Abstract
Background. Treatment initiation rapidly kills most drug-susceptible Mycobacterium tuberculosis, but a bacterial subpopulation tolerates prolonged drug exposure. We evaluated drug-tolerant bacilli in human sputum by comparing messenger RNA (mRNA) expression of drug-tolerant bacilli that survive the early bactericidal phase with treatment-naive bacilli.
Methods. M. tuberculosis gene expression was quantified via reverse-transcription polymerase chain reaction in serial sputa from 17 Ugandans treated for drug-susceptible pulmonary tuberculosis.
Results. Within 4 days, bacterial mRNA abundance declined >98%, indicating rapid killing. Thereafter, the rate of decline slowed >94%, indicating drug tolerance. After 14 days, 16S ribosomal RNA transcripts/genome declined 96%, indicating slow growth. Drug-tolerant bacilli displayed marked downregulation of genes associated with growth, metabolism, and lipid synthesis and upregulation in stress responses and key regulatory categories—including stress-associated sigma factors, transcription factors, and toxin-antitoxin genes. Drug efflux pumps were upregulated. The isoniazid stress signature was induced by initial drug exposure, then disappeared after 4 days.
Conclusions. Transcriptional patterns suggest that drug-tolerant bacilli in sputum are in a slow-growing, metabolically and synthetically downregulated state. Absence of the isoniazid stress signature in drug-tolerant bacilli indicates that physiological state influences drug responsiveness in vivo. These results identify novel drug targets that should aid in development of novel shorter tuberculosis treatment regimens.
Keywords: gene expression profiling, Mycobacterium tuberculosis/genetics, Mycobacterium tuberculosis/physiology, pulmonary/epidemiology, sputum/microbiology, tuberculosis
Drug-tolerant bacterial phenotypes play a central role in tuberculosis, a disease that kills 1.5 million people annually [1]. In patients treated for drug-susceptible tuberculosis, >99% of the Mycobacterium tuberculosis population is killed within the first 3–5 days [2–4]. Thereafter, the rate of killing decreases by >80% [2–4], indicating that surviving bacilli are drug tolerant. Unlike drug resistance, drug tolerance is a reversible phenotypic state that occurs in the absence of resistance-conferring mutations [5].
Drug tolerance is a central challenge for drug development and tuberculosis control. Most forms of tuberculosis require at least 6 months of treatment to eradicate slowly killed, drug-tolerant M. tuberculosis populations and minimize risk of relapse [6, 7]. This duration burdens patients and health systems and contributes to medication nonadherence, enabling further transmission and acquired drug resistance [8].
Little is known about drug-tolerant M. tuberculosis in human tuberculosis. Genetically homogeneous bacterial populations are hypothesized to include heterogeneous bacterial phenotypes [9, 10]. Phenotypes that survive prolonged antibiotic exposure despite genetic susceptibility have been called persisters [5]. However, it is unknown whether there is a single M. tuberculosis persister phenotype or a spectrum of drug-tolerant phenotypes. Drug tolerance is observed under a variety of experimental conditions [11–17], but it is unknown whether these models recapitulate drug-tolerant phenotypes in humans [5]. There is currently no method of identifying distinct phenotypes within an isogenic M. tuberculosis population. A better understanding of the clinically relevant drug-tolerant population that emerges in tuberculosis patients during treatment is essential to developing novel, shorter-treatment regimens.
We used multiplex reverse-transcription polymerase chain reaction (RT-PCR) gene expression profiling to characterize the metabolic state and adaptive mechanisms of drug-tolerant M. tuberculosis remaining in sputum after the early bactericidal phase of treatment. Messenger RNA (mRNA) expression provides a contemporaneous measure of bacterial physiologic state because the estimated half-life of M. tuberculosis mRNA is 9.5 to 50 minutes [18] and mRNA is not detectable from dead bacilli [19]. To identify the transition from rapid to slow killing that is the hallmark of a drug-tolerant bacterial population, we evaluated change in the abundance of M. tuberculosis genomic DNA, 16S ribosomal RNA, and mRNA over time. We analyzed gene expression profiles, identifying change in functional gene categories that reflect the physiologic state of the drug-tolerant population. We examined the expression of genes previously associated with drug tolerance, including efflux pumps, drug-activating enzymes, drug targets, and the isoniazid stress signature. Finally, we compared M. tuberculosis expression patterns in tuberculosis patients undergoing treatment with expression patterns in experimental models of drug tolerance [11, 13, 15–17].
METHODS
Enrollment and Specimen Collection
A random sample of consecutive adults with pulmonary tuberculosis were enrolled at Mulago National Referral Hospital in Kampala, Uganda (details in online Supplementary Materials). Before tuberculosis treatment (day 0) and after 2, 4, 7, 14, 28, and 56 days, patients expectorated into a sterile cup containing guanidine thiocyanate solution for immediate RNA preservation [20]. Within 2 hours, sputa were needle-sheared, centrifuged at 9000 × g, and stored in Trizol at −80°C. Ugandan and US institutional review boards approved the study. All patients provided written informed consent.
Expression Profiling
Total RNA was extracted using a phenol/chloroform protocol described in the Supplementary Materials. After first-strand complementary DNA (cDNA) synthesis, cDNA underwent controlled multiplex amplification using M. tuberculosis-specific flanking primers as previously described [21]. A total of 2411 M. tuberculosis transcripts were quantified via multiplex quantitative RT-PCR with a LightCycler 480 (Roche, Indianapolis, Indiana) using TaqMan primers.
