Abstract
Monosomy of chromosome 17 may affect the assessment of HER2 amplification. Notably, the prevalence ranges from 1% up to 49% due to lack of consensus in recognition. We sought to investigate the impact of monosomy of chromosome 17 to interpretation of HER2 gene status. 201 breast carcinoma were reviewed for HER2 gene amplification and chromosome 17 status. FISH analysis was performed by using double probes (LSI/CEP). Absolute gene copy number was also scored per each probe. HER2 FISH test was repeated on serial tissue sections, ranging in thickness from 3 to 20 µm. Ratio was scored and subsequently corrected by monosomy after gold control test using the aCGH method to overcome false interpretation due to artefactual nuclear truncation. HER2 immunotests was performed on all cases. 26/201 cases were amplified (13%). Single signals per CEP17 were revealed in 7/201 (3.5%) cases. Five out of 7 cases appeared monosomic with aCGH (overall, 5/201, 2.5%) and evidenced single signals in >60% of nuclei after second-look on FISH when matching both techniques. Among 5, one case showed amplification with a pattern 7/1 (HER2/CEP17>2) of copies (3+ at immunotest); three cases revealed single signals per both probes (LSI/CEP=1) and one case revealed a 3:1 ratio; all last 4 cases showed 0/1+ immunoscore. We concluded that: 1) monosomy of chromosome 17 may be observed in 2.5% of breast carcinoma; 2) monosomy of chromosome 17 due to biological reasons rather than nuclear truncation was observed when using the cut-off of 60% of nuclei harboring single signals; 3) the skewing of the ratio due to single centromeric 17 probe may lead to false positive evaluation; 4) breast carcinomas showing a 3:1 ratio (HER2/CEP17) usually show negative 0/1+ immunoscore and <6 gene copy number at FISH.
Keywords: Breast carcinoma, monosomy, chromosome 17, HER 2 amplification, FISH analysis, double probes, aCGH method, single signals, ratio (HER2/CEP 17), false positive
Introduction
The presence of monosomy of chromosome 17 among breast carcinoma has been already documented [1,2]. Differently to the biological patterns of gains and/or polysomies of chromosome 17 [3], the impact of monosomy of chromosome 17 during the HER2 gene testing, has not been extensively studied [4]. At clinical level, the features of monosomy of chromosome 17 is still a matter of debate, with some arguing a worse prognostic value and less responsiveness to anti-HER2 targeted therapy [1]. The most recent guidelines (ASCO/CAP 2013) addressed appropriate HER2 scoring among common and uncommon patterns of HER2 values [5,6] and about the monosomy of chromosome 17 it has been observed that this pattern may lead to overall false interpretation, caused by the skewing of the ratio due to single centromeric 17 probe loss [4]. Some reports also compared the presence of single signals due to the artefactual nuclear truncation versus those being as true biological pattern [7], however no consensus is actually achieved. The recognition of gains/polysomy of chromosome 17, was changed along time and the overall interpretation was gradually modified through consensus and guidelines [8,9], differently from the recognition of monosomy of chromosome 17. We sought to evaluate the incidence and impact of monosomy of chromosome 17 to HER2 testing, focusing at technical level the analytical phase, to avoid misinterpretation due to nuclear truncation for appropriate HER2 gene interpretation.
Material and methods
Samples
201 consecutive cases were recruited from the Database of the Fluorescent Molecular Lab Verona, Italy. Cases represented breast carcinoma with matched immunophenotypical and molecular analysis for HER2. All cases were tested for HER2 by immunohistochemistry and by FISH technique (not only score 2+ cases).
Immunophenotypical analysis
All cases were tested with the Hercept test (Dako Glostrup, Denmark) and scored according to 2013 ASCO/CAP [5].
Molecular interphase cytogenetic FISH analysis
Analysis was performed on formalin-fixed, paraffin embedded tissues. Cytogenetic analysis was performed using commercially available locus-specific probes mapping the chromosome 17 centromere and HER2 hot locus (Abbott-Vysis, Milan). FISH procedure was developed according to standard protocol from the company. Five-micron sections were cut from paraffin-embedded blocks. The slides were examined using an Olympus BX61 (Olympus, Hamburg, Germany) with appropriate filters for Spectrum Orange, Spectrum Green and the UV filter for the DAPI nuclear counterstain. The signals were recorded with a CCD camera (CytoVysion, Olympus Berlin, Germany).
Controls
To avoid false interpretation of single fluorescent signals due to nuclear truncation at serial sections of the paraffin blocks, we retested cases showing single signals for HER2 by using additional thickness ranging from 3 to 5, 10, 15 and up to 20 µm per case. Monosomy of chromosome 17 was assessed when at least 60% of nuclei harbored single fluorescent centromeric signals. Ratio was initially scored and subsequently corrected by monosomy. Again, HER2 immunotests (Hercept Test) were matched for all cases.
