Abstract
The bacteria involved in organic pollutant degradation in pelagic deep-sea environments are largely unknown. In this report, the diversity of polycyclic aromatic hydrocarbon (PAH)-degrading bacteria was analyzed in deep-sea water on the Southwest Indian Ridge (SWIR). After enrichment with a PAH mixture (phenanthrene, anthracene, fluoranthene, and pyrene), nine bacterial consortia were obtained from depths of 3946–4746 m. While the consortia degraded all four PAHs when supplied in a mixture, when PAHs were tested individually, only phenanthrene supported growth. Thus, degradation of the PAH mixture reflected a cometabolism of anthracene, fluoranthene, and pyrene with phenanthrene. Further, both culture-dependent and independent methods revealed many new bacteria involved in PAH degradation. Specifically, the alpha and gamma subclasses of Proteobacteria were confirmed as the major groups within the communities. Additionally, Actinobacteria, the CFB group and Firmicutes were detected. Denaturing Gradient Gel Electrophoresis (DGGE) analysis showed that bacteria closely affiliated with Alcanivorax, Novosphingobium, and Rhodovulum occurred most frequently in different PAH-degrading consortia. By using general heterotrophic media, 51 bacteria were isolated from the consortia and of these 34 grew with the PAH mixture as a sole carbon source. Of these, isolates most closely related to Alterierythrobacter, Citricella, Erythrobacter, Idiomarina, Lutibacterium, Maricaulis, Marinobacter, Martelella, Pseudidiomarina, Rhodobacter, Roseovarius, Salipiger, Sphingopyxis, and Stappia were found to be PAH degraders. To the best of our knowledge, this is the first time these bacteria have been identified in this context. In summary, this report revealed significant diversity among the PAH-degrading bacteria in the deep-sea water column. These bacteria may play a role in PAH removal in deep-sea environments.
Keywords: Southwest Indian Ridge, deep-sea water column, PAHs, biodegradation, bacterial biodiversity, pelagic ocean
Introduction
Polycyclic aromatic hydrocarbons (PAHs) are a group of cytotoxic, mutagenic and carcinogenic compounds with two or more fused aromatic rings. They can be found in deep-sea sediments, such as those found 2.7 m beneath the bottom surface at a water depth of 3,962 m on the Middle Atlantic Ridge. This locations has a total PAH concentration (ΣPAHs) of 260 ∼440 ng/g dw (dry weight) (Cui et al., 2008; Wang et al., 2008). Recently, even higher PAH concentrations were detected in the hydrothermal vent area of the South Mid-Atlantic Ridge (Huang et al., 2014). There, the ΣPAHs ranged from 2768 to 9826 ng/g dry sediment at different sites (Huang et al., 2014). Among the PAHs detected in these reports, phenanthrene was most abundant. However, the origin and fate of PAHs in this extreme environment are not well understood.
Biodegradation of PAHs by bacteria has been investigated for a long time in coastal and gulf areas (Kanaly and Harayama, 2000; Harayama et al., 2004; Head et al., 2006; Yakimov et al., 2007; Kappell et al., 2014). Various PAH-degrading bacteria have been found in coastal environments (Head et al., 2006; Yakimov et al., 2007; Lamendella et al., 2014). Of these, bacteria of the genus Cycloclasticus are among the most wide-spread PAH degraders in marine systems (Dyksterhouse et al., 1995; Wang et al., 1996; Kasai et al., 2002; Yakimov et al., 2005; McKew et al., 2007; Teira et al., 2007). In deep-sea sediment, Cycloclasticus also can be found functioning as a PAH degrader (Cui et al., 2008; Wang et al., 2008).
In deep-sea water conditions, PAHs are thought to be absorbed on suspended particles and sink to the floor. As labile carbon sources in deep water are limited, PAHs in these environments could serve as valuable carbon and energy sources for bacterial growth. However, little is known about the diversity of PAH-degrading bacteria therein. In the present report, PAH degraders were examined at water depths ranging from 3946 to 4746 m on the Southwest Indian Ridge (SWIR). These bacteria displayed significant diversity and novelty, indicating the prevalence of PAH-degrading bacteria in the deep water system of the open sea.
Materials and Methods
Medium
For enrichments, a mineral salt medium (NH) was used that contained: 1.0 g NH4NO3, 0.5 g KH2PO4, 2.8 mg FeSO4, 1000 ml filtered sea water, adjusted to pH 7.5. PAHs were used as the sole carbon and energy source and included phenanthrene (>97%, Fluka), anthracene (>96%, Fluka), fluoranthene (≥98.5%, Fluka), pyrene (98%, Sigma).
Bacteria were isolated from enrichments by using two types of heterotrophic growth media, 216L and M2. The former medium contained: 1.0 g CH3COONa, 10.0 g Tryptone, 2.0 g Yeast extract, 0.5 g Sodium citrate, 0.2 g NH4NO3, 1000 ml sea water, adjusted to pH 7.5. The latter medium contained: 5.0 g CH3COONa, 0.5 g Tryptone, 0.5 g Yeast extract, 0.5 g Glucose, 0.5 g Sucrose, 0.05 g Sodium citrate, 0.05 g Malic acid, 1.0 g NH4NO3, 0.2 g NH4Cl, 0.5 g KH2PO4, 1000 ml sea water, adjusted to pH 7.6.
Sampling and PAH Enrichment on Board
Water samples were collected from the site IR-CTD5 (31°0710′S, 58°9945′E) on the SWIR during the DY105-13 cruise of R/V “Da-Yang Yi-Hao” on December 7, 2005 (Supplementary Figure S1). Niskin bottles attached to a CTD (conductivity, temperature, and depth) circular rosette were used to collect water samples. The bottles were purchased from OceanTest Equipment, INC (Davie, FL, USA). The CTD instrument (SEB, Model: 13-02-B) was obtained from Seabird Electronics, INC (Bellevue, WA, USA). In each bottle, 20 L water was loaded at depths of 4746, 4696, 4546, 4396, 4296, 4196, 4146, 4096 and 3946 m. These depths are 20 to 820 m above the sea bottom, and the samples were numbered IR5-1 to IR5-9, respectively. The parameters recorded by the CTD at 3946 m and 4746 m were 0.9785°C, 11.2262 mg/liter DO, 3.47% salinity and 0.9316°C, 11.2415 mg/liter DO, 3.47% salinity, respectively.
To enrich the PAH-degrading bacteria, 400 ml water from each layer was drained into autoclaved bottles directly from the sampler. The enrichment process was initiated on board the boat at 20°C with PAHs as the carbon and energy sources. Samples were kept for 2 months until laboratory analysis.
PAH Enrichment in the Laboratory
Polycyclic aromatic hydrocarbons dissolved in chloroform were added to flasks containing 150 ml autoclaved NH medium, with final concentrations of 100 mg/liter phenanthrene, 10 mg/liter of anthracene, 10 mg/liter fluoranthene, and 10 mg/liter pyrene. To evaporate the chloroform, flasks were shaken at 160 rpm for 2 days. Then, enrichment was initiated by adding 5.0 ml primary enriched cultures from each layer in the flasks, followed by shaking at 160 rpm in the dark at 25°C for 20 days. For a second round of enrichment in the lab, a 2.0 ml culture of each sample was transferred into the same medium in triplicate. In addition, each PAH was tested individually for degradation in parallel to the PAH mixture treatments. At days 4, 8, 12, and 20, 2 ml cultures of treatments with obvious bacterial growth were used for community genomic DNA extraction. At day 30, the residual PAHs in each flask quantified as described below.
Bacterial Isolation from the PAH Enriched Consortia
After the samples were enriched by being transferred twice in lab, bacteria were isolated from PAH-degrading consortia by serial dilutions ranging from 10-3 to 10-7. These dilutions were then spread on both M2 agar and 216L agar plates and incubated at 18°C for ∼2 weeks. A total of 238 isolates were purified and then subjected to Rep-PCR analysis to eliminate redundant strains, as described previously (Ma et al., 2006). The isolates were then identified by 16S rRNA gene sequencing and analyzed using BLASTN at http://www.ncbi.nlm.nih.gov/BLAST. Nearly full length sequence of 16 rRNA gene was obtained by universal primer set 27f (forward) 5′-AGAGTTTGATCCTGGCTCAG-3′/and 1502r (reverse):5′ -ACGGCTACCTTGTTACGACT-3′. The ability of these bacteria to degrade PAH was tested with a mixture containing phenanthrene, anthracene, fluoranthene, and pyrene.
