Abstract
The relevance of superinfection as a model to identify correlates of protection against human immunodeficiency virus (HIV) depends on whether the superinfecting transmission resembles primary infection, which has not been established. Here, we characterize the genetic bottleneck in superinfected individuals for the first time. In all 3 cases, superinfection produced a spike in viral load and could be traced to a single, C-C chemokine receptor 5–tropic founder virus with shorter, less glycosylated variable regions than matched chronic viruses. These features are consistent with primary HIV transmission and provide support for the use of superinfection as a model to address correlates of protection against HIV.
Keywords: HIV, superinfection, T/F, transmitted, founder, correlates of protection
A significant challenge facing the development of a human immunodeficiency virus (HIV) vaccine is that correlates of protection against HIV are not clearly defined. Vaccine efficacy trials are time consuming and expensive, and models that increase our understanding of correlates of protection can potentially accelerate vaccine development. By definition, in HIV superinfection a secondary acquisition of HIV occurs in the face of an ongoing anti-HIV immune response and therefore provides a unique opportunity to assess whether the immune responses typically elicited during natural HIV infection provide any protection against subsequent HIV infection. Indeed, several studies have compared immunological markers in superinfected participants to those who were presumably exposed but not superinfected [1–4]. However, the utility of superinfection as a challenge model depends on whether superinfection resembles primary transmission, which has not yet been established.
Primary HIV sexual transmission results in a severe genetic bottleneck associated with the acquisition of HIV. It is estimated that during the sexual transmission of HIV, approximately 80% of infections are the result of a single founder variant, despite exposure to hundreds to millions of virions in the inoculum [5–7]. These transmitted/founder (T/F) viruses are consistently C-C chemokine receptor 5 (CCR5)–tropic, and it has been shown elsewhere that subtype C has shorter, less glycosylated envelopes than viruses collected during chronic infection [8]. However, the genetic bottleneck associated with superinfection and the question of whether the superinfecting viruses have phenotypes typical of primary viruses have not been studied, to our knowledge. Here, we characterize the genetic bottleneck in three superinfected individuals, and characterize the features of the T/Fs of superinfection.
Methods
Longitudinal samples were provided by participants in the CAPRISA [Centre for the AIDS Programme of Research in South Africa] 002 Acute Infection Study (n = 62). Plasma samples were taken weekly for the first 3 weeks after primary infection, fortnightly until approximately 3 months after primary infection, monthly until approximately 1 year after primary infection, and quarterly thereafter (described in detail in [9]). Written informed consent was obtained from all participants, and ethical approval for the study was granted by the University of KwaZulu-Natal (E013/04), University of Cape Town (025/2004), and University of the Witwatersrand (M040202).
Putative cases of superinfection were identified retrospectively by the presence of divergent variants (DNA distance >5%) in previously generated sequences. If contamination could not adequately explain the divergent sequences, additional env sequences were generated from longitudinal plasma samples by single-genome amplification (SGA) and, in the case of CAP256, strain-specific polymerase chain reaction (PCR).
Plasma viral RNA was extracted from 200L or 400 µL of plasma using either the Roche MagNApure (Roche Applied Science) or Qiagen viral RNA extraction kits (Qiagen). RNA was reversed-transcribed to complementary DNA (cDNA), using Superscript III (Invitrogen, Life Technologies), as described elsewhere [7].
