Abstract
Background
Apurinic/apyrimidinic endonuclease-1 (APE1) is a rate-limiting enzyme in DNA base excision repair and has been implicated in carcinogenesis. In this study, we summarize available data to examine the susceptibility of APE1 gene Asp148Glu variant to digestive cancer via a meta-analysis.
Material/Methods
Study selection and data abstraction were conducted independently by 2 authors. Random-effects model was utilized to pool effect estimates. Heterogeneity and publication bias were addressed.
Results
Sixteen articles involving 4916 digestive cancer patients and 7748 controls were qualified for this meta-analysis. Overall association showed an indicative association between Asp148Glu variant and digestive cancer under allelic (odds ratio or OR=1.11; 95% confidence interval or CI: 0.99–1.25; P=0.074) and dominant (OR=1.18; 95% CI: 1.00–1.40; P=0.056) models, with strong evidence of heterogeneity. Deviation from Hardy-Weinberg equilibrium was an obvious source of heterogeneity. In subgroup analyses by cancer sites, this variant was significantly associated with the increased risk for hepatocellular cancer under allelic (OR=1.50; 95% CI: 1.25–1.80; P<0.001) and homozygous genotypic (OR=1.55; 95% CI: 1.02–2.29; P=0.028) models. There were low probabilities of publication bias for the above comparisons.
Conclusions
The results of this meta-analysis collectively suggest that APE1 gene Asp148Glu variant is not a risk-conferring factor for digestive cancer. Further large and well-designed studies are required.
MeSH Keywords: Digestive System Neoplasms, Genetic Association Studies, Meta-Analysis
Background
DNA damage refers to an alteration in the chemical structure of DNA, and usually gives rise to mutations and epimutations [1,2]. In the body, damaged DNA or inappropriate bases can be identified and properly repaired by some enzymes, such as apurinic/apyrimidinic endonuclease-1 (APE1) [3]. APE1 is a rate-limiting enzyme in DNA base excision repair and is increasingly recognized to play an important role in cancer cell growth and tumorigenicity [4]. For example, in pancreatic cancer, APE1 has been implicated in anticancer properties via inhibiting pancreatic tumor growth, as well as cancer cell migration and invasion [5,6]. Moreover, APE1 was observed to be implicated in sustaining cell variability and proliferation of colon cancer and breast cancer cells [7]. It is therefore reasonable to conjecture that APE1 might play a contributory role in unraveling the molecular mechanisms of cancer.
The gene encoding APE1 is mapped on chromosome 14q11.2–14q12 and consists of 5 exons spanning approximate 2.21 kb. APE1 has a DNA-repairing domain and a redox domain, and its carboxy-terminus contains the endonuclease activity required for DNA repair [8]. A non-synonymous exonic variant, Asp148Glu (rs1130409), that resides in the carboxy-terminus of APE1 has attracted special attention in genetic cancer research. Many association studies have examined the relationship between APE1 gene Asp148Glu variant and cancer [9–11]; however, the results of most studies remain inconclusive, with no consensus on their implications, possibly due to the insufficient power of individual studies, the genetic diversity of ethnic populations, and the potentially uncontrolled confounding effects [12]. To systematically address this uncertainty, we undertook a meta-analysis by summarizing available data on the association between Asp148Glu variant and digestive cancer risk. Digestive cancer is a family of malignancies that originate from digestive organs, such as the stomach, colon, and liver, and has a strong inherited basis. For example, family members who have a mutation in a mismatch repair gene are observed to have a much higher rate of colorectal cancer than those who do not have the mutation [13].
Material and Methods
Article search
An attempt to find all original articles on the association between APE1 gene Asp148Glu variant and digestive cancer risk was conducted in the electronic databases PubMed and Embase up to December 2014. The following medical subject headings and key words were used: “apurinic/apyrimidinic or APE1 or APEX1”, “gastric or stomach or colorectal or colon or rectal or esophageal or liver or hepatic or hepatocellular or pancreatic or gallbladder or biliary”, “cancer or carcinoma or tumor or sarcoma or leiomyoma”, along with “polymorphism or genetic or variant or mutation or allele or genotype”. The bibliographies of primarily retrieved articles and previous meta-analyses were manually searched to identify citations that were not identified initially.