Quantification of PAH Compounds
To assay the biodegradation capacity of the consortia, PAHs from the mixture were analyzed with Gas Chromatography Mass Spectrometry (GC-MS) after 1 month of incubation. The residual PAHs in each flask were extracted according to Guo et al. (2005). Simultaneously, 5 mg/liter fluorene was added as an internal standard of extraction. Then, degradation percentages were determined using GC-MS QP2010 (SHIMADZU) with a 30-m RTS-5MS capillary column (0.25 mm inside diameter and 0.25 μm film thickness). Details of thermal program and injection conditions were according to our previous report (Cui et al., 2008). Selected ion monitoring (SIM) mode was used for quantification of PAH, and full-scan mode mode was used for identification of PAH with the GC-MS Postrun Analysis software (Version 2.10, SHIMADZU).
DNA Preparation
Total community DNA was collected from 2 mL culture of each sample on days 4, 8, 12, and 20 in sterile 2.0 ml Eppendorf tubes, and then immediately extracted. Genomic DNA of both consortia and isolates were extracted using the modified SDS-CTAB method (Sambrook et al., 1989).
16S rRNA PCR for DGGE Analysis
Polymerase chain reaction amplification of the V3 variable region of 16S rRNA fragments prior to Denaturing Gradient Gel Electrophoresis (DGGE) was performed as described by (Muyzer et al., 1993). To amplify the variable V3 region of bacterial 16S rDNA genes, the primer set was used: DGGEf (forward), 5′-CGCCCGCCGCGCGCGGCGGGCG GGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG-3′, which contained a GC clamp linked to the 5′end; DGGEr (reverse), 5′-ATTACCGCGGCTGCTGG-3′.
All PCRs were performed with a Mastercycler (Eppendorf, Hamburg, Germany). Reaction mixtures contained 1.0 U of rTaq (Takara), each primer at a concentration of 0.5 μM, and 0.6 μl of template DNA in a total volume of 50 μl. After an initial 6 min denaturation at 95°C, a touchdown thermal profile protocol was used. The annealing temperature was decreased by 0.5°C per cycle from 65 to 55°C, followed by 20 additional cycles at 55°C. Amplification was carried out with 1 min of denaturation at 95°C, 1 min of primer annealing, and 1.0 min of primer extension at 72°C, followed by a final extension at 72°C for 10 min. PCRs were carried out in replicates of four using the consortium DNA, then the mixtures were combined PCR amplifications were checked on 1% (w/v) agarose gel prior to DGGE analysis.
Denaturing Gradient Gel Electrophoresis
The purified PCR product was loaded onto 8% (w/v) polyacrylamide gels (ratio of acrylamide to bisacrylamide, 37.5:1). The denaturing gradients used ranged from 30 to 70% denaturant [100% denaturant contained 7 M urea and 40% (v/v) formamide]. The gels were electrophoresed using a D-Code instrument (Bio-Rad) in 1 × TAE buffer (40 mM Tris-acetate, 1 mM Na-EDTA, pH 8.0) running at 30 V for 15 min first and then 130 V for 4.5 h at 60°C.
After electrophoresis, the gel was stained with ethidium bromide (10 μg/ml) for 15 min, and images were captured using an Alpha-Imager Imaging System with AlphaEase FC image software 4.1.0 (Alpha Innotech).
Each visible band on the DGGE gels containing the PAH-degrading consortia was excised manually, and the DNA was extracted using the method described by Muyzer et al. (1993). Using 5 μl of the extracted DNA as template, PCR was carried out to amplify target DNA for cloning. Positive PCR products were purified using the E.Z.N.A Cycle-Pure Kit (OMEGA Bio-tek, USA) and cloned into the pMD19-T Vector (Takara). After being confirmed using another DGGE gel, they were sequenced with an ABI model 3730 DNA sequencer (Invitrogen Shanghai).
Nucleotide Sequence Deposit
The 16S rRNA sequences of bacteria isolated from the nine consortia have been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST) under accession No. EU440952∼EU441002. The DGGE band sequences determined in this study have also been submitted to the GenBank database and have been assigned accession numbers EU441003∼EU441036.
Results
PAH Degradation by Deep Water Layers
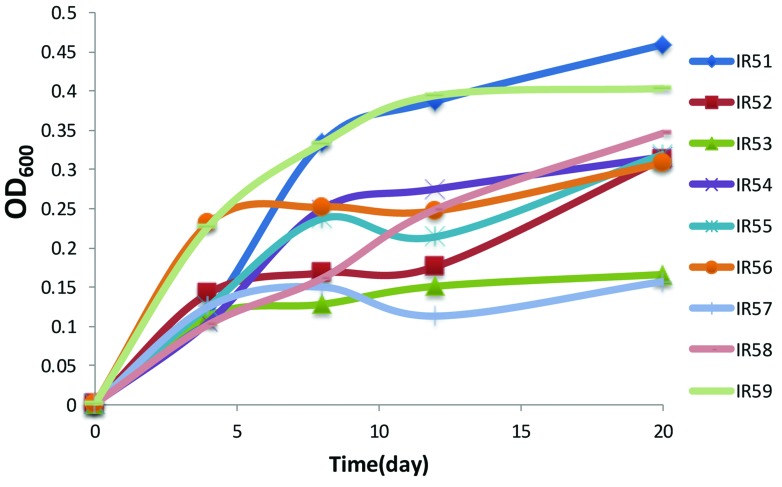
After the initial 2 months enrichment with a PAH mixture of phenanthrene, anthracene, fluoranthene, and pyrene as the sole carbon and energy sources, PAH degradation occurred in all treatment conditions of nine layers of water samples acquired from 3946 to 4746 m. Obvious bacterial growth was observed in all the enrichment cultures with the color changing to dark brown. In the lab, the initial enrichment cultures were transferred to fresh NH medium with PAHs as the sole carbon source (with 5% inoculum). The resulting final nine consortia were numbered IR51–IR59 (IR5, representing the sampling site IR-CTD-5; 1-9, the layer number based on the water depths listed above). The growth curves of all cultures during the second round of enrichment in the lab are shown in Figure 1. Of these cultures, layer 1 (IR51, 4746 m) showed the best growth with the highest cell concentration (OD600 = 0.459) at day 20, followed by layer 9 (IR59, 3946 m) (OD600 = 0.400) (Figure 1).
FIGURE 1.
The growth of the nine consortia on a polycyclic aromatic hydrocarbon (PAH) mixture of phenanthrene (100 mg/liter), fluoranthene (10 mg/liter), anthracene (10 mg/liter), and pyrene (10 mg/liter). Cell growth was determined by OD600 from 4 to 20 days. Incubation was at 28°C in the dark in a constant shaker.
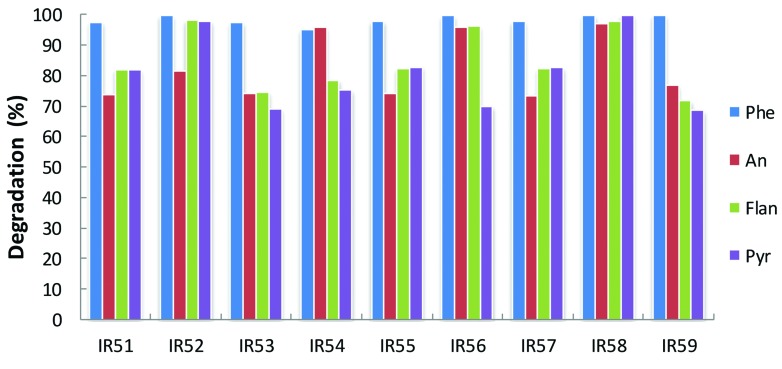
Meanwhile, PAH degradation was further confirmed by GC-MS quantification. As shown in Figure 2, phenanthrene was almost completely degraded in all cases after 30 days incubation, while anthracene, fluoranthene, and pyrene showed degradation ranging from 69 to 99%.
FIGURE 2.
Degradation of the 4-PAH mixture by nine consortia from the deep sea water column of the Indian Ocean. PAH degradation experiments were conducted in 100 ml Erlenmeyer flasks containing 30 ml NH medium with a mixture of 4-PAHs as carbon sources (using the same concentrations in Figure 1). The residual PAHs were quantified with GC-MS; the degradation percentage was the average of triplicate repeats at 30 days. Phe, An, Flan, and Pyr are abbreviations of phenanthrene, anthracene, fluoranthene, and pyrene, respectively.
