Abstract
MicroRNAs (miRNAs) are a class of small non-coding RNAs that play key roles in cancer development and progression. Therefore, the discovery of miRNAs may provide a new and powerful tool for understanding the mechanism of carcinogenesis. In the present study, we aimed to investigate the functional significance of miR-630 and to identify its possible target genes in human non-small cell lung cancer (NSCLC). Our results showed that miR-630 was significantly down-regulated in NSCLC tissues and cell lines. The enforced expression of miR-630 was able to inhibit cell proliferation, migration, and invasion of NSCLC cells. Moreover, our results further revealed that LMO3, a nuclear LIM-only proteins, was identified as a target of miR-630. Restoration of LMO3 remarkably reversed the tumor-suppressive effects of miR-630 on cell proliferation, migration, and invasion in NSCLC cells. Therefore, we demonstrated that miR-630 suppressed the proliferation, migration, and invasion of NSCLC cells by down-regulating LMO3 expression, suggesting miR-630 as a potential therapeutic target for the treatment of human NSCLC in the future.
Keywords: Non-small cell lung cancer, miR-630, LMO3, proliferation, migration, invasion
Introduction
Lung cancer is the leading cause of cancer-related mortality, with 1.4 million deaths worldwide annually. Non-small cell lung cancer (NSCLC) accounts for approximately 80% of primary lung cancer [1]. The prognosis for NSCLC remains poor despite recent advances in the diagnosis and chemotherapies used for this cancer, and the 5-year overall survival rate of NSCLC is a dismal 11% [2]. Thus, the elucidation of the molecular mechanisms that control NSCLC tumor growth and metastasis is urgently needed.
MicroRNAs (miRNAs) are a class of small non-coding RNAs(ncRNA) that negatively regulate the expression of their target genes by binding to the 3’-untranslated regions (3’-UTRs) of target mRNAs that leads to mRNA degradation or translational suppression [3]. The miRNAs regulate the expression of multiple target genes involved in various biological processes, including cell apoptosis, proliferation, differentiation and migration [4-6]. Recently, mounting evidence indicated that aberrant changes in miRNA expression correlates with a wide range of cancers and that miRNAs act as oncogenes and tumor suppressors [7,8]. In NSCLC, multiple miRNAs, such as miR-186, miR-205 and miR-21 were found to promote NSCLC carcinogenesis [9-11]. However, the detailed role of miR-630 in NSCLC carcinogenesis remains unclear.
In the present study, we found that miR-630 was significantly decreased in NSCLC tissues and cell lines. Enforced expression of miR-630 inhibited cell proliferation, migration, and invasion of NSCLC cells. Furthermore, we identified LIM-only proteins 3 (LMO3) as a target of miR-630 in NSCLC cells. Finally, we found that miR-630 suppressed cell proliferation, migration and invasion of NSCLC cells partially by down-regulating LMO3 expression. Together, our results suggested that miR-630 could be a therapeutic target for the treatment of NSCLC.
Materials and methods
Patients and specimens
Twenty-two paired NSCLC tissues and matched adjacent normal tissues were obtained from The Fuzhou General Hospital of Nanjing Military Command (Fuzhou, China) between March 2007 and March 2009. All patients recruited in this study were not subjected to preoperative radiotherapy and/or chemotherapy and were diagnosed as infiltrating carcinoma by pathology. Tumor tissues and matched adjacent normal tissues were collected and stored in liquid nitrogen until use. The study was approved by the Medical ethics committee of Xiamen University. Written informed consent was obtained from each patient.