Study selection
The eligibility of all retrieved articles was independently ascertained by 2 of us (He Li and Jing Zou) according to the predefined criteria through scanning the titles and abstracts. As a prerequisite, only articles written in English and performed in humans were considered. Inclusion criteria for selection were: (1) all eligible articles should be original investigations; (2) clinical endpoints should be digestive cancer, including esophageal cancer, gastric cancer, colorectal cancer, hepatocellular carcinoma, biliary tract cancer and pancreatic cancer; (3) all studies should be retrospective or nested case-control studies; and (4) the genotype counts of APE1 gene Asp148Glu variant should be provided in both digestive cancer patients and controls. Abstracts and conference posters or proceedings were not included in this meta-analysis due to insufficient information of interest. All eligible articles were reported to have received approval from the local Institutional Review Board (IRB) committees.
Data abstraction
The 2 authors who were responsible for study selection independently abstracted data from each qualified article according to a standardized collection form, including the first author’s last name, year of publication, ethnicity of study population, type of digestive cancer, study design, genotyping platform, matched condition, sample size, and the genotype counts of APE1 gene Asp148Glu variant between digestive cancer patients and controls, as well as the average levels of study characteristics, if available, including age, sex (the percentage of males), body mass index (BMI), and the percentages of smoking, drinking, and family history of cancer between the 2 groups. Discrepancies in data abstraction were resolved by consensus through discussion with other investigators of the present meta-analysis or through reference to the original or indexed articles. Study authors were contacted if necessary for additional information.
Statistical analysis
For the association of APE1 gene Asp148Glu variant with digestive cancer risk, 3 genetic models of inheritance including allelic (148Glu versus 148Asp), homozygous genotypic (148Glu/Glu versus 148Asp/Asp), and dominant (148Glu/Glu plus 148Asp/Glu versus 148Asp/Asp) models were calculated, and the risk effects were expressed as odds ratio (OR) and its corresponding 95% confidence interval (95% CI). Assessment of Hardy-Weinberg equilibrium for Asp148Glu variant was conducted only among controls using the chi-squared test at a significance level of 5%.
Heterogeneity among studies was examined for risk effects using the I2 statistic, a transformation of the Q statistic (I2=100%×(Q-df)/Q, where DF denotes degrees of freedom) that estimates the percentage of the variation in effect sizes that is due to heterogeneity rather than due to chance. The I2 statistic takes values between 0 and 100% with higher values (>50%) indicating the existence of heterogeneity.
In the absence of between-study heterogeneity, fixed- and random-effects models yielded similar estimates, while in view of significant heterogeneity for several comparisons, only results from the random-effects model using the DerSimonian & Laird method [14] are presented in the present meta-analysis.
To seek potential sources of heterogeneity, both subgroup analyses and meta-regression analyses were conducted. Subgroup analyses were predefined according to the test results of Hardy-Weinberg equilibrium, different sites of digestive cancer, ethnicities, study designs, genotyping platforms, matched conditions and sample sizes. Continuous variables including age, gender, body mass index (BMI), and the percentages of smoking, drinking, family history of cancer were incorporated into a meta-regression model. The probability of publication bias was inspected by the visual Begg’s funnel plots and was quantified by both Begg’s and Egger’s tests at a significance level of 10% [15]. In addition, the trim and fill method was adopted to estimate the number and outcomes of potentially missing studies resulting from publication bias. Statistical calculations were completed by the STATA software (StataCorp, Texas, USA, version 12.0 for Windows).
Results
Description of studies
Initial search yielded 294 potentially relevant articles according to the predefined subject headings and key words. After reviewing these articles, 278 articles were excluded with specified reasons and a total of 16 qualified articles involving 4916 digestive cancer patients and 7748 controls were left for final analysis [10,16–30].
Tables 1 and 2 show the baseline characteristics of study populations and the genotype distributions of APE1 gene Asp148Glu variant of each qualified study. Out of 16 eligible studies, 8 studies analyzed the association of this variant with colorectal cancer, 3 studies with gastric cancer, 2 studies for pancreatic cancer, and 1 study respectively for cancer of esophageal, gallbladder and hepatocellular. Eight studies involved populations of Caucasian descent, 6 studies of Asian descent and 2 studies of mixed descents. Nine studies enrolled controls from hospitals and 7 from general populations. Age or gender was reported to be matched in thirteen studies, unavailable in 2 studies, and unmatched in only 1 study. For the genotype distributions of Asp148Glu variant, Hardy-Weinberg equilibrium was satisfied in 13 studies and was not in 3 studies. Seven studies had genotypes determined by restriction fragment length polymorphism (RFLP) method, and the other 9 studies by Taqman or array method. There were twelve of 16 studies with total sample size of less than 1000. The average frequency of 148Glu allele was 45.35% in digestive cancer patients and 42.57% in controls.