Isolation of PAH-Degrading Bacteria
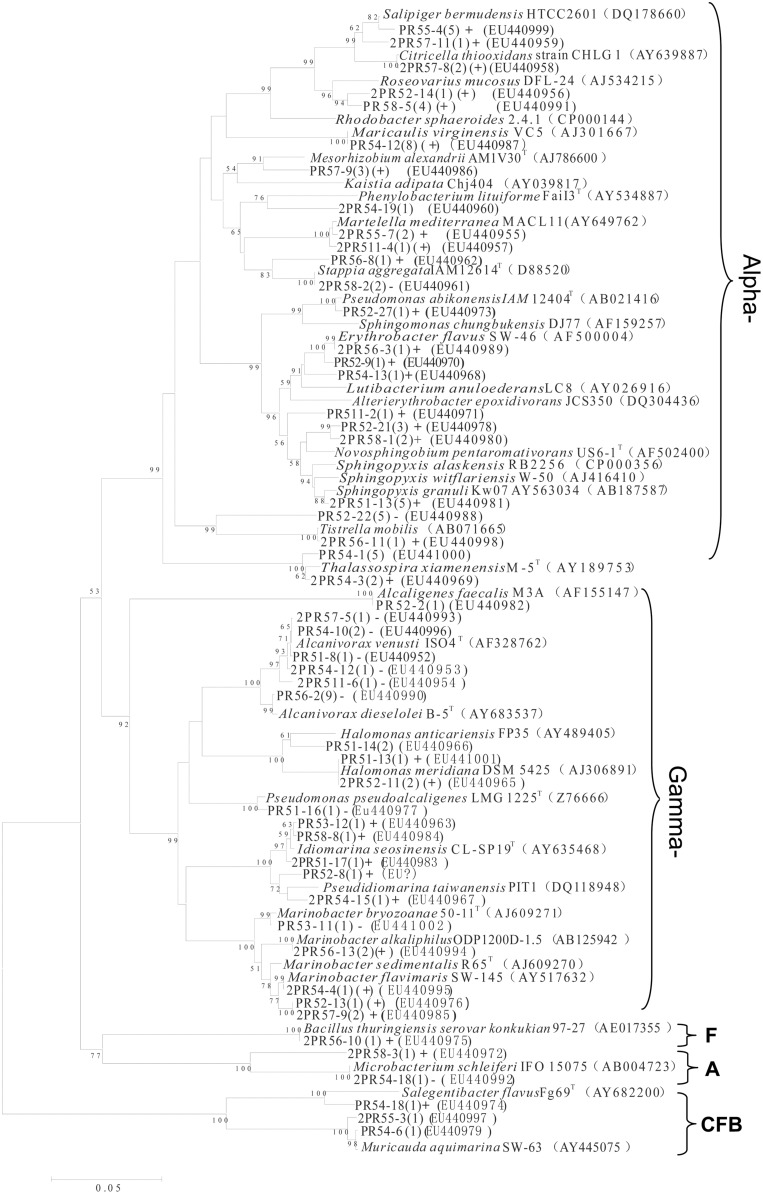
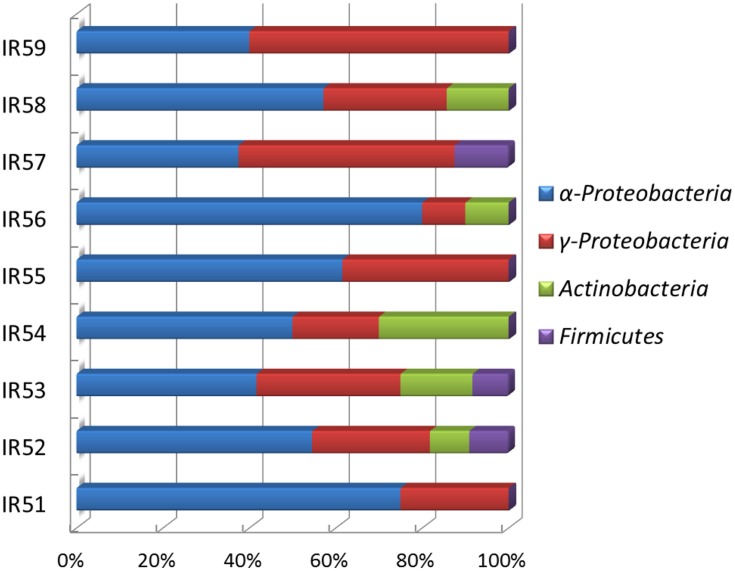
From the nine consortia, 238 isolates were purified. Of these, 168 strains showed a unique Rep-PCR pattern (not shown), 51 of which varied in their 16S rRNA gene sequences (Table 1). Phylogenetic analysis showed that they belonged to 29 genera. As shown in the phylogenetic tree, most belonged to Proteobacteria (88% of the total), in addition to Actinobacteria (4.0%), the CFB (Cytophaga-Flexibacter-Bacteroides) group (6%), and Firmicutes (2%). The largest subgroup was the Alphaproteobacteria, containing 18 genera of Erythrobacter, Thalassospira, Martelella, Novosphingobium, Pseudaminobacter, Tistrella, Salipiger, Sphingomonas, Sphingopyxis, and Stappia. The Gammaproteobacteria we detected contained Alcaligenes, Alcanivorax, Halomonas, Idiomarina, Marinobacter, Pseudidiomarina, and Pseudomonas. In addition, isolates closely related to Muricauda and Salegentibacte in CFB group, a bacterium belonging to the genus Microbacterium in the Actinobacteria group and an isolate of Bacillus were also found. Among these, 13 appear to be novel species, with similarities to the 16S rRNA sequences of known species ranging from 88.6 to 97.3%. In GenBank, the closest relatives of these 13 species belonged to Alterierythrobacter, Halomonas, Kaistia, Pseudaminobacter, Phenylobacterium, Pseudidiomarina, Roseovarius, Rhodobacter, Salegentibacte, Sphingomonas, Sphingopyxis, and Stappia (Figure 3).
Table 1.
The bacteria detected by Denaturing Gradient Gel Electrophoresis (DGGE) in PAHs- and Phe-degrading consortia.