Cell culture
The human lung cell lines NCI-H23, A549, H157, H1299 and normal bronchial epithelial cell line 16HBE were purchased from the American Type Culture Collection (ATCC, USA). All cell lines were routinely maintained in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U/ml penicillin sodium, and 100 mg/ml streptomycin sulfate at 37°C in a humidified air atmosphere containing 5% CO2. Cell transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
Plasmids and luciferase activity assay
MiR-630 and the scramble control mimics were obtained from Ribio Bio (Guangzhou, China). LMO3 over-expression plasmid, pcDNA3-LMO3 was purchased from Promega. The LMO3 3’-UTR fragment containing putative miR-630 binding sites was amplified from complementary DNA (cDNA) of NCI-H23 cells. The 3’-UTR was inserted into pGL3 vector (Invitrogen), and named pGL3-LMO3 3’-UTR (WT). Mutation of the miR-630 target sites in pGL3-LMO3 3’-UTR (Mut) was performed using a Strata gene mutation kit (Stratagene).
HEK293 cells were cultured in 24-well plates, and each well was co-transfected with 100 ng WT or Mut and 100 ng miR-630 mimics using Lipofectamine 2000. Renilla luciferase pRL-SV40 (Promega) was used as a control. Cells were collected 48 h after transfection and the dual-luciferase activity was examined with a Dual-Luciferase Reporter Assay (Promega) according to the manufacturer’s instructions.
Cell proliferation assay
To determine cell proliferation, cells were plated in 96-well plates overnight and transfected with miR-630 mimic or/and LMO3 plasmid. The cell proliferation was determined with 3-(4,5-dimethylthiazol-2-yl)-2-5 diphenyltetrazolium bromide (MTT, Sigma) assay at 24, 48, 72, and 96 h after transfection respectively, and the absorbance of samples was measured with a spectrophotometer reader (Molecular Deviced) at 570 nm. All experiments were performed in six replicates and were repeated three times independently.
In vitro migration and invasion assays
Cell migration and invasion assays were performed in a 24-well plate with 8 mm pore size chamber inserts (Corning). For migration assays, 5×104 cells were placed into the upper chamber per well with the non-coated membrane. For invasion assays, 1×105 cells were placed into the upper chamber per well with the Matrigel-coated membrane, which was diluted with serum-free culture medium. In both assays, cells were suspended in 200 μL of DMEM without FBS when they were seeded into the upper chamber. In the lower chamber, 800 μL of DMEM supplemented with 10% FBS was added. After incubation for 48 h at 37°C and 5% CO2, the membrane inserts were removed from the plate, and non-invading cells were removed from the upper surface of the membrane. Cells that moved to the bottom surface of the chamber were fixed with 100% methanol for 20 min and stained with 0.1% crystal violet for 30 min. Then, cells were imaged and counted in at least 5 random fields using a microscope (Olympus). The assays were conducted three independent times.
RNA extraction and quantitative real-time PCR
Total RNA from tissue samples and cell lines were isolated by the TRIZOL reagent (Invitrogen) according to the manufacturer’s protocol. Quantitative real-time PCR (qRT-PCR) was performed with an Applied Biosystems 7900HT system (Applied Biosystems) using SYBR Premix Ex Taq (Takara). PCR volume was 20 μL, containing 1 μL reverse transcript product. Cycling conditions were 1 cycle of 95°C for 30 seconds and 40 cycles of 95°C for 5 seconds and 60°C for 30 seconds. PCR was performed in triplicate. For measurement of the LMO3 transcript from total RNA, total cDNA was synthesized using a Takara reverse transcription kit (Takara). qRT-PCR was performed using SYBR Pre-mix ExTaq (Takara). The U6 and GAPDH were used as endogenous control for miRNA and mRNA, respectively. The ΔΔCt method was used to determine relative quantitation of miRNA and mRNA expression, and fold change was determined as 2-ΔΔCt. Primer sequences used were shown in Table 1.
Table 1.