Table 1.
Baseline characteristics of the study populations in this meta-analysis.
| Author (year) | Cancer type | Ethnicity | Design | Matched | Genotyping | Sample size | Age (years) | Male | BMI (kg/m2) | Smoking | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | ||||||
| Moreno V. et al. (2006) | Colorectal | Caucasian | Hospital | NA | Array | 359 | 312 | NA | NA | NA | NA | NA | NA | NA | NA |
| Jiao L. et al. (2006) | Pancreatic | Mixed | Hospital | YES | PCR-ASG | 367 | 330 | NA | NA | 0.557 | 0.515 | NA | NA | 0.612 | 0.535 |
| Berndt S. et al. (2007) | Colorectal | Mixed | Population | YES | TaqMan | 739 | 757 | NA | NA | 0.696 | 0.692 | NA | NA | 0.663 | 0.595 |
| Tse D. et al. (2008) | Esophageal | Caucasian | Hospital | YES | TaqMan | 311 | 454 | 64.0 | 64.0 | 0.894 | 0.874 | 23.00 | 22.00 | 0.800 | 0.683 |
| Pardini B. et al. (2008) | Colorectal | Caucasian | Hospital | YES | PCR-RFLP | 531 | 530 | 58.5 | 57.4 | 0.553 | 0.553 | NA | NA | 0.268 | 0.283 |
| Kasahara M. et al. (2008) | Colorectal | Asian | Hospital | YES | PCR-RFLP | 68 | 121 | 67.3 | 67.4 | 0.544 | 0.612 | NA | NA | 0.471 | 0.545 |
| Huang W.Y. et al. (2008) | Gallbladder | Asian | Population | YES | Array | 236 | 734 | NA | NA | 0.274 | 0.388 | NA | NA | 0.271 | 0.302 |
| Palli D. et al. (2010) | Gastric | Caucasian | Population | YES | TaqMan | 298 | 546 | 68.8 | 55.5 | 0.564 | 0.493 | NA | NA | 0.558 | 0.586 |
| Jelonek K. et al. (2010) | Colorectal | Caucasian | Hospital | YES | PCR-RFLP | 113 | 153 | NA | NA | NA | NA | NA | NA | NA | NA |
| Brevik A. et al. (2010) | Colorectal | Caucasian | Population | NA | TaqMan | 304 | 359 | NA | NA | NA | NA | NA | NA | NA | NA |
| Canbay E. et al. (2010) | Gastric | Caucasian | Population | YES | PCR-RFLP | 40 | 247 | 60.1 | 52.8 | NA | NA | NA | NA | 0.625 | 0.368 |
| Gu D. et al. (2011) | Gastric | Asian | Hospital | YES | PCR-RFLP | 338 | 362 | 61.8 | 62.5 | 0.657 | 0.660 | NA | NA | 0.461 | 0.362 |
| Canbay E. et al. (2011) | Colorectal | Caucasian | Population | YES | PCR-RFLP | 79 | 247 | 60.2 | 59.7 | 0.646 | 0.526 | 28.50 | 27.70 | 0.380 | 0.368 |
| Nakao M. et al. (2012) | Pancreatic | Asian | Population | YES | TaqMan | 185 | 1465 | NA | NA | 0.687 | 0.749 | NA | NA | NA | NA |
| Zeng X. et al. (2012) | Hepatocellular | Asian | Hospital | YES | TaqMan | 497 | 500 | NA | NA | 0.787 | 0.742 | NA | NA | 0.328 | 0.096 |
| Li Y. et al. (2013) | Colorectal | Asian | Hospital | NO | PCR-RFLP | 451 | 631 | 59.4 | 57.0 | 0.583 | 0.577 | 22.92 | 23.58 | 0.419 | 0.475 |
BMI – body mass index; ASG – allele-specific genotyping; PCR – polymerase chain reaction; RCLP – restriction fragment length polymorphism; NA,– not available.
Table 2.