Analysis
We plotted mRNA, 16S ribosomal RNA (rRNA), and DNA abundance over time relative to pretreatment values. We fitted change in geometric mean percentage with 95% confidence intervals using saturated linear mixed models. mRNA expression data were normalized using a minimum-variance data-driven method [22]. We identified altered gene expression at days 2, 4, 7, and 14 relative to day 0 via paired t tests with Benjamini-Hochberg [23] multiple testing correction using a false discovery rate of 0.05. To identify highly expressed genes, expression values for each sample were ranked from 1 (gene with lowest expression) to 100 (highest expression). The median percentile rank of each gene at each day was calculated.
To identify bacterial functions that changed with treatment, we summarized the proportion of genes in previously described functional categories with significantly increased or decreased expression from day 0 with an unadjusted P value <.05. A modified 1-way Fisher exact test [24] was used to identify enriched functional categories (P-value threshold <.05).
Data from days 28 and 56 were not normalized because of low transcript detection. To evaluate similarity in expression across days 14–56, we quantified between-day overlap in the genes consistently detected in >50% of samples.
Nonspecific changes were defined as genes that were significantly induced or suppressed both immediately after drug exposure (day 2) and at all subsequent time points. Genes significantly altered between days 2 and 14 were considered likely specific to drug-tolerant bacilli. Hierarchical clustering using a Euclidean distance was performed on the median fold change from day 0 for tolerance-associated genes. A static tree cut at height 4 was used to create 5 distinct clusters.
Enrichment of functional gene categories was compared between sputum and 5 experimental models. For sputum and each model, the ratio of genes up- and downregulated was calculated for each functional category and log2 transformed. Pearson's correlation coefficients were calculated between each model pair using the log2-transformed values. Analysis used the R statistical suite, version 3.0.1 except as noted in the Supplementary Materials. Minimum information about a microarray experiment (MIAME)-compliant data are available at http://www.tbdb.org/rtpcrData.shtml. Supplementary Table 1 shows median fold-change for all genes assayed.
RESULTS
Seventeen Ugandan adults with drug-susceptible, sputum acid-fast bacillus smear-positive, culture-positive pulmonary tuberculosis were enrolled (Supplementary Figure 1, Supplementary Table 2). Nine were human immunodeficiency virus–infected. All were treated with isoniazid, rifampicin, pyrazinamide, and ethambutol. After 2 months, 4 remained sputum M. tuberculosis-culture positive. One patient had recurrent tuberculosis 13 months after completing treatment.
Change in Nucleic Acid Abundance
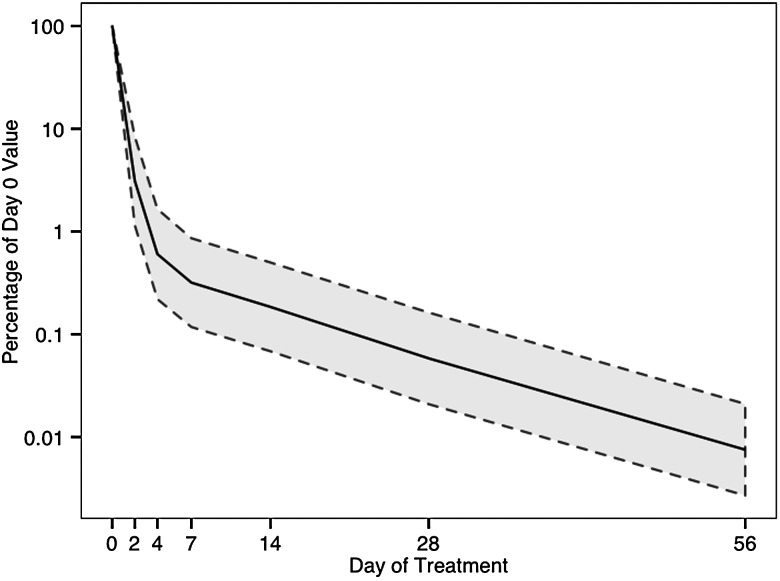
Mycobacterium tuberculosis mRNA abundance declined 0.55 log/day during days 0–4, consistent with rapid killing during the early bactericidal phase (Figure 1). After day 7, the decline in mRNA abundance slowed to 0.03 log/day (slowing 94% relative to day 0–4), consistent with transition to a drug-tolerant population. 16S rRNA and DNA declined more slowly (Supplementary Figure 2). 16S rRNA transcripts per genome equivalent of DNA declined 96%–99% from day 0 to days 14 through 56 (Supplementary Table 3), consistent with a slowing of replication. All samples, including those that were culture negative after 56 treatment days, had greater than 100 M. tuberculosis mRNA transcripts detected, indicating the presence of a viable, but not culturable population, as previously reported [25–28].
Figure 1.
Decline in Mycobacterium tuberculosis mRNA abundance among 17 patients during the first 56 days of tuberculosis treatment. The solid line represents the geometric mean percentage of mRNA abundance remaining at each day. The shaded area between dotted lines indicates 95% confidence intervals. Abbreviation: mRNA, messenger RNA.
M. tuberculosis Transcriptional Alteration During Treatment
Treatment markedly altered the bacterial transcriptome. Relative to pretreatment expression, 452 (19%) to 741 (31%) transcripts had significantly altered expression on treatment days 2, 4, 7, and 14 (Supplementary Table 4).