Sequences of Primers
| Gene | Primers | Sequence (5’-3’) |
|---|---|---|
| miR-630 | forward | TAAAGGAGGAAGATAAGG |
| reverse | GTAGCAGTGATAGGCATT | |
| LMO3 | forward | ATGCTCTCAGTCCAGCCAGA |
| reverse | TCAGCGAACCTGGGGTGCAT | |
| U6 | forward | CCTGCTTCGGCAGCACA |
| reverse | TGGAACGCTTCACGAA | |
| GAPDH | forward | CCACTCCTCCACCTTTGAC |
| reverse | ACCCTGTTGCTGTAGCCA |
Western blotting
Cultured cells were harvested and lysed with RIPA buffer (Beyotime). Separated on 10% SDS-PAGE, and then transferred to PVDF membranes (Millipore). Membranes were blocked with 3% non-fat dried milk solution for 0.5 h at room temperature and then probed with primary antibodies overnight at 4°C. The membranes were further developed with HRP-conjugated secondary antibodies for 1 h at 37°C. Blots were visualized using an ECL detection system (Amersham) and analyzed by Kodak Digital Science 1D software (Eastman Kodak).
Statistical analysis
All statistical analyses were performed using SPSS version 18.0 software. The data are shown as the mean ± SD from at least three independent experiments. Differences between groups were analyzed using Student’s t test or one-way ANOVA analysis. P<0.05 was considered statistically significant.
Results
miR-630 was decreased in NSCLC tissues and cell lines
The expression of miR-630 in 22 pairs of NSCLC tissues and their matched normal tissues was measured using qRT-PCR. Our results showed that miR-630 expression was significantly down-regulated in NSCLC tissues compared with matched adjacent normal tissues (P<0.05, Figure 1A). In addition, the expression level of miR-630 in four NSCLC cell lines (NCI-H23, A549, H157 and H1299) were determined. Our data showed that the relative expression of miR-630 in NSCLC cells was strikingly decreased compared to the normal lung bronchus epithelial cell line 16HBE (P<0.05, Figure 1B). These results indicated that miR-630 may play a critical role in the progression of NSCLC.
Figure 1.
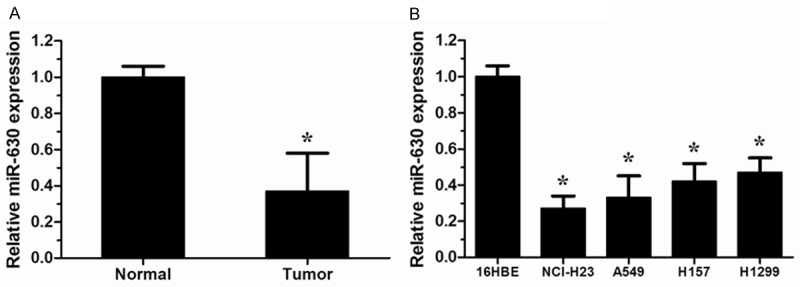
miR-630 was down-regulated in human non-small cell lung cancer (NSCLC) tissues and cell lines. A. Expression of miR-630 in 22 paired NSCLC tissues and their matched normal tissues was measured by qRT-PCR. U6 was used as an internal control. B. The expression of miR-630 in the human bronchial epithelial cell line 16HBE and four NSCLC cell lines (NCI-H23, A549, H157 and H1299 ). *P<0.05.
miR-630 inhibited NSCLC cell proliferation, migration, and invasion
We further investigated the function of miR-630 in the progression of NSCLC, miR-630 mimics and control mimics was transfected into NCI-H23 cells, and the effect of miR-630 mimics was confirmed by qRT-PCR (P<0.05, Figure 2A), MTT assay revealed that over-expression of miR-630 significantly inhibited proliferation ability of NCI-H23 cells (P<0.05, Figure 2B). Similarly, in vitro migration and invasion assays showed that enforced expression of miR-630 could inhibit the migration and invasion ability of NCI-H23 cells (P<0.05, Figure 2C and 2D). These data indicated that miR-630 was able to inhibit the development and progression of NSCLC.
Figure 2.
