcon tube containing 20 ml preheated RPMI 1640 media (BioWhittaker; Lonza/Biowest) containing 10% heat inactivated FBS (Life Technologies) and 1% l-glutamine (Life Technologies; Medium+). The falcon tubes were centrifuged at 350 g for 10 min at 24°C. Fibroblasts were obtained from a skin biopsy from the patient and cultured in RPMI 1640.
Lentiviral vector production and transduction of fibroblasts and THP1-derived, IRF3-deficient monocytes.
Lentiviral vector constructs pCCL/PGK-IRF3 and pCCL/PGK-IRF3(R285Q) were generated by insertion of PCR-amplified IRF3 or IRF3(R285Q) cDNA sequences, amplified from pCDNA3_V5-IRF3 or pCDNA3_V5-IRF3(R285Q), respectively, into BamHI–XhoI-digested pCCL/PGK-eGFP (Jakobsen et al., 2009). Lentiviral vectors were produced as previously described (Jakobsen et al., 2009). In brief, packaging plasmids pMD2.G, pRSV-Rev, and pMDIg/pRRE were calcium phosphate-transfected together with either pCCL/MCS, pCCL/PGK-eGFP, pCCL/PGK-IRF3, or pCCL/PGK-IRF3(R285Q) into HEK293T cells. Vector-containing supernatants were harvested by filtration through a 0.45-µm filter, and polybrene was added to a final concentration of 8 µg/ml. For transduction of patient fibroblasts and normal human dermal fibroblast control cells, filtered supernatants were added to fibroblasts seeded at 1 × 106 cells/dish in 6-cm dishes in RPMI 1640 1 d before transduction. The cells were then passaged for 4 d before being subjected to further analysis. The transduction efficiency as assessed by eGFP expression was between 60 and 70%. Transduction of THP-1–derived IRF3-deficient cells was done in a similar manner as described above for fibroblasts. The cells were passaged for 5 d before being differentiated into macrophages by addition of 100 nM phorbol myristate acetate (PMA). After 24 h, the media was changed and the cells were left resting for another 24 h before being subjected to further analysis.
In vitro stimulations.
PBMCs and fibroblasts were seeded in 24-well tissue culture plates at a concentration of 106 and 1 × 105 cells/well, respectively, in 300 µl media and incubated overnight at 37°C and 5% CO2. For stimulation with PRR agonists, the following reagents and concentrations were used: extracellular poly(I:C) (50 µg/ml), R848 (1 µg/ml), and ODN2216 (5 µM), transfected poly(dA:dT) (4 µg/ml; all from InvivoGen), HSV-1–derived dsDNA 60mer (DNA Technology; 4 µg/ml). For infection with viruses, the following viruses and infection doses were used: SeV (strain Cantell, 20 HA U/well), HSV-1 (strain 17+, MOI 9), HSV-2 (strain 333, MOI 9), HHV8 (30 genomes/cell), and IAV (PR8, MOI 3). At the end of the incubation period, adherent and nonadherent cells were lysed and the pooled lysates were subjected to RNA purification.
RNA purification.
High Pure RNA Isolation kit (Roche) was used for RNA purification following instructions of the manufacturer. cDNA was synthesized from RNA using QuantiTect Reverse Transcription kit (QIAGEN) following the manufacturers recommendations.
DNA purification.
Genomic DNA was isolated from 1 ml EDTA stabilized blood using MagNA Pure Compact Nucleic Acid Isolation kit I – Large Volume (Roche).
WES and bioinformatics.