Figure 2 shows change in key functional categories during treatment (categories defined in Supplementary Table 5; change within each functional category is shown in Supplementary Figures 3–23). Expression of genes associated with metabolic processes was predominantly downregulated. Expression of genes involved in aerobic respiration, the tricarboxylic acid (TCA) cycle, and adenosine triphosphate (ATP) synthesis decreased immediately and remained suppressed through day 14. By contrast, expression of genes associated with anaerobic respiration was not downregulated; expression of narG and frdA increased significantly by day 14. Genes of the glyoxylate bypass, an alternative to the TCA cycle, were downregulated yet remained among the most highly expressed genes.
Figure 2.
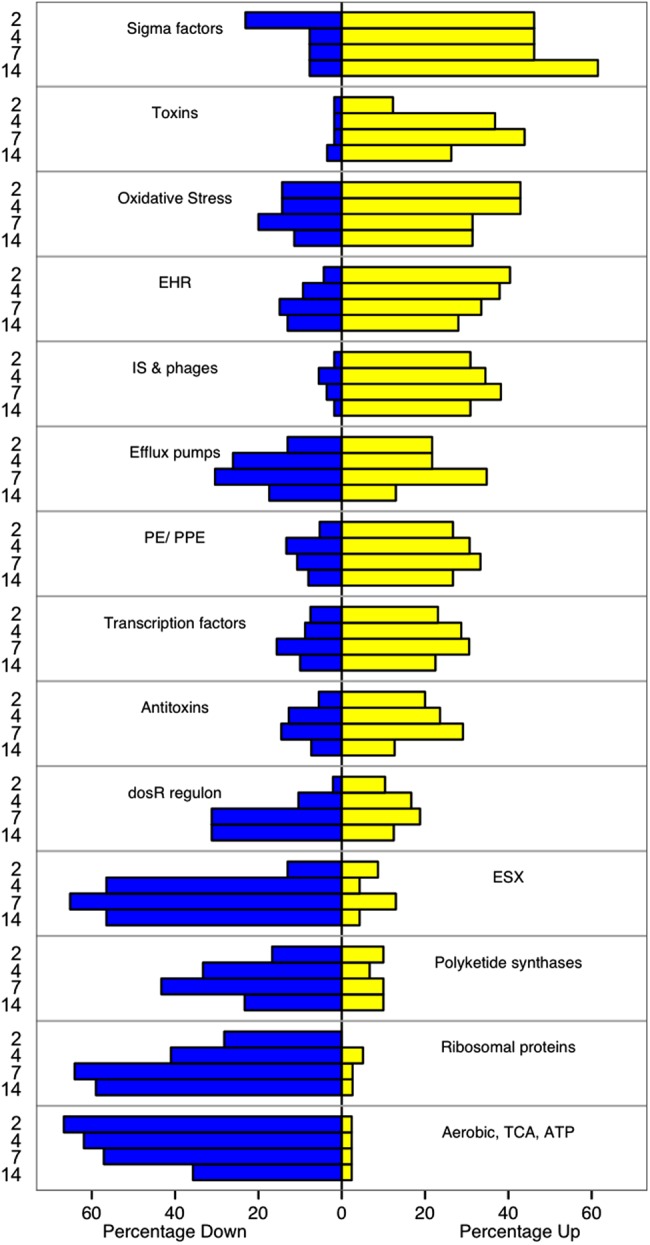
Change in expression of selected functional categories of genes after initiation of tuberculosis treatment. The percentage of genes in each category significantly up- (yellow) or down- (blue) regulated at each day relative to pretreatment expression (Day 0) is illustrated. Genes that did not display statistically significant change are not shown. Categories are ordered from most upregulated (top) to most downregulated (bottom). Category references are provided in the Supplementary Materials, Supplementary Table 5. Abbreviations: ATP, adenosine triphosphate; EHR, enduring hypoxic response; ESX, ESX/Type VII secretion system; IS & phages, insertion sequences and phage-related genes; TCA, tricarboxcylic acid cycle.
Expression of genes coding for ribosomal proteins and other genes such as translation elongation factor (tuf, Rv0685) was rapidly downregulated, suggesting a global decrease in protein translation. Downregulation of polyketide synthase gene expression suggested diminished production of cell envelope lipids. Genes coding for the ESX/Type VII secretion system (ESX), which secretes highly immunogenic virulence factors (including ESAT-6), were highly expressed prior to treatment but were downregulated with treatment, particularly at days 7–14. Expression of genes central to DNA replication, including DNA gyrase subunit A (gyrA), subunit B (gyrB), and DNA topoisomerase I (topA), was significantly downregulated during days 7–14.
Genes typically induced by stress had increased expression, including: oxidative stress response, the immunogenic PE/PPE genes, and insertion sequences and phages. There was induction of genes for the enduring hypoxic response, a response induced in vitro by prolonged hypoxia and other stressors. In contrast, expression of DosR regulon genes that are induced in vitro by hypoxia and other conditions that inhibit aerobic respiration was significantly downregulated in sputum on day 14. Expression of the gene for RelA, the regulator of the starvation-associated stringent response, increased significantly on day 2 but not on subsequent days. The stringent response expression signature includes induction of specific stress-associated gene sets and suppression of ribosomal protein mRNA and other genes [29]. Genes classically induced during the stringent response did not have significantly increased expression in sputum during days 2–14. By contrast, genes previously described as suppressed during the stringent response did have significantly decreased expression during days 2–14.