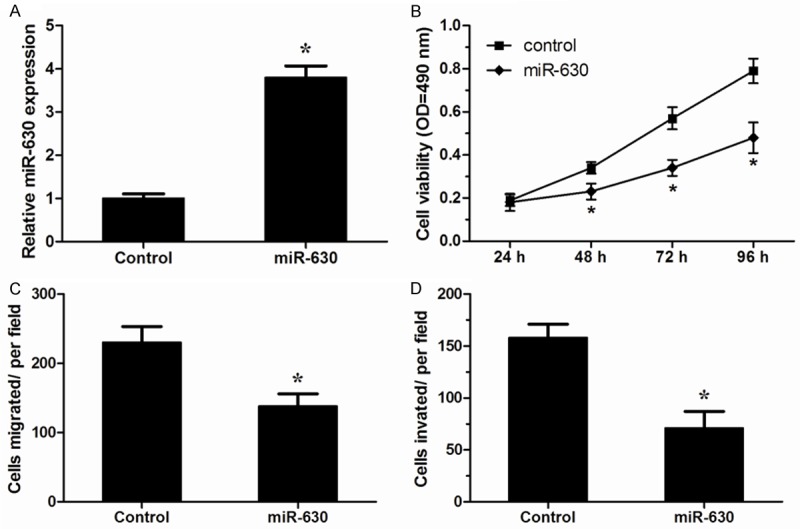
miR-630 inhibited proliferation, migration and invasion of NSCLC cells. A. Expression of miR-630 in NCI-H23 cells transfected with miR-630 mimics or control mimics was detected by qRT-PCR. B. MTT assay was performed to analyze the effect of miR-630 on proliferation of NCI-H23 cells. C, D. Transwell migration and invasion assays were utilized to analyze the effect of miR-630 on migration and invasion of NCI-H23 cells. *P<0.05.
LMO3 was a direct target of miR-630
We further explored the downstream molecular target of miR-630 in NSCLC. We used TargetScan 6.2 (http://www.targetscan.org), a widely used miRNA target prediction website, LMO3 was found to be a potential target of miR-630 (Figure 3A). Luciferase activity assay showed that miR-630 significantly inhibited the luciferase activity of the wild-type (WT) 3’-UTR of LMO3, without effect on its mutant (Mut) (P<0.05, Figure 3B). Moreover, qRT-PCR and Western blotting confirmed that miR-630 suppressed the expression of LMO3 in NCI-H23 cells (P<0.05, Figure 3C and 3D). These data suggested that LMO3 was a target of miR-630 in NSCLC cells.
Figure 3.
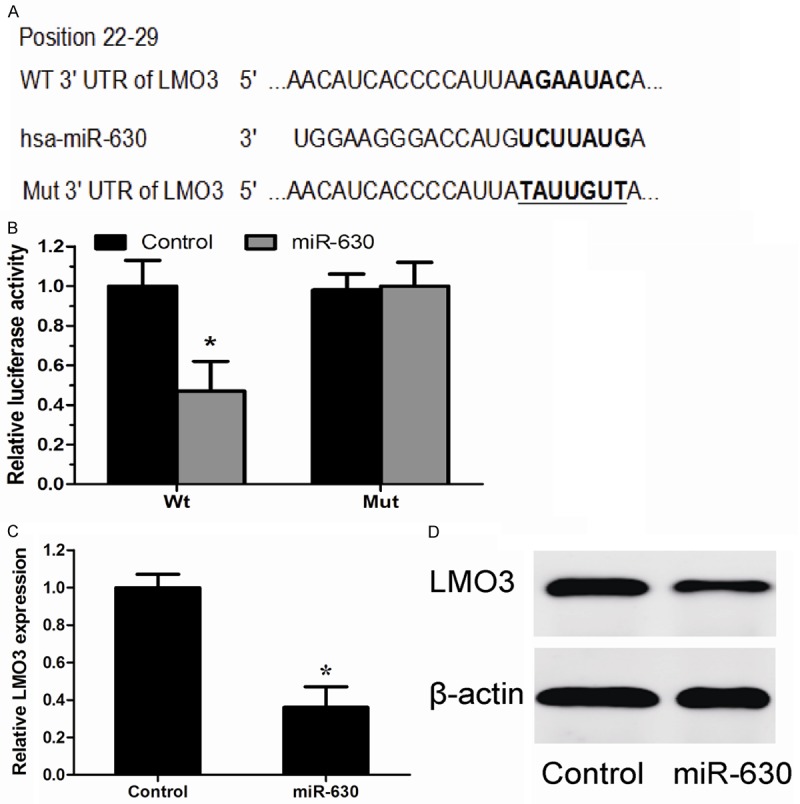
LMO3 was a direct target of miR-630. A. The potential miR-630 binding sites of LMO3 3’-UTR. B. A luciferase reporter assay showed the inhibitory effect of miR-630 on LMO3 3’UTR luciferase activity in HEK293 cells. C. Expression of LMO3 mRNA was detected by qRT-PCR in NCI-H23 cells transfected with miR-630 mimics or control mimics. D. Protein levels were detected by western blotting in NCI-H23 cells transfected with miR-630 mimics or control mimics. *P<0.05.