Baseline characteristics of the study populations in this meta-analysis.
| Author (year) | Drinking | Family cancer history | Cases | Controls | P for HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | 148Asp/Asp | 148Asp/Glu | 148Glu/Glu | 148Asp/asp | 148Arg/Glu | 148Glu/Glu | ||
| Moreno V. et al. (2006) | NA | NA | NA | NA | 95 | 177 | 87 | 99 | 147 | 66 | 0.406 |
| Jiao L. et al. (2006) | NA | NA | NA | NA | 108 | 180 | 79 | 85 | 174 | 71 | 0.305 |
| Berndt S. et al. (2007) | NA | NA | NA | NA | 186 | 387 | 166 | 222 | 357 | 178 | 0.140 |
| Tse D. et al. (2008) | 0.890 | 0.820 | NA | NA | 75 | 162 | 74 | 123 | 228 | 103 | 0.892 |
| Pardini B. et al. (2008) | NA | NA | NA | NA | 140 | 261 | 130 | 157 | 267 | 106 | 0.696 |
| Kasahara M. et al. (2008) | NA | NA | NA | NA | 23 | 45 | 0 | 70 | 51 | 0 | 0.003 |
| Huang W.Y. et al. (2008) | 0.152 | 0.206 | NA | NA | 76 | 118 | 42 | 221 | 358 | 155 | 0.653 |
| Palli D. et al. (2010) | NA | NA | 0.166 | 0.089 | 103 | 147 | 48 | 208 | 243 | 95 | 0.102 |
| Jelonek K. et al. (2010) | NA | NA | NA | NA | 49 | 59 | 5 | 38 | 87 | 28 | 0.079 |
| Brevik A. et al. (2010) | NA | NA | NA | NA | 102 | 137 | 65 | 108 | 167 | 84 | 0.215 |
| Canbay E. et al. (2010) | 0.675 | 0.146 | NA | NA | 14 | 18 | 8 | 151 | 63 | 33 | 0.000 |
| Gu D. et al. (2011) | 0.373 | 0.287 | NA | NA | 69 | 185 | 84 | 110 | 183 | 69 | 0.645 |
| Canbay E. et al. (2011) | 0.241 | 0.146 | NA | NA | 28 | 43 | 8 | 151 | 63 | 33 | 0.000 |
