Abstract
Many studies have suggested the significance of glycosyltransferase-mediated macromolecule glycosylation in the regulation of pluripotent states in human pluripotent stem cells (hPSCs). Here, we observed that the sialyltransferase ST6GAL1 was preferentially expressed in undifferentiated hPSCs compared to non-pluripotent cells. A lectin which preferentially recognizes α-2,6 sialylated galactosides showed strong binding reactivity with undifferentiated hPSCs and their glycoproteins, and did so to a much lesser extent with differentiated cells. In addition, downregulation of ST6GAL1 in undifferentiated hPSCs led to a decrease in POU5F1 (also known as OCT4) protein and significantly altered the expression of many genes that orchestrate cell morphogenesis during differentiation. The induction of cellular pluripotency in somatic cells was substantially impeded by the shRNA-mediated suppression of ST6GAL1, partially through interference with the expression of endogenous POU5F1 and SOX2. Targeting ST6GAL1 activity with a sialyltransferase inhibitor during cell reprogramming resulted in a dose-dependent reduction in the generation of human induced pluripotent stem cells (hiPSCs). Collectively, our data indicate that ST6GAL1 plays an important role in the regulation of pluripotency and differentiation in hPSCs, and the pluripotent state in human cells can be modulated using pharmacological tools to target sialyltransferase activity.
The glycosylation of macromolecules, including proteins and lipids, is critically involved in the regulation of numerous biochemical and physiological activities in eukaryotic cells1. It is known that glycosylation plays essential roles to ensure normal cell differentiation and embryogenesis2,3,4, but its function in human pluripotent stem cells (hPSCs) remains largely unclear. A recent report demonstrated that O-linked glycosylation influences cellular pluripotency by acting on core components of the pluripotency signaling network in mouse embryonic stem cells (mESCs)5, revealing a direct link between protein glycosylation and pluripotency regulation that is highly likely to exist in human cells.
Indeed, evidence suggests that protein modification by sialic acid-containing glycans and sialyltransferases may be important in the regulation of cellular pluripotency in human cells6,7,8,9. Characterization of significant differences between the glycomic composition of undifferentiated hPSCs and their differentiated derivatives revealed that sialylation is a type of glycomodification that is typically altered when hPSCs lose their pluripotency6,7. In human cells, the sialylation of macromolecules is regulated by many sialyltransferases, including ST6GAL1 (β-galactoside alpha-2,6-sialyltransferase 1)10,11. ST6GAL1 is a type II membrane protein that is typically found in the Golgi apparatus and catalyzes the transfer of sialic acid monosaccharide to galactose-containing substrates12. Using lectin microarrays and global gene expression profiling, we confirmed that there is a significant change in protein sialylation during differentiation in a large panel of hPSCs, and discovered the preferential expression of ST6GAL1 in undifferentiated hPSCs8,13. These results suggested that ST6GAL1 may be functionally important in hPSCs, and that changes in ST6GAL1 expression may impact the regulation of pluripotency and cell differentiation. Interestingly, another recent study has provided an additional indication that the loss of terminal sialylation on the cell surface of hPSCs may lead to neuronal differentiation14.
In this study, we set out to determine whether and how the sialyltransferase ST6GAL1 may be involved in the modulation of pluripotency in human cells. We used biochemical and molecular biology approaches to demonstrate that modifying ST6GAL1 expression levels in human cells induced phenotypic changes in the maintenance and establishment of pluripotency. In addition, systems biology tools were used to further dissect the signaling networks that are potentially modulated by ST6GAL1 in controlling the pluripotent state of hPSCs and cellular reprogramming.
Materials and Methods
Cell Culture
HDF51 (HDF-f; ScienCell Research Laboratories, Carlsbad, CA), normal mouse embryonic fibroblast (MEF), and DR4 MEF (ATCC, Manassas, VA) cells were cultured in DMEM containing 10% (15% for DR4 MEF cells) fetal bovine serum (FBS) at 37°C. HM (HEMl; ScienCell Research Laboratories, Carlsbad, CA) cells were cultured in melanocyte medium (MelM; ScienCell Research Laboratories, Carlsbad, CA). Methods for culturing undifferentiated hPSC lines used in our study were previously described8. WA07 and WA09 hESCs were obtained from the WiCell Stem Cell Bank (WiCell Research Institute, Madison, WI). MEL1 hESCs15 were provided by Dr. Andrew Laslett at CSIRO, Australia. The cells used in our study were free of mycoplasma, tested using a MycoAlertTM mycoplasma detection kit (Lonza, Walkersville, MD). Additional information with regard to the cells used in our study is provided in Supplementary Table 1.
ST6GAL1 Knockdown in hPSCs
pLKO.1 lentiviral vectors which express ST6GAL1 shRNA (shRNA2 targeting sequence: CGTGTGCTACTACTACCAG_TRCN0000035432, shRNA5 targeting sequence: CGCTGCTCTATGAGAAGAA_TRCN0000035429; Sigma Aldrich, St. Louis, MO) were used to knock down ST6GAL1 protein expression in undifferentiated hPSCs. Cells (~30% confluency) cultured in feeder-free conditions were transduced using the lentiviral vectors with and without ST6GAL1 shRNA. The transduced cells were allowed to recover in StemPro® hESC serum- and feeder-free medium (Life Technologies, Carlsbad, CA) for 24 hours. StemPro® hESC medium containing puromycin (2 μg/ml) was used to culture the transduced cells for an additional 3 days prior to sample collection.