Array comparative genomic hybridization (aCGH) analysis
Genomic DNA was isolated using QIAamp DNA mini kit (Qiagen, Valensia, CA, Usa) and quantified on the NanoDrop-1000 Spectrophotometer (NanoDrop Technologies Inc., DE, USA). The quantities and qualities of the DNA allowed us to continue with the analyses. With Agilent Human 244K array format/platform containing around 244000 oligonucleotide probes, covering the coding and noncoding genome regions (Agilent Technologies, Santa Clara, CA), we tested tumors for assessment of the genomic profile, previously screened by FISH analysis. Briefly, 1.5 µg of tumor DNA and gender-matched reference DNA were digested, labeled, hybridized and washed according to Agilent Oligonucleotide Array-Based CGH for Genomic DNA analysis protocol version 6.1 (Agilent Technologies, Santa Clara, CA, USA). The digested DNAs were labeled by random priming with Cy3-dUTP (reference DNA) and Cy5-dUTP (patient DNA) by use of the Agilent Complete SureTag Complete DNA Labeling Kit (Agilent Technologies, Santa Clara, CA, USA), after which the labeled DNAs were purified. The resolution of the fragments analyzed varied from several Kb to 1 Mb depending upon the case selected. The array images obtained after scanning (high resolution Agilent scanner) were processed with the Feature Extraction software (v11.5.1.1.), and the output data files were analyzed with the Agilent CytoGenomics v.2.0.6.0 Software. To identify copy number alterations we used Aberration Detection Method 2 (ADM-2) algorithm with sensitivity threshold of 6.0. (and in our lab the default aberration filter with minimum number or probes in region is 3 and minimum absolute average log ratio of region is 0.25) and to exclude the small variances in the data we set up a custom aberration filter identifying an alterations in copy number if minimum of 8 probes gained or lost to be present and with minimum absolute average log ratio for region to be 0.5. The region with small copy number variations were excluded by comparing and visualizing the copy number variant regions tool of the CytoGenomics software.
Interpretation
We primarily scored the absolute numbers of HER2 spots and the centromeric ones. For the purpose of the actual study we initially categorized a case as “amplified” when the presence of absolute number of HER2 set >6 and if the ratio (HER2/CEP17) set ≥2. We also categorized the presence of single signals for chromosome 17 reporting the percentage and the interpretation of monosomic status of the chromosome. We secondly matched the score of the analytical phase to interpretation of aCGH findings and corrected the false positive and negative interpretation.
Digital imaging analysis
FISH slides were also digitalized by D-Sight/Fluo instrument (Visia Imaging, Florence, Italy). FISH digital analysis was performed as additional control for scoring, due to the capacity of the instruments to assess hundred of nuclei.
Ethical issues
Our internal Institutional review board approved the design and aim of the actual study (07012015 request).
Results
Clinico-pathological and molecular findings are summarized in Tables 1 and 2.
Table 1.
Histopathological and immunophenotypical features of 201 consecutive breast carcinomas
| Breast cancers, n (%) | |
|---|---|
| Histotype | |
| Ductal | 159 (79.1) |
| Lobular | 31 (14.4) |
| Special types | 11 (5.5) |
| Grade | |
| G1 | 58 (28.9) |
| G2 | 113 (56.2) |
| G3 | 30 (14.9) |
| pT | |
| pT1 | 142 (70.6) |
| pT2 | 49 (23.4) |
| pT3 | 10 (5) |
| pN | |
| N+ | 49 (24.4) |
| N- | 152 (75.6) |
| Molecular subtype | |
| Luminal A | 96 (47.8) |
| Luminal B | 54 (26.9) |
| HER2-enriched | 23 (11.4) |
| Triple negative | 28 (13.9) |
Table 2.
Breast carcinoma with monosomy of chromosome 17: relevant clinico-pathological and molecular characters
| Case | Age | pT | Molecular subtype | HER2 IHC | FISH Nuclei with loss CEP17 (%) | HER2 status by FISH | aCGH status on chr. 17 | Interpretation losses of CEP17 |
|---|---|---|---|---|---|---|---|---|
| 1 | 77 | pT1 | Luminal A | 1 | 70 | Not amplified | Whole loss | Monosomy |
| 2 | 63 | pT2 | Triple negative | 1 | 80 | Not amplified | Loss 17p and gain 17q | Monosomy |
| 3 | 38 | pT1 | Luminal A | 0 | 70 | Not amplified | Loss part of chr 17q | Monosomy |
| 4 | 34 | pT1 | Triple negative | 0 | 75 | Not amplified | Loss 17p and part of 17q | Monosomy |
| 5 | 53 | pT1 (bilateral) | Luminal B | 3 | 80 | Amplified | Loss part of 17q | Monosomy |
| 6 | 42 | pT1 | Luminal A | 0 | 30 | Not amplified | Flat | Nuclear truncation |
| 7 | 80 | pT3 | Triple negative | 1 | 20 | Not amplified | Flat | Nuclear truncation |
Pathological findings
159/201 (79.1%) breast carcinomas showed ductal differentiation (Figure 1), 31/201 (14.4%) lobular and the remaining minor subtypes. 58/201 (28.9%) revealed G1, 113 (56.2%) G2 and 30 (14.9%) G3 grading of differentiation. 142/201 (70.6%) staged pT1, 49/201 (23.4%) pT2 and 10/201 (5%) pT3.
