Abstract
Background: Personalization of cancer therapy requires molecular evaluation of tumor tissue. Traditional tissue preservation involves formalin fixation, which degrades the quality of nucleic acids. Strategies to bank frozen prostate tissue can interfere with diagnostic studies. PAXgene is an alternative fixative that preserves protein and nucleic acid quality. Methods: Portions of prostates obtained from autopsy specimens were fixed in either 10% buffered formalin or PAXgene, and processed and embedded in paraffin. Additional sections were immediately embedded in OCT and frozen. DNA and RNA were extracted from the formalin-fixed, PAXgene-fixed, or frozen tissue. Quantitative PCR was used to compare the quality of DNA and RNA obtained from all three tissue types. In addition, 5 μm sections were cut from specimens devoid of cancer and from prostate cancer specimens obtained at prostatectomy and fixed in PAXgene. They were either stained with hematoxylin and eosin or interrogated with antibodies for p63, PSA and p504. Results: Comparable tissue morphology was observed in both the formalin and PAXgene-fixed specimens. Similarly, immunohistochemical expression of the P63, PSA and P504 proteins was comparable between formalin and PAXgene fixation techniques. DNA from the PAXgene-fixed tissue was of similar quality to that from frozen tissue. RNA was also amplified with up to 8-fold greater efficiency in the PAXgene fixed tissue compared to the formalin-fixed tissue. Conclusions: Prostate specimens fixed with PAXgene have preserved histologic morphology, stain appropriately, and have preserved quality of nucleic acids. PAXgene fixation facilitates the use of prostatectomy tissue for molecular biology techniques such as next-generation sequencing.
Keywords: Prostate cancer, PAXgene, formalin, fixation, next-generation sequencing, immunohistochemistry
Introduction
Prostate cancer (PCa) is the most common malignancy in many Western countries, affecting over 230,000 men each year in the U.S. alone, and it is the second leading cause of cancer death among American men [1]. Localized PCa is a clinically heterogeneous disease with significant variation in patient outcome. Determining the most beneficial management strategy requires risk stratification based on clinical factors and diagnostic/prognostic biomarkers [2]. Though advanced PCa is also clinically heterogeneous, most men are treated with the same therapies. Biomarker-driven therapies are just now beginning to enter into clinical trials, and none have entered into routine clinical practice.
It is essential to identify better predictors that could guide therapeutic actions and help reach the target of personalized medicine in PCa. In this challenging context, next-generation sequencing (NGS) technologies constitute powerful tools to identify potential prognostic and predictive biomarkers based on each patient’s mutational landscape and expression signature. The recent rise of sequencing of PCa genomes has significantly increased our understanding of the molecular basis of the disease [3-6]. Efforts are being made towards translating these new findings into clinical care [7,8].
The development of biomarkers through NGS or other methods requires access to high quality specimens. In the case of PCa, this resource is particularly difficult to acquire. PCa is rarely visible on gross examination due to its small size or the intermingling of benign and malignant glands, making sampling difficult and often poorly representative of the tumor. Moreover, standard prostatectomy specimen processing requires that the gland should be fully fixed in formaldehyde solution before sectioning to guarantee the accurate assessment of surgical margins [9]. Others have proposed methods to freeze a larger fraction of the specimen, but these methods compromise the “capsule” [10]. Formalin fixation causes the formation of chemical cross links of nucleic acids and proteins, thus resulting in degraded, fragmented DNA and RNA [11]. The current study sought to evaluate an alternative, non-crosslinking fixation reagent for prostate specimens. We demonstrate that fixation of prostates with PAXgene results in preservation of high quality DNA and RNA with preserved tissue architecture and staining characteristics.
Materials and methods
Tissue preparation
Four random prostates devoid of PCa were obtained at autopsy and prepared by the Human Tissue Resource Center (HTRC) at the University of Chicago. Before fixation, a punch biopsy was immediately embedded in OCT compound and frozen. The remainder of each specimen was bisected. Half was fixed in 10% buffered formalin for 24 hours using standard fixation techniques and embedded in paraffin. The other half was placed in PAXgene Tissue FIX, 150 mL, or adjusted to ~4 mL per gram of tissue, for either 24 or 48 hours (Figure 1). Specimens were then incubated in ethanol-diluted PAXgene Tissue STABILIZER, 150 mL, or adjusted to ~4 mL per gram of tissue, for at least three hours. PAXgene-fixed specimens were embedded in paraffin using standard ethanol-based techniques, avoiding all exposure to formalin.