LMO3 over-expression partially attenuated the tumor suppressive effect of miR-630
We further investigated whether LMO3 over-expression could attenuate tumor suppressive effects of miR-630. The effect of pcDNA-LMO3 was confirmed by qRT-PCR and western blot (P<0.05, Figure 4A and 4B). MTT assay, migration assay and invasion assay revealed that over-expression of LMO3 significantly reversed the tumor suppressive effects of miR-630 on NCI-H23 cells (P<0.05, Figure 4C-E). These results demonstrated that restoration of LMO3 significantly attenuated the tumor suppressive effect of miR-630 in NSCLC.
Figure 4.
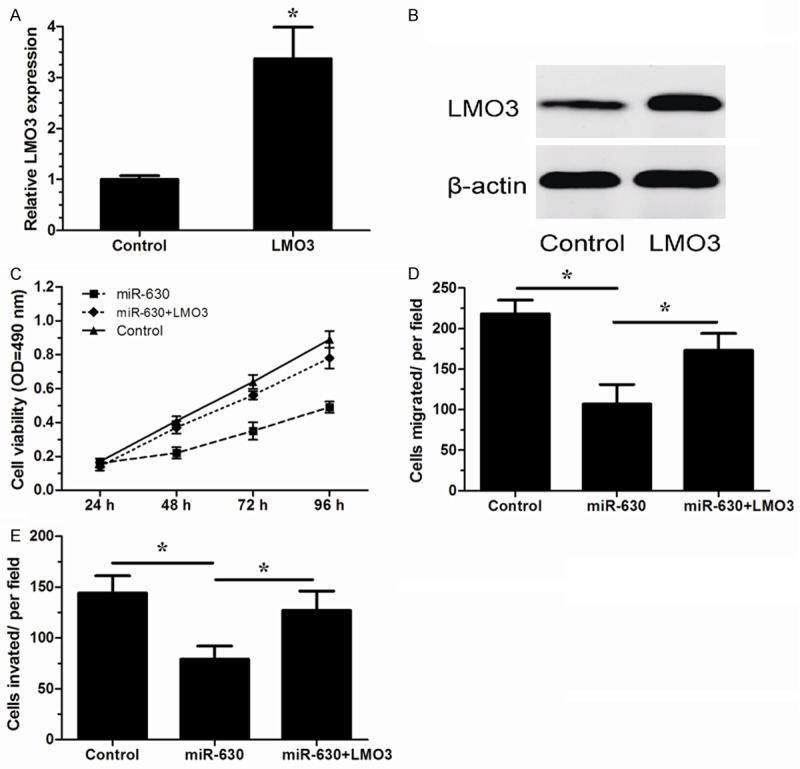
LMO3 over-expression partially attenuated the tumor suppressive effect of miR-630. (A, B) LMO3 expression in NCI-H23 cells transfected with an pcDNA-LMO3 or empty vector (control) was measured by qRT-PCR (A) and western blotting (B). (C-E) NCI-H23 cells were transfected with miR-630 mimics or control mimics (control) with LMO3 over-expression plasmid. MTT assay (C), in vitro migration assay (D) and in vitro invasion assay (E) were performed. *P<0.05.