TruSeq DNA sample preparation was performed according to the manufacturer’s recommendations (Illumina) on a Caliper Sciclone robot (Perkin Elmer). Targeting of exomes with SeqCap EZ Human Exome Library v3.0 (Roche) and purification of libraries was performed using a Caliper Zephyr robot. Libraries were quantified using Kapa quantification (Kapabiosystems) and sequencing was performed on HiSeq, paired-end 2X101 bp indexed. Adapters were identified and removed, and reads were mapped to hg19 using BWA mem. PCR and optical duplicates were identified and marked. The alignment file (bam) was realigned using GATK to refine the alignment, especially around indels at the ends of reads. The alignment was recalibrated using GATK. Single nucleotide polymorphisms were called using HaplotypeCaller from the GATK package. Variant call files (VCF) were uploaded to Cartagenia and filtered using a list of 204 genes known to be involved in immunodeficiency or involved in relevant pathways. Variants of interest were selected on the basis of frequency in the NHLBI Exome Sequencing Project (ESP) Exome Variant Server (ESP6500) and 1000 Genomes. The list was made on the basis of genes present in the IDBases (Piirilä et al., 2006), along with a search in the KEGG Pathway Database (Kanehisa et al., 2014). Variants were selected on the basis of both rarity (<0.001) and evaluation by prediction tools. The R285Q mutation is predicted to be probably damaging or damaging by PolyPhen-2 and SIFT software, respectively. In addition, the CADD software predicts a score of 21.8, indicating that the mutation belongs to the 1% most deleterious mutations. By using Ingenuity Variant Analysis, we analyzed for the existence of rare homozygous and compound heterozygous mutations. The identified mutation in IRF3 was analyzed by the Integrative Genomics Viewer (IGV) based on BAM files, and further confirmed by Sanger sequencing. No other homozygous or compound heterozygous mutations were identified. There are other rare nonsynonymous mutations reported for IRF3; from a total of 29 rare mutations, 13 are predicted to be damaging by Polyphen2. The identified mutation was not present in 1,000 genomes or WES datasets from 60 controls of Danish origin. Data from the Exome Aggregation Consortium server shows that of 207 missense and loss-of-function mutations identified in IRF3, only 8 have a frequency >0.001, suggesting that purifying selection may be acting on the gene. The R285Q mutation in IRF3 has also been reported in this database with a frequency of <0.0001. In addition to the IRF3 mutation, 7 other rare heterozygous (not compound heterozygous) mutations were identified based on the list of 204 genes, of which mutations in only FEN1 and AIP were predicted to be possibly damaging. However, these proteins are not linked to antiviral immunity to HSV, and thus we did not pursue the function of these any further.
Sanger sequencing.
Confirmation of sequence data were obtained by Sanger sequencing by using the following primers (IRF3-2360F, 5′-GAAGCCCCTGTCTCACTCAC-3′; IRF3-3100R, 5′-TCCAGAAAAGGATATGGAAATGCC-3′). PCR was performed using AmpliTaq Gold (Life Technologies), and PCR products were sequenced on an ABI 3130 XL.
Whole-cell lysate.
PBMCs used for Western blotting were thawed, washed in PBS, and lysed in a Triton-based lysis buffer (Cell Signaling Technologies) supplemented with Complete-mini protease inhibitor cocktail following the manufacturer’s instructions (Roche). The cells were lysed on ice for 30 min and centrifuged at 17,000 g to remove cellular debris.
Western blotting.
The lysates were mixed with SDS loading buffer (Sigma-Aldrich) and heated for 5 min at 95°C. The samples were subjected to 10% SDS-PAGE and then transferred to a PVDF-membrane (Applichem). The amount of IRF3 present in the cell lysates was detected using a rabbit anti-IRF3 antibody (Santa Cruz Biotechnology, Inc.) and the amount of GAPDH was detected using a rabbit anti-GAPDH (Santa Cruz Biotechnology, Inc.) antibody. Both primary antibodies were followed by a secondary HRP-conjugated swine anti–rabbit antibody (Dako). The proteins on the membrane were visualized on x-ray film (Konica Minolta) using the SuperSignal West Dura chemiluminescence system (Thermo Fisher Scientific).
TaqMan RT-qPCR.
Expression levels of IFN-β, CXCL10, and TNF and the two household genes GAPDH and TBP were analyzed by RT-qPCR using TaqMan probes. RT-qPCR was done in 2-steps (1-RT, 2-qPCR); the second step in the qPCR was performed on the synthesized cDNA using PerfeCTa ToughMix II (Quanta BioSciences) and the following TaqMan probes purchased from Life Technologies: were GAPDH (Hs02758991), TBP (Hs00427620), IFNB1 (Hs01077958), CXCL10 (Hs01124251), IFNα2 (Hs00265051_s1), TNF (Hs01113624). IFN-λ1 was amplified using CYBR Green kit (Life Technologies) and the following primers: forward, 5′-GGAAGCAGTTGCGATTTAGCC-3′; reverse, 5′-GACTCTTCCAAGGCGTCCCT-3′. Samples were analyzed in duplicates. Relative mRNA levels were calculated using the following formula: 2ΔCt(control − sample). Control Ct values were generated as a mean of GAPDH and TBP Ct values.
Cell lines.