Figure 1.
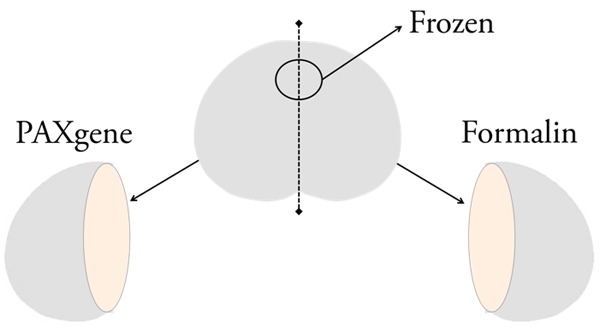
Autopsy prostate processing. Mirrored sections from four prostate specimens from autopsies were fixed either in PAXgene or formalin. Two specimens were fixed in PAXgene for 24 hours, and two specimens were fixed in PAXgene for 48 hours. For each prostate a frozen sample was taken before fixation.
Similarly, as part of an Internal Review Board-approved protocol, specimens with PCa from four patients undergoing radical prostatectomy were fixed in PAXgene Tissue FIX for 48 hours and in PAXgene Tissue STABILIZER for at least three hours. They were then embedded in paraffin using standard ethanol-based techniques.
Hematoxylin and eosin (H&E) staining and immunohistochemistry (IHC)
Five μm sections from PAXgene-fixed paraffin embedded (PAXPE) and formalin-fixed paraffin embedded (FFPE) sample blocks were stained with hematoxylin and eosin according to standard procedures. Two pathologists with expertise in genitourinary malignancies (T.A., G.P.) performed independent morphology assessment including overall morphology and nuclear, cytoplasmic and cell membrane details of the prostatic gland and stroma, while blinded to the fixation method.
For IHC, tissue sections were treated and stained using the automated VentanaBenchMark XT System (PSA) or the Leica Bond III System (P63 and P504). For PSA detection, anti-PSA antibody (DAKO, cat#A0562, Rabbit IgG, polyclonal, 1:1500) was applied on tissue sections and the antigen-antibody binding was detected with Ultraview detection kit from Ventana. For P63 detection, tissue sections were treated with Bond Epitope Retrieval Solution 2 (AR9640, Leica, Biosystems, EDTA, pH 9) for 20 minutes. Anti-P63 antibody (Biocaremedical, cat#CM163, clone: 4A4, mouse IgG2a, 1:100) was applied on tissue sections for 25 minutes. For P504 detection, tissue sections were treated with Bond Epitope Retrieval Solution 1 (AR9961, Leica, Biosystems, citrate buffer, pH 6) for 30minutes. Anti-P504S antibody (Biocaremedical, cat#ACA200BK, Rabbit IgG, polyclonal, 1:50) was applied on tissue sections for 25 minutes. For P63 and P504 the antigen-antibody binding was detected using Leica Bond Refine polymer detection system (Leica Biosystems, DS9800). Tissue sections were briefly immersed in hematoxylin for counterstaining. Images were obtained on a DM6000B microscope (Leica) at 10x magnification.
Nucleic acids extraction
Genomic DNA and total RNA from PAXgene-fixed samples were isolated using PAXgene Tissue DNA kit (PreAnalytiX) and PAXgene Tissue miRNA kit (PreAnalytiX), respectively. Genomic DNA and total RNA from formalin-fixed samples were isolated using QIAamp DNA FFPE Tissue kit (Qiagen) and miRNeasy FFPE kit (Qiagen), respectively. Genomic DNA and total RNA from frozen OCT embedded samples were isolated using DNeasy Blood and Tissue kit (Qiagen) and miRNeasy Mini kit (Qiagen), respectively. All extractions were performed using the manufacturer’s recommended protocols. Extracted DNA concentration was measured by Qbit® fluorometric quantification (Life Technologies). DNA fragmentation and RNA integrity number (RIN) were evaluated by Screentape® electrophoresis system (Agilent Technologies). After extraction, DNA was stored at 4°C and RNA at -80°C.