LMO3 expression was inversely correlated with that of miR-630 in NSCLC tissues
To further explore the relationship between LMO3 and miR-630 in vivo, we examined the expression of LMO3 in 22 pairs of NSCLC tissues and their matched normal tissues using qRT-PCR. We found that LMO3 expression was significantly increased in NSCLC tissues relative to their matched normal tissues (P<0.05, Figure 5A). Moreover, LMO3 was negatively correlated with miR-630 expression in the same NSCLC tissues (P<0.05, Figure 5B). These data further indicated that LMO3 was a target of miR-630.
Figure 5.
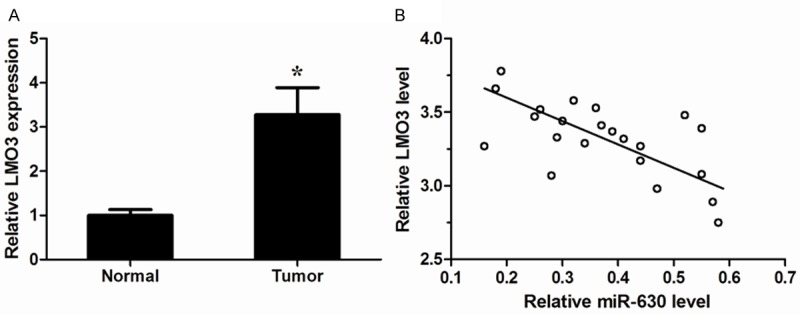
LMO3 levels were inversely correlated with miR-630 in NSCLC tissues. A. LMO3 expression levels in 22 pairs of NSCLC tissues and their matched normal tissues were measured by qRT-PCR. GAPDH was used as an internal control. B. Correlation analysis between LMO3 mRNA levels and miR-630 expression in NSCLC tissues (Spearman’s correlation analysis, r=-0.5592; P<0.05). *P<0.05.
Discussion
According to recent studies, more than 1,900 human miRNAs have been identified, which are estimated to regulate over 60% of genes in mammals [12]. And miRNAs have been reported to play important roles in carcinogenesis and tumor progression [13,14]. In addition, acting as either tumor suppressors or oncogenes, miRNAs are involved in several aspects of cancer biology including cell proliferation, apoptosis, migration and invasion. In this study, we focused on miR-630. Numerous studies have revealed that miR-630 potentially act as oncogenes or tumor suppressors in different cancers. For example, Chu showed miR-630 was significantly elevated in gastric cancer and associated with gastric cancer invasion, lymph node metastasis, distant metastasis and TNM stage [15]. Zhao found that miR-630 was markedly increased in renal cancer and associated with renal cancer histologic grade, lymph node metastasis, distant metastasis and TNM stage, which indicated that miR-630 could play an important role in the progression of renal cancer [16]. However, Farhana et al. proved that up-regulation of miR-630 in pancreatic cancer cells could induce apoptosis by targeting IGF-1R [17]. Corcoran showed that decreased expression of miR-630 could induce the cellular motility by increasing the migration and invasion ability of breast cancer cells. Conversely, up-regulation of miR-630 expression in breast cancer cells could decrease the cellular motility [18]. miR-630 was also found to be up-regulated in head and neck squamous cell carcinoma after cisplatin treatment and can modulate the protein expression of ATG5, ATG6/BECN1, ATG10, ATG12, ATG16L1 and UVRAG [19,20].