| Closest relatives in GenBank∗ | Accession No. | Similarity (%) | Corresponding bands# |
Corresponding isolates | |
|---|---|---|---|---|---|
| IR51-59 | IR51P-59P | ||||
| Alcanivorax dieselolei B-5 | AY683537 | 99.47 | 1-2, 2-3, 3-3, 4-4, 5-5, 7-3, 9-3 | P1-3, P2-4, P4-1, P5-2, P6-3, P7-1, P8-3, P9-1 | / |
| A. dieselolei B-5 | AY683537 | 96.87 | / | P2-2 | / |
| A. dieselolei B-5 | AY683537 | 98.43 | / | P2-5, P6-4 | / |
| Alcanivorax sp. EPR 7 | AY394866 | 100.00 | 3-4, 8-2 | P8-4, P9-2 | / |
| A. venusti ISO4 | AF328762 | 98.09 | 9-5 | P5-5, P9-3 | 2PR511-6 |
| A. venusti ISO4 | AF328762 | 100.00 | 5-9 | / | PR51-8 |
| Arthrobacter sp. 255-8a | AY444852 | 90.68 | 3-7, 4-8 | P4-5 | / |
| Arthrobacter sp. 255-8a | AY444852 | 91.27 | 3-6, 4-7 | P4-4 | / |
| Arthrobacter sp. 255-8a | AY444852 | 96.46 | 8-4 | P8-5, P9-4 | / |
| Arthrobacter sp. ADG1 | AY651318 | 91.27 | 2-5, 6-7 | P1-5, P2-8, P5-6, P6-7 | / |
| Azospirillum sp. 5C | AF413109 | 95.21 | 1-8, 3-10, 4-10, 5-14 | / | / |
| Bacillus subtilis B-2009 | AM110937 | 99.40 | 2-8, 3-9, 7-8 | P3-9, P7-10 | / |
| Bartonella sp. AD273 | DQ113447 | 98.79 | 1-4, 6-8, 7-6, 8-5 | P6-8, P7-8, P8-6, P9-5 | / |
| Citricella thiooxidans CHLG 1 | AY639887 | 98.81 | 2-7, 3-8 | P1-6, P3-7, P4-6 | / |
| C. spirillensus M4-6 | AY026915 | 100.00 | 2-1, 3-1, 5-2 | P1-1, P2-1 | / |
| Erythrobacter sp.G265 | AY371411 | 99.26 | / | P3-3 | / |
| Erythrobacter sp.G265 | AY371411 | 100.00 | 6-4 | P6-2 | 2PR56-3 |
| Halomonas aquamarina DSM 30161 | AJ306888 | 100.00 | 9-6 | / | PR51-13 |
| Halomonas meridiana DSM 5425 | AJ306891 | 99.88 | 8-6 | / | 2PR52-11 |
| Halomonas sp. 3029 | AM110984 | 100.00 | 7-7 | P7-9, P8-7 | 2PR52-11 |
| Maricaulis virginensis VC5 | AJ301667 | 100.00 | 8-7 | / | PR54-12 |
| Marinobacter aquaeolei OC-11 | AY669171 | 99.19 | 2-2 | P2-3, P3-2 | / |
| Microbacterium schleiferi DSM 20489 | Y17237 | 100.00 | 4-9 | / | 2PR54-18 |
| Novosphingobium aromaticivorans DSM 12444 | CP000248 | 98.79 | 5-8, 6-5, 8-3 | P3-5 | / |
| N. aromaticivorans DSM 12444 | CP000248 | 100.00 | 9-4 | P5-4, P6-5, P7-5 | 2PR58-1 |
| N. aromaticivorans5-8B | DQ768707 | 100.00 | 4-2 | / | / |
| Novosphingobium hassiacum W-51 | AJ416411 | 95.81 | 2-4 | P1-4, P2-6 | / |
| Novosphingobium pentaromativorans US6-1 | AF502400 | 98.64 | 1-3 | / | / |
| Novosphingobium sp. MG36 | AJ746093 | 98.77 | 3-5, 4-6, 6-6 | P2-7, P4-3 | / |
| Novosphingobium sp. MG36 | AJ746093 | 100.00 | / | P6-6, P7-6 | PR52-21 |
| Porphyrobacter sp. J3-AN66 | DQ454121 | 98.79 | 7-1 | / | / |
| Pseudaminobacter salicylatoxidans BN12 | AF072542 | 95.60 | 1-5, 2-9, 3-11, 5-11 | P1-7, P3-10 | / |
| Pseudomonas sp. BJC3 | DQ834358 | 100.00 | 1-1 | P1-2 | / |
| Rhizobiales bacterium RR47 | AB174816 | 93.08 | 4-5, 5-7 | P3-4, P4-2, P5-3, P7-4 | / |
| Rhizobium undicola Liujia-81 | DQ648579 | 92.90 | 4-1, 5-1, 6-1, 8-1, 9-1 | P8-1 | / |
| Rhodospirillaceae bacterium PH30 | AF513476 | 99.30 | / | P3-8 | / |
| Rhodovulum sp. AR2002 | AJ401214 | 95.21 | 1-7, 2-11, 3-12, 5-13 | P1-8, P3-11 | / |
| Rhodovulum sp. AR2002 | AJ401214 | 96.75 | 1-6, 2-10, 5-12, 6-10, 9-7 | P5-8, P6-10, P9-6 | / |
| Rhodovulum sp. AR2002 | AJ401214 | 95.81 | 5-10 | P5-7 | / |
| Shewanella oneidensis 84F | AB182078 | 91.09 | 3-2, 4-3, 5-3, 6-3, 7-2 | P5-1, P6-1 | / |
| Sinorhizobium medicae M58 | DQ423248 | 100.00 | 6-9 | P6-9 | PR57-9 |
| Novosphingobium sp. FND-3 | DQ831000 | 99.40 | 5-4 | / | / |
| Sphingomonas sp. PXM | AY232825 | 98.41 | 7-4 | P7-3 | / |
| Sphingomonas sp. SAFR-042 | AY167827 | 99.30 | / | P3-6 | / |
| Sphingomonas sp.MD-1 | AB110635 | 98.82 | 6-2 | P7-2 | / |
| Thalassospira lucentensis DSM 14000 | AM294944 | 99.39 | 7-5 | P7-7 | PR54-1, 2PR54-3 |
| Zymomonas mobilis ATCC29191 | AF281034 | 97.00 | 2-6 | / | / |
∗Reference bacteria listed in order of alphabet. #The boldface bands in blue were the predominant member in their own consortium.
FIGURE 3.
Phylogenetic tree of culturable strains isolated from the nine PAH-degrading bacterial consortia. The dendrogram was constructed from a matrix by the neighbor-joining method using the MEGA3.1 program. Thousands of trial bootstrap analysis was used to provide confident estimates for phylogenetic tree topologies. The scale bar represented 0.05 substitutions per nucleotide site. The number in the bracket shows the frequency with which the bacterium appeared in the nine PAH-degrading consortia. “+” means the bacterium can grow well in the medium with PAHs as carbon sources. “(+)” means the bacterium can grow but not very well in the medium with PAHs as carbon sources. “-” means the bacterium cannot grow in the medium with PAHs as the carbon sources. Subgroups: Alpha-, Gammaproteobacteria; F, Firmicutes; A, Actinobacteria. Isolates named as PR and 2PR were isolated on agar plates of M2 agar and 216L, respectively.
In addition to phylogenic analysis, these 51 isolates were also tested for PAH-degradation. We found that 34 isolates could grow with PAHs as the sole carbon and energy sources (labeled with “+” in Figure 3). Among them, 20 bacteria belonged to Alphaproteobacteria. These 34 degraders contained some bacteria that have been previously reported, such as Bacillus (Hunter et al., 2005), Novosphingobium (Sohn et al., 2004), Lutibacterium (Chung and King, 2001), Sphingomonas (Bastiaens et al., 2000), and Marinobacter (Hedlund et al., 2001). However, many others had not been reported to be PAH degraders prior to this study. This latter group included Alterierythrobacter, Citricella, Erythrobacter, Idiomarina, Maricaulis, Martelella, Pseudidiomarina, Rhodobacter, Roseovarius, Salipiger, Sphingopyxis, and Stappia.
Bacterial Community Composition of the PAH-Degrading Consortia
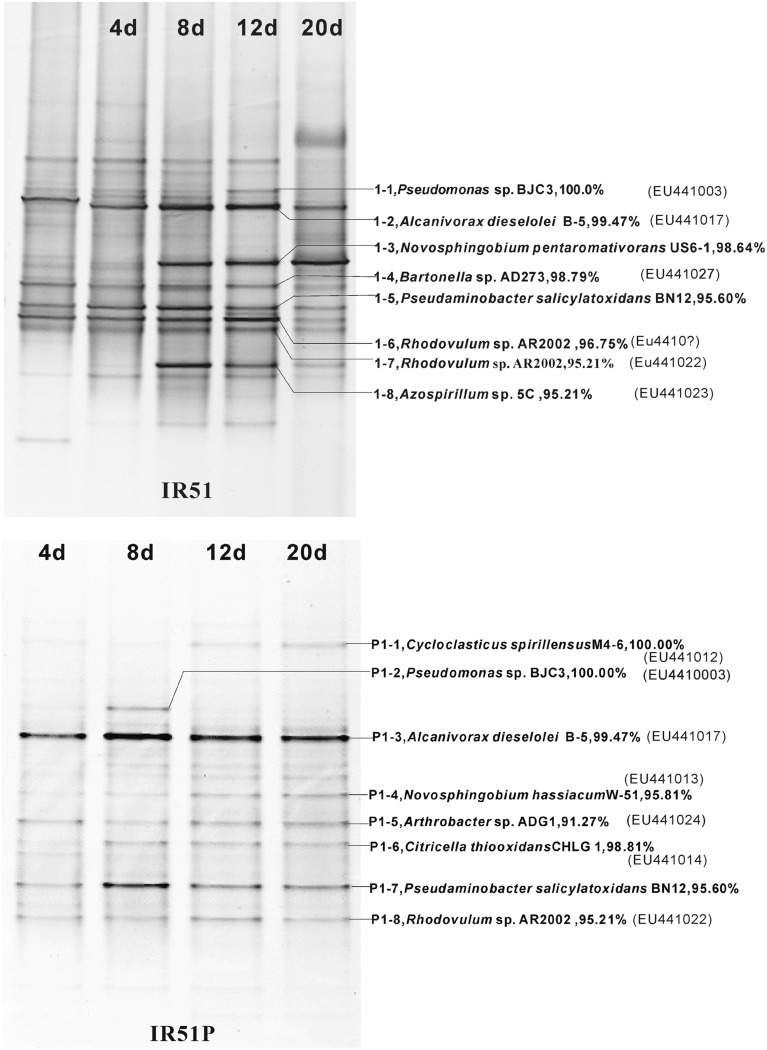
The bacterial composition of all consortia changed from the first day to the 20th day of the incubation process and also varied from layer to layer (Figure 4, Supplementary Figures S2 and S3). For an example, the consortium of IR51 that was generated from the first layer contained at least seven major bands (Band 2 to Band 8), as shown in the DGGE profile in Figure 4. These bands belonged to the genera Alcanivorax, Novosphingobium, Bartonella, Pseudaminobacter, Rhodovulum, and Azospirillum. Two bacteria, Novosphingobium pentaromativorans US6-1T (98.64%; Band1-3, with “1-” indicating the first layer) and Azospirillum sp. 5C (95.21%; Band1-8), became predominant at day 8. The bacterium of Band1-3 remained a predominant member throughout the time course, while the abundance of Band1-8 decreased after day 8. At day 20, Novosphingobium (Band1-3) turned out to be the most predominant member, indicating a key role in PAH degradation. In addition, Band1–2, closely related to Alcanivorax dieselolei B-5 (99.47%), was also an important member and was a major band from days 8 to 12 (Figure 4). This bacterium reoccurred as the most dominant member in the community with phenanthrene as the sole carbon and energy source (Figure 4); however, it failed to degrade any PAHs by itself.