Reverse transcription assay
For each sample, 1 μg of RNA was transcribed into cDNA using SuperScript VILO MasterMix (Life Technologies) according to the manufacturer’s instructions. After reverse transcription, cDNA was stored at -20°C.
Real time PCR assay
For testing DNA integrity, one forward primer and six reverse primers (Supplementary Table 1) were designed to amplify six different transcript sequences with lengths of 88 to 817 base-pairs (bp) of the GAPDH gene. For testing cDNA integrity, one forward primer and five reverse primers (Supplementary Table 1) were designed to amplify five different transcript sequences with lengths of 95 to 983 bp of the β-Actin (ACTB) gene. Real time quantitative PCR assays were performed with 10 ng of genomic DNA and one microliter of 1:4 cDNA dilution, using iTaq Universal SYBR Green Supermix (Bio-rad) according to the manufacturer’s instructions. Reactions were performed on a ViiA 7 Real-Time PCR System (Life Technologies). Thermal cycling conditions were as followed: an activation step of 15 min at 95°C and 40 cycles of 15 s at 94°C, 30 s at 62°C and 30 s at 70°C. PCR reactions were run in triplicate. The acceptance criteria for single reactions were that the cycle threshold (CT) must be below 40 and the standard deviation in CT between triplicate reactions must be below 0.15. Melting curves had to be free of extraneous peaks. The significance of the difference between PAXPE and FFPE samples was tested using a paired Student’s T-test.
Results
Preservation of tissue morphology and immunohistochemistry characteristics
To evaluate the preservation of tissue morphology under PAXgene fixation, sections of formalin-fixed, paraffin-embedded (FFPE) and PAXgene-fixed, paraffin-embedded (PAXPE) specimens were stained with hematoxylin and eosin. On pathology assessment, the preservation of tissue morphology in specimens was similar regardless of fixation agent used (Figure 2A). It was noted that using equivalent staining protocols, PAXgene-fixed tissue exhibits stronger eosinophilic stain, which does not affect the nuclear detail. Some compromise in morphology was noted in the autopsy sections fixed using either technique, which was likely due to extended post-mortem time prior to fixation.
Figure 2.
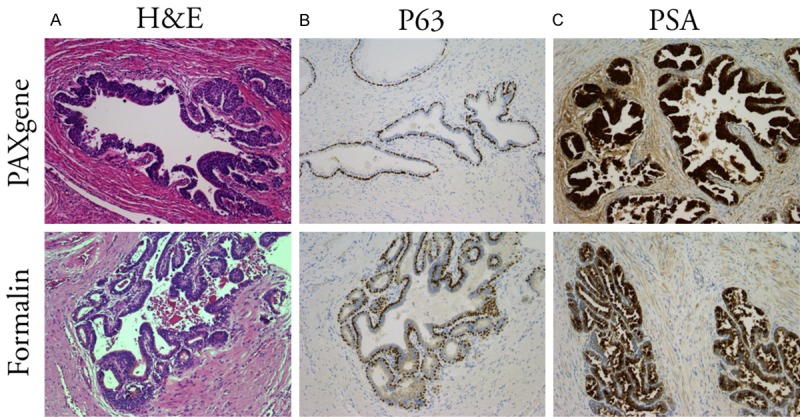
Preservation of morphology and antigenicity depending on fixation method. H&E staining (left), and P63 (center) and PSA (right) immunohistochemistry of prostate tissues from autopsy specimens fixed in PAXgene (top) for 48 hours or in formalin (bottom). Magnification X10.