Cellular Reprogramming and Differentiation
To generate induced pluripotency in HDFs, we primarily used retroviral vectors to deliver four reprogramming factors (POU5F1, SOX2, KLF4, and MYC genes) into HDF51 cells. Methods for retrovirus-mediated cell reprogramming were previously described8. The transgene-free hiPSCs used in this study were generated through Sendai virus-mediated cell reprogramming. The recombinant DNA work in this study was performed according to the National Institutes of Health guidelines. To test the effect of ST6GAL1 knockdown on the establishment of pluripotency, HDFs were co-transduced with the ST6GAL1 shRNA lentiviral expression vector and the retroviral vectors for reprogramming. The transduced cells were placed onto radiation-inactivated DR4 (multiple drug resistant) MEF feeder cells at a density of 1 × 104 cells per well of a six well plate and cultured for 14 days with puromycin selection (1 μg/ml for 4 days followed by 0.5 μg/ml for the rest of the culture period). To test the effect of a sialyltransferase inhibitor on the establishment of pluripotency, HDFs transduced with the retroviral vectors for reprogramming were placed onto radiation-inactivated MEF feeder cells at a density of 1 × 104 cells per well of a six well plate and cultured for 14 days with 3Fax-peracetyl Neu5Ac, a cell-permeable sialic acid analog (Millipore, Billerica, MA). The reprogramming efficiency was evaluated using an alkaline phosphatase (AP) staining kit II (Stemgent, Cambridge, MA). To test the effect of ST6GAL1 knockdown during reprogramming, the transduced cells were placed onto Geltrex® (Life Technologies, Carlsbad, CA)-coated wells at a density of ~3.8 × 105 (a quarter of the original cell number for transduction) cells per well of a six well plate and cultured for the indicated periods with puromycin selection. For non-directed differentiation of hPSCs by embryoid body (EB) formation, hPSCs grown on a MEF feeder layer were incubated with pre-warmed (37 °C) 300 U/ml Collagenase I (Worthington Biomedical Corp., Lakewood, NJ) in DMEM/F12 (Life Technologies, Carlsbad, CA), typically for 60–75 minutes, to yield small hPSC colony clumps in suspension and leave most of the feeder cells behind. The cell clumps were collected with minimal trituration into bFGF-deficient DMEM/F12 medium with L-glutamine containing 20% KnockOut™ Serum Replacement, 100 μM non-essential amino acids, and 100 μM ß-mercaptoethanol (hESC medium; all components from Life Technologies, Carlsbad, CA) and left to sediment by gravity for 20–30 minutes in an incubator, to enable the removal of residual MEFs from the supernatant fraction. The cells were washed, pelleted at low centrifugation speed (50 g for 2 minutes), and plated into non-adherent polystyrene petri dishes (Simport, Beloeil, Canada) in hESC medium containing 10 ng/ml bFGF and left undisturbed in an incubator for 24–48 hours to establish viable aggregate cultures before changing to differentiation culture conditions. Aggregates were collected into 25 ml conical skirt tubes (Greiner, Monroe, NC), left to sediment by gravity for ~30 minutes in an incubator, removing initial single cell debris in the supernatant, and replated to low adherence petri dishes in EB differentiation medium comprised of high glucose DMEM, 2 mM Glutamax, 1% v/v non-essential amino acids (all from Life Technologies, Carlsbad, CA) and 10% v/v fetal bovine serum (FBS), (Sigma-Aldrich, St. Louis, MO). Suspension cultures were subsequently replenished with EB differentiation medium each 3–4 days. EBs were collected into 50 ml conical tubes (BD Biosciences, San Jose, CA) following 7, 14, and 28 days of differentiation, washed twice with PBS and dissociated to single cell suspensions usingTrypLETM Express (Life Technologies, Carlsbad, CA) and a 15–30 minute incubation and gentle pipetting to assist breaking up the EB structures for the ease of flow cytometry analysis and cell sorting. The protocol used to generate melanocytic differentiated derivatives of hPSCs was reported in a previous study16.
Western and Lectin-mediated Blotting
Methods for Western blotting were described in our previously published report16. The primary antibodies used in this study were purchased from R&D Systems (ST6GAL1; cat# AF5924), Cell Signaling (POU5F1; cat# 2840), Millipore (NANOG; cat# MABD24) and MP Biomedicals (ACTIN; cat# 08691001). HRP-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). For SNA lectin-mediated blotting, 10 ug of total proteins from each sample were separated by SDS-PAGE and transferred onto nitrocellulose membranes. The membranes with transferred proteins were blocked using a polyvinyl alcohol solution to prevent non-specific binding. After blocking, the membranes were reacted with PBS containing Triton X-100 (0.1%) and biotinylated SNA lectin (2 μg/ml; Vector Laboratories, Burlingame, CA) at 4 °C for 2 hours. After thorough washing, the membranes were reacted with HRP-conjugated streptavidin (Jackson ImmunoResearch Laboratories, West Grove, PA) in PBS for 30 minutes. After thorough washing, the chemiluminescence of an ECL substrate catalyzed by HRP was detected using film exposure.
Flow Cytometry and Cell Sorting
To measure SNA binding, 5 × 105 cells per sample were harvested using Accutase (Life Technologies, Carlsbad, CA), pelleted, resuspended in Hank′s balanced salt solution (HBSS; Life Technologies, Carlsbad, CA) containing fluorescein-labeled SNA lectin (4 μg/ml; Vector Laboratories, Burlingame, CA), and incubated at 4°C with mild agitation for 45 minutes. At the end of the reaction, the cells were pelleted and resuspended in fresh HBSS. The fluorescence intensity of samples was analyzed using a FACSCalibur flow cytometer equipped with CellQuest Pro software (BD Biosciences, San Jose, CA). For measuring UEA-I binding and the expression of NANOG and POU5F1, aliquots of harvested cell samples were fixed using PBS containing 4% paraformaldehyde, perforated using PBS containing 0.1% Triton X-100, labeled with biotinylated UEA-I lectin (6.5 μg/ml; Vector Laboratories, Burlingame, CA) and specific antibodies targeting NANOG and POU5F1 (Millipore, Billerica, MA), and stained with fluorescein-conjugated streptavidin and secondary antibodies. The unstained, isotype and single color fluorophore controls were included for each cell sample. All tubes were kept on ice during lectin and antibody labeling procedures, and prior to multiple color analyses on a LSRII flow cytometry analyzer (BD Biosciences, San Jose, CA). A routine hPSC maintenance culture was harvested with TrypLETM Express to provide a Day 0 control for each analysis. For the subsequent collection of UEA-I positive and negative live cell populations from Day 0, 7,14 and 28 cultures of EB differentiation, cell suspensions were labeled only with the biotinylated UEA-I lectin and detected with fluorescein-conjugated streptavidin. Two populations of cells in each sample were gated and sorted on a FACSVantage Diva cell sorter (BD Biosciences, San Jose, CA). UEA-I positive and negative cell fractions of 100,000–700,000 (relative to differentiation state) were collected and pelleted for global gene expression profiling. For detecting apoptosis, 5 × 105 cells from each sample were stained using a dead cell apoptosis Kit (Alexa Fluor 488-conjugated annexin V and propidium iodide; Life Technologies, Carlsbad, CA) according to the manufacturer’s instruction. The fluorescence intensity indicating apoptotic cell death was analyzed using a LSRII flow cytometry analyzer (BD Biosciences, San Jose, CA).
Global Gene Expression and qRT-PCR Analysis
Global gene expression profiling was performed using HT-12v4 Human Gene Expression BeadChips (Illumina, Hayward, CA), according to the manufacturer’s instructions. Data were filtered for detection P value <0.01 in GenomeStudio (Illumina, Hayward, CA), and normalized using the LUMI package with RSN (Robust spline normalization) algorithm in R. Qlucore Omics Explorer was used to perform differential gene expression analysis and hierarchical clustering. The ontology analysis of differentially expressed genes was performed using Genomic Regions Enrichment of Annotations Tool (GREAT, http://bejerano.stanford.edu/great/public/html/). To validate the expression of genes that were differentially expressed in distinct samples, cDNA of each RNA sample was first synthesized using the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA). Multiplex qRT-PCR was performed using Taqman® assays for ST6GAL1, PITX2 and KAT6A genes (cat# Hs00949382_m1, Hs04234069_mH and Hs01063029_m1; Life Technologies, Carlsbad, CA), according to the manufacturer’s instructions. To measure the expression of endogenous POU5F1, SOX2, KLF4 and MYC genes, qRT-PCR was performed after cDNA synthesis using SYBR® Green PCR Master Mix (Life Technologies, Carlsbad, CA) and specific primers that target untranslated regions of four endogenous gene transcripts (absent in exogenously expressed reprogramming factors). The specific primers that target endogenous transcripts included: KLF4 3′UTR_For (5′TATGACCCACACTGCCAGAA3′), KLF4 3′UTR_Rev (5′ATCCAGTCACAGACCCCATC3′), SOX2 endo_S1430 (5′GGGAAATGGGAGGGGTGCAAA3′), SOX2 endo_AS1555 (5′TTGCGTGAGTGTGGATGGGAT3′), MYC 3′UTR_For (5′CGGAACTCTTGTGCGTAAGG3′), MYC 3′UTR_Rev (5′CTCAGCCAAGGTTGTGAGGT3′), POU5F1 3′UTR_For (5′GTACTCCTCGGTCCCTTTCC3′), POU5F1 3′UTR_Rev (5′CAAAAACCCTGGCACAACT3′).