Figure 1.
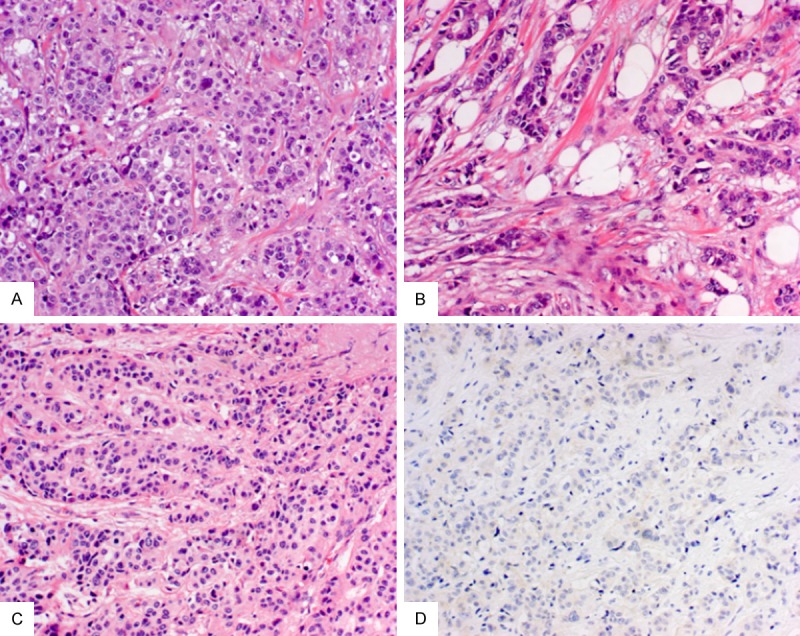
Breast carcinoma with monosomy of chromosome 17. Ductal phenotype, luminal A young age (2/5 cases <38 years) (A); triple negative breast carcinoma (2/5 cases) (B); a case of bilateral breast carcinoma (1/5) (C). Four out of five displayed a negative (0 or 1+) HER2 immunoscore (D).
Immunophenotypical findings
96/201 (47.8%) of breast carcinomas were Luminal A, 54/201 (26.9%) Luminal B, 23/201 (11.4%) HER2-enriched and 28/201 (13.9%) triple negative (Figure 1).
FISH cytomolecular findings
26/201 cases were amplified (13%). Single signals per CEP17 were revealed in 7/201 (3.5%) cases. Five out of 7 cases (overall 2/201, 2.5%) appeared monosomic after matching with aCGH analysis and evidenced single signals in >60% of nuclei after second-look on FISH analysis (Figure 2; Table 2). Among 5, one case showed amplification with a pattern 7/1 (HER2/CEP17>2) of copies (3+ at immunotest); three cases revealed single signals per both probes (LSI/CEP=1) and one case revealed a 3:1 (LSI/CEP>2) count ratio; importantly all last 4 cases showed 0/1+ score at immunotest. All cases except one (6/7) exhibited <6 absolute HER2 gene copy number. One case showed seven signals.
Figure 2.
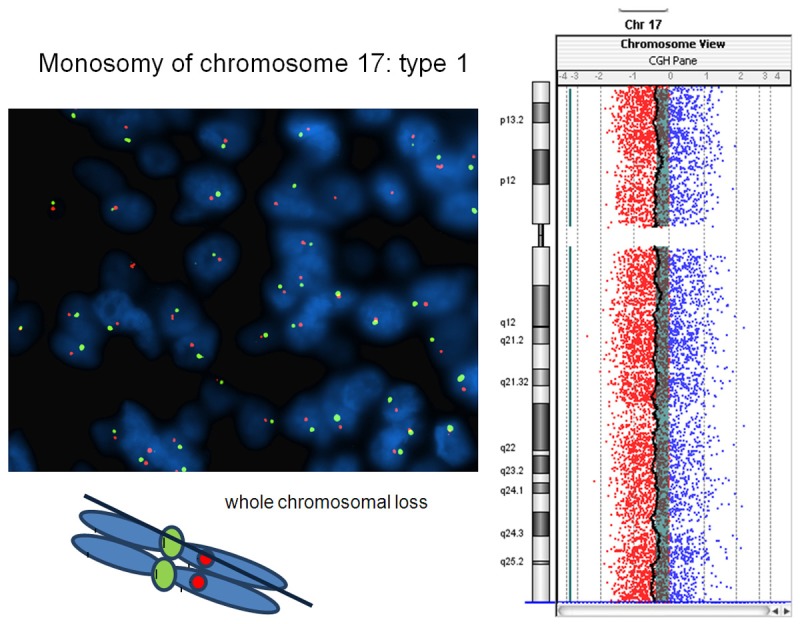
Breast carcinoma with monosomy of chromosome 17. The tumor harbors single signals in >60% of neoplastic nuclei per both LSI Her-2/neu and CEP17 loci at FISH analysis and reveals loss of whole chromosome 17 (type 1 monosomy) after aCGH molecular profiling.