Envelope cassettes were amplified from the full-length cDNA in nested PCR by SGA using a Platinum Taq High Fidelity kit (Invitrogen, Life Technologies), as described elsewhere [7]. Briefly, cDNA was serially diluted until <30% of the reactions were PCR positive, such that >80% of the amplicons, according to a Poisson distribution would probably have been amplified from a single template. This approach prevents template resampling and PCR-induced recombination that can occur if multiple templates are amplified in a reaction. Primers used in the outer reaction were Vif1 (5′-GGGTTTATTACAGGGACAGCAGAG-3′; HXB2 nucleotides 4900–4923) and OFM19 (5′-GCACTCAAGGCAAGCTTTATTGAGGCTTA-3′; HXB2 nucleotides 9604–9632). Inner primers used were ENV1A-Rx (5′-CACCGGCTTAGGCATCTCCTATAGCAGGAAGAA-3′; HXB2 nucleotides 5954–5982) and EnvN (5′-TTGCCAATCAAGGAAGTAGCCTTGTGT-3′; HXB2 nucleotides 9145–9171). For strain-specific SGA the CAP256 superinfecting virus at select time points, reverse transcription was performed using a primer specific for the superinfecting variant (256spR 5′-CTCCCTCTGCTGTTGGCTGCGCTCGCGC-3′; HXB2 nucleotides 8856–8884) as the antisense primer in both rounds of amplification [10]. Outer reaction thermal cycling conditions were as follows: initial denaturation at 94°C for 2 minutes, followed by 35 cycles (94°C for 15 seconds, 55°C for 30 seconds, 68°C for 4 minutes), followed by a final extension for 1 cycle (68°C for 20 minutes). Inner reaction thermal cycling conditions were the same as above, for 45 cycles.
To refine the estimates of the timing of superinfection and differentiate the transmission of one virus or multiple viruses, we estimated the time to the MRCA of multiple closely related sequences generated from the first time point when superinfection was detectable. The time to the MRCA was then estimated with Poisson Fitter software (http://www.hiv.lanl.gov/content/sequence/poisson_fitter/poisson_fitter/html), using a mutation rate of 2.16 × 10–05 mutations per base pair (bp) per cycle and a generation time of 2 days. Sequences were determined to have originated from a single T/F virus if they displayed a star phylogeny and the number of mutations conformed to a Poisson distribution (described in detail in [6]).
Variable region lengths and the number of potential asparagine (N)–linked glycosylation sites were calculated using an in-house script. The V1, V2, V3, V4, and V5 regions were measured from HXB2 positions 126–157, 158–196, 296–331, 385–418 and 460–471 respectively, and summed. Potential N-linked glycosylation sites were identified within the variable regions by the presence of sequons (N-X-S/T, where X is any amino acid except proline). Overlapping sequons (NNST, NNSS, NNTT, and NNTS) were counted as a single N-linked glycosylation site.
The length and glycosylation of the variable regions in superinfecting viruses were compared with those in viruses from primary infection and consensus sequences from 3 years after infection (chronic infection), using an unpaired, 2-tailed t test. The acute and chronic infection groups represented all participants in the CAPRISA 002 cohort for whom env SGA sequences were already available from both acute infection and 3 years after infection. Consensus sequences for chronic infection were derived according to “majority rules” from a median of 10 sequences from each individual (range, 5–24). The length and glycosylation of the variable regions in matched acute and chronic sequences were compared using a 2-tailed, paired t test implemented in Prism 5 software (GraphPad Software).
To determine whether the superinfecting sequences harbored recombinant regions inherited from primary viruses, we calculated Smith-Waterman similarities over a 100-bp sliding window (5-bp step size), using an in-house script. Similarities of the superinfecting virus to the primary infecting viruses were compared with similarities to viruses from a panel of geographically matched env sequences (n = 68). A similarity score of ≥98% for the primary infecting virus but not for any of the panel viruses (over ≥3 consecutive windows) was considered evidence of recombination. An example of a detectable case of recombination is shown in Supplementary Figure 1. Coreceptor tropism was predicted using the Geno2pheno [11] Web server (http://coreceptor.geno2pheno.org/), with a false-positive rate cutoff of 10%.