To generate HEK293T and THP1 cells deficient in IRF3, cells were plated at a density of 2 × 104 cells per 96-well. The next day, CRISPR plasmids were transfected using GeneJuice transfection reagent (Merk Millipore) according to the manufacturer’s protocol. pRZ-mCherry-Cas9 and pLenti-gRNA constructs (Ablasser et al., 2013) were transfected at a ratio of 3:1 (i.e., 150 ng:50 ng). THP1 cells were electroporated with the same plasmid combinations (2.5 × 106 cells with 5 µg of DNA) and sorted for mCherry-Cas9-positive cells. Exon 4 of IRF3 was targeted in the following region: 5′-GGGGGTCCCGGATCTGGGAGTGG-3′. Subsequently, limiting dilution cloning was performed and, after 10 d, growing monoclones were selected by bright field microscopy. Identified clones were trypsinized and expanded in two separate wells. One well was used to recover gDNA and, subsequently, the target region of interest was amplified in a two-step PCR and subjected to deep sequencing, as previously described (Schmid-Burgk et al., 2014). Knockout cell clones were identified as cell clones harboring all-allelic frame shift mutations.
IRF3-deficient HEK293T cells and murine L929 cells were cultured in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Sigma-Aldrich), 100 U/ml penicillin (Sigma-Aldrich), and 100 mg/ml streptomycin (Sigma-Aldrich), whereas IRF-deficient THP-1 cells were cultured in RPMI 1640 supplemented with 10% FBS (Sigma-Aldrich), 200 mM l-glutamine, 100 U/ml penicillin (Sigma-Aldrich), and 100 mg/ml streptomycin (Sigma-Aldrich). All cell lines were and maintained at 37°C with 5% CO2.
Plasmids.
The IRF3 R285Q construct was generated by site-directed mutagenesis using pfuUltra II Fusion HS DNA polymerase according to the manufacturer’s instructions (Agilent). The IRF3 R285Q construct was generated on the human V5-IRF3-pcDNA3 vector (Addgene ID, 32713) using the forward primer 5′-CTGGGCCCAGCAGCTGGGGCACT-3′ and the reverse primer 5′-AGTGCCCCAGCTGCTGGGCCCAG-3′.
Luciferase assay.
The IRF3-deficient HEK293T cells were seeded in 12-well plates at a density of 4 × 105 cells/well and cultured for 24 h. The cells were transiently transfected with a transfection mixture consisting of DNA and the transfection agent polyethylenimine (PEI) in a ratio of 1:3. For all experiments, the DNA mixture contained 970 ng reporter plasmid containing firefly luciferase under the control of the IFN-β promoter, 30 ng reporter plasmid containing Renilla luciferase under the control of the β-actin promoter and 50 ng of the IRF3 WT plasmid or the IRF3 R285Q plasmid except for the experiments presented in Fig. 4 f, where the IRF3 R285Q plasmid was transfected in increasing doses from 0 to 200 ng. For the experiments presented in Fig. 4 b, the cells were also transfected with either 250 ng MAVS, STING, or TRIF. In all cases, empty plasmid was added to give a total amount of 2 µg of DNA in a total volume of 200 µl DMEM/well. The DNA and PEI mixtures were mixed and incubated 15 min at room temperature before gently being applied to the cells. In Fig. 4 (a and f), the cells were incubated for 24 h before infection with 20 HAU of SeV for 16 h. The cells were lysed with Passive Lysis Buffer (Promega) and the luciferase activity was measured with Dual-Luciferase Reporter Assay system (Promega) according to the manufactures instructions. In Fig. 4 b, luciferase activity was measured in lysates isolated 40 h after transfection.
IRF3 dimerization assay.
L929 cells were transfected with empty vector, IRF3 WT or IRF3 R285Q. The cells were stimulated by cotransfection with vectors encoding human TBK1 or TRIF, or infected with Newcastle Disease Virus. Cells were lysed in lysis buffer (1% NP-40, 50 mM Tris-Cl pH8.0, 150 mM NaCl, 1 mM Vanadate, 1 mM PMSF, and 0.1 µg/ml leupeptin) and subjected to native-PAGE followed by immunoblotting with antibodies against human IRF3 (18781) and human IRF3 S386-P (18783) from Immuno-Biological Laboratories Co. Ltd. (Mori et al., 2004).
Statistics.
Experiments were performed on biological duplicates and each experiment was repeated two to three times. The nonparametric Mann-Whitney ranked sum test was used to establish statistical significance.
Ethics.
The National Committee on Health Research Ethics and the Danish Data Protection Agency approved the study (project # 1–10-72-586-12). The patient provided written consent before inclusion. All personal information is protected as required by the Data Protection Agency and the relevant Danish laws.