aCGH molecular findings
Two cases revealed complete loss of whole chromosome 17 (Figure 2). One case revealed a complete loss of chromosome 17q and part of chromosome 17p (Figure 3). One case evidenced a loss of chromosome 17q with a partial gains of the telomeric chromosome 17p region. One case showed a complex chromosome rearrangement and imbalance on chromosome 17 (possible loss of chromosome 17 with the formation of an isochromosome for long arm of chromosome 17 and an unbalanced translocation with retention of the derivative chromosome containing 17q material and the HER2 locus). All these cases revealed >60% of neoplastic nuclei harboring single signals per CEP17 at FISH analysis (Table 2; Figure 4). Type 1 chr17 monosomy reveals loss of whole chromosome 17 whereas type 2 chr17 monosomy show partial losses of chromosome 17 (i.e. loss of 17q, CEP17 and part of 17p). Two cases revealed a flat profile with absence of chromosome 17 aberrations (Table 2). These cases revealed <60% nuclei harboring single signals. At Hercept test, three cases showed score 0, three cases showed score 1+, one case scored 3+ (Figure 1). The case showing 3+ at immunotest showed a pattern 7/1 (HER2/CEP17>2) at FISH test. The remaining showed respectively 1/1 in four cases and 3:1 in one case.
Figure 3.
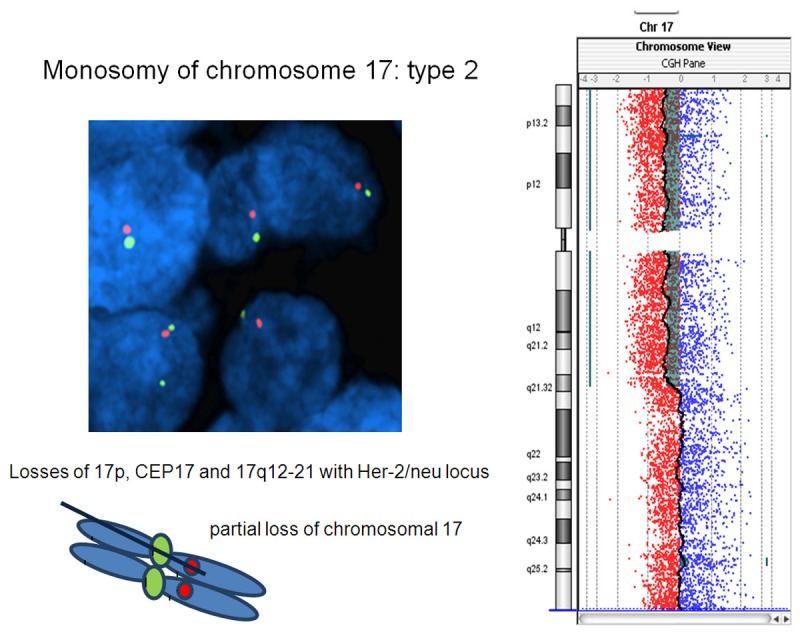
Breast carcinoma with partial monosomy of chromosome 17. The tumor harbors single signals per both LSI Her-2/neu and CEP17 probes in >60% of neoplastic nuclei and evidenced losses of chromosome 17p, CEP17 and the partial chromosome 17q12-21 (type 2 monosomy).
Figure 4.
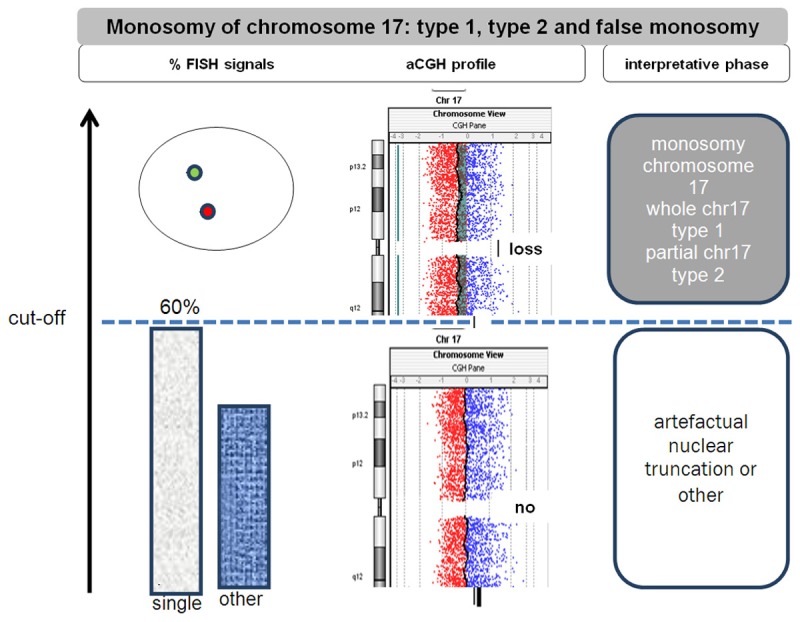
Recommendations’ on Her-2/neu FISH testing when single signals is observed at FISH analysis; when monosomy of chromosome 17 is observed (>60% nuclei with single CEP17 probe), use the single absolute Her-2/neu gene copy number (>6) when distinguishing a case of breast carcinoma with amplification versus not amplified. Type 1 chr17 monosomy reveals loss of whole chromosome 17 whereas type 2 chr17 monosomy show partial losses of chromosome 17 (i.e. loss of 17q, CEP17 and part of 17p).