Analysis of SNA lectin binding affinity and kinetics
Methods for functionalizing microfluidic channels with lectins and using a microfluidic device to examine the affinity and kinetics of specific lectin binding to cells were described in a previously published report17.
Results
ST6GAL1 is highly expressed and active in undifferentiated hPSCs
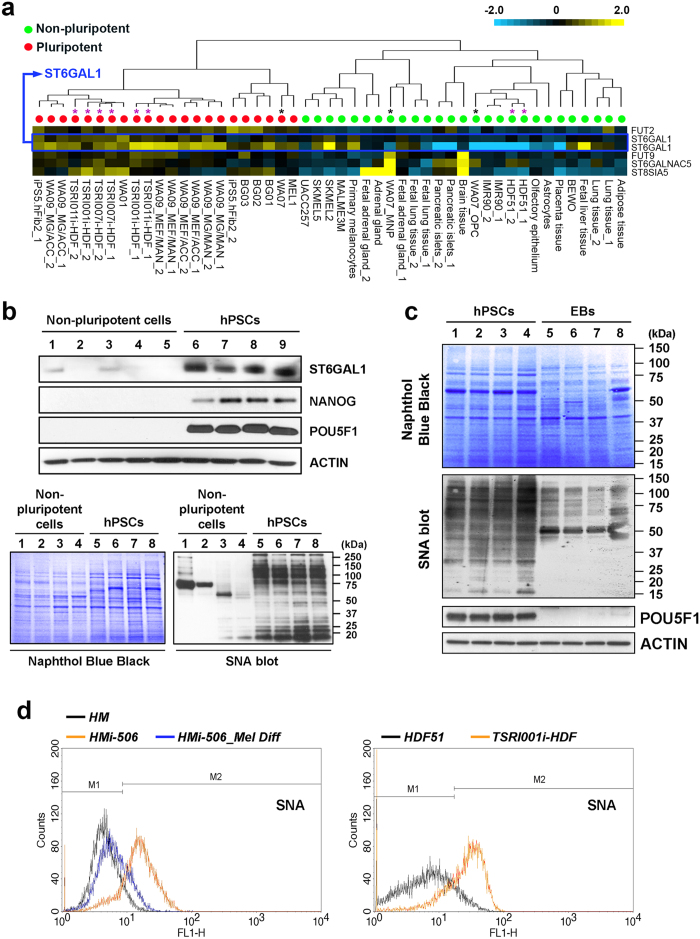
To begin to address the significance of ST6GAL1 expression in the regulation of pluripotent states in human cells, we analyzed ST6GAL1 mRNA and protein expression in multiple undifferentiated hPSC lines [including human embryonic stem cells (hESCs) and induced pluripotent stem cells (hiPSCs)], their differentiated derivatives, and somatic cells that were used for generating the hiPSCs. In particular, we examined several hESC and hiPSC lines paired with their differentiated derivatives. Analysis of gene expression using both microarrays and quantitative RT-PCR (qRT-PCR) showed that cells in the pluripotent state generally had higher expression of the ST6GAL1 transcript (Fig. 1a and Supplementary Figure 1), despite a noticeable variation of ST6GAL1 mRNA expression among different hPSC lines. This higher expression of ST6GAL1 in undifferentiated hPSCs was also reflected at the protein level (Fig. 1b upper panel), consistent with a previous finding18. Since ST6GAL1 catalyzes the terminal addition of sialic acid to β-galactosides to form α-2,6 sialylated glycoconjugates that can be selectively bound by the SNA lectin19, we used SNA as a probe to detect enzymatic products of ST6GAL1 in protein samples isolated from hPSCs and non-pluripotent cells. The SNA-mediated blotting revealed that proteins in undifferentiated hPSCs had distinguishably higher reactivity to SNA, compared to those in non-pluripotent cells (Fig. 1b lower panel and 1c). At the cellular level, hPSCs also showed higher reactivity to SNA binding, compared to their non-pluripotent counterparts (Fig. 1d and Supplementary Figure 2). These results indicate that not only is ST6GAL1 highly expressed but it is also enzymatically active in undifferentiated hPSCs.
Figure 1. ST6GAL1 is preferentially expressed and active in hPSCs.
(a) Heatmap representation of ST6GAL1 transcript expression, as well as other glycosyltransferases, measured by global gene expression analysis in hPSCs and non-pluripotent cell samples. The expression of ST6GAL1 in each cell sample, detected by two independent probes targeting different regions of the ST6GAL1 transcript is shown. Black asterisk: WA07 hESCs and their isogenic differentiated derivatives. Purple asterisk: HDF51 cells and hiPSCs generated from HDF51 cells. (b) Upper panel: Western blotting analysis of ST6GAL1 in cell samples showed that undifferentiated hPSCs had higher ST6GAL1 protein expression, compared to differentiated cells. Lane 1: HMi-506_Mel Diff16, 2: HDF68i-505_Mel Diff16, 3: HM (HEMl), 4: HDF68, 5: HDF51, 6: WA09, 7: HDF68i-50516, 8: HMi-50616, 9: TSRI001i-HDF8. Lower panel: SNA-mediated blotting showed that protein samples extracted from undifferentiated hPSCs had higher reactivity to SNA binding. Lane 1: HM, 2: HDF68i-505_Mel Diff, 3: HDF51, 4: HDF68, 5: WA09, 6: HMi-506, 7: TSRI001i-HDF, 8: HDF68i-505. (c) SNA-mediated blotting showed that protein samples extracted from derivatives of undirected differentiation (embryoid bodies, EBs) had less reactivity to SNA, compared to those from paired, undifferentiated hPSCs. Lane 1: WA09, 2: WA07, 3: TSRI001i-HDF, 4: HMi-506, 5: WA09_EBs, 6: WA07_EBs, 7: TSRI001i-HDF_EBs, 8: HMi-506_EBs. (d) Flow cytometric analysis demonstrated that live human cells in the pluripotent state were more reactive to SNA at the cellular level.