Three categories of regulators—sigma factors, transcription factors, and toxin-antitoxin (TA) modules—were markedly altered. Expression of the gene for SigA, the principal growth-associated sigma factor, was suppressed. Conversely, expression of genes for stress-associated sigma factors, including SigB, SigE, SigF, SigH, SigI, and SigJ, increased significantly. Expression of genes coding for toxins were upregulated—particularly of the VapBC family, which cleave existing mRNA to rapidly remodel the transcriptome in response to stress.
Transcriptome analysis after day 14 was limited because mRNA abundance was insufficient. However, transcripts consistently detected (present in >50% of samples) at days 14, 28, and 56 overlapped almost entirely (Supplementary Figure 21), suggesting a stable expression pattern during the sterilizing phase.
Transcriptional Patterns Specific to the Drug-tolerant Population
The M. tuberculosis transcriptome is massively altered by drug exposure. However, expression changes apparent immediately after treatment initiation (eg, day 2) may represent nonspecific stress responses rather than adaptations of drug-tolerant populations [11]. To elucidate drug-tolerant phenotypes that emerge later in treatment, we distinguished alterations characteristic of drug-tolerant populations from nonspecific stress responses based on the following established definitions: Nonspecific responses were genes significantly induced or suppressed immediately after drug exposure (day 2) with changes sustained at all subsequent time points. Transcriptional adaptations specific to drug-tolerant bacilli were genes whose expression changed significantly between day 2 and day 14 of drug treatment [11].
Of 104 nonspecific stress-response genes, 64 were consistently induced and 40 were consistently suppressed. Induced genes were statistically enriched for the enduring hypoxic response (P = .003). Suppressed genes were enriched for metabolism genes involved in the TCA cycle, aerobic respiration, and ATP synthesis (P = .006).
We identified 82 genes that appeared to be specifically associated with emergence of a drug-tolerant population (Figure 3). Clustering by treatment day showed progressive expression change with increasing duration of treatment. Days 7 and 14 clustered closely together, suggesting transition to a stable drug-tolerant phenotype. Clustering of genes over time identified subsets with similar patterns of expression change, suggesting a coordinated role in development or maintenance of drug-tolerant bacilli. Clusters A and B (35 genes) demonstrated little to no change between days 0 and 2, but significant induction by days 7 and 14. These included triacylglycerol synthases, and ATP-binding cassette transporter and toxin molecules. Clusters C and D (9 genes) were strongly induced immediately following antibiotic exposure (day 2) and were notable for 3 genes in the KAS operon required for mycolic acid synthesis. Cluster E (38 genes) was significantly downregulated at days 7 and 14; it consisted primarily of ESX and ribosomal genes.
Figure 3.
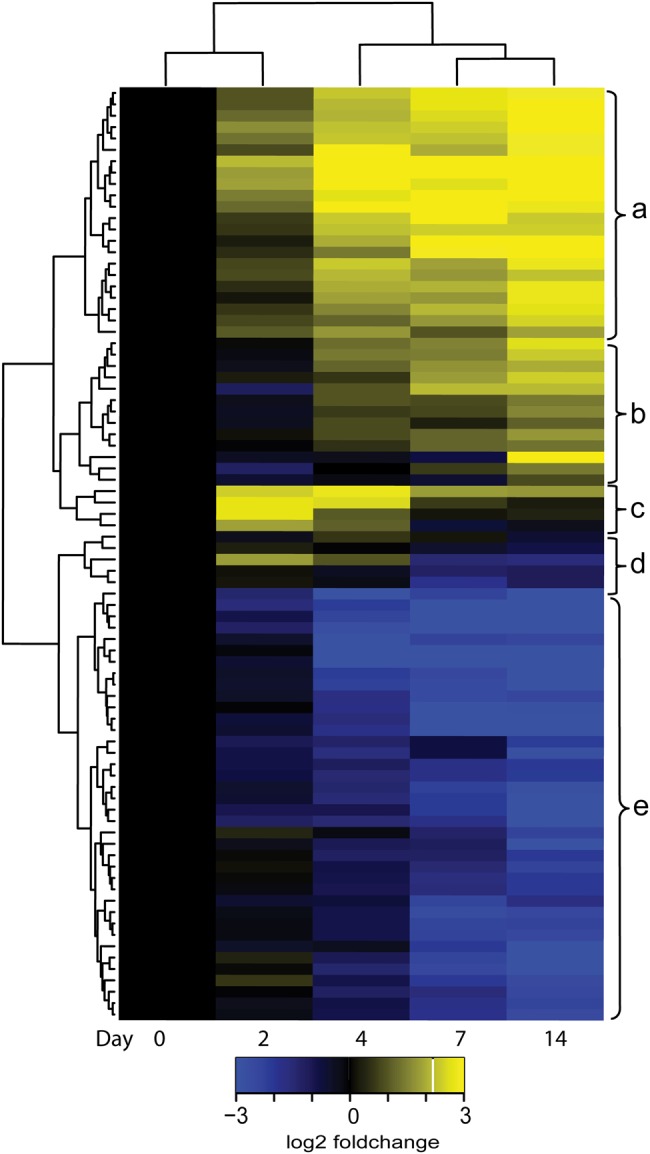
Heat map of median expression fold-change of 82 genes differentially expressed (adjusted P value <.05) between the early bactericidal phase (Day 2) and sterilizing phase (Day 14). Yellow indicates increased expression relative to day 0; blue indicates decreased expression. Clustering by day (row) indicates similarity between expression patterns at different days. Clustering by gene (column) identifies 5 clusters of genes (a–e) with similar expression patterns over time.