Discussion
In the present study we show that: 1) monosomy of chromosome 17 may be observed in 2.5% of breast carcinomas; 2) monosomy due to biological reasons rather than nuclear truncation is observed when the cut-off set to 60% of nuclei; 3) the skewing of the ratio due to single centromeric 17 probe may lead to false positive evaluation of HER2 gene amplification; 4) when dealing with cases showing a 3:1 ratio (HER2/CEP17) the interpretation should be “HER2 negative”, being Her-2 negative at immunolevel and characterized by <6 Her-2/neu signals. It is well known that chromosome 17 in breast cancer shows very complex abnormalities and aneusomy of chromosome 17 can lead to troublesome Her2 gene assessment in routine diagnostic practice [10]. It has been demonstrated that chromosome 17 polysomy is rare, but few data on monosomy are present in the literature, especially when dealing with interphase nuclei assessable on formalin fixed paraffin embedded samples. In the actual study, we show an analytical phase for the assessment of monosomy of chromosome 17 in a series of consecutive breast carcinomas. Based on our data, monosomy of chromosome 17 may be observed in about 2.5% of breast carcinomas. Among the Literature few data are recruitable due to the fact that most previous researches focused on gene amplification, polisomy and/or heterogeneity. Moreover there are no strictly standard guidelines on how to assess monosomy of chromosome 17 in interphase nuclei. The recent ASCO/CAP 2013 guidelines pose attention to the identification of unusual HER2 genotypic abnormalities, like aneusomy of chromosome 17 (i.e monosomy), that affect HER2/CEP17 ratio in dual signal in situ hybridization assays. From cytogenetists it is known that the skewing of the ratio due to single centromeric 17 probe may bring to false positive evaluation of gene amplification. All cases with monosomy of chromosome 17 except one did not show HER2 immunoexpression. In the literature a subset of such a cases may harbor pitfalls when the copy number of HER2 duplicate, showing a 3:1 ratio. These scenarios happen when i.e. uniparental disomy and polyploidy nuclei replicate or when breast carcinoma BRCA positive progresses. Definitions of aneusomy 17 differ with the threshold criteria for monosomy. Setting the thresholds is complicated not only by genomic heterogeneity and proliferative activity of tumors, but also by the substantial nuclear truncation resulting from tissue sectioning. Pathologists and biologists routinely dealing with molecular tests such as HER2 FISH assay are well aware of the presence of nuclear truncation. We know that up to 30% of cells may display monosomy due to artefactual reasons. To overcome this gap, we retested all seven cases by using different thickness of tissue sections ranging from 3 to 5, 10, 15 and up to 20 µm. All 5 cases showing the entire or partial loss of the chromosome 17 at aCGH analysis maintained the pattern of single signals in more than 1000 nuclei counted per case. Moreover, the digitalization of the images permitted the count of thousands of nuclei in at least five spots of the neoplastic tissue. The two cases not confirmed at aCGH analysis as having a potential monosomic status, revealed at aCGH presence of single signals per chromosome 17 in no more than 45% of nuclei at 5 µm thickness tissue sections. Cases that were confirmed as monosomic at aCGH analysis harbored at least 60% of nuclei with single fluorescent orange signals on tissue section evaluation. Type 1 chr17 monosomy reveals loss of whole chromosome 17 whereas type 2 chr17 monosomy shows partial losses of chromosome 17 (i.e. loss of 17q, CEP17 and part of 17p). When considering the cut-off of 60% at FISH test, the cases matched with those characterized by monosomy at aCGH analysis. The aCGH technique is not biased by nuclear truncation and may be considered as gold standard. Reinholtz et al. reported similar findings (cut-off of 60% of nuclei) when evaluating thresholds for the recognition of monosomic cases (one centromeric 17 signals). The higher the threshold is set, the lower is the possibility to face artefactual monosomy due to nuclear truncation; Nakopoulou et al., Reinholz et al. and Jemenez et al. respectively set the cut-off at least 40% of neoplastic nuclei, to 60%, up to 80% for the monosomic status [11,12]. Kouvaras et al. focused on nuclear truncation during breast carcinoma HER2 assessment. The HER2/CEP 17 ratios were compared in routine (4 μm) vs. thicker (15 μm) tissue sections. HER2 and CEP 17 number of signals increased in thick sections; however, HER2/CEP 17 ratios were decreased [7]. They concluded that at least a subset