| Nakao M. et al. (2012) | 0.694 | 0.663 | 0.043 | 0.040 | 77 | 75 | 33 | 542 | 681 | 242 | 0.257 |
| Zeng X. et al. (2012) | 0.396 | 0.116 | 0.095 | 0.006 | 66 | 198 | 440 | 56 | 203 | 241 | 0.186 |
| Li Y. et al. (2013) | NA | NA | 0.183 | 0.154 | 123 | 247 | 81 | 186 | 335 | 110 | 0.052 |
HWE – Hardy-Weinberg equilibrium; NA – not available.
APE1 gene Asp148Glu variant and digestive cancer risk
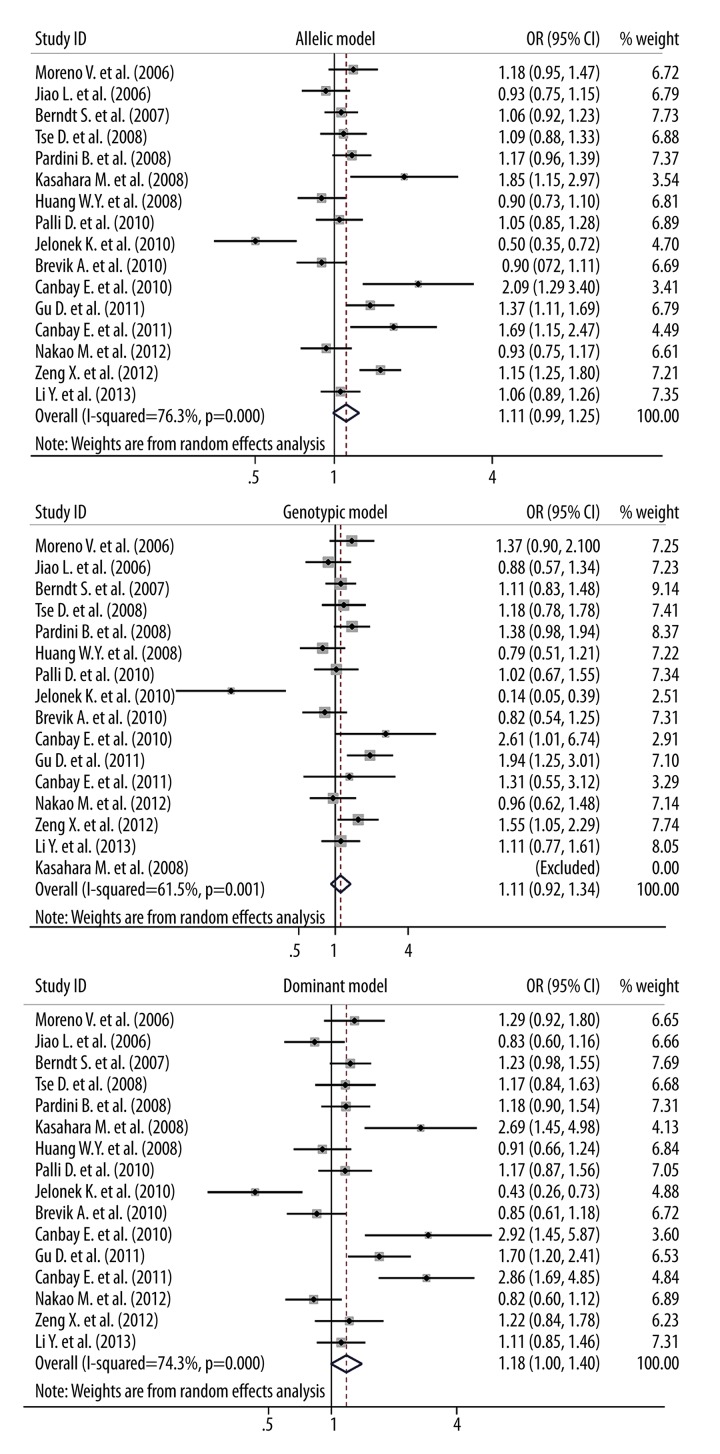
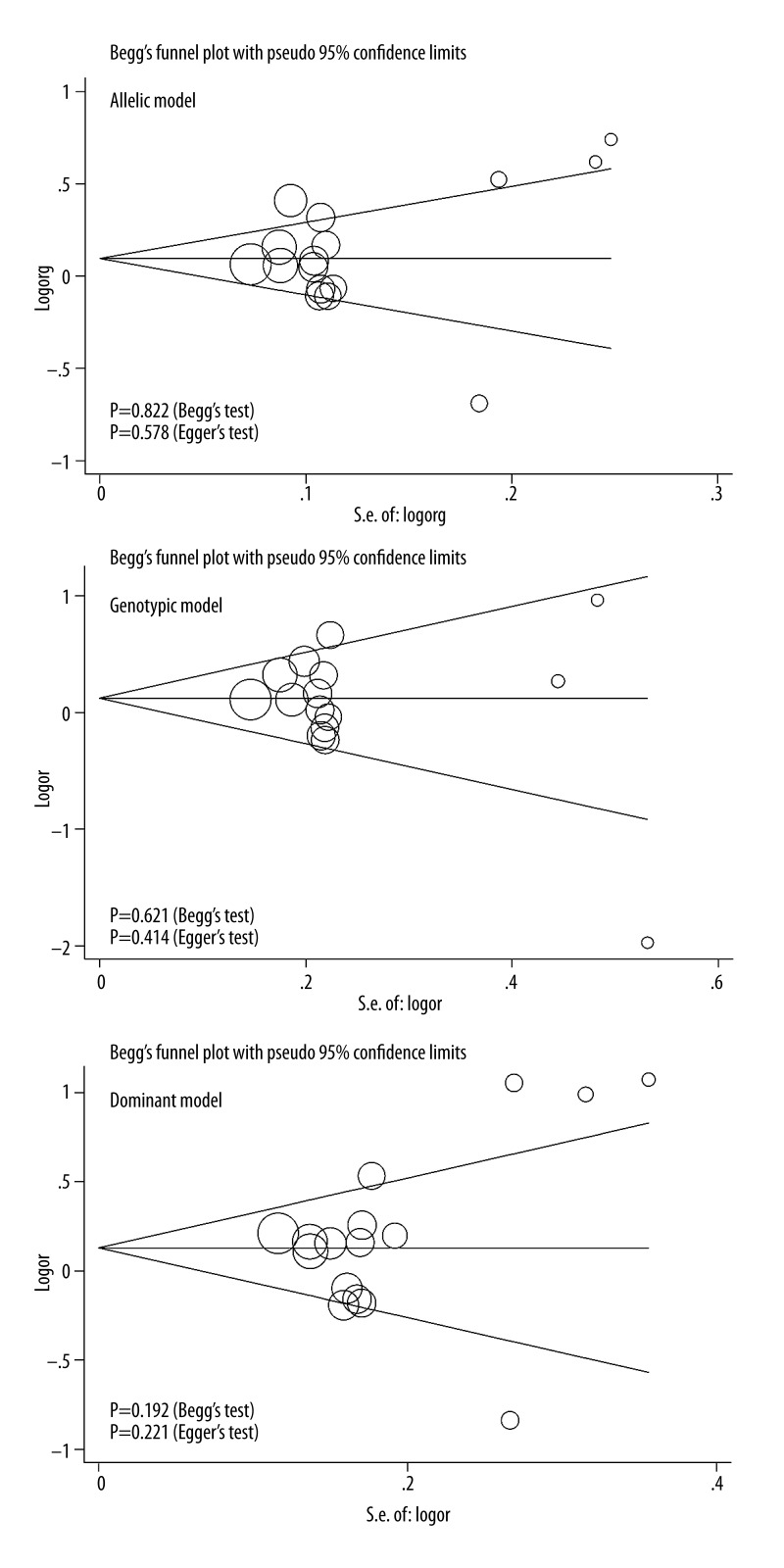
When all qualified studies were analyzed together, significance was indicative for the association between Asp148Glu variant and digestive cancer risk under allelic (OR=1.11; 95% CI: 0.99–1.25; P=0.074) and dominant (OR=1.18; 95% CI: 1.00–1.40; P=0.056) models (Figure 1). There was strong evidence of heterogeneity for all 3 genetic models (I2=76.3%, 61.5% and 74.3% for allelic, homozygous genotypic and dominant models, respectively), while low probabilities of publication bias were observed (Figure 2). In addition, as reflected by the trim and fill method, 1 study for allelic model and 2 studies for dominant model were required to make filled funnel plots symmetrical (Supplementary Figure 1). Adjusting for the missing studies still failed to attain statistical significance for both genetic models of inheritance (data not shown).
Figure 1.
Forest plots of APE1 gene Asp148Glu variant for digestive cancer risk under 3 genetic models.
Figure 2.
Begg’s funnel plots of APX1 gene Asp148Glu variant with digestive cancer risk under 3 genetic models.