RESULTS
Longitudinal gag (n = 365), nef (n = 289), and/or env (n = 2011) single-genome sequences were available from 32 participants in the CAPRISA 002 cohort over a total of 54.9 person-years of follow-up [7, 12–14] (R. Thebus, B. Lambson, unpublished data). These sequences were screened retrospectively for superinfection, defined as infection after seroconversion with a second variant that is separated phylogenetically from the primary infecting virus by epidemiologically unlinked viruses (phylogenetically unlinked). For 3 participants (CAP237, CAP256, and CAP281), a phylogenetically unlinked virus could be detected after seroconversion that was not detectable at or before seroconversion (Figure 1A–D). We performed SGA and strain-specific PCR on sequential plasma samples and sequencing to confirm superinfection and to identify the timing.
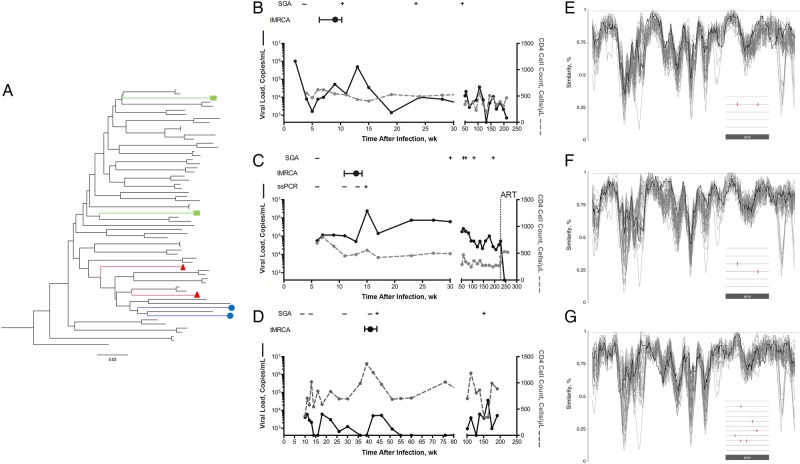
Figure 1.
Identification of the transmitted/founder superinfecting envelopes, before recombination with the primary virus. A, Maximum likelihood phylogeny depicting the presence of 2 strains separated phylogenetically by epidemiologically unlinked viruses in participants CAP237 (circles), CAP256 (triangles), and CAP281 (squares). The phylogeny was reconstructed using env sequences from CAP237, CAP256, CAP281 along with 58 subtype C sequences from 30 individuals, including 24 participants from the CAPRISA [Centre for the AIDS Programme of Research in South Africa] 002 cohort. B–D, Viral load (solid black lines) and CD4 cell counts (dotted gray lines) over time are shown for the 3 superinfected participants: CAP237 (B), CAP256 (C), and CAP281 (D). The limit of detection for the viral load assay was 400 copies/mL. In each case, the detection (+) or absence (−) of the superinfecting virus in sequences from single-genome amplification (SGA) and strain-specific polymerase chain reaction (ssPCR) is indicated. The estimated window of superinfection is shown, based on the 95% confidence interval for the estimated time since a most recent common ancestor (tMRCA) for the early (nonrecombinant) superinfecting viral sequences. ART, initiation of antiretroviral therapy. E–G, Similarity of the consensus of the early superinfecting env sequences from CAP237 (E), CAP256 (F), and CAP281 (G), to sequences from an alignment of subtype C sequences (including sequences from the CAPRISA 002 cohort and the primary infecting viruses). Over all windows (100–base pair [bp] windows with a 5-bp shift), the superinfecting viruses are not more similar to the primary virus (black line) than would be expected for unlinked sequences (gray lines), indicating the absence of recombinant regions inherited from the primary virus. Insets depict highlighter plots of the early single-genome env sequences of the superinfecting viral populations indicative of a “star” phylogeny of near identical sequences in each case.
In all 3 cases, the time between sequential plasma samples that tested negative and then positive for the superinfecting virus could be narrowed to within 2–6 weeks. To determine the multiplicity of superinfection and to accurately identify the founder(s) of superinfection, we analyzed 5–11 superinfecting single-genome env sequences generated from the first plasma sample with evidence of the superinfecting virus. For CAP237 and CAP281, these were generated without the need for strain-specific primers. In all 3 cases, the early sampled superinfecting viral population was highly homogenous, with single-genome env sequences differing from each other by ≤2 mutations (Figure 1E–G: see insets). The superinfecting sequences in all 3 participants conformed to a star phylogeny with a Poisson distribution of mutations, consistent with superinfection being the result of a single T/F virus.