Analysis of prostate tissue relies on immunohistochemical analysis using antibodies to Prostate Specific Antigen (PSA), the basal cell marker P63, and P504, a marker supporting a diagnosis of malignancy. Using standard IHC workflows, sections of PAXPE and FFPE prostates devoid of cancer were evaluated for PSA and P63. Comparable staining characteristics and staining intensity were observed between PAXPE and FFPE specimen for PSA and P63, (Figure 2B, 2C). PSA staining was detected as a very intense stain in luminal cells of prostate glands whereas P63 showed as strong nuclear staining in the basal cells.
To confirm that there was minimal effect of PAXgene fixation on immunohistochemical analysis of tissue containing PCa, four prostatectomy specimens were fixed in PAXgene reagent for 48 hours. As seen with non-PCa tissue, H&E staining demonstrated that tissue morphology and architecture was well preserved (Figure 3A). PCa specimens underwent immunohistochemical evaluation for P504, in addition to PSA and P63. All three stained strongly, demonstrating high levels of immune reactivity on PAXPE sections (Figure 3B-D).
Figure 3.
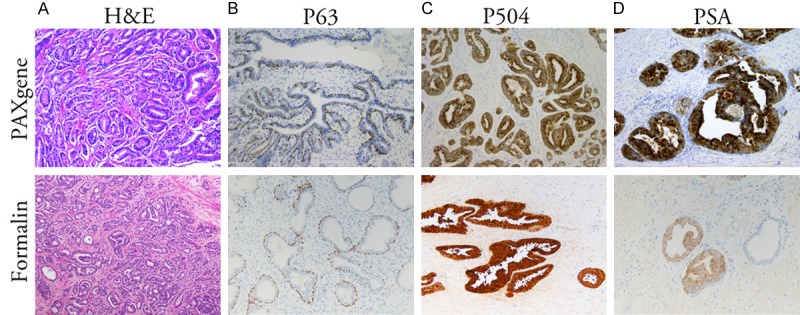
Prostate cancer staining. H&E staining (A) and P63 (B), P504S (C) and PSA (D) immunohistochemical staining of prostatectomy tissues fixed in PAXgene for 48 hours (top) or formalin (bottom). Magnification X10.
Preservation of nucleic acid quality
To investigate the quality of nucleic acids from the different fixation methods, total DNA and RNA were extracted from each autopsy prostate specimen. Sample yield varied highly between specimens but did not show significant differences between the preservation methods (data not shown). For DNA from both types of tissue the average fragment length was 5Kb or more. PAXgene-fixed samples were ~25% longer than formalin-fixed ones (Figure 4). For RNA samples, RIN scores were below 2, and there was no significant difference between PAXgene- and Formalin-fixed tissues. Quality of the RNA extracted from frozen tissue sample from each autopsy specimen ranged from RIN 1.5 to 3.
Figure 4.
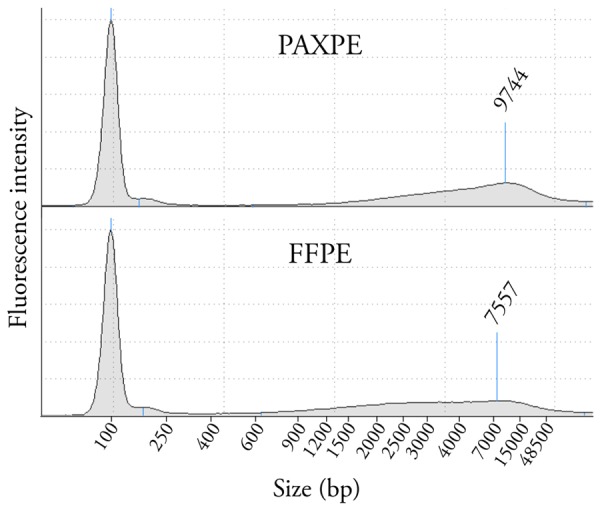
DNA fragment length depending on fixation method. Representative results of DNA fragments length between prostate tissue samples fixed in PAXgene for 48 hours or in formalin evaluated by Screentape® electrophoresis system. The electrophoregram for the FFPE sample shows smaller more degraded DNA as compared to the PAXPE sample.