Expression of Antibiotic-associated Genes
We evaluated genes for proteins previously associated with drug tolerance or resistance, including efflux pumps, drug-activating enzymes, drug targets, and the isoniazid stress signature. As a category, efflux pump genes were not significantly enriched following treatment (Supplementary Figure 22). However, a subset of efflux genes shown in vitro to export drugs—including the Tap protein coded for by Rv1258c, bacA (Rv1819c), and mmr (Rv3065)—were induced >3-fold during days 7–14.
Decreased expression of genes coding for drug-activating enzymes—katG (isoniazid), pncA (pyrazinamide), and ethA (ethionamide)—would be predicted to increase drug tolerance. However, after 7–14 days’ treatment, expression of these genes was not suppressed.
Drug tolerance could also result from increased expression of drug targets. Expression of the gene for rifampicin target DNA-dependent RNA polymerase (RpoB) was significantly induced at all intervals after treatment initiation. Expression of the gene for isoniazid target InhA did not change significantly. In contrast, genes for DNA gyrase subunits (GyrA, GyrB), which are targeted by fluoroquinolones (drugs these patients did not receive), had progressively decreased expression, consistent with decreased replication. Similarly, expression of the gene for bedaquiline target AtpE was suppressed at all intervals, consistent with downregulation of aerobic metabolism.
We examined the effects of drug treatment on the isoniazid stress response—a set of mycolic acid synthesis and stress response genes previously shown to be induced and inversely proportional to growth rate during in vitro isoniazid exposure (Supplementary Table 7) [30–33]. After 2 treatment days, the isoniazid stress signature was robustly induced in sputum, including mycolic acid biosynthesis genes (fabD, acpM, kasA, kasB, accD6), an efflux gene (efpA), and an oxidative stress-response gene (aphC). After 7 days, this signature was no longer upregulated, consistent with the known poor sterilizing activity of isoniazid [34].
Comparison With Experimental Models of M. tuberculosis Drug Tolerance
To test whether existing experimental models recapitulate drug-tolerant M. tuberculosis in humans, we compared sputum with 5 widely cited models (details in Supplementary Table 8) [11, 13, 15–17]. Similar to sputum, all models identified downregulation of ribosomal proteins and TCA cycle, aerobic respiration, and ATP synthesis genes, indicating metabolic slowdown (Figure 4). Most found downregulation of ESX genes. All reported upregulation of the enduring hypoxic response, and most found upregulation of oxidative stress responses. Notably, the oxygen-responsive DosR regulon was induced in models that applied hypoxic stress [15–17, 35] but not in sputum, the antibiotic selection model [11], or the nutrient starvation model [13]. The stringent response regulator relA was induced in the nutrient starvation model [13], the hollow-fiber granuloma model [32], and the oxygen deprivation model [16]. The initial transient induction of relA at day 2 in sputum is similar to the antibiotic selection model [11] in which relA was significantly induced after 1 day but then significantly downregulated after 7 and 14 days (Supplementary Table 9). Drug-tolerant M. tuberculosis in sputum had greater upregulation of regulatory genes—TA modules, sigma factors, and transcription factors—than did several models [13, 15, 35]. Sputum M. tuberculosis also had greater induction of PE/PPE genes and insertion sequences and phage genes than most models [11, 15, 16]. Overall, expression in sputum was most similar to the antibiotic selection model [11] (Pearson correlation coefficient, 0.73) (Supplementary Table 10).
Figure 4.
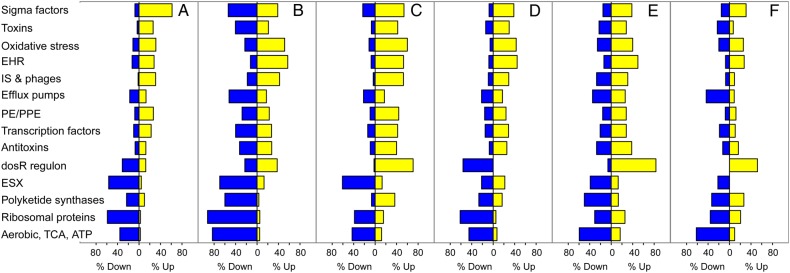
Comparison of enrichment of functional gene categories between drug-tolerant Mycobacterium tuberculosis in sputum and selected experimental models. A, Sputum after 14 days of drug treatment. B, Antibiotic selection model [11]. C, Hollow fiber artificial granuloma model [16]. D, Nutrient starvation model [13]. E, Oxygen deprivation nonreplicating persistence model [16], (F) Multiple-stress model [15]. Abbreviations: ATP, adenosine triphosphate; EHR, enduring hypoxic response; ESX, ESX/Type VII secretion system; IS & phages, insertion sequences and phage-related genes; TCA, tricarboxcylic acid cycle.
DISCUSSION
This study provides the first direct characterization of drug-tolerant M. tuberculosis during human tuberculosis treatment, a refractory population that dictates the components and duration of tuberculosis treatment. Gene expression of drug-tolerant bacilli suggests slowing of growth and metabolic and synthetic downregulation. Nonetheless, the drug-tolerant population is not transcriptionally dormant. Rather, upregulation of genes involved in stress responses, drug efflux, and regulatory functions point to mechanisms that enable survival under drug stress. Understanding the transcriptional state of drug-tolerant bacilli in vivo should aid development of novel shorter tuberculosis treatment regimens.