To ensure that the inferred T/F represented the superinfecting virus before any recombination with the primary virus, we looked for regions within the superinfecting sequences that were more similar to the primary virus than they were to a panel of geographically matched sequences using sliding window similarity measures. In all 3 cases, there was no significant evidence of recombinant regions inherited from the primary virus (Figure 1E–G). The consensus/MRCA was therefore considered an accurate representation of the superinfecting T/F virus. The 3 superinfecting viruses were all classified as subtype C, were computationally predicted by Geno2pheno to be CCR5 tropic, and differed from the primary infecting variants (also all subtype C) across env by 12.53% (CAP237), 11.92% (CAP256), and 13.99% (CAP281).
To refine the likely window of superinfection, we estimated the time from a most recent common ancestor of the early (highly homogenous, nonrecombinant) superinfecting sequences. The timing of superinfection for CAP237, CAP256, and CAP281 was estimated as 9 (range, 6–11), 13 (11–14), and 42 (40–44) weeks after infection, respectively. In all 3 cases, this inferred time of superinfection preceded a >10-fold spike in viral load (Figure 1B–D). Similar spikes in viral load (>10-fold increase from the previous visit), outside of acute infection, were observed only infrequently in the other 29 participants (1 spike per 13 person-years of follow-up).
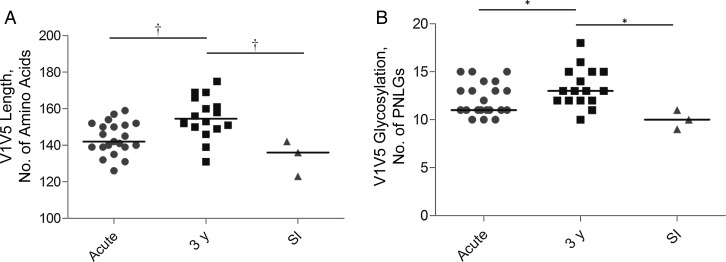
Recently transmitted subtype C viruses seem to possess shorter, less glycosylated variable regions than viruses from chronic infection [8]. To establish whether superinfecting founder viruses had similar characteristics, we compared the length and potential N-linked glycosylation of the variable regions with a panel of matched acute and chronic subtype C sequences from the same cohort, sampled over a similar period. There was no variance in variable region length or number of N-linked glycosylation sites among the sampled early superinfecting quasispecies within participants. env SGA sequences for acute infection were available from 21 participants in the CAPRISA 002 cohort, of whom 16 had matched SGA sequences available 3 years after infection. Consistent with previous observations, viruses sampled 3 years after infection had longer (P = .001), potentially more glycosylated (P = .01) variable regions compared with those sampled shortly after transmission. Superinfecting viruses had characteristics typical of recently transmitted viruses, with length and potential N-linked glycosylation significantly lower than in viruses sampled 3 years after infection (P = .007 and P = .01, respectively; Figure 2).
Figure 2.
Superinfecting (SI) transmitted/founder viruses have envelopes with shorter (A), less glycosylated (B) variable regions than do viruses sampled 3 years after infection. *P < .05; †P < .01. Abbreviation: PNLGs, potential N-linked glycosylation sites.
DISCUSSION
Since the first case report in 2002 [15], individuals with HIV superinfection are increasingly being identified. The implication is that the natural immune response to HIV infection is frequently not sufficient to provide protection from infection with another HIV variant, raising concerns for vaccine design. Even if the immune responses generated during natural infection provide no protection from HIV infection, quantifying these responses at the time of superinfection will still address an important question, namely: what is not sufficient to provide protection? However, the utility of superinfection as a model of vaccine protection depends on the ability of the superinfection event to accurately reflect primary infection, though this has not previously been demonstrated, to our knowledge.