of the equivocal cases could represent an artefactual increase of the ratio related to nuclear truncation and loss of peripheral CEP 17 signals in routine sections. In our study we assessed the cut-off percentage of nuclei harboring single signals due to nuclear truncation (<60%) versus those being characterized by biological pattern of monosomy of chromosome 17 (>60%). Gunn et al. matched aCGH and FISH analysis on 20 formalin fixed paraffin embedded tissue samples from newly diagnosed cases of invasive ductal carcinoma referred to their laboratory with unresolved HER2 status and revealed two false positives and one false negative by FISH due to “ratio skewing” caused by chromosomal gains and losses in the centromeric region [13]. Marchiò et al. elegantly evidenced the presence of part of chromosome 17 such as the single centromeric region to be gained in apparent polysomic cases. The use of aCGH is useful in this contex of aneusomy of chromosome 17. In our cohort of cases the gold standard aCGH revealed in 4 cases the complete loss of the entire chromosome 17 (p and q arms and the centromeric region) and in one case a partial loss of two regions on the opposite arms. Reported incidences ranged from 0% to 49% for monosomy [2]. Some Authors reported the presence of cases having monosomy of chromosome 17 when a mean of one single molecular signal was assessed per nuclei [14,15]. Other Authors performed the assessment of fine needle aspiration or touch preparation and interpreted the monosomic status not only when dealing with one signals per chromosome 17 per nuclei but also when no signals were observed [12,16-20]; a similar interpretation was scored on formalin-fixed and paraffin embedded tissues [11,21]. A modulation of the score to assess the monosomic status of chromosome 17 ranged from 1.23 to 1.75 [22-26]. Salido et al. reported the incidence of 2% of monosomy without specification of the cut-off [27]. Kokate et al. evaluated monosomy 17 and found a 7.3% incidence and was more preponderant in FISH negative cases [28]. Our findings are also important for quality scheme of HER2 assessment and external quality control. Most of these cases are considered negative by using a single probe approach (i.e <6 absolute copy number in 6/7 cases from the actual study) and do show 0 and or 1+ at immunohistochemistry. Taking into account the ratio score by dual color ISH, the two cases showing analytically ratio >2 (3:1 LSI/CEP17 pattern) do not match with interpretation by using a single probe approach (three signals are <6). These contradictory findings are not due to pre-analytical problematic and agreement for interpretation is needed. At clinical level, preliminary findings suggest that patients with metastatic breast cancer with HER2 amplification and chromosome 17 monosomy did not respond to trastuzumab [1]. Monosomy 17 also has been associated with nodal metastasis [11]. Results from N9831 further suggest that patients with primary breast cancer with HER2/CEP17 ratios greater than 15, most of whom displayed monosomy 17, did not benefit from adjuvant trastuzumab (hazard ratio 1.01) [29]. Interestingly, breast carcinoma with monosomy of chromosome 17 from our study, respectively showed a ductal phenotype, luminal A with young age in 2 out of 5 cases (<38 years), a triple negative phenotype in 2/5 cases and one case the patient was affected bilaterally by cancer. HER2 gene final interpretation is the results of a strong analytical phase. The analytical phase proposed from our study in such a cases is as follows: firstly, number the absolute copy number of HER2 and the centromeric color probes; secondly, assess the cases having the monosomic status if the centromeric probe show single signals in major than 60% of neoplastic nuclei (biological pattern rather than nuclear artefactual truncation); thirdly, the overall interpretation has to be posed to the absolute copy number of HER2 gene (>6 HER2 gene amplification versus <6 HER2 not amplified). The subset 3:1 HER2/CEP17 patterns does merit a distinct evaluation at clinical level in order to test its predictiveness to the efficacy of actual targeted therapies already available or available in the near future. Breast carcinomas showing a 3:1 ratio (HER2/CEP17) usually show negative 0 or 1+ immunoscore and <6 overall gene copy number at FISH analysis by single probe approach.
Acknowledgements
Supported by 1) Internal Funding (MB FUR 2013), Department of Pathology and Diagnostic, University of Verona, 2) Italian Association for Cancer Research AIRC (MFAG13310) 3) Cancer Society of Finland and 4) Sigrid Jusélius Foundation, Finland.
Disclosure of conflict of interest
None.