FIGURE 4.
Community structures of the PAH-degrading consortia from 4746 m water depth (20 m to the sea floor). Numbers (4–20 days) above each lane indicate the number of days inoculated. The first lane is the initial inoculum of the last transfer. The closest described relatives in GenBank of the main bands in profiles are labeled at the right. (Top), IR51grown with 4-PAH mixture; (Bottom), IR51P grown with phenanthrene.
The community structures of the other water layers (IR52–IR59) were also examined (Supplementary Figures S2 and S3). Consistent with the results above, Alcanivorax and Novosphingobium were the most common bacteria in all the consortia. For an instance, an isolate closely related to A. dieselolei B-5 was a predominant member of almost all consortia. Bacteria of the genus Novosphingobium were also primary members of consortia from all depths and were closely related to Novosphingobium aromaticivorans, N. hassiacum, N. pentaromativorans, and Novosphingobium sp. MG36.
Interestingly, some novel species were detected as key members of the PAH-degrading consortia. For example, those with only 90.7–96.5% similarity to Arthrobacter sp. 255-8a (AY444852), as well as the bacteria with only 95.2–96.7% similarity to Rhodovulum sp. AR2002 and the bacterium with only 93.1% similarity to Rhizobiales bacterium RR47, occurred as a strong band in consortia IR53, 54, and 58. These bacteria have never been reported to be PAH degraders. Conversely, Cycloclasticus spirillensus (100.00%), the most famous PAH-degrading bacterium in marine environments (Head et al., 2006), occurred in consortia IR52, IR53, and IR55 (Supplementary Figures S2 and S3), but was not as important here as Novosphingobium.
Single PAH Degradation
To detect the degradation of a single PAH, phenanthrene, pyrene, anthracene, and fluoranthene were used separately as the sole carbon source. Unexpectedly, none of the nine consortia could grow with anthracene, fluoranthene, or pyrene. However, all consortia could grow with phenanthrene.
During the degradation of phenanthrene, the community structure was relatively stable and not as dynamic as communities with the PAH mixture. Additionally, the bacterial composition was notably simplified with phenanthrene alone. The DGGE profile of consortium IR51P is shown in Figure 4, and other consortia are displayed in Supplementary Figure S3 (IR52P–IR59P). Consistent with results in IR51 (Figure 4), the predominant band in the other consortia was A. dieselolei B-5 (99.47%; Band P1-3). However, another predominant member, N. pentaromativorans US6-1T (98.64%; IR51, Band1-3, Figure 4), was replaced with N. hassiacum (95.81%; IR51P, Band P1-4, Figure 4). In addition, C. spirillensus (100%; Band P1-1) was present. The bacterium of Pseudomonas sp. BJC3 (100.0%; Band P1-2) occurred with both phenanthrene alone and the consortia grown with the PAH mixture.
Some bacteria occurred only in the phenanthrene consortia, such as Erythrobacter sp.G265 (99.26%; Band P3-3), Sphingomonas sp. SAFR-042 (100%; P3-6) and Novosphingobium sp. MG36 (100%; P6-6) (Table 1). The most prevalent bacteria involved in phenanthrene degradation also belonged to Novosphingobium and Alcanivorax.
Discussion
Oil spilling into the Gulf of Mexico from the Deepwater Horizon blowout stimulated indigenous bacteria of the order Oceanospirillales in the Gammaproteobacteria, which are likely involved in alkane degradation in the deep-water column from 1099 to 1219 m (Hazen et al., 2010). However, bacteria involved in organic pollutant degradation in deep-sea environments are largely unknown, especially in the open sea (Deming, 1998). In deep-sea water columns of the open Mediterranean Sea, various types of PAHs have been found sinking with suspended particles (Bouloubassi et al., 2006). The source and fate of PAHs in deep-sea environments are poorly understood. One possible source is hydrothermal activity. A variety of organic compounds, including PAHs and long-chain hydrocarbons, were detected in the hydrothermal fluids (McCollom and Seewald, 2007). These hydrocarbons may serve as organic carbon sources that are otherwise scarce in the deep-sea environments but needed for heterotrophic bacterial growth.
Along the SWIR, hydrothermal vent fields were recently found (German et al., 1998; Tao et al., 2012). The microbial diversity of this region remains poorly characterized, and the bacterial diversity with regards to PAH degradation is unclear (Chen and Shao, 2009; Cao et al., 2014; Li et al., 2014). In this report, a variety of PAH-degrading bacteria were found in the deep water column on the Ridge at depths ∼4766 m. Among the discovered bacterial phylotypes, Proteobacteria dominated all the consortia, which was constituted of mainly Alphaproteobacteria and Gammaproteobacteria (Figures 3 and 5). Interestingly, the predominant PAH degraders obtained from the deep-sea water were significantly different from those in marine sediments. Cycloclasticus, thought to be the key PAH-degrading bacterium in marine environments, was not as important as previously reported (Dyksterhouse et al., 1995; Wang et al., 1996, 2008; Kasai et al., 2002; Yakimov et al., 2005; McKew et al., 2007; Teira et al., 2007; Cui et al., 2008). Instead, Novosphingobium seemed to be most important genus in this report. Bacteria of this genus were also previously detected as PAH degraders in the deep-sea hydrothermal environment of the Lau Basin of the West Pacific (Dong et al., 2011). They are closely related to those detected in this report, belonging to N. indicum, which we isolated here from sample CTD-5 (Yuan et al., 2008, 2009). This bacterium can degrade many types of aromatic hydrocarbons including biphenyl, naphthalene, acenaphthene, 2-methylnaphthalene, dibenzofuran, dibenzothiophene, 2,6-dimethylnaphthalene, 4- methyldibenzothiophene, phenanthrene, anthracene, chrysene, and fluoranthene (Yuan et al., 2008). However, contrary to expectations, only phenanthrene could be degraded alone, while the three other PAHs (anthracene, fluoranthene, and pyrene) could not be individually utilized by any of the consortia, as evidenced by the lack of clear bacterial growth within 1 month. We propose that phenanthrene metabolism induced the bacterial cells to utilize the three other PAHs more rapidly. This possibility is supported by a previous report, which found that pyrene degradation relies on the presence of phenanthrene in the case of the isolate Cycloclasticus sp. N3 (Geiselbrecht et al., 1998).
FIGURE 5.
Phylogenetic distributions of bacteria detected by DGGE in nine PAH-degrading consortia.