However, the role of miR-630 in NSCLC carcinogenesis remains unclear. A recent report by Kuo et al showed that miR-630 was decreased in lung cancer and inversely correlated with advanced stage, higher lymph node metastasis, higher grade invasion, poor overall survival, and poor disease free survival in lung cancer [21]. Patnaik found that miR-630 could predict recurrence of localized stage I non-small cell lung cancer after surgical resection [22]. It was previously shown that miR-630 conferred robust cytoprotection against CDDP and carboplatin, resulting from decreased proliferation coupled to upstream inhibition of the signaling cascades that emanate from damaged DNA and converge on p53 activation [23]. In this study, we found that miR-630 was markedly reduced in NSCLC tissues and cell lines. The ectopic over-expression of miR-630 effectively inhibited NSCLC cell proliferation, migration and invasion. These results suggested that miR-630 was a novel tumor suppressive miRNA in NSCLC progression.
To elucidate the molecular mechanisms involved in miR-630 induced inhibition of NSCLC growth and metastasis, we used TargetScan 6.2 to predict miR-630 target genes. LMO3 was identified as a target of miR-630 in NSCLC. The restoration of LMO3 markedly attenuated the tumor suppressive effects of miR-630 on NSCLC cells. Furthermore, LMO3 levels were increased and were negatively correlated with miR-630 levels in NSCLC tissues. The miRNAs can induce mRNA degradation or translational suppression, and down-regulated mRNA levels may result in decreased protein levels. Thus, the observed decrease in LMO3 protein levels induced by miR-630 might be because of the degradation of LMO3 mRNA. Taken together, these data indicated that miR-630 inhibited NSCLC growth and metastasis by inhibiting LMO3 expression. In various cancers, LMO3 acts as an essential regulator of cell growth. Aoyama reported that high levels of LMO3 mRNA were found in aggressive neuroblastomas with a poor prognosis as compared with more favorable subtypes [24]. Larsen found that LMO3 in highly expressing tumors may simultaneously permit activation of the p53 pathway and inhibit LMO3-mediated pro-survival mechanisms [25]. Hui suggested that over-expression of LMO3 promoted cell growth and the effect of LMO3 on proliferation may be explained by its role in cell-cycle progression [26]. Isogai demonstrated that oncogenes LMO3 collaborates with HEN2 to enhance neuroblastoma cell growth through transactivation of Mash1 [27]. Here, our work further investigated the role of LMO3 in NSCLC.
In summary, our data revealed that miR-630 was dramatically down-regulated in NSCLC tissues and cell lines, and enforced expression of miR-630 could suppress NSCLC cell proliferation, migration and invasion. Moreover, we found that the tumor suppressor function of miR-630 was mediated by the down-regulation of its downstream target gene LMO3. These results suggested that miR-630 deregulation might play important roles in tumor progression and miR-630 could be a potential therapeutic strategy for the treatment of NSCLC.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Verdecchia A, Francisci S, Brenner H, Gatta G, Micheli A, Mangone L, Kunkler I. Recent cancer survival in Europe: a 2000-02 period analysis of EUROCARE-4 data. Lancet Oncol. 2007;8:784–796. doi: 10.1016/S1470-2045(07)70246-2. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Chen CZ. MicroRNAs as oncogenes and tumor suppressors. N Engl J Med. 2005;353:1768–1771. doi: 10.1056/NEJMp058190. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 9.Cui G, Cui M, Li Y, Liang Y, Li W, Guo H, Zhao S. MiR-186 targets ROCK1 to suppress the growth and metastasis of NSCLC cells. Tumour Biol. 2014;35:8933–7. doi: 10.1007/s13277-014-2168-6. [DOI] [PubMed] [Google Scholar]