Effects of loss of ST6GAL1 function in undifferentiated hPSCs
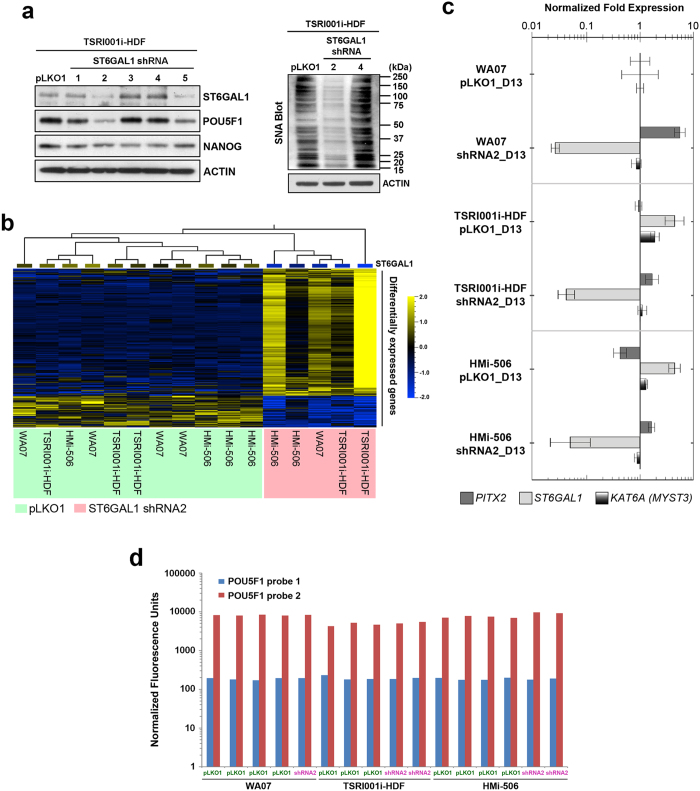
We identified two independent shRNA sequences (shRNA2 and shRNA5) that reduced ST6GAL1 protein expression in hPSCs by more than 60% (Fig. 2a). Despite the fact that NANOG protein levels were relatively unaffected by ST6GAL1 knockdown in hiPSCs after 72 hours, the reduction of ST6GAL1 induced by both shRNA sequences was paralleled by decreases in POU5F1 (a.k.a. OCT4) protein and SNA reactivity (Fig. 2a) in the cells. This suggests that the pluripotency signaling network in hPSCs can be partially modulated by ST6GAL1. Using global gene expression profiling followed by differential expression analysis, we further identified a group of genes (~400 genes) that were significantly upregulated or downregulated in hPSCs in which ST6GAL1 had been knocked down (Fig. 2b,c and Supplementary Figure 3a). Notably, many genes (e.g., COL6A3, S100A4, SNAI2, HOXC8, PITX2, and TWIST220,21,22,23,24,25) known to be involved in organ development or cell morphogenesis during differentiation were upregulated by ST6GAL1 knockdown (Table 1), while the transcripts of genes (e.g., KAT6A (MYST326)) known to be associated with pluripotency were downregulated. Genes including PTPRM and GNG11 that appeared as potential regulators of cellular pluripotency in a genome-wide RNAi screening27 were differentially expressed in hPSCs with ST6GAL1 knockdown (Supplementary Table 2). The ontology analysis of the differentially expressed genes showed that they are highly enriched in the regulation of biological processes relevant to cell differentiation and organogenesis (Supplementary Figure 4). This suggests that ST6GAL1 knockdown may bias cellular pluripotency and differentiation capacity in hPSCs through its global impact on the expression of genes involved in multiple cell lineages. Although POU5F1 protein was reduced in hPSCs with successful knockdown of ST6GAL1, we did not find similarly reduced expression of the POU5F1 gene at the transcriptional level (Fig. 2d), suggesting that the ST6GAL1 knockdown-induced alteration of POU5F1 protein may work through a post-transcriptional or post-translational mechanism.
Figure 2. Downregulation of ST6GAL1 in hPSCs has an impact on signaling networks involved in pluripotency regulation and embryogenesis.
(a) Left Panel: Western blotting analysis showed that two independent shRNA sequences (shRNA2 and shRNA5) that target ST6GAL1 transcripts led to effective downregulation of ST6GAL1 protein 72 hours after transduction. While the protein level of NANOG was relatively unaffected, the protein level of POU5F1 was decreased in hPSCs that received shRNA2 and shRNA5. Right Panel: SNA-mediated blotting showed that protein samples extracted from hPSCs which received shRNA2 had lower reactivity to SNA, indicating a decreased amount of α-2,6 sialylated glycoconjugates in the cells. pLKO1: the empty factor control for the transduction of shRNA expression vectors. (b) Global gene expression profiling followed by differential gene expression analysis revealed a group of genes (~400 genes) that were differentially expressed (P < 0.01, F-test) in multiple lines of hPSCs with and without ST6GAL1 knockdown. (c) Quantitative RT-PCR was used to validate the expression of a selection of differentially expressed genes, confirming the upregulation of the PITX2 gene and the downregulation of KATA6A and ST6GAL1 genes due to ST6GAL1 knockdown. D13: 13 days after the beginning of puromycin selection (14 days post transduction). (d) The expression level of POU5F1 transcripts reflected by normalized fluorescence units in expression array analysis indicated that the POU5F1 gene expression was relatively unchanged at the transcriptional level in multiple lines of hPSCs with ST6GAL1 knockdown.
Table 1. Genes that are involved in cell or organ morphogenesis and differentially expressed in hPSCs with ST6GAL1 knockdown.