The relative quality of DNA and RNA was evaluated by real-time PCR and reverse transcription real-time PCR, respectively. To measure the DNA template quality, as a function of the fixation method, six primer pairs were designed to amplify fragments of increasing length of the GAPDH gene, from 88 bp to 817 bp. Consistently, DNA from PAXPE samples demonstrated 4-8 fold increased amplification efficiency (corresponding to 2-3 lower cycle threshold value) compared to FFPE samples (Figure 5A).
Figure 5.
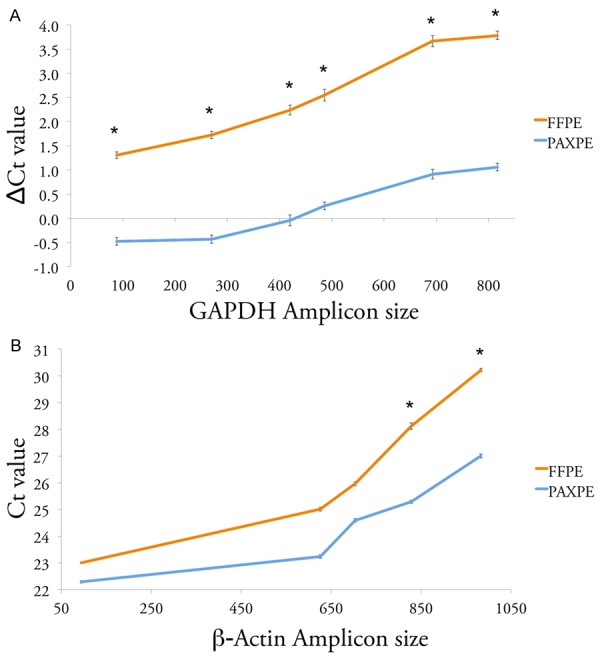
Preservation of DNA and RNA depending on fixation method. A. Preservation of DNA. Line plot showing the difference in cycle threshold (Ct) in real-time PCR targeting GAPDH gene DNA, between PAXgene-fixed and formalin-fixed prostate tissue samples, and the corresponding Frozen tissue. Each line represents the average values from 2 prostate specimens from autopsies fixed for 48 hours. *: statistical significance, P-value < 0.05. B. Preservation of RNA. Line plot showing threshold cycle (Ct) of real-time RT-PCR targeting β-Actin gene cDNA from PAXgene-fixed or formalin-fixed prostate tissue samples. Each line represents the average values from 2 prostate specimens from autopsies fixed for 48 hours. *: statistical significance, P-value < 0.05.
Five primer pairs were used to evaluate cDNA quality, producing amplicons of increasing length of the β-Actin house-keeping gene, from 95 bp to 983 bp. While RIN scores were low in both formalin and PAXgene tissues, there was a significant difference in cDNA template quality, with up to 8-fold increased amplification efficiency in PAXPE samples (Figure 5B).
Fixation with PAXgene for 48 hours requires alterations to the typical tissue processing workflow and a delay in availability of tissue for diagnostic studies. Therefore we also evaluated the quality of DNA and RNA from PAXPE prostate specimens fixed for 24 hours. As seen in Figure 6, quality of both DNA and RNA from these specimens was slightly inferior to that fixed for 48 hours.
Figure 6.