References
- 1.Risio M, Casorzo L, Redana S, Montemurro F. HER2 gene-amplified breast cancers with monosomy of chromosome 17 are poorly responsive to trastuzumab-based treatment. Oncol Rep. 2005;13:305–9. [PubMed] [Google Scholar]
- 2.Reinholz MM, Bruzek AK, Visscher DW, Lingle WL, Schroeder MJ, Perez EA, Jenkins RB. Breast cancer and aneusomy 17: implications for carcinogenesis and therapeutic response. Lancet Oncol. 2009;10:267–77. doi: 10.1016/S1470-2045(09)70063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marchio C, Lambros MB, Gugliotta P, Di Cantogno LV, Botta C, Pasini B, Tan DS, Mackay A, Fenwick K, Tamber N, Bussolati G, Ashworth A, Reis-Filho JS, Sapino A. Does chromosome 17 centromere copy number predict polysomy in breast cancer? A fluorescence in situ hybridization and microarray-based CGH analysis. J Pathol. 2009;219:16–24. doi: 10.1002/path.2574. [DOI] [PubMed] [Google Scholar]
- 4.Starczynski J, Atkey N, Connelly Y, O’Grady T, Campbell FM, di Palma S, Wencyk P, Jasani B, Gandy M, Bartlett JM. HER2 gene amplification in breast cancer: a rogues’ gallery of challenging diagnostic cases: UKNEQAS interpretation guidelines and research recommendations. Am J Clin Pathol. 2012;137:595–605. doi: 10.1309/AJCPATBZ2JFN1QQC. [DOI] [PubMed] [Google Scholar]
- 5.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, Spears PA, Vance GH, Viale G, Hayes DF. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J. Clin. Oncol. 2013;31:3997–4013. doi: 10.1200/JCO.2013.50.9984. [DOI] [PubMed] [Google Scholar]
- 6.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, Hanna W, Jenkins RB, Mangu PB, Paik S, Perez EA, Press MF, Spears PA, Vance GH, Viale G, Hayes DF. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138:241–56. doi: 10.5858/arpa.2013-0953-SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kouvaras E, Papandreou CN, Daliani DD, Athanasiadis A, Koukoulis GK. Comparative study of spatial localization of HER-2 and CEP17 signals and of HER-2/CEP17 ratios, in “thin” and “thick” tissue sections. Breast. 2012;21:34–9. doi: 10.1016/j.breast.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. Arch Pathol Lab Med. 2007;131:18–43. doi: 10.5858/2007-131-18-ASOCCO. [DOI] [PubMed] [Google Scholar]
- 9.Hanna WM, Ruschoff J, Bilous M, Coudry RA, Dowsett M, Osamura RY, Penault-Llorca F, van de Vijver M, Viale G. HER2 in situ hybridization in breast cancer: clinical implications of polysomy 17 and genetic heterogeneity. Mod Pathol. 2014;27:4–18. doi: 10.1038/modpathol.2013.103. [DOI] [PubMed] [Google Scholar]
- 10.Rondon-Lagos M, Verdun Di Cantogno L, Rangel N, Mele T, Ramirez-Clavijo SR, Scagliotti G, Marchio C, Sapino A. Unraveling the chromosome 17 patterns of FISH in interphase nuclei: an in-depth analysis of the HER2 amplicon and chromosome 17 centromere by karyotyping, FISH and M-FISH in breast cancer cells. BMC Cancer. 2014;14:922. doi: 10.1186/1471-2407-14-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakopoulou L, Giannopoulou I, Trafalis D, Gakiopoulou H, Keramopoulos A, Davaris P. Evaluation of numeric alterations of chromosomes 1 and 17 by in situ hybridization in invasive breast carcinoma with clinicopathologic parameters. Appl Immunohistochem Mol Morphol. 2002;10:20–8. doi: 10.1097/00129039-200203000-00004. [DOI] [PubMed] [Google Scholar]
- 12.Jimenez RE, Wallis T, Tabasczka P, Visscher DW. Determination of Her-2/Neu status in breast carcinoma: comparative analysis of immunohistochemistry and fluorescent in situ hybridization. Mod Pathol. 2000;13:37–45. doi: 10.1038/modpathol.3880007. [DOI] [PubMed] [Google Scholar]
- 13.Gunn S, Yeh IT, Lytvak I, Tirtorahardjo B, Dzidic N, Zadeh S, Kim J, McCaskill C, Lim L, Gorre M, Mohammed M. Clinical array-based karyotyping of breast cancer with equivocal HER2 status resolves gene copy number and reveals chromosome 17 complexity. BMC Cancer. 2010;10:396. doi: 10.1186/1471-2407-10-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merola R, Mottolese M, Orlandi G, Vico E, Cognetti F, Sperduti I, Fabi A, Vitelli G, Cianciulli AM. Analysis of aneusomy level and HER-2 gene copy number and their effect on amplification rate in breast cancer specimens read as 2+ in immunohistochemical analysis. Eur J Cancer. 2006;42:1501–6. doi: 10.1016/j.ejca.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Farabegoli F, Ceccarelli C, Santini D, Baldini N, Taffurelli M, Marrano D, Trere D, Derenzini M. c-erbB-2 over-expression in amplified and non-amplified breast carcinoma samples. Int J Cancer. 1999;84:273–7. doi: 10.1002/(sici)1097-0215(19990621)84:3<273::aid-ijc13>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Fehm T, Morrison L, Saboorian H, Hynan L, Tucker T, Uhr J. Patterns of aneusomy for three chromosomes in individual cells from breast cancer tumors. Breast Cancer Res Treat. 2002;75:227–39. doi: 10.1023/a:1019901010758. [DOI] [PubMed] [Google Scholar]