| Gene Name | Fold Change (knockdown/control) | P value (F-test) |
|---|---|---|
| ANTXR1 | 1.358615 | 0.004427 |
| COL3A1 | 18.10201 | 3.27E-07 |
| COL6A2 | 1.877785 | 0.002152 |
| COL6A3 | 4.273398 | 0.00015 |
| DAB2 | 4.075949 | 0.000344 |
| DKK1 | 1.911963 | 0.004 |
| FN1 | 2.018123 | 0.001211 |
| MEG3 | 2.525871 | 0.000155 |
| NTF3 | 1.498846 | 0.003855 |
| PRSS23 | 3.534211 | 0.001322 |
| RPS6KA2 | 1.768516 | 0.00018 |
| S100A4 | 6.758956 | 1.93E-05 |
| S100A6 | 3.890132 | 0.000399 |
| SLIT3 | 2.238777 | 0.001546 |
| SNAI2 | 4.19717 | 6.73E-05 |
| TRAK1 | 1.606 | 0.001001 |
| TSPO | 1.745324 | 0.004938 |
| ANGPT1 | 1.608027 | 0.000904 |
| ARID5B | 3.490712 | 0.000275 |
| CD44 | 3.305001 | 0.004976 |
| COL1A2 | 5.251568 | 3.90E-06 |
| COL5A1 | 5.055658 | 0.000118 |
| COL8A1 | 6.030956 | 0.001054 |
| COMP | 2.422019 | 0.000522 |
| CRYGB | 1.326064 | 8.74E-05 |
| DCN | 58.4075 | 6.95E-10 |
| ENG | 1.674513 | 0.001303 |
| FZD1 | 1.20207 | 0.001537 |
| GAS1 | 5.137223 | 0.000236 |
| GREM1 | 9.592929 | 9.73E-05 |
| HEG1 | 2.404444 | 0.000406 |
| HOXA5 | 1.164782 | 0.003494 |
| HOXC4 | 1.773052 | 0.001589 |
| HOXC8 | 4.19109 | 0.000182 |
| ID2 | 2.850354 | 0.003447 |
| IGFBP5 | 2.114798 | 5.94E-05 |
| MEF2D | 0.813538 | 0.003326 |
| MSX1 | 3.0921 | 0.004755 |
| MYLK | 1.946184 | 0.004114 |
| NNMT | 7.347762 | 4.22E-05 |
| NRP1 | 2.751723 | 0.000539 |
| OSR1 | 2.245728 | 0.001449 |
| PDGFRA | 2.562541 | 0.000506 |
| PHACTR3 | 2.016834 | 0.002722 |
| PITX2 | 1.47441 | 0.000332 |
| PTPRM | 1.444159 | 0.004339 |
| RDH10 | 1.565426 | 0.002461 |
| RTN4 | 0.857992 | 0.00114 |
| TGFBR2 | 1.583883 | 0.002067 |
| TSHZ1 | 1.545548 | 0.003036 |
| TWIST2 | 2.225789 | 0.001767 |
| TXNDC5 | 1.195257 | 0.003299 |
| VEGFC | 1.696232 | 0.001064 |
Effects of loss of ST6GAL1 function during cellular reprogramming
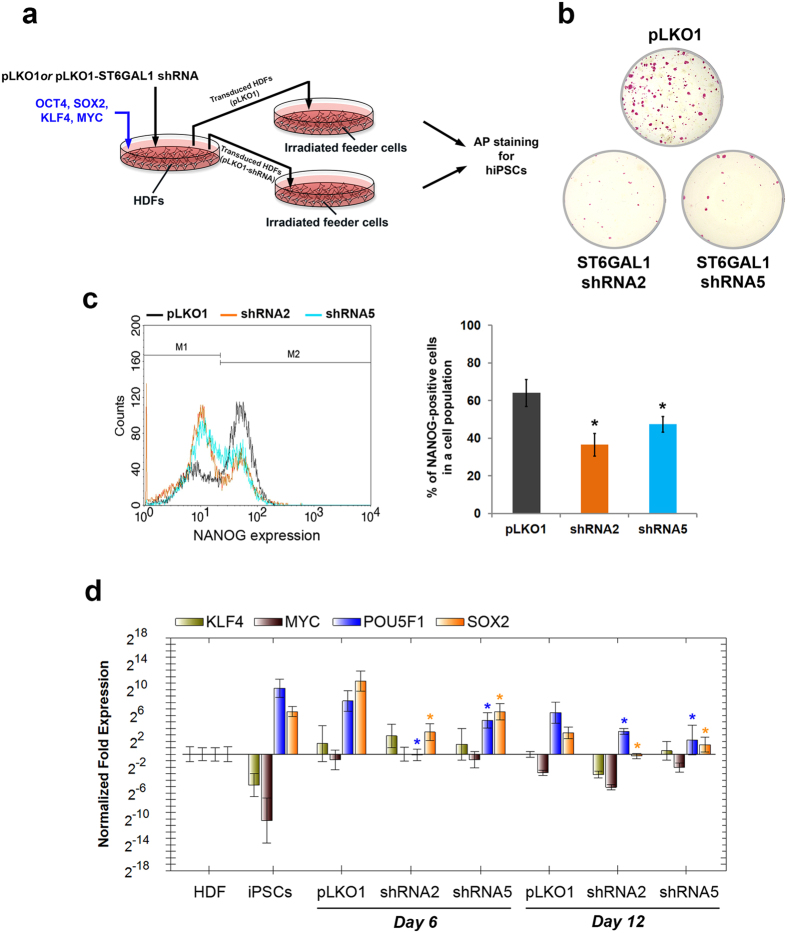
Having demonstrated that ST6GAL1 is preferentially expressed in bona fide pluripotent cells and functionally involved in the regulation of their pluripotency signaling, we wanted to understand whether ST6GAL1 also plays a role in the establishment of induced pluripotency in somatic cells. We performed cell reprogramming in human dermal fibroblasts (HDFs) using four transcription factors (POU5F1, SOX2, KLF4, and MYC) with and without shRNA-mediated ST6GAL1 knockdown (Fig. 3a). Reprogramming efficiency was measured by alkaline phosphatase staining to detect hiPSC colonies and flow cytometric analysis of NANOG expression. As shown in Fig. 3b, the shRNA targeting ST6GAL1 dramatically diminished the number of hiPSC colonies induced by the reprogramming factors. Also, the induction of endogenous NANOG expression in the reprogrammed cells was significantly reduced by the shRNA (Fig. 3c). These results suggest that ST6GAL1 is crucial for the establishment of pluripotency, and that induced pluripotency may be impaired if ST6GAL1 activity is not present during reprogramming. Consistent with the reduced induction of NANOG expression, the expression of endogenous POU5F1 and SOX2 at the initial stage of reprogramming was greatly suppressed by ST6GAL1 knockdown (Fig. 3d), providing a possible mechanistic explanation for the inefficiency of generating hiPSCs in the absence of ST6GAL1. Interestingly, POU5F1 and SOX2 have been shown to counteract lineage specification signaling and interact with each other and other factors to orchestrate self-renewal and pluripotency in hPSCs28,29,30. Our results therefore suggest that ST6GAL1 may regulate the establishment of induced pluripotency by optimizing the balance of POU5F1 and SOX2 during the reprogramming process.
Figure 3. ST6GAL1 knockdown impedes cellular reprogramming and establishment of induced pluripotency in human somatic cells.
(a) Schematic illustration of the experimental strategy to examine the influence of ST6GAL1 knockdown on cellular reprogramming. Twenty-four hours after HDFs received POU5F1, SOX2, KLF4, and MYC with or without ST6GAL1 shRNA, the transduced HDFs were seeded on X-ray irradiated feeder cells (mouse embryonic fibroblasts, MEFs). Fourteen days later, alkaline phosphatase (AP) staining was used to examine hiPSC colonies formed by transduced HDFs on the feeder cells. (b) AP staining showed that dramatically fewer hiPSC colonies with AP activity were obtained from cellular reprogramming under ST6GAL1 knockdown mediated by shRNA2 and shRNA5. (c) Quantitative analysis of NANOG expressing cells in the reprogrammed cell population showed that shRNA2 and shRNA5 both led to a significant reduction in NANOG expressing cells in the analyzed cell populations. Left Panel: The histogram representation of flow cytometry analysis. Right panel: the quantitative result of flow cytometry analysis (n = 3; *P < 0.05, t-test). (d) The expression of endogenous POU5F1, SOX2, KLF4 and MYC genes in cells that underwent reprogramming with or without ST6GAL1 knockdown was measured by qRT-PCR with primer sets that target untranslated regions of the endogenous gene transcripts. The induction of endogenous POU5F1 and SOX2 gene expression at the early stage of cellular reprogramming in HDFs was substantially suppressed by shRNA-mediated ST6GAL1 knockdown (n = 3; *P < 0.05, t-test). Day 6: cell samples collected at 6 days after the initial transduction (2 days after the beginning of puromycin selection). Day 12: cell samples collected at 12 days after the initial transduction (8 days after the beginning of puromycin selection).