Online supplemental material.
The medical history of the patient is provided as supplemental text. Table S1 lists demography, clinical symptoms, and findings. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20142274/DC1.
Supplementary Material
Acknowledgments
This work was supported by The Lundbeck Foundation (R151-2013-14668 and R144-2013-13436; T.H. Mogensen), The Foundation for the Advancement of Medical Sciences (T.H. Mogensen), the Danish Research Council (12-124330 [S.R. Paludan], 11-107588 [T.H. Mogensen], 4004-00047 [R. Hartmann], and 4004-00220 [J.G. Mikkelsoe]), Region Midt (E. Kofod-Olsen, L. Østergaard, and T.H. Mogensen), and The Danish Cancer Society (grant R20-A927; R. Hartmann).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AD
- autosomal dominant
- AR
- autosomal recessive
- HSE
- herpes simplex encephalitis
- HHV8
- human herpesvirus 8
- IAV
- influenza A virus
- IRF
- IFN regulatory factor
- PAMP
- pathogen associated molecular pattern
- PRR
- pattern recognition receptor
- RLR
- RIG-like receptor
- SeV
- Sendai virus
- WES
- whole-exome sequencing
References
- Abel L., Plancoulaine S., Jouanguy E., Zhang S.Y., Mahfoufi N., Nicolas N., Sancho-Shimizu V., Alcaïs A., Guo Y., Cardon A., et al. 2010. Age-dependent Mendelian predisposition to herpes simplex virus type 1 encephalitis in childhood. J. Pediatr. 157:623–629: e1. 10.1016/j.jpeds.2010.04.020 [DOI] [PubMed] [Google Scholar]
- Ablasser A., Schmid-Burgk J.L., Hemmerling I., Horvath G.L., Schmidt T., Latz E., and Hornung V.. 2013. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 503:530–534. 10.1038/nature12640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audry M., Ciancanelli M., Yang K., Cobat A., Chang H.H., Sancho-Shimizu V., Lorenzo L., Niehues T., Reichenbach J., Li X.X., et al. 2011. NEMO is a key component of NF-κB- and IRF-3-dependent TLR3-mediated immunity to herpes simplex virus. J. Allergy Clin. Immunol. 128:610–617: e1–e4. 10.1016/j.jaci.2011.04.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L., and Abel L.. 2007. Primary immunodeficiencies: a field in its infancy. Science. 317:617–619. 10.1126/science.1142963 [DOI] [PubMed] [Google Scholar]
- Casanova J.L., Conley M.E., Seligman S.J., Abel L., and Notarangelo L.D.. 2014. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. J. Exp. Med. 211:2137–2149. 10.1084/jem.20140520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casrouge A., Zhang S.Y., Eidenschenk C., Jouanguy E., Puel A., Yang K., Alcais A., Picard C., Mahfoufi N., Nicolas N., et al. 2006. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 314:308–312. 10.1126/science.1128346 [DOI] [PubMed] [Google Scholar]
- Griffin D.E. 2005. Encephalitis, myelitis, and neuritis. Principles and practice in infectious diseases. Mandell G.L., Bennett J.E., and Dolin R., Elsevier, Philadelphia: pp. 1143–1150. [Google Scholar]
- Guo Y., Audry M., Ciancanelli M., Alsina L., Azevedo J., Herman M., Anguiano E., Sancho-Shimizu V., Lorenzo L., Pauwels E., et al. 2011. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J. Exp. Med. 208:2083–2098. 10.1084/jem.20101568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman M., Ciancanelli M., Ou Y.H., Lorenzo L., Klaudel-Dreszler M., Pauwels E., Sancho-Shimizu V., Pérez de Diego R., Abhyankar A., Israelsson E., et al. 2012. Heterozygous TBK1 mutations impair TLR3 immunity and underlie herpes simplex encephalitis of childhood. J. Exp. Med. 209:1567–1582. 10.1084/jem.20111316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., and Taniguchi T.. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644–658. 10.1038/nri1900 [DOI] [PubMed] [Google Scholar]