By comparing the bacterial structure of these consortia, a trend in bacterial distribution from 20 to 820 m above the bottom can be observed. Consortia from neighboring layers were similar in bacterial structure. For example, in phenanthrene-enriched consortia, the community structures of the upper two layers were similar to each other but different from those of the other layers (Supplementary Figure S3). Some bacteria occurred only in specific consortia. For instance, the bacterium closely related to N. aromaticivorans DSM 12444 (100%) occurred from 470 to 620 m above the bottom as bands P5-4, P6-5 and P7-5, but was not found in other layers (Table 1). The bacterium similar to Azospirillum sp. 5C (95.21%) occurred below 470 m from the bottom as bands 1–8, 3–10, 4–10, and 5–14, as did the bacterium similar to Rhodovulum sp. AR2002 (95.21%), which frequently occurred as bands 1–7, 2–11, 3–12, 5–13, P1–8 and P3–11 (Table 1). However, the strain that displayed 96.75% similarity with Rhodovulum sp. AR2002 was detected in a wider range of 20–820 m above the bottom, presenting as bands 1–6, 2–10, 5–12, 9–7, 6–10, P6–10, and P9–6 (Table 1). Additionally, the bacterium similar to N. hassiacum W-51 (95.81%) only occurred in the first and second layers above the bottom as bands 2–4, P1–4, and P2–6, while the bacterium similar to N. pentaromativorans US6-1 (98.64%) only occurred in the bottom layer as a key member of the PAH mixture consortia. However, the bacterium similar to A. dieselolei B-5 (99.47%) frequently occurred in all layers, and the bacterium similar to Bartonella sp. AD273 (98.79%) occurred in many consortia regardless of layer. Among these bacteria, Alcanivorax, and Novosphingobium have been reported in deep-sea sediments involved in PAH degradation (Cui et al., 2008; Wang et al., 2008; Cui and Shao, 2009). Recently, we found that PAH-degrading bacteria, including Cycloclasticus, Pseudomonas, Pseudoalteromonas, Halomonas, Marinomonas, and Dietzia, were widespread in the deep-sea sediments of the Arctic Ocean (Dong et al., 2015). In addition, Alcanivorax frequently occurred in the Arctic PAH-degrading consortia, while bacteria Novosphingobium did not.
Among the bacteria detected in this report, the bacterium similar to A. dieselolei B-5 (99.47%) was a key member of all consortia. The predominance of A. dieselolei was further confirmed by 16S rRNA gene library construction of two communities, IR58 and IR59 (data not shown). From the library, we obtained a nearly full length 16S rRNA, which showed 99.8% (1502/1505) similarity with the type strain A. dieselolei B-5. Moreover, Alcanivorax bacteria appeared in both the PAH mixture and the phenanthrene-degrading consortia. It has been demonstrated that A. dieselolei is an obligate alkane-degrading bacterium (Liu and Shao, 2005; Wang et al., 2010). However, bacteria of this genus have never been shown to be PAH degraders. Iwabuchi et al. (2002) suggested that Alcanivorax may enhance PAH degradation by utilizing the alkyl side-chains on methylated PAHs within crude oil (Iwabuchi et al., 2002). However, in this study, such alkyl side-chain-substituted PAHs were not used. Why Alcanivorax coexist with other bacteria in these communities remains an intriguing question.
Some bacteria grew better with the PAH mixture, while others preferred phenanthrene alone (Table 1). For example, the bacterium similar to Zymomonas mobilis (97.00%; Band2-6), the bacterium similar to Halomonas aquamarina DSM 30161 (100; Band9-6), the bacterium similar to Microbacterium schleiferi DSM 20489(100%; Band4–9), the bacterium similar to Azospirillum sp. 5C (95.21%), and the bacterium similar to N. pentaromativorans US6-1 (98.64%; Band1-3) occurred as major bands only in the PAH mixture. However, some were found only in the phenanthrene treatments, such as the bacterium similar to Erythrobacter sp.G265 (99.26%; Band P3–3), the bacterium similar to Novosphingobium sp. MG36 (100%; Band P6–6) and the bacterium similar to Sphingomonas sp. SAFR-042 (100%; Band P3-6) (Table 1). Recently, some of these bacteria have been reported to be PAH degraders in marine environments. For example, an isolate of Microbacterium from mangrove sediments (Wongwongsee et al., 2013) and Erythrobacter strain N3, isolated from south Atlantic deep-sea sediment (unpublished data from our group), can utilize phenanthrene to grow. A bacterium of Sphingomonas has also been reported in both soils and marine sediment involved PAH degradation (Nadalig et al., 2002; Uyttebroek et al., 2007). However, bacteria of Zymomonas and Azospirillum have not been confirmed yet in PAH degradation.
Conversely, some bacteria in this study occurred as major bands in both types of enrichment, including the bacteria similar to N. aromaticivorans (98.79, 100%), Novosphingobium sp. MG36 (98.77%), Alcanivorax sp. EPR 7 and Alcanivorax dieselolei B-5 (99.47%), and Arthrobacter sp. ADG1 (91.27%). Others occurred in both treatments but only appeared as a major band in the PAH mixture. These included the bacteria similar to Arthrobacter sp. 255-8a (96.46%), Thalassospira lucentensis (99.39%), Novosphingobium hassiacum W-51 (95.81%), Rhodovulum sp. AR2002 (95.21 and 96.75%), S. oneidensis (91.09%), and C. spirillensus (100%). Bacteria of different Thalassospira species have been previously detected, and then later confirmed, in PAH-degrading consortia (Liu et al., 2007; Kodama et al., 2008; Zhao et al., 2010). Many of these bacteria have not been successfully isolated on agar plates, such as Cycloclasticus spirillensus, and potentially novel species such as Arthrobacter, Azospirillum, and the Rhizobiales and Rhodovulum with 16S rRNA sequences 90–97% similar to related strains. Azospirillum, Porphyrobacter, Shewanella, and Bartonella also fall into this category. More efforts toward pure culture cultivation are needed to confirm their role in PAH degradation.
Among the PAHs used, phenanthrene is probably the easiest to degrade, as the resulted PAH-degrading consortia failed to show obvious bacterial growth when using pyrene, anthracene, or fluoranthene as the sole carbon source. However, when used in form a mixture, all of them can de degraded, suggesting a cometabolism between phenanthrene and other PAHs. This phenomenon has been found in other reports (Geiselbrecht et al., 1998; Kanaly and Harayama, 2000). In the case of pure cultures, 34 of the isolates have been confirmed to grow with the PAH mixture. By now, their degradation ability on each kind of PAHs remains undetermined except one novel species of N. indicum mentioned above (Yuan et al., 2008, 2009).
In respect of temperatures and hydrostatic pressures in situ that might select bacteria different to the above described, obligate psychrophilic and barophilic microbes involved in PAH degradation probably exist in the deep sea water column. Bacterial isolates in this report ever tested showed temperature growth ranges from 10 to 41°C mostly, with optimal growth temperatures of 20 to 25°C, such as Alcanivorax marinus, N. indicum, Oceanibaculum indicum, and Stappia indica et al., which are the novel species isolated from the deep sea water of SWIR (Lai et al., 2009a,b, 2010, 2011a,b, 2013; Yuan et al., 2009). They did not show an obvious growth under 4°C within 1 week, which is higher than the temperature (about 1°C) in situ. Probably, they actually grow slightly under temperature at 4°C and below, or need long time incubation. Anyhow, we believe that microbes in situ would possess some taxa that are psychrophilic and barophilic, but remain uncultivated by now.
In summary, this report describes the discovery of diverse PAHs-degrading bacteria in the deep-sea water column of the SWIR at a depth of 4766 m. Bacteria affiliated with Alcanivorax, Novosphingobium, and Rhodovulum frequently occurred in different PAH-degrading consortia, and 34 isolates were confirmed to grow with our PAH mixture. These included novel PAH-degrading bacteria belonging to Alterierythrobacter, Citricella, Erythrobacter, Idiomarina, Lutibacterium, Maricaulis, Marinobacter, Martelella, Pseudidiomarina, Rhodobacter, Roseovarius, Salipiger, Sphingopyxis, and Stappia. These bacteria are thought to play a role in PAH degradation in deep-sea water. More efforts are needed to examine the diversity of extremophiles in situ of deep sea that involved in PAH degradation.
Author Contributions
ZS: conceived and designed the project, oceanic sampling and on board enrichment. JY, QL: bacterial isolation, identification, DGGE and GS-MS analysis. FS: bacteria management. ZS, YJ, and TZ: writing the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the scientists cruise DY105-17A of the “Da-Yang Yi-Hao”. Thanks to Professor I. J. Hodgkiss from University of Hong Kong for his assistance with English. This report was supported by the China Ocean Mineral Resources R&D Association Program (COMRA No. DY125-15-R-01; DYXM115-02-2-05), Xiamen Marine Economy Development Project (14CZP034HJ08), and the National Infrastructure of Microbial Resources of China (NIMR-2014-9; NIMR-2015-9).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00853
Sampling site IR-CTD5 (The map based on NOAA topography). The solid red circles are hydrothermal vent fields previously detected. IR-CTD5 was in the B-B’ section, where plume 2 was found by anomalies (German et al., 1998). Edmond and Kairei are confirmed vent sites.