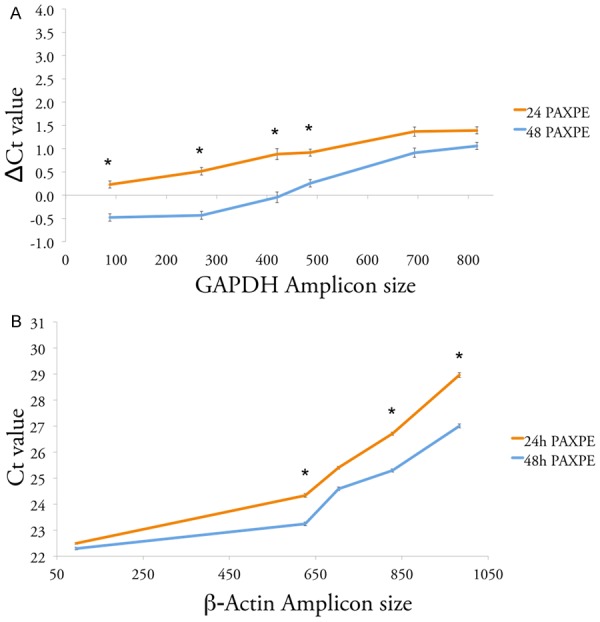
Preservation of DNA and RNA depending on PAXgene fixation time. A. Preservation of DNA. Line plot showing the difference in cycle threshold (Ct) in real-time PCR targeting GAPDH gene DNA, between prostate tissue samples fixed in PAXgene for 24 or 48 hours, and the corresponding Frozen tissue. Each line represents the average values from 2 prostate specimens from autopsies. *: statistical significance, P-value < 0.05. B. Preservation of RNA. Line plot showing threshold cycle (Ct) of real-time RT-PCR targeting β-Actin gene cDNA from prostate tissue samples fixed in PAXgene for 24 or 48 hours. Each line represents the average values from 2 prostate specimens from autopsies. *: statistical significance, P-value < 0.05.
Discussion
We report here that PAXgene fixation and stabilization reagents preserve morphology and immunoreactivity equivalent to formalin fixation, but with superior preservation of nucleic acid integrity. This technique can facilitate collection of tissue with high quality DNA and RNA, without compromising tissue architecture by disrupting the capsule prior to fixation, and without limiting tissue collection to small samples obtained without prior full pathological review. This represents a particular advantage for studies evaluating the entire gland or correlating morphologic or staining features with molecular characteristics.
Hematoxylin and eosin stained PAXPE prostate tissues provided good representation of histologic features, similar to that of formalin. As previously described [12,13] an EC FP7 project aimed to improve pre analytic procedures, the PAXgene Tissue System (PAXgene) both hematoxylin and eosin were modestly more intense in PAXPE samples than in the corresponding FFPE samples. Immunohistochemical analysis revealed that fixation with PAXgene did not alter immunoreactivity of antigens relevant for PCa as compared to formalin fixation. Staining and counterstaining were intense for all markers tested. Importantly, immunohistochemical analysis for this study did not require specific optimization of routine staining procedures for PAXPE samples, in contrast to what has been previously reported [12].
Consistent with previous studies, we found that PAXgene reagents preserved both DNA and RNA quality significantly better than formalin [14-16]. Using real-time PCR on DNA and RNA obtained from prostate autopsy specimens, we showed that PAXPE samples have lower CT values compared to FFPE samples. This difference increased with amplicon length, indicating that PAXgene allows efficient amplification even of long fragments. Chromosomal rearrangements and fusion genes appear to play an important role in prostate cancer development and progression [4,17-19]. Identification of these structural variants using next-generation sequencing is improved by sequencing across longer DNA inserts, thus increasing the physical coverage of the genome [20]. Sequencing libraries made from FFPE require short inserts to allow for amplification across cross-linked DNA. DNA from PAXPE tissue facilitates use of sequencing libraries more amenable to identifying the rearrangements characteristic of prostate cancer.
We demonstrate here significant differences in downstream assays using RNA extracted from PAXPE versus FFPE prostatectomy samples, though RNA from both tissue types were of low quality. It should be noted that RNA is likely to have substantially degraded during post-mortem time before fixation, which is supported by the low quality of RNA from matched frozen tissue.
A limitation of this study is that a portion of the study used autopsy specimens, which have already undergone some degree of post-mortem degradation prior to PAXgene and formalin fixation. This strategy was undertaken to avoid compromising the capsule of clinical samples prior to fixation. Prostatectomy specimens fixed wholly in PAXgene also demonstrated preserved morphology and staining characteristics. Although we demonstrate superior preservation of nucleic acids with PAXgene, the question of how long those features remain stable is still to be answered.
The prolonged duration of fixation (48 hours) may be problematic for the workflow of some centers. Though 24 hour fixation is inferior to 48 hour fixation in preserving DNA quality, it is superior to formalin and preserves staining characteristics and therefore is a suitable alternative. In addition, the routine use of PAXgene fixation increases financial costs. If prohibitively expensive for routine use, PAX gene fixation is an excellent method for preserving specimens of high research interest.