- 10.Larzabal L, de Aberasturi A, Redrado M, Rueda P, Rodriguez MJ, Bodegas ME, Montuenga LM, Calvo A. TMPRSS4 regulates levels of integrin α5 in NSCLC through miR-205 activity to promote metastasis. Br J Cancer. 2014;110:764–774. doi: 10.1038/bjc.2013.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC) Clin Chim Acta. 2010;411:846–852. doi: 10.1016/j.cca.2010.02.074. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 13.Mueller DW, Bosserhoff AK. Role of miRNAs in the progression of malignant melanoma. Br J Cancer. 2009;101:551–556. doi: 10.1038/sj.bjc.6605204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nature Reviews Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 15.Chu D, Zhao Z, Li Y, Li J, Zheng J, Wang W, Zhao Q, Ji G. Increased MicroRNA-630 Expression in Gastric Cancer Is Associated with Poor Overall Survival. PLoS One. 2014;9:e90526. doi: 10.1371/journal.pone.0090526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao JJ, Chen PJ, Duan RQ, Li KJ, Wang YZ, Li Y. Up-regulation of miR-630 in clear cell renal cell carcinoma is associated with lower overall survival. Int J Clin Exp Pathol. 2014;7:3318–3323. [PMC free article] [PubMed] [Google Scholar]
- 17.Farhana L, Dawson MI, Murshed F, Das JK, Rishi AK, Fontana JA. Upregulation of miR-150* and miR-630 induces apoptosis in pancreatic cancer cells by targeting IGF-1R. PLoS One. 2013;8:e61015. doi: 10.1371/journal.pone.0061015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corcoran C, Rani S, Breslin S, Gogarty M, Ghobrial IM, Crown J, O’Driscoll L. miR-630 targets IGF1R to regulate response to HER-targeting drugs and overall cancer cell progression in HER2 over-expressing breast cancer. Mol Cancer. 2014;13:71. doi: 10.1186/1476-4598-13-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Chuang A, Hao H, Talbot C, Sen T, Trink B, Sidransky D, Ratovitski E. Phospho-ΔNp63α is a key regulator of the cisplatin-induced microRNAome in cancer cells. Cell Death Differ. 2011;18:1220–1230. doi: 10.1038/cdd.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Guerrero-Preston R, Ratovitski EA. Phospho-ΔNp63α-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle. 2012;11:1247. doi: 10.4161/cc.11.6.19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo TC, Tan CT, Chang YW, Hong CC, Lee WJ, Chen MW, Jeng YM, Chiou J, Yu P, Chen PS, Wang MY, Hsiao M, Su JL, Kuo ML. Angiopoietin-like protein 1 suppresses SLUG to inhibit cancer cell motility. J Clin Invest. 2013;123:1082–1095. doi: 10.1172/JCI64044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36–45. doi: 10.1158/0008-5472.CAN-09-3153. [DOI] [PubMed] [Google Scholar]
- 23.Galluzzi L, Morselli E, Vitale I, Kepp O, Senovilla L, Criollo A, Servant N, Paccard C, Hupé P, Robert T. miR-181a and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010;70:1793–1803. doi: 10.1158/0008-5472.CAN-09-3112. [DOI] [PubMed] [Google Scholar]
- 24.Aoyama M, Ozaki T, Inuzuka H, Tomotsune D, Hirato J, Okamoto Y, Tokita H, Ohira M, Nakagawara A. LMO3 interacts with neuronal transcription factor, HEN2, and acts as an oncogene in neuroblastoma. Cancer Res. 2005;65:4587–4597. doi: 10.1158/0008-5472.CAN-04-4630. [DOI] [PubMed] [Google Scholar]
- 25.Larsen S, Yokochi T, Isogai E, Nakamura Y, Ozaki T, Nakagawara A. LMO3 interacts with p53 and inhibits its transcriptional activity. Biochem Biophys Res Commun. 2010;392:252–257. doi: 10.1016/j.bbrc.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 26.Hui L, Ji C, Hui B, Lv T, Ha X, Yang J, Cai W. The oncoprotein LMO3 interacts with calcium-and integrin-binding protein CIB. Brain Res. 2009;1265:24–29. doi: 10.1016/j.brainres.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 27.Isogai E, Ohira M, Ozaki T, Oba S, Nakamura Y, Nakagawara A. Oncogenic LMO3 collaborates with HEN2 to enhance neuroblastoma cell growth through transactivation of Mash1. PLoS One. 2011;6:e19297. doi: 10.1371/journal.pone.0019297. [DOI] [PMC free article] [PubMed] [Google Scholar]