Several lines of evidence indicate that M. tuberculosis surviving in sputum after 14 days of treatment is drug tolerant. In vitro [11] and human data [36] indicate that 7 days of drug exposure clears rapidly killed bacilli, leaving a drug-tolerant population. We identified a biphasic decline of M. tuberculosis mRNA characteristic of early bactericidal activity studies [2–4]. The slowdown in killing after the bactericidal phase is the clinical hallmark of drug-tolerance [2–4].
Whether drug-tolerant M. tuberculosis phenotypes continue replication or become nonreplicating is intensely debated [5, 37, 38]. Our gene expression data do not provide a direct measure of growth rate, but the following observations suggest decreased replication: 16S rRNA per genome declined 96%–99% from day 0 to days 14–56, genes coding for ribosomal proteins were strongly downregulated, and expression of DNA gyrases and topoisomerases was suppressed. Conversely, the continued detection of transcripts for ribosomal proteins and DNA synthesis functions suggests that replication may not cease entirely.
Drug-tolerant M. tuberculosis are metabolically downregulated but are not transcriptionally dormant. Induction of multiple stress responses and of genes with regulatory functions suggest adaptations that enable survival of drug-tolerant phenotypes. Our data support the role of growth-inhibiting TA modules in mediating transition to and maintenance of drug tolerance through posttranscriptional regulation [39, 40].
We evaluated mechanisms hypothesized to contribute to drug tolerance. Slow growth decreases drug effectiveness [15–17, 32], likely because antibiotics generally target replication-associated functions. Drug-tolerant M. tuberculosis had increased expression of genes for efflux pumps, which export rifampicin and isoniazid [41, 42]. This supports the proposal that pharmacologic efflux pump inhibition might shorten treatment regimens [43]. Conversely, drug tolerance is not readily explained by regulation of drug-activating enzymes. In vitro data suggest that one tolerance mechanism is suppression of katG expression, leading to diminished isoniazid activation [37, 44]. We did not identify downregulation of katG in vivo. However, this does not rule out the possibility that decreased katG expression occurs in a subset of drug-tolerant bacilli or fosters the initial formation of drug tolerance after isoniazid exposure. Drug tolerance was not readily explained by altered expression of drug targets. Induction of rpoB in drug-tolerant M. tuberculosis could be a compensatory adaptation for rifampicin's inactivation of RNA polymerase. Alternatively, it could indicate a general stress-induced increase in transcriptional reprogramming. In either case, rpoB overexpression has not been associated with resistance [45]. Increased expression of the gene for isoniazid target InhA causes low-level resistance [34], but, as in experimental models [11, 13, 30, 46], inhA was not induced in drug-tolerant M. tuberculosis in sputum. Drug-tolerant M. tuberculosis had decreased expression of genes for fluoroquinolone targets GyrA and GyrB and bedaquiline target AtpE. Because patients were not administered these drugs, this may reflect downregulation of replication and aerobic metabolism as part of a broader regulatory program rather than drug-specific responses.
These results have clinical implications. We have shown that monitoring bacterial physiology during human tuberculosis treatment is feasible through M. tuberculosis expression profiling. Practical applications—including monitoring response to novel drugs or as biomarkers predictive of treatment failure or relapse—should be evaluated [47]. Detection of transcriptionally active bacilli after 2 months of treatment—including in culture-negative samples—underscores existing evidence that culture detects a small proportion of viable bacilli [48]. Importantly, our findings indicate that standard doses of rifampicin—an inhibitor of transcription initiation—fail to fully interrupt bacterial gene expression. Evidence suggests higher rifampicin doses may improve killing and shorten treatment durations [49].
Prior to this study, it was not possible to test whether experimental models that inform current understanding of drug tolerance successfully recapitulate M. tuberculosis phenotypes in human tuberculosis. Gene expression of drug-tolerant M. tuberculosis in sputum was most similar to an in vitro model of drug tolerance based on antibiotic exposure alone in the absence of hypoxia or other adverse conditions [11]. Drug-tolerant M. tuberculosis in sputum did not have increased expression of the DosR regulon that is characteristic of models that use hypoxia [15, 16, 35] or hypoxic tissue conditions [17]. Genes of the DosR regulon were highly expressed in sputum prior to drug exposure in our data and in a previous study [20], but were not further induced by treatment. This indicates that drug-tolerant bacilli are not necessarily in a more hypoxic environment than drug-susceptible bacilli. The transient induction of relA we observed in sputum is consistent with the antibiotic selection model but not with nutrient-limited models. RelA with consequent ppGpp synthesis may direct the initial formation of drug-tolerant phenotypes. Alternatively, it is possible that the stringent response is not robustly expressed in drug-tolerant M. tuberculosis in sputum because antibiotic exposure may not alter nutrient availability.
Treatment failure and relapse are surmised to result from drug-tolerant M. tuberculosis persister bacilli. However, there is no direct evidence or scientific consensus regarding the nature of the M. tuberculosis persister phenotype [5] or even whether a single persister phenotype exists [8, 10]. The drug-tolerant M. tuberculosis population we studied in sputum meets a proposed functional definition of persisters: bacteria that survive prolonged antibiotic exposure despite genetic susceptibility [5]. Nonetheless, because it is not currently possible to distinguish between bacterial phenotypes or track individual bacilli in humans, we cannot prove that the drug-tolerant M. tuberculosis present in sputum represents the hypothesized persister phenotype.