In this study we thus demonstrate the novel use of PAXgene for fixation of prostatectomy specimens. Its use improves the quality of molecular analysis without compromising histopathological analysis. PAXgene fixation may prove especially useful for assays for which longer amplicons are required, such as for chromosomal rearrangements, or for studies making use of more tissue than can be obtained with a simple punch.
Conclusions
Given the difficulty of identifying PCa on gross examination and the potential for compromise of the prostate “capsule” if tissue is divided prior to fixation, banking significant amounts of PCa tissue samples is challenging. The alternative, non-crosslinking fixation agent PAXgene preserves the quality of DNA and RNA isolated from fixed tissue without compromising the quality of tissue histology or staining characteristics. Incorporation of PAXgene fixation into banking protocols should facilitate the wider availability of high quality PCa tissues for next-generation sequencing and other studies.
Acknowledgements
This work was supported in part by the University of Chicago Cancer Center Support Grant (CA014599), and some PAXgene reagents were provided by Qiagen.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Mohler J, Bahnson RR, Boston B, Busby JE, D’Amico A, Eastham JA, Enke CA, George D, Horwitz EM, Huben RP, Kantoff P, Kawachi M, Kuettel M, Lange PH, Macvicar G, Plimack ER, Pow-Sang JM, Roach M 3rd, Rohren E, Roth BJ, Shrieve DC, Smith MR, Srinivas S, Twardowski P, Walsh PC. NCCN clinical practice guidelines in oncology: prostate cancer. J Natl Compr Cancer Netw JNCCN. 2010;8:162–200. doi: 10.6004/jnccn.2010.0012. [DOI] [PubMed] [Google Scholar]
- 3.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, Onofrio R, Carter SL, Park K, Habegger L, Ambrogio L, Fennell T, Parkin M, Saksena G, Voet D, Ramos AH, Pugh TJ, Wilkinson J, Fisher S, Winckler W, Mahan S, Ardlie K, Baldwin J, Simons JW, Kitabayashi N, MacDonald TY, Kantoff PW, Chin L, Gabriel SB, Gerstein MB, Golub TR, Meyerson M, Tewari A, Lander ES, Getz G, Rubin MA, Garraway LA. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, Nickerson E, Chae SS, Boysen G, Auclair D, Onofrio RC, Park K, Kitabayashi N, MacDonald TY, Sheikh K, Vuong T, Guiducci C, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Hussain WM, Ramos AH, Winckler W, Redman MC, Ardlie K, Tewari AK, Mosquera JM, Rupp N, Wild PJ, Moch H, Morrissey C, Nelson PS, Kantoff PW, Gabriel SB, Golub TR, Meyerson M, Lander ES, Getz G, Rubin MA, Garraway LA. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ, Chinnaiyan AM, Tomlins SA. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, Wang Y, Sheikh KL, Terry S, Tagawa ST, Dhir R, Nelson JB, De La Taille A, Allory Y, Gerstein MB, Perner S, Pienta KJ, Chinnaiyan AM, Wang Y, Collins CC, Gleave ME, Demichelis F, Nanus DM, Rubin MA. Molecular Characterization of Neuroendocrine Prostate Cancer and Identification of New Drug Targets. Cancer Discov. 2011;1:487–495. doi: 10.1158/2159-8290.CD-11-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H, Yu J, Palanisamy N, Siddiqui J, Yan W, Cao X, Mehra R, Sabolch A, Basrur V, Lonigro RJ, Yang J, Tomlins SA, Maher CA, Elenitoba-Johnson KSJ, Hussain M, Navone NM, Pienta KJ, Varambally S, Feng FY, Chinnaiyan AM. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19:664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samaratunga H, Montironi R, True L, Epstein JI, Griffiths DF, Humphrey PA, van der Kwast T, Wheeler TM, Srigley JR, Delahunt B, Egevad L. International Society of Urological Pathology (ISUP) Consensus Conference on Handling and Staging of Radical Prostatectomy Specimens. Working group 1: specimen handling. Mod Pathol. 2011;24:6–15. doi: 10.1038/modpathol.2010.178. [DOI] [PubMed] [Google Scholar]