Community structures of the PAH-degrading consortia growing with 4-PAH mixture in 20 days. Derived from water samples of IR5-2 to IR5-9 after PAH enrichment, with water depths of 4696, 4546, 4396, 4296, 4196, 4146, 4096 and 3946 m, and 40–820 m above the sea floor.
Community structures of the PAH-degrading consortia growing with phenanthrene in 20 days. Treatments were in parallel with those in Figure S2, except using phenanthrene as a sole carbon source.
References
- Bastiaens L., Springael D., Wattiau P., Harms H., deWachter R., Verachtert H.et al. (2000). Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Appl. Environ. Microbiol. 66 1834–1843. 10.1128/AEM.66.5.1834-1843.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloubassi I., Mejanelle L., Pete R., Fillaux J., Lorre A., Point V. (2006). PAH transport by sinking particles in the open Mediterranean Sea: a 1 year sediment trap study. Mar. Pollut. Bull. 52 560–571. 10.1016/j.marpolbul.2005.10.003 [DOI] [PubMed] [Google Scholar]
- Cao H., Wang Y., Lee O. O., Zeng X., Shao Z., Qian P. Y. (2014). Microbial sulfur cycle in two hydrothermal chimneys on the Southwest Indian Ridge. MBio 5:e00980-13. 10.1128/mBio.00980-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Shao Z. (2009). Isolation and diversity analysis of arsenite-resistant bacteria in communities enriched from deep-sea sediments of the Southwest Indian Ocean Ridge. Extremophiles 13 39–48. 10.1007/s00792-008-0195-1 [DOI] [PubMed] [Google Scholar]
- Chung W. K., King G. M. (2001). Isolation, characterization, and polyaromatic hydrocarbon degradation potential of aerobic bacteria from marine macrofaunal burrow sediments and description of Lutibacterium anuloederans gen. nov., sp. nov., and Cycloclasticus spirillensus sp. nov. Appl. Environ. Microbiol. 67 5585–5592. 10.1128/AEM.67.12.5585-5592.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Lai Q., Dong C., Shao Z. (2008). Biodiversity of polycyclic aromatic hydrocarbon-degrading bacteria from deep sea sediments of the Middle Atlantic Ridge. Environ. Microbiol. 10 2138–2149. 10.1111/j.1462-2920.2008.01637.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Shao Z. (2009). [Predominant strains of polycyclic aromatic hydrocarbon-degrading consortia from deep sea of the Middle Atlantic Ridge]. Wei Sheng Wu Xue Bao 49 902–909. [PubMed] [Google Scholar]
- Deming J. W. (1998). Deep ocean environmental biotechnology. Curr. Opin. Biotechnol 9 283–287. 10.1016/S0958-1669(98)80060-8 [DOI] [PubMed] [Google Scholar]
- Dong C. M., Bai X. H., Sheng H. F., Jiao L. P., Zhou H. W., Shao Z. Z. (2015). Distribution of PAHs and the PAH-degrading bacteria in the deep-sea sediments of the high-latitude Arctic Ocean. Biogeosciences 12 2163–2177. 10.5194/bg-12-2163-2015 [DOI] [Google Scholar]
- Dong C., Chen L., Liao Y., Shao Z. (2011). [Phylogenetic and degrading genes analysis of a PAH-degrading bacterium TVG9-VII from deep-sea hydrythermal environment]. Wei Sheng Wu Xue Bao 51 1548–1554. [PubMed] [Google Scholar]
- Dyksterhouse S. E., Gray J. P., Herwig R. P., Lara J. C., Staley J. T. (1995). Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int. J. Syst. Bacteriol. 45 116–123. 10.1099/00207713-45-1-116 [DOI] [PubMed] [Google Scholar]
- Geiselbrecht A. D., Hedlund B. P., Tichi M. A., Staley J. T. (1998). Isolation of marine polycyclic aromatic hydrocarbon (PAH)-degrading Cycloclasticus strains from the Gulf of Mexico and comparison of their PAH degradation ability with that of puget sound Cycloclasticus strains. Appl. Environ. Microbiol. 64 4703–4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German C. R., Baker E. T., Mevel C., Tamaki K. The FUJI Science Team. (1998). Hydrothermal activity along the southwest Indian ridge. Nature 395 490–493. 10.1038/26730 [DOI] [Google Scholar]
- Guo C. L., Zhou H. W., Wong Y. S., Tam N. F. Y. (2005). Isolation of PAH-degrading bacteria from mangrove sediments and their biodegradation potential. Mar. Pollut. Bull. 51 1054–1061. 10.1016/j.marpolbul.2005.02.012 [DOI] [PubMed] [Google Scholar]
- Harayama S., Kasai Y., Hara A. (2004). Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 15 205–214. 10.1016/j.copbio.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Hazen T. C., Dubinsky E. A., DeSantis T. Z., Andersen G. L., Piceno Y. M., Singh N.et al. (2010). Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330 204–208. 10.1126/science.1195979 [DOI] [PubMed] [Google Scholar]
- Head I. M., Jones D. M., Roling W. F. M. (2006). Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4 173–182. 10.1038/nrmicro1348 [DOI] [PubMed] [Google Scholar]
- Hedlund B. P., Geiselbrecht A. D., Staley J. T. (2001). Marinobacter strain NCE312 has a Pseudomonas-like naphthalene dioxygenase. FEMS Microbiol. Lett. 201 47–51. 10.1111/j.1574-6968.2001.tb10731.x [DOI] [PubMed] [Google Scholar]
- Huang X., Zeng Z., Chen S., Yin X., Wang X., Ma Y.et al. (2014). Component characteristics of polycyclic aromatic compounds in sediments of the South Mid-Atlantic Ridge. Acta Oceanol. Sinica 33 150–154. 10.1007/s13131-014-0564-9 [DOI] [Google Scholar]
- Hunter R. D., Ekunwe S. I., Dodor D. E., Hwang H. M., Ekunwe L. (2005). Bacillus subtilis is a potential degrader of pyrene and benzo[a]pyrene. Int. J. Environ. Res. Public Health 2 267–271. 10.3390/ijerph2005020010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi N., Sunairi M., Urai M., Itoh C., Anzai H., Nakajima M.et al. (2002). Extracellular polysaccharides of Rhodococcus rhodochrous S-2 stimulate the degradation of aromatic components in crude oil by indigenous marine bacteria. Appl. Environ. Microbiol. 68 2337–2343. 10.1128/AEM.68.5.2337-2343.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaly R. A., Harayama S. (2000). Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182 2059–2067. 10.1128/JB.182.8.2059-2067.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappell A. D., Wei Y., Newton R. J., Van Nostrand J. D., Zhou J., McLellan S. L.et al. (2014). The polycyclic aromatic hydrocarbon degradation potential of Gulf of Mexico native coastal microbial communities after the Deepwater Horizon oil spill. Front. Microbiol. 5:205 10.3389/fmicb.2014.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai Y., Kishira H., Harayama S. (2002). Bacteria belonging to the genus Cycloclasticus play a primary role in the degradation of aromatic hydrocarbons released in a marine environment. Appl. Environ. Microbiol. 68 5625–5633. 10.1128/AEM.68.11.5625-5633.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama Y., Stiknowati L. I., Ueki A., Ueki K., Watanabe K. (2008). Thalassospira tepidiphila sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 58 711–715. 10.1099/ijs.0.65476-0 [DOI] [PubMed] [Google Scholar]
- Lai Q., Qiao N., Wu C., Sun F., Yuan J., Shao Z. (2010). Stappia indica sp. nov., isolated from deep seawater of the Indian Ocean. Int. J. Syst. Evol. Microbiol. 60 733–736. 10.1099/ijs.0.013417-0 [DOI] [PubMed] [Google Scholar]
- Lai Q., Wang J., Gu L., Zheng T., Shao Z. (2013). Alcanivorax marinus sp. nov., isolated from deep sea water of Indian Ocean. Int. J. Syst. Evol. Microbiol. 3(Pt 12), 4428–4432. 10.1099/ijs.0.049957-0 [DOI] [PubMed] [Google Scholar]