Here, we characterized the multiplicity of infection for a superinfecting strain for the first time. Frequent sampling allowed us to show that 3 cases of superinfection were all probably established by a single virus. This is consistent with primary HIV infection, in which clinical infection is established by a single founder virus in approximately 80% of individuals [6, 7]. With the limited sequencing depth, it remains possible that we failed to detect minority superinfecting variants. It is also possible that the use of strain-specific primers for CAP256 failed to amplify other superinfecting variants. Nevertheless, our data are consistent with a severe genetic bottleneck after superinfection. We also showed that superinfection was followed by a spike in viral load and that the superinfecting viruses were predicted to be CCR5 tropic and had shorter, less glycosylated variable regions than variants from chronic infection; characteristics of recently transmitted viruses. These data indicate that superinfection transmission resembles primary sexual transmission, providing support for superinfection as a model to address correlates of protection against HIV.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank the participants in the Centre for the AIDS Programme of Research in South Africa (CAPRISA) acute infection cohort for providing specimens and the clinical and laboratory staff at CAPRISA for the outstanding management of the cohort.
Financial support. This work was funded by CAPRISA, through the National Institutes of Health (NIH) (HIV Vaccine Research and Design grant AI088610) and by the National Research Foundation of South Africa (grant 78747). CAPRISA is part of the Comprehensive International Program of Research on AIDS and is supported by the National Institute of Allergy and infectious Disease, NIH (grant AI51794). D. J. S. has received financial support from the University of Cape Town, the Poliomyelitis Research Foundation, and the German Academic Exchange Service (DAAD).
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Smith DM, Strain MC, Frost SDW, et al. Lack of neutralizing antibody response to HIV-1 predisposes to superinfection. Virology 2006; 355:1–5. [DOI] [PubMed] [Google Scholar]
- 2.Basu D, Kraft CS, Murphy MK, et al. HIV-1 subtype C superinfected individuals mount low autologous neutralizing antibody responses prior to intrasubtype superinfection. Retrovirology 2012; 9:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blish CA, Dogan OC, Derby NR, et al. Human immunodeficiency virus type 1 superinfection occurs despite relatively robust neutralizing antibody responses. J Virol 2008; 82:12094–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu D, Xiao P, Ende Z, et al. Low antibody-dependent cellular cytotoxicity responses in Zambians prior to HIV-1 intrasubtype C superinfection. Virology 2014; 462–463:295–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta P, Mellors J, Kingsley L, et al. High viral load in semen of human immunodeficiency virus type 1-infected men at all stages of disease and its reduction by therapy with protease and nonnucleoside reverse transcriptase inhibitors. J Virol 1997; 71:6271–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 2008; 105:7552–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrahams M-R, Anderson JA, Giorgi EE, et al. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol 2009; 83:3556–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derdeyn CA, Decker JM, Bibollet-Ruche F, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 2004; 303:2019–22. [DOI] [PubMed] [Google Scholar]
- 9.van Loggerenberg F, Mlisana K, Williamson C, et al. Establishing a cohort at high risk of HIV infection in South Africa: challenges and experiences of the CAPRISA 002 acute infection study. PLoS One 2008; 3:e1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore PL, Sheward D, Nonyane M, et al. Multiple pathways of escape from HIV broadly cross-neutralizing V2-dependent antibodies. J Virol 2013; 87:4882–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol 2007; 25:1407–10. [DOI] [PubMed] [Google Scholar]
- 12.Moore PL, Ranchobe N, Lambson BE, et al. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog 2009; 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore PL, Gray ES, Wibmer CK, et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med 2012; 18:1688–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wibmer CK, Bhiman JN, Gray ES, et al. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS Pathog 2013; 9:e1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jost S, Bernard M-C, Kaiser L, et al. A patient with HIV-1 superinfection. N Engl J Med 2002; 347:731–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.