ST6GAL1 knockdown impedes cellular reprogramming through multiple mechanisms
To further dissect the signaling networks modulated by ST6GAL1 in cells undergoing reprogramming, we analyzed the global gene expression profiles of HDFs that were reprogrammed while ST6GAL1 was knocked down. Differential expression analysis revealed that a group of genes (~570 genes) were differentially expressed in the reprogrammed cells with and without ST6GAL1 knockdown (Supplementary Figure 5a and 5b). Many of these genes are involved in ribosomal biogenesis and the post-transcriptional regulation of gene expression (e.g., RNA processing and translation; Supplementary Figure 5c), indicated by gene ontology analysis. It has been reported that alterations in ribosome synthesis (ribosomal stress) and decreases in ribosome levels may cause cell cycle arrest through both TP53-dependent and TP53-independent mechanisms31,32. In addition, cell cycle arrest is a well-documented barrier to the induction of cellular pluripotency33,34,35,36. Thus, our findings suggest that ribosomal stress-mediated inhibition of cell proliferation may also contribute to the decreased reprogramming efficiency caused by ST6GAL1 knockdown.
A sialyltransferase inhibitor reduces the efficiency of cellular reprogramming
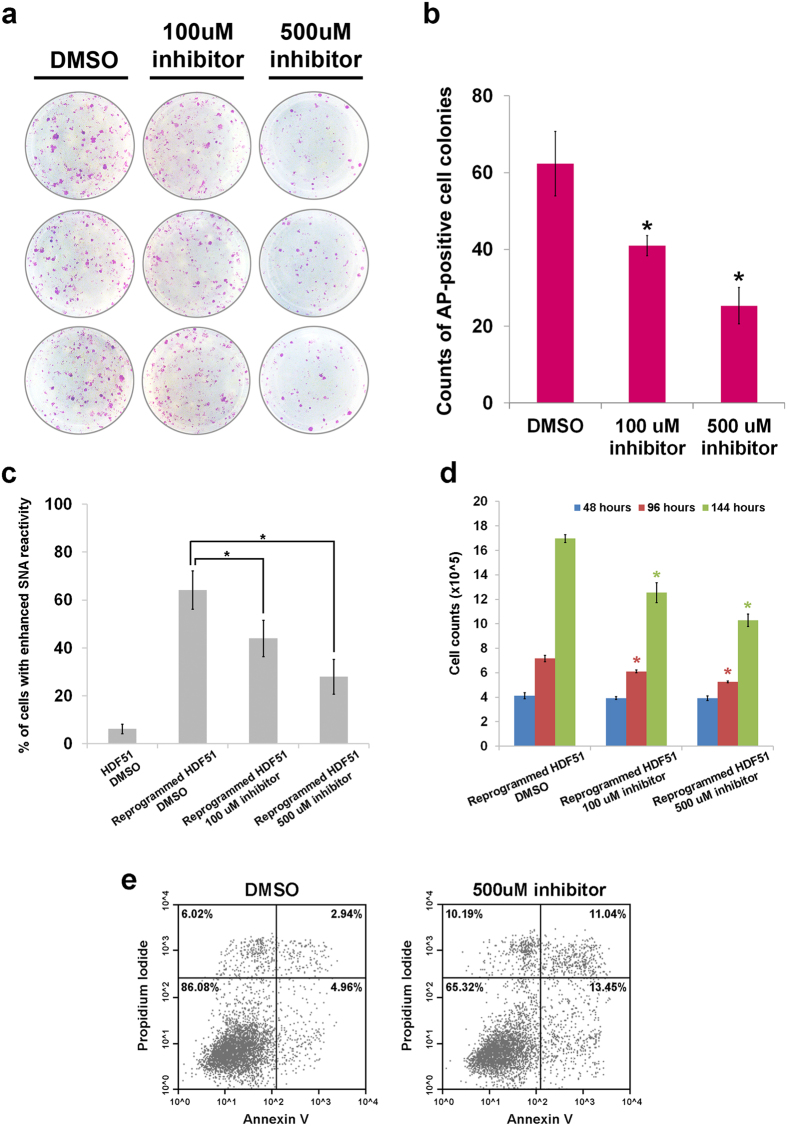
In light of the reduction in reprogramming efficiency due to ST6GAL1 knockdown, we used a cell-permeable sialyltransferase inhibitor, 3Fax-peracetyl Neu5Ac, to test whether it would also suppress the establishment of cellular pluripotency in somatic cells. Rillahan et al. have reported significant inhibition of ST6GAL1-mediated sialylation in HL-60 cells treated with 200 uM 3Fax-peracetyl Neu5Ac37. As shown in Fig. 4a,b, treatment with the sialyltransferase inhibitor during cell reprogramming reduced the number of AP-positive cell colonies in a dose-dependent manner. The inhibitor-induced reduction in reprogramming efficiency was concordant with the decreased SNA reactivity (Fig. 4c). In addition, treatment with the inhibitor led to a slight but significant reduction in proliferative capacity and viability of the cells (Figs 4d,e). Although the shRNA-mediated ST6GAL1 knockdown repressed induction of pluripotency to a greater extent than the drug treatment (Fig. 3b and Supplementary Figure 3b), our results suggest that the modulation of cellular pluripotency can be achieved using pharmacological tools to target sialyltransferase activity.
Figure 4. Treatment of a sialyltransferase inhibitor suppresses cellular reprogramming and establishment of induced pluripotency in human somatic cells.
(a) AP staining showed that fewer hiPSC colonies with AP activity were obtained from cellular reprogramming under the treatment of 3Fax-peracetyl Neu5Ac. (b) The numbers of AP-positive colonies with a size equal or larger than 1.0 mm in diameter were counted at the end of reprogramming experiments, which showed a dose-dependent reduction of hiPSC formation due to the treatment of 3Fax-peracetyl Neu5Ac (n = 3; *P < 0.05, t-test). (c) The percentages of cells with enhanced SNA reactivity in reprogrammed HDF51 cells were significantly reduced by the treatment of 3Fax-peracetyl Neu5Ac for 6 days in a dose-dependent manner (n = 3; *P < 0.05, t-test), indicating decreased sialylation on the cell surface. (d) The cell counts of reprogrammed HDF51 cells at different time points over 6 days were marginally but significantly reduced by the treatment of 3Fax-peracetyl Neu5Ac in a dose-dependent manner (n = 3; *P < 0.05, t-test), indicating a slight suppression of cell proliferation. (e) The number of apoptotic cells in reprogrammed HDF51 cells was slightly increased by the treatment of 500 μM 3Fax-peracetyl Neu5Ac for 6 days.
Discussion
The pivotal roles of sialic acid and sialyltransferase-mediated macromolecule sialylation in many physiological and pathological processes, including the development of embryos, cancer progression, the regulation of immune systems, and host-pathogen interactions, have been identified by numerous studies38,39,40. In our study, we demonstrated that the downregulation of ST6GAL1 in undifferentiated hPSCs biases the core signaling of cellular pluripotency and significantly alters the expression of many genes that orchestrate cellular differentiation and organ development. Most interestingly, the induction of cellular pluripotency in somatic cells was dramatically impeded by ST6GAL1 suppression, likely through the perturbation of endogenous POU5F1 and SOX2 expression as well as inhibition of cell proliferation.