- 10.Esgueva R, Park K, Kim R, Kitabayashi N, Barbieri CE, Dorsey PJ Jr, Abraham C, Banerjee S, Leung RA, Tewari AK, Terry S, Shevchuk MM, Rickman DS, Rubin MA Weill Cornell Medical College. Next-generation prostate cancer biobanking: toward a processing protocol amenable for the International Cancer Genome Consortium. Diagn Mol Pathol. 2012;21:61–68. doi: 10.1097/PDM.0b013e31823b6da6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Beers EH, Joosse SA, Ligtenberg MJ, Fles R, Hogervorst FBL, Verhoef S, Nederlof PM. A multiplex PCR predictor for aCGH success of FFPE samples. Br J Cancer. 2005;94:333–337. doi: 10.1038/sj.bjc.6602889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kap M, Smedts F, Oosterhuis W, Winther R, Christensen N, Reischauer B, Viertler C, Groelz D, Becker KF, Zatloukal K, Langer R, Slotta-Huspenina J, Bodo K, de Jong B, Oelmuller U, Riegman P. Histological Assessment of PAXgene Tissue Fixation and Stabilization Reagents. PLoS One. 2011;6:e27704. doi: 10.1371/journal.pone.0027704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gündisch S, Slotta-Huspenina J, Verderio P, Ciniselli CM, Pizzamiglio S, Schott C, Drecoll E, Viertler C, Zatloukal K, Kap M, Riegman P, Esposito I, Specht K, Babaryka G, Asslaber M, Bodó K, den Bakker M, den Hollander J, Fend F, Neumann J, Reu S, Perren A, Langer R, Lugli A, Becker I, Richter T, Kayser G, May AM, Carneiro F, Lopes JM, Sobin L, Höfler H, Becker KF. Evaluation of colon cancer histomorphology: a comparison between formalin and PAXgene tissue fixation by an international ring trial. Virchows Arch. 2014;465:509–519. doi: 10.1007/s00428-014-1624-4. [DOI] [PubMed] [Google Scholar]
- 14.Ergin B, Meding S, Langer R, Kap M, Viertler C, Schott C, Ferch U, Riegman P, Zatloukal K, Walch A, Becker KF. Proteomic Analysis of PAXgene-Fixed Tissues. J Proteome Res. 2010;9:5188–5196. doi: 10.1021/pr100664e. [DOI] [PubMed] [Google Scholar]
- 15.Belloni B, Lambertini C, Nuciforo P, Phillips J, Bruening E, Wong S, Dummer R. Will PAXgene substitute formalin? A morphological and molecular comparative study using a new fixative system. J Clin Pathol. 2013;66:124–135. doi: 10.1136/jclinpath-2012-200983. [DOI] [PubMed] [Google Scholar]
- 16.Viertler C, Groelz D, Gündisch S, Kashofer K, Reischauer B, Riegman PHJ, Winther R, Wyrich R, Becker KF, Oelmüller U, Zatloukal K. A New Technology for Stabilization of Biomolecules in Tissues for Combined Histological and Molecular Analyses. J Mol Diagn. 2012;14:458–466. doi: 10.1016/j.jmoldx.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 18.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, Van Allen E, Kryukov GV, Sboner A, Theurillat JP, Soong TD, Nickerson E, Auclair D, Tewari A, Beltran H, Onofrio RC, Boysen G, Guiducci C, Barbieri CE, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Ramos AH, Winckler W, Cipicchio M, Ardlie K, Kantoff PW, Berger MF, Gabriel SB, Golub TR, Meyerson M, Lander ES, Elemento O, Getz G, Demichelis F, Rubin MA, Garraway LA. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubin MA, Maher CA, Chinnaiyan AM. Common Gene Rearrangements in Prostate Cancer. J. Clin. Oncol. 2011;29:3659–3668. doi: 10.1200/JCO.2011.35.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