This study has limitations. The sample size did not permit correlation of bacterial expression with clinical outcomes. Although phenotypic drug-susceptibility testing was not performed, strains profiled in this study were likely drug-susceptible because rates of primary drug resistance in Uganda are low [50]. No rifampicin resistance–conferring mutations were detected with GeneXpert MTB/RIF. Our analysis cannot differentiate between transcriptional reprogramming of individual bacilli and alteration in the M. tuberculosis population structure. A component of the transcriptional change we observed is likely due to selective elimination of rapidly growing nontolerant subpopulations. However, regardless of the relative balance of transcriptional reprogramming versus population restructuring, transcriptional profiling of the bacilli that survive prolonged drug exposure provides important insights into drug tolerance.
CONCLUSIONS
This first description of the M. tuberculosis transcriptome during human tuberculosis treatment suggests a shift to a slowly replicating, metabolically downregulated phenotype that no longer displays signals of isoniazid-induced stress. Our findings reinforce the critical need for novel antituberculosis drugs to target the recalcitrant drug-tolerant bacterial population, leading to shorter, more effective drug regimens.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We wish to acknowledge the patients who participated in the study and the staff and administration of Mulago Hospital and the Makerere University and University of California, San Francisco Research Collaboration. We thank Eric Vittinghoff for analytic support.
Financial support. This work was supported by National Institutes of Health (grant numbers UL1 RR025780, 2T15LM009451-06 [to B. J. G.], K23HL094141 [to A. C.], R21AI101714 [to J. L. D.], K23AI080147 [to J. L. D.], K24HL087713 [to L. H.], R01HL090335 [to L. H.], 5R01AI104589 [to P. N.], 2RO1 AI061505 [to M. I. V.]) and Veteran's Administration (CDA1IK2CX000914-01A1 [to N. D. W.]), and a Boettcher Foundation Webb-Waring Award (to M. S.).
Potential conflicts of interest. All authors: No reported.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. Global tuberculosis control: WHO report 2014. 2014.
- 2.Mitchison DA, Selkon JB. The bactericidal activities of antituberculosis drugs. Am Rev Respir Dis 1956; 74:109–16. [DOI] [PubMed] [Google Scholar]
- 3.Jindani A, Doré CJ, Mitchison DA. Bactericidal and sterilizing activities of antituberculosis drugs during the first 14 Days. Am J Resp Crit Care 2003; 167:1348–54. [DOI] [PubMed] [Google Scholar]
- 4.Davies GR, Brindle R, Khoo SH, Aarons LJ. Use of nonlinear mixed-effects analysis for improved precision of early pharmacodynamic measures in tuberculosis treatment. Antimicrob Agents Chemother 2006; 50:3154–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balaban NQ, Gerdes K, Lewis K, McKinney JD. A problem of persistence: still more questions than answers? Nat Rev Micro 2013; 11:587–91. [DOI] [PubMed] [Google Scholar]
- 6.Balaban NQ, Gerdes K, Lewis K, McKinney JD. American thoracic society/centers for disease control and prevention/infectious diseases society of America: treatment of tuberculosis. Am J Resp Crit Care 2003; 167:603–62. [DOI] [PubMed] [Google Scholar]
- 7.Gillespie SH, Crook AM, McHugh TD, et al. Four-month moxifloxacin-based regimens for drug-sensitive tuberculosis. N Engl J Med 2014; 371:1577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipsitch M, Levin BR. Population dynamics of tuberculosis treatment: mathematical models of the roles of non-compliance and bacterial heterogeneity in the evolution of drug resistance. Int J Tuberc Lung Dis 1998; 2:187–99. [PubMed] [Google Scholar]
- 9.Balaban NQ. Persistence: mechanisms for triggering and enhancing phenotypic variability. Curr Opin Genet Dev 2011; 21:768–75. [DOI] [PubMed] [Google Scholar]
- 10.Manina G, Dhar N, McKinney JD. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 2015; 17:32–46. [DOI] [PubMed] [Google Scholar]
- 11.Keren I, Minami S, Rubin E, Lewis K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2011; 2:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun 1996; 64:2062–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 2002; 43:717–31. [DOI] [PubMed] [Google Scholar]
- 14.Rifat D, Bishai WR, Karakousis PC. Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J Infect Dis 2009; 200:1126–35. [DOI] [PubMed] [Google Scholar]
- 15.Deb C, Lee C-M, Dubey VS, et al. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLOS One 2009; 4:e6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis 2004; 84:218–27. [DOI] [PubMed] [Google Scholar]
- 17.Karakousis PC, Yoshimatsu T, Lamichhane G, et al. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med 2004; 200:647–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rustad TR, Minch KJ, Brabant W, et al. Global analysis of mRNA stability in Mycobacterium tuberculosis. Nucleic Acids Res 2013; 41:509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Mangan JA, Dhillon J, et al. Detection of mRNA transcripts and active transcription in persistent Mycobacterium tuberculosis induced by exposure to rifampin or pyrazinamide. J Bacteriology 2000; 182:6358–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garton NJ, Waddell SJ, Sherratt AL, et al. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculosis sputum. PLOS Med 2009; 5:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galagan JE, Minch K, Peterson M, et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 2013; 499:178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia B, Walter ND, Dolganov G, et al. A minimum variance method for genome-wide data-driven normalization of qRT-PCR expression data. Anal Biochem 2014; 458:11–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995; 57:289–300. [Google Scholar]