To assess variation among different hPSC lines41,42,43,44, the expression of ST6GAL1 was analyzed in a broad spectrum of pluripotent and non-pluripotent cell samples including a variety of hPSC lines paired with isogenic differentiated derivatives and the primary cells used for reprogramming. Despite variation in levels of ST6GAL1 mRNA expression among different hPSC lines, the undifferentiated pluripotent cells consistently showed higher ST6GAL1 expression compared with the differentiated derivatives and primary cells. In addition, protein samples extracted from differentiated cells have relatively low binding reactivity with the SNA lectin, compared with those from isogenic undifferentiated hPSCs. Our results suggest that high levels of ST6GAL1 expression and enzymatic activity are specifically associated with pluripotency in human cells.
We sought to determine what mechanisms may underlie the effect of ST6GAL1 on pluripotency. Using global gene expression profiling followed by differential gene expression analysis, we identified a group of genes that are upregulated in response to ST6GAL1 knockdown in undifferentiated hPSCs. The gene ontology analysis indicated that many of these genes are involved in the regulation of developmental processes and appear to influence multiple cell lineages. Although additional study is needed to determine whether these changes in gene expression are primarily mediated through biasing the pluripotency core signaling, our data suggest a role of ST6GAL1 in the negative regulation of differentiation signaling that maintains pluripotency in hPSCs. Published work in other systems has shown that the α-2,6 sialylation mediated by ST6GAL1 is associated with a less differentiated phenotype in mouse mammary tumors12 and enhanced PI3K/AKT signaling in human cancer cells45,46. Since PI3K/AKT activity plays an important role in the maintenance of self-renewal in hPSCs by restraining prodifferentiation signaling47, it appears possible that the suppression of ST6GAL1 activity may promote differentiation by interfering with PI3K/AKT signaling in hPSCs.
Our findings regarding the ST6GAL1 knockdown-mediated downregulation of POU5F1 protein that occurred without an obvious decrease of POU5F1 mRNA indicate that a post-transcriptional or post-translational mechanism controlling POU5F1 expression may be regulated through ST6GAL1. The expression of genes involved in protein synthesis or quality control mechanisms in undifferentiated hPSCs did not seem particularly affected by ST6GAL1 knockdown. However, we have observed the expression of genes highly relevant to ribosomal biogenesis and the post-transcriptional regulation of gene expression are particularly disturbed by ST6GAL1 knockdown in somatic cells undergoing cell reprogramming. Catalyzed by another glycosyltransferase (O-GlcNAc transferase), the post-translational O-GlcNAcylation of POU5F1 significantly influences the activity of POU5F1 in mESCs5. Although additional investigation would be needed to elucidate mechanistic details, our data together with other studies indeed support the likelihood of post-transcriptional or post-translational mechanisms mediated by the sialyltransferase ST6GAL1 in controlling the protein level and function of POU5F1 in hPSCs.
ST6GAL1 knockdown affected cellular reprogramming, reducing the number of reprogrammed cells. This was accompanied by the altered expression of genes involved in ribosomal biogenesis and RNA processing in addition to the repression of endogenous POU5F1 and SOX2 expression in the reprogrammed somatic cells. Consistent with our results, several recent reports have described the finding that interference with ribosome homeostasis (ribosomal stress) led to growth delay, apoptosis and differentiation in murine pluripotent stem cells48,49. Thus, our results suggest that ST6GAL1 activity may help to stabilize pluripotency by preventing ribosomal stress during the reprogramming process in hPSCs.
To determine the effects of blocking the enzymatic activity of ST6GAL1 without altering its expression, we used a newly-developed sialic acid analog (3Fax-peracetyl Neu5Ac) that has been extensively characterized in biochemical and cell-based analyses37. The inhibitor did reduce reprogramming efficiency, but not as much as the effect observed by knocking down ST6GAL1 expression. This may be due to the incomplete suppression of ST6GAL1 activity and the inactivation of another enzyme, ST3GAL1, which is known to occur in live cells treated with 3Fax-peracetyl Neu5Ac37. Our previous study revealed that ST3GAL1 is preferentially expressed in many differentiated cells, including the HDFs that we used for the reprogramming studies8. Thus, it seems possible that suppression of ST3GAL1 may counteract the effects of the loss of ST6GAL1, allowing more cells to be reprogrammed. A more specific and potent small-molecule inhibitor may better mimic the effect of shRNA targeting ST6GAL1 in the regulation of cellular pluripotency.
Conclusions
In this study, we have shown that ST6GAL1 is critically involved in the regulation of cellular pluripotency and essential for efficient induction of pluripotency in somatic cells, potentially through multiple mechanisms. To our knowledge, this work provides the first direct evidence for the functional significance of a sialyltransferase and macromolecule sialylation in regulating the pluripotent state in human cells. Targeting glycosyltransferases and protein glycosylation could be a useful strategy for manipulating hPSCs for research and regenerative medicine.
Additional Information
How to cite this article: Wang, Y.-C. et al. Glycosyltransferase ST6GAL1 contributes to the regulation of pluripotency in human pluripotent stem cells. Sci. Rep. 5, 13317; doi: 10.1038/srep13317 (2015).
Supplementary Material
Acknowledgments
Y.C.W is supported by NIH (R33 MH087925), CIRM (CL1-00502) and UNTHSC Start-up Fund for Stem Cell Laboratory. J.W.S. is supported by CIRM (TB1-01186). C.Y.L. is supported by CIRM (TG2-01165). R.C. is supported by Ken and Karen Craven Graduate Fellowship and CIRM (RM1-01717 and TR3-05603). A.L.L. is supported by CIRM (TR1-01250, TR3-05603) and a partner investigator on the Australian Research Council (ARC) Special Research Initiative in Stem Cell Science, Stem Cells Australia. C.M.O’B. and H.S.C. are supported by CIRM (TR1-01250, TR3-05603). S.K.M. is supported by NIH (R01 EB009327) and NSF (CAREER grant 0747166). S.E.P. and J.F.L. are supported by CIRM (RT1-01108, TR1-01250, TR3-05603, and CL1-00502), NIH (R33 MH087925) and the Esther O’Keeffe Foundation.
Footnotes
Author Contributions Y.C.W. supervised the study and contributed to study concept and design, data collection and analysis, and manuscript writing. J.W.S., C.L.L., H.T.T, C.Y.L., R.C., A.H, V.G.A., H.S.C., C.O’.B., S.K.M. and A.L.L. contributed to data collection, analysis, and interpretation. S.E.P. contributed to data interpretation and manuscript writing. J.F.L. provided conceptual advice and supervised the study.