- Jakobsen M., Stenderup K., Rosada C., Moldt B., Kamp S., Dam T.N., Jensen T.G., and Mikkelsen J.G.. 2009. Amelioration of psoriasis by anti-TNF-alpha RNAi in the xenograft transplantation model. Mol. Ther. 17:1743–1753. 10.1038/mt.2009.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., and Tanabe M.. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42:D199–D205. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaille F.G., Pessach I.M., Zhang S.Y., Ciancanelli M.J., Herman M., Abhyankar A., Ying S.W., Keros S., Goldstein P.A., Mostoslavsky G., et al. 2012. Impaired intrinsic immunity to HSV-1 in human iPSC-derived TLR3-deficient CNS cells. Nature. 491:769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim H.K., Seppänen M., Hautala T., Ciancanelli M.J., Itan Y., Lafaille F.G., Dell W., Lorenzo L., Byun M., Pauwels E., et al. 2014. TLR3 deficiency in herpes simplex encephalitis: high allelic heterogeneity and recurrence risk. Neurology. 83:1888–1897. 10.1212/WNL.0000000000000999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y.T., Grishin N.V., and Chen Z.J.. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 347:aaa2630 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- Menachery V.D., Pasieka T.J., and Leib D.A.. 2010. Interferon regulatory factor 3-dependent pathways are critical for control of herpes simplex virus type 1 central nervous system infection. J. Virol. 84:9685–9694. 10.1128/JVI.00706-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen T.H. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22:240–273. 10.1128/CMR.00046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Yoneyama M., Ito T., Takahashi K., Inagaki F., and Fujita T.. 2004. Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 279:9698–9702. 10.1074/jbc.M310616200 [DOI] [PubMed] [Google Scholar]
- Osterlund P., Veckman V., Sirén J., Klucher K.M., Hiscott J., Matikainen S., and Julkunen I.. 2005. Gene expression and antiviral activity of alpha/beta interferons and interleukin-29 in virus-infected human myeloid dendritic cells. J. Virol. 79:9608–9617. 10.1128/JVI.79.15.9608-9617.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan S.R., Bowie A.G., Horan K.A., and Fitzgerald K.A.. 2011. Recognition of herpesviruses by the innate immune system. Nat. Rev. Immunol. 11:143–154. 10.1038/nri2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez de Diego R., Sancho-Shimizu V., Lorenzo L., Puel A., Plancoulaine S., Picard C., Herman M., Cardon A., Durandy A., Bustamante J., et al. 2010. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 33:400–411. 10.1016/j.immuni.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piirilä H., Väliaho J., and Vihinen M.. 2006. Immunodeficiency mutation databases (IDbases). Hum. Mutat. 27:1200–1208. 10.1002/humu.20405 [DOI] [PubMed] [Google Scholar]
- Rieux-Laucat F., and Casanova J.L.. 2014. Immunology. Autoimmunity by haploinsufficiency. Science. 345:1560–1561. 10.1126/science.1260791 [DOI] [PubMed] [Google Scholar]
- Sancho-Shimizu V., Perez de Diego R., Jouanguy E., Zhang S.Y., and Casanova J.L.. 2011a. Inborn errors of anti-viral interferon immunity in humans. Curr Opin Virol. 1:487–496. 10.1016/j.coviro.2011.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho-Shimizu V., Pérez de Diego R., Lorenzo L., Halwani R., Alangari A., Israelsson E., Fabrega S., Cardon A., Maluenda J., Tatematsu M., et al. 2011b. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J. Clin. Invest. 121:4889–4902. 10.1172/JCI59259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk J.L., Schmidt T., Gaidt M.M., Pelka K., Latz E., Ebert T.S., and Hornung V.. 2014. OutKnocker: a web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome Res. 24:1719–1723. 10.1101/gr.176701.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K., Horiuchi M., Fujii K., Nakamura S., Noda N.N., Yoneyama M., Fujita T., and Inagaki F.. 2010. Ser386 phosphorylation of transcription factor IRF-3 induces dimerization and association with CBP/p300 without overall conformational change. Genes Cells. 15:901–910. [DOI] [PubMed] [Google Scholar]
- Whitley R.J., and Lakeman F.. 1995. Herpes simplex virus infections of the central nervous system: therapeutic and diagnostic considerations. Clin. Infect. Dis. 20:414–420. 10.1093/clinids/20.2.414 [DOI] [PubMed] [Google Scholar]
- Zhang S.Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., et al. 2007. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 317:1522–1527. 10.1126/science.1139522 [DOI] [PubMed] [Google Scholar]
- Zhang S.Y., Herman M., Ciancanelli M.J., Pérez de Diego R., Sancho-Shimizu V., Abel L., and Casanova J.L.. 2013. TLR3 immunity to infection in mice and humans. Curr. Opin. Immunol. 25:19–33. 10.1016/j.coi.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.