- 17.Tsukamoto F, Miyoshi Y, Egawa C, Kasugai T, Takami S, Inazawa J, Noguchi S. Clinicopathologic analysis of breast carcinoma with chromosomal aneusomy detected by fluorescence in situ hybridization. Cancer. 2001;93:165–70. doi: 10.1002/cncr.9024. [DOI] [PubMed] [Google Scholar]
- 18.McManus DT, Patterson AH, Maxwell P, Humphreys MW, Anderson NH. Fluorescence in situ hybridisation detection of erbB2 amplification in breast cancer fine needle aspirates. Mol Pathol. 1999;52:75–7. doi: 10.1136/mp.52.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichikawa D, Hashimoto N, Hoshima M, Yamaguchi T, Sawai K, Nakamura Y, Takahashi T, Abe T, Inazawa J. Analysis of numerical aberrations of specific chromosomes by fluorescent in situ hybridization as a diagnostic tool in breast cancer. Cancer. 1996;77:2064–9. doi: 10.1002/(SICI)1097-0142(19960515)77:10<2064::AID-CNCR15>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 20.Takehisa M, Sasa M, Bando Y, Hirose T, Morimoto T, Nagao T, Tangoku A. Chromosomal aneusomy (chr 1, 11, 17) detected by fluorescence in situ hybridization may be a prognostic factor in breast cancer. Anticancer Res. 2007;27:1073–8. [PubMed] [Google Scholar]
- 21.Bose S, Mohammed M, Shintaku P, Rao PN. Her-2/neu gene amplification in low to moderately expressing breast cancers: possible role of chromosome 17/Her-2/neu polysomy. Breast J. 2001;7:337–44. doi: 10.1046/j.1524-4741.2001.21018.x. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, Hossein Saboorian M, Frenkel EP, Haley BB, Siddiqui MT, Gokaslan S, Hynan L, Ashfaq R. Aneusomy 17 in breast cancer: its role in HER-2/neu protein expression and implication for clinical assessment of HER-2/neu status. Mod Pathol. 2002;15:137–45. doi: 10.1038/modpathol.3880505. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y, Lespagnard L, Durbecq V, Paesmans M, Desmedt C, Gomez-Galdon M, Veys I, Cardoso F, Sotiriou C, Di Leo A, Piccart MJ, Larsimont D. Polysomy 17 in HER-2/neu status elaboration in breast cancer: effect on daily practice. Clin Cancer Res. 2005;11:4393–9. doi: 10.1158/1078-0432.CCR-04-2256. [DOI] [PubMed] [Google Scholar]
- 24.Watters AD, Going JJ, Cooke TG, Bartlett JM. Chromosome 17 aneusomy is associated with poor prognostic factors in invasive breast carcinoma. Breast Cancer Res Treat. 2003;77:109–14. doi: 10.1023/a:1021399923825. [DOI] [PubMed] [Google Scholar]
- 25.Hyun CL, Lee HE, Kim KS, Kim SW, Kim JH, Choe G, Park SY. The effect of chromosome 17 polysomy on HER-2/neu status in breast cancer. J Clin Pathol. 2008;61:317–21. doi: 10.1136/jcp.2007.050336. [DOI] [PubMed] [Google Scholar]
- 26.Beser AR, Tuzlali S, Guzey D, Dolek Guler S, Hacihanefioglu S, Dalay N. HER-2, TOP2A and chromosome 17 alterations in breast cancer. Pathol Oncol Res. 2007;13:180–5. doi: 10.1007/BF02893497. [DOI] [PubMed] [Google Scholar]
- 27.Salido M, Tusquets I, Corominas JM, Suarez M, Espinet B, Corzo C, Bellet M, Fabregat X, Serrano S, Sole F. Polysomy of chromosome 17 in breast cancer tumors showing an overexpression of ERBB2: a study of 175 cases using fluorescence in situ hybridization and immunohistochemistry. Breast Cancer Res. 2005;7:R267–73. doi: 10.1186/bcr996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kokate P, Sawaimoon S, Bhatia S, Mandava S. Evaluation of genetic status of HER-2/neu and aneusomy 17 by fluorescence in situ hybridization and comparison with immunohistochemistry assay from Indian breast cancer patients. Genet Test Mol Biomarkers. 2012;16:239–45. doi: 10.1089/gtmb.2011.0125. [DOI] [PubMed] [Google Scholar]
- 29.Perez EA, Reinholz MM, Hillman DW, Tenner KS, Schroeder MJ, Davidson NE, Martino S, Sledge GW, Harris LN, Gralow JR, Dueck AC, Ketterling RP, Ingle JN, Lingle WL, Kaufman PA, Visscher DW, Jenkins RB. HER2 and chromosome 17 effect on patient outcome in the N9831 adjuvant trastuzumab trial. J. Clin. Oncol. 2010;28:4307–15. doi: 10.1200/JCO.2009.26.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]