- Lai Q., Wang L., Liu Y., Yuan J., Sun F., Shao Z. (2011a). Parvibaculum indicum sp. nov., isolated from deep-sea water. Int. J. Syst. Evol. Microbiol. 61 271–274. 10.1099/ijs.0.021899-0 [DOI] [PubMed] [Google Scholar]
- Lai Q., Yu Z., Yuan J., Sun F., Shao Z. (2011b). Nitratireductor indicus sp. nov., isolated from deep-sea water. Int. J. Syst. Evol. Microbiol. 61 295–298. 10.1099/ijs.0.022962-0 [DOI] [PubMed] [Google Scholar]
- Lai Q., Yuan J., Gu L., Shao Z. (2009a). Marispirillum indicum gen. nov., sp. nov., isolated from a deep-sea environment. Int. J. Syst. Evol. Microbiol. 59 1278–1281. 10.1099/ijs.0.003889-0 [DOI] [PubMed] [Google Scholar]
- Lai Q., Yuan J., Wu C., Shao Z. (2009b). Oceanibaculum indicum gen. nov., sp. nov., isolated from deep seawater of the Indian Ocean. Int. J. Syst. Evol. Microbiol. 59 1733–1737. 10.1099/ijs.0.004341-0 [DOI] [PubMed] [Google Scholar]
- Lamendella R., Strutt S., Borglin S., Chakraborty R., Tas N., Mason O. U.et al. (2014). Assessment of the Deepwater Horizon oil spill impact on Gulf coast microbial communities. Front. Microbiol. 5:130 10.3389/fmicb.2014.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Peng X., Zhou H., Sun Z., Chen S. (2014). Microbial communities in semi-consolidated carbonate sediments of the Southwest Indian Ridge. J. Microbiol. 52 111–119. 10.1007/s12275-014-3133-1 [DOI] [PubMed] [Google Scholar]
- Liu C., Shao Z. (2005). Alcanivorax dieselolei sp. nov., a novel alkane-degrading bacterium isolated from sea water and deep-sea sediment. Int. J. Syst. Evol. Microbiol. 55 1181–1186. 10.1099/ijs.0.63443-0 [DOI] [PubMed] [Google Scholar]
- Liu C., Wu Y., Li L., Ma Y., Shao Z. (2007). Thalassospira xiamenensis sp. nov. and Thalassospira profundimaris sp. nov. Int. J. Syst. Evol. Microbiol. 57 316–320. 10.1099/ijs.0.64544-0 [DOI] [PubMed] [Google Scholar]
- Ma Y., Wang L., Shao Z. (2006). Pseudomonas, the dominant polycyclic aromatic hydrocarbon-degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ. Microbiol. 8 455–465. 10.1111/j.1462-2920.2005.00911.x [DOI] [PubMed] [Google Scholar]
- McCollom T. M., Seewald J. S. (2007). Abiotic synthesis of organic compounds in deep-sea hydrothermal environments. Chem. Rev. 107 382–401. 10.1021/cr0503660 [DOI] [PubMed] [Google Scholar]
- McKew B. A., Coulon F., Osborn A. M., Timmis K. N., McGenity T. J. (2007). Determining the identity and roles of oil-metabolizing marine bacteria from the Thames estuary, UK. Environ. Microbiol. 9 165–176. 10.1111/j.1462-2920.2006.01125.x [DOI] [PubMed] [Google Scholar]
- Muyzer G., de Waal E. C., Uitterlinden A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadalig T., Raymond N., Ni’matuzahroh, Gilewicz M., Budzinski H., Bertrand J. C. (2002). Degradation of phenanthrene, methylphenanthrenes and dibenzothiophene by a Sphingomonas strain 2mpII. Appl. Microbiol. Biotechnol. 59 79–85. 10.1007/s00253-002-0960-5 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, 2nd Edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Sohn J. H., Kwon K. K., Kang J. H., Jung H. B., Kim S. J. (2004). Novosphingobium pentaromativorans sp. nov., a high-molecular-mass polycyclic aromatic hydrocarbon-degrading bacterium isolated from estuarine sediment. Int. J. Syst. Evol. Microbiol. 54 1483–1487. 10.1099/ijs.0.02945-0 [DOI] [PubMed] [Google Scholar]
- Tao C., Lin J., Guo S., Chen Y. J., Wu G., Han X.et al. (2012). First active hydrothermal vents on an ultraslow-spreading center: Southwest Indian Ridge. Geology 40 47–50. 10.1130/G32389.1 [DOI] [Google Scholar]
- Teira E., Lekunberri I., Gasol J. M., Nieto-Cid M., Alvarez-Salgado X. A., Figueiras F. G. (2007). Dynamics of the hydrocarbon-degrading Cycloclasticus bacteria during mesocosm-simulated oil spills. Environ. Microbiol. 9 2551–2562. 10.1111/j.1462-2920.2007.01373.x [DOI] [PubMed] [Google Scholar]
- Uyttebroek M., Spoden A., Ortega-Calvo J. J., Wouters K., Wattiau P., Bastiaens L.et al. (2007). Differential responses of eubacterial, Mycobacterium, and Sphingomonas communities in polycyclic aromatic hydrocarbon (PAH)-contaminated soil to artificially induced changes in PAH profile. J. Environ. Qual. 36 1403–1411. 10.2134/jeq2006.0471 [DOI] [PubMed] [Google Scholar]
- Wang B., Lai Q., Cui Z., Tan T., Shao Z. (2008). A pyrene-degrading consortium from deep-sea sediment of the West Pacific and its key member Cycloclasticus sp. P1. Environ. Microbiol. 10 1948–1963. 10.1111/j.1462-2920.2008.01611.x [DOI] [PubMed] [Google Scholar]
- Wang L., Wang W., Lai Q., Shao Z. (2010). Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean. Environ. Microbiol. 12 1230–1242. 10.1111/j.1462-2920.2010.02165.x [DOI] [PubMed] [Google Scholar]
- Wang Y., Lau P. C., Button D. K. (1996). A marine oligobacterium harboring genes known to be part of aromatic hydrocarbon degradation pathways of soil pseudomonads. Appl. Environ. Microbiol. 62 2169–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongwongsee W., Chareanpat P., Pinyakong O. (2013). Abilities and genes for PAH biodegradation of bacteria isolated from mangrove sediments from the central of Thailand. Mar. Pollut. Bull. 74 95–104. 10.1016/j.marpolbul.2013.07.025 [DOI] [PubMed] [Google Scholar]
- Yakimov M. M., Denaro R., Genovese M., Cappello S., D’Auria G., Chernikova T. N.et al. (2005). Natural microbial diversity in superficial sediments of Milazzo Harbor (Sicily) and community successions during microcosm enrichment with various hydrocarbons. Environ. Microbiol. 7 1426–1441. 10.1111/j.1462-5822.2005.00829.x [DOI] [PubMed] [Google Scholar]
- Yakimov M. M., Timmis K. N., Golyshin P. N. (2007). Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 18 257–266. 10.1016/j.copbio.2007.04.006 [DOI] [PubMed] [Google Scholar]
- Yuan J., Lai Q., Zheng T., Shao Z. (2008). [Polycyclic aromatic hydrocarbon-degrading bacterium Novosphingobium sp. H25 isolated from deep sea and its degrading genes]. Wei Sheng Wu Xue Bao 48 1208–1213. [PubMed] [Google Scholar]
- Yuan J., Lai Q., Zheng T., Shao Z. (2009). Novosphingobium indicum sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from a deep-sea environment. Int. J. Syst. Evol. Microbiol. 59 2084–2088. 10.1099/ijs.0.002873-0 [DOI] [PubMed] [Google Scholar]
- Zhao B., Wang H., Li R., Mao X. (2010). Thalassospira xianhensis sp. nov., a polycyclic aromatic hydrocarbon-degrading marine bacterium. Int. J. Syst. Evol. Microbiol. 60 1125–1129. 10.1099/ijs.0.013201-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sampling site IR-CTD5 (The map based on NOAA topography). The solid red circles are hydrothermal vent fields previously detected. IR-CTD5 was in the B-B’ section, where plume 2 was found by anomalies (German et al., 1998). Edmond and Kairei are confirmed vent sites.
Community structures of the PAH-degrading consortia growing with 4-PAH mixture in 20 days. Derived from water samples of IR5-2 to IR5-9 after PAH enrichment, with water depths of 4696, 4546, 4396, 4296, 4196, 4146, 4096 and 3946 m, and 40–820 m above the sea floor.
Community structures of the PAH-degrading consortia growing with phenanthrene in 20 days. Treatments were in parallel with those in Figure S2, except using phenanthrene as a sole carbon source.