- 24.Hosack D, Dennis G, Sherman B, Lane H, Lempicki R. Identifying biological themes within lists of genes with EASE. Genome Biol 2003; 4:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hellyer TJ, DesJardin LE, Hehman GL, Cave MD, Eisenach KD. Quantitative analysis of mRNA as a marker for viability of Mycobacterium tuberculosis. J Clin Microbiol 1999; 37:290–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desjardin L, Perkins M, Wolski K, et al. Measurement of sputum Mycobacterium tuberculosis messenger RNA as a surrogate for response to chemotherapy. Am J Respir Crit Care Med 1999; 160:203–10. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Mahan CS, Palaci M, et al. Sputum Mycobacterium tuberculosis mRNA as a marker of bacteriologic clearance in response to antituberculosis therapy. J Clin Microbiol 2010; 48:46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honeyborne I, McHugh TD, Phillips PPJ, et al. Molecular bacterial load assay, a culture-free biomarker for rapid and accurate quantification of sputum Mycobacterium tuberculosis bacillary load during treatment. J Clin Microbiol 2011; 49:3905–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahl JL, Kraus CN, Boshoff HIM, et al. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Nat Acad Sci USA 2003; 100:10026–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson M, DeRisi J, Kristensen H-H, et al. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA 1999; 96:12833–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betts JC, McLaren A, Lennon MG, et al. Signature gene expression profiles discriminate between isoniazid-, thiolactomycin-, and triclosan-treated Mycobacterium tuberculosis. Antimicrob Agents Chemother 2003; 47:2903–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karakousis PC, Williams EP, Bishai WR. Altered expression of isoniazid-regulated genes in drug-treated dormant Mycobacterium tuberculosis. J Antimicrob Chemother 2008; 61:323–31. [DOI] [PubMed] [Google Scholar]
- 33.Fu LM. Exploring drug action on Mycobacterium tuberculosis using Affymetrix oligonucleotide genechips. Tuberculosis 2006; 86:134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Jacobs WR., Jr Mechanisms of drug action, drug resistance and drug tolerance in Mycobacterium tuberculosis: Expected phenotypes from evolutionary pressures from a highly sucessful pathogen. In: Kaufmann SHE, Rubin E, eds. Handbook of tuberculosis: molecular biology and biochemistry. Vol. I Weinheim: Wiley-VCH, 2008:323–58. [Google Scholar]
- 35.Muttucumaru DGN, Roberts G, Hinds J, Stabler RA, Parish T. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis 2004; 84:239–46. [DOI] [PubMed] [Google Scholar]
- 36.Chigutsa E, Patel K, Denti P, et al. A time-to-event pharmacodynamic model describing treatment response in patients with pulmonary tuberculosis using days to positivity in automated liquid mycobacterial culture. Antimicrob Agents Chemother 2013; 57:789–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakamoto Y, Dhar N, Chait R, et al. Dynamic persistence of antibiotic-stressed mycobacteria. Science 2013; 339:91–5. [DOI] [PubMed] [Google Scholar]
- 38.Boshoff HIM, Barry CE. Tuberculosis metabolism and respiration in the absence of growth. Nat Rev Micro 2005; 3:70–80. [DOI] [PubMed] [Google Scholar]
- 39.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLOS Genet 2009; 5:e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fasani RA, Savageau MA. Molecular mechanisms of multiple toxin–antitoxin systems are coordinated to govern the persister phenotype. Proc Natl Acad Sci USA 2013; 110:E2528–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams Kristin N, Takaki K, Connolly Lynn E, et al. Drug tolerance in replicating mycobacteria mediated by a macrophage-induced efflux mechanism. Cell 2011; 145:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta AK, Chauhan DS, Srivastava K, et al. Estimation of efflux mediated multi-drug resistance and its correlation with expression levels of two major efflux pumps in mycobacteria. J Commun Dis 2006; 38:246–54. [PubMed] [Google Scholar]
- 43.Louw GE, Warren RM, Gey van Pittius NC, McEvoy CR, Van Helden PD, Victor TC. A balancing act: efflux/influx in mycobacterial drug resistance. Antimicrob Agents Chemother 2009; 53:3181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niki M, Niki M, Tateishi Y, et al. A novel mechanism of growth phase-dependent tolerance to isoniazid in mycobacteria. J Biol Chem 2012; 287:27743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tupin A, Gualtieri M, Roquet-Baneres F, Morichaud Z, Brodolin K, Leonetti J-P. Resistance to rifampin: at the crossroads between ecological, genomic and medical concerns. Int J Anntimicrob Agents 2010; 35:519–23. [DOI] [PubMed] [Google Scholar]
- 46.Tudó G, Laing K, Mitchison DA, Butcher PD, Waddell SJ. Examining the basis of isoniazid tolerance in nonreplicating Mycobacterium tuberculosis using transcriptional profiling. Future Med Chem 2010; 2:1371–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nahid P, Saukkonen J, Kenzie WRM, et al. Tuberculosis biomarker and surrogate endpoint research roadmap. Am J Resp Crit Care 2011; 184:972–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. Resuscitation-promoting factors reveal an occult population of tubercle bacilli in sputum. Am J Resp Crit Care 2010; 181:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Steenwinkel JEM, Aarnoutse RE, de Knegt GJ, et al. Optimization of the rifampin dosage to improve the therapeutic efficacy in tuberculosis treatment using a murine model. Am J Resp Crit Care 2013; 187:1127–34. [DOI] [PubMed] [Google Scholar]
- 50.Lukoye D, Cobelens FGJ, Ezati N, et al. Rates of anti-tuberculosis drug resistance in Kampala-Uganda are low and not associated with HIV infection. PLOS One 2011; 6:e16130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.