References
- Wang Y. C., Peterson S. E. & Loring J. F. Protein post-translational modifications and regulation of pluripotency in human stem cells. Cell Res. 24, 143–160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzkopf M. et al. Sialylation is essential for early development in mice. P Natl Acad Sci USA. 99, 5267–5270 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A., Machingo Q. J., Fritz A. & Shur B. D. Core Fucosylation Is Required for Midline Patterning During Zebrafish Development. Dev Dynam. 239, 3380–3390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q. et al. O-fucose modulates Notch-controlled blood lineage commitment. Am J Pathol. 176, 2921–2934 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H. et al. O-GlcNAc Regulates Pluripotency and Reprogramming by Directly Acting on Core Components of the Pluripotency Network. Cell Stem Cell. 11, 62–74 (2012). [DOI] [PubMed] [Google Scholar]
- Hasehira K. et al. Structural and Quantitative Evidence for Dynamic Glycome Shift on Production of Induced Pluripotent Stem Cells. Mol Cell Proteomics. 11, 1913–1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satomaa T. et al. The N-glycome of human embryonic stem cells. BMC Cell Biol. 10, 10.1186/1471-2121-10-42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. C. et al. Specific lectin biomarkers for isolation of human pluripotent stem cells identified through array-based glycomic analysis. Cell Res. 21, 1551–1563 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S. et al. Possible linkages between the inner and outer cellular states of human induced pluripotent stem cells. BMC Syst Biol. 5 Suppl 1, S17; 10.1186/1752-0509-5-S1-S17 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harduin-Lepers A. Comprehensive Analysis of Sialyltransferases in Vertebrate Genomes. Glycobiol Insights. 2, 29–61; 10.4137/GBI.S3123 (2010). [DOI] [Google Scholar]
- Harduin-Lepers A. et al. The human sialyltransferase family. Biochimie. 83, 727–737 (2001). [DOI] [PubMed] [Google Scholar]
- Hedlund M., Ng E., Varki A. & Varki N. M. alpha 2-6-Linked sialic acids on N-glycans modulate carcinoma differentiation in vivo. Cancer Res. 68, 388–394 (2008). [DOI] [PubMed] [Google Scholar]
- Tateno H. et al. Glycome Diagnosis of Human Induced Pluripotent Stem Cells Using Lectin Microarray. J Biol Chem. 286, 20345–20353 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alisson-Silva F. et al. Evidences for the involvement of cell surface glycans in stem cell pluripotency and differentiation. Glycobiology. 24, 458–468 (2014). [DOI] [PubMed] [Google Scholar]
- International Stem Cell I. et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 25, 803–816 (2007). [DOI] [PubMed] [Google Scholar]
- Jones J. C. et al. Melanocytes Derived from Transgene-Free Human Induced Pluripotent Stem Cells. J Invest Dermatol. 133, 2104–2108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers D. A. et al. Lectin-functionalized microchannels for characterizing pluripotent cells and early differentiation. Biomicrofluidics. 6, 24122–2412210, 10.1063/1.4719979 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindall A. F. et al. ST6Gal-I Protein Expression Is Upregulated in Human Epithelial Tumors and Correlates with Stem Cell Markers in Normal Tissues and Colon Cancer Cell Lines. Cancer Res. 73, 2368–2378 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings R. D. & Etzler M. E. Antibodies and Lectins in Glycan Analysis in Essentials of Glycobiology 2nd edn (eds. Varki A. et al.) Ch. 45 (Cold Spring Harbor Laboratory Press, 2009). [PubMed] [Google Scholar]
- Bell C. E. & Watson A. J. SNAI1 and SNAI2 Are Asymmetrically Expressed at the 2-Cell Stage and Become Segregated to the TE in the Mouse Blastocyst. PLoS One. 4, 10.1371/journal.pone.0008530 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir E. et al. Mutations in COL6A3 cause severe and mild phenotypes of Ullrich congenital muscular dystrophy. Am J Hum Genet. 70, 1446–1458 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherzi R. et al. Akt2-mediated phosphorylation of Pitx2 controls Ccnd1 mRNA decay during muscle cell differentiation. Cell Death Differ. 17, 975–983 (2010). [DOI] [PubMed] [Google Scholar]
- Kiryushko D. et al. Molecular mechanisms of Ca2+ signaling in neurons induced by the S100A4 protein. Mol Cell Biol. 26, 3625–3638 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D. et al. Hox genes are involved in vascular wall-resident multipotent stem cell differentiation into smooth muscle cells. Sci Rep-Uk. 3, 10.1038/srep02178 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade C. et al. Twist2 contributes to termination of limb bud outgrowth and patterning through direct regulation of Grem1. Dev Biol. 370, 145–153 (2012). [DOI] [PubMed] [Google Scholar]
- Scheper W. & Copray S. The Molecular Mechanism of Induced Pluripotency: A Two-Stage Switch. Stem Cell Rev Rep. 5, 204–223 (2009). [DOI] [PubMed] [Google Scholar]
- Chia N. Y. et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 468, 316–U207 (2010). [DOI] [PubMed] [Google Scholar]
- Montserrat N. et al. Reprogramming of Human Fibroblasts to Pluripotency with Lineage Specifiers. Cell Stem Cell. 13, 341–350 (2013). [DOI] [PubMed] [Google Scholar]
- Wang Z. et al. Distinct Lineage Specification Roles for NANOG, OCT4, and SOX2 in Human Embryonic Stem Cells. Cell Stem Cell. 10, 440–454 (2012). [DOI] [PubMed] [Google Scholar]
- Thomson M. et al. Pluripotency Factors in Embryonic Stem Cells Regulate Differentiation into Germ Layers. Cell. 145, 875–889 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati G., Montanaro L. & Derenzini M. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res. 72, 1602–1607 (2012). [DOI] [PubMed] [Google Scholar]
- Iadevaia V. et al. PIM1 kinase is destabilized by ribosomal stress causing inhibition of cell cycle progression. Oncogene. 29, 5490–5499 (2010). [DOI] [PubMed] [Google Scholar]
- Hong H. et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 460, 1132–1135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T. et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 460, 1140–1144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof D. et al. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 5, 214–226 (2009). [DOI] [PubMed] [Google Scholar]
- Utikal J. et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 460, 1145–1148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rillahan C. D. et al. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat Chem Biol. 8, 661–668 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. & Varki A. Advances in the biology and chemistry of sialic acids. ACS Chem Biol. 5, 163–176 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzkopf M. et al. Sialylation is essential for early development in mice. Proc Natl Acad Sci USA. 99, 5267–5270 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angata T. & Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chemical Rev. 102, 439–469 (2002). [DOI] [PubMed] [Google Scholar]
- Bock C. et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 144, 439–452 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K. H. et al. Diverse hematopoietic potentials of five human embryonic stem cell lines. Exp Cell Res. 314, 2930–2940 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetty S. et al. A simple tool to improve pluripotent stem cell differentiation. Nat Methods. 10, 553–556 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K. et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 26, 313–315 (2008). [DOI] [PubMed] [Google Scholar]
- Ma H. et al. Reversal effect of ST6GAL 1 on multidrug resistance in human leukemia by regulating the PI3K/Akt pathway and the expression of P-gp and MRP1. PLoS One. 9, 10.1371/journal.pone.0085113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao Y. et al. Modification of sialylation mediates the invasive properties and chemosensitivity of human hepatocellular carcinoma. Mol Cell Proteomics. 13, 520–536 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Singh A. M. et al. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell. 10, 312–326 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgado-Palacin L., Llanos S. & Serrano M. Ribosomal stress induces L11- and p53-dependent apoptosis in mouse pluripotent stem cells. Cell Cycle. 11, 503–510 (2012). [DOI] [PubMed] [Google Scholar]
- Watanabe-Susaki K. et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells. 32, 3099–3111 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.