After grouping studies by the degree of Hardy-Weinberg equilibrium test at a significance level of 5%, it was of interest to note that the corresponding effect estimates were exceedingly overestimated in studies with Asp148Glu genotypes deviating from Hardy-Weinberg equilibrium across 3 genetic models, especially under dominant model (OR=2.82; 95% CI: 1.99–3.99; P<0.001), without heterogeneity. In contrast, conformity to Hardy-Weinberg equilibrium greatly attenuated the risk estimates, yet with significant heterogeneity. In view of this divergence and to avoid biased estimates, the following subgroup analyses were restricted to the studies with Asp148Glu genotypes in Hardy-Weinberg equilibrium (Table 3).
Table 3.
Subgroup analyses of all qualified studies under 3 genetic models.
| Subgroups | No. of studies (cases/controls), n (n/n) | Allelic model | Genotypic model | Dominant model | |||
|---|---|---|---|---|---|---|---|
| OR; 95% CI; P | I2 (P) | OR; 95% CI; P | I2 (P) | OR; 95% CI; P | I2 (P) | ||
| HWE test | |||||||
| Yes | 13 (4729/7133) | 1.04; 0.94–1.16; 0.449 | 74.0% (<0.001) | 1.08; 0.89–1.30; 0.450 | 63.8% (0.001) | 1.05; 0.92–1.21; 0.472 | 60.8% (0.002) |
| No | 3 (187/615) | 1.84; 1.43–2.36; <0.001 | 0.0% (0.791) | 1.80; 0.91–3.55; 0.089 | 10.6% (0.290) | 2.82; 1.99–3.99; <0.001 | 0.0% (0.982) |
| Cancer site (HWE=YES) | |||||||
| Colorectal cancer | 6 (2497/2742) | 0.99; 0.84–1.16; 0.858 | 76.0% (0.001) | 0.98; 0.70–1.37; 0.909 | 74.8% (0.001) | 1.02; 0.81–1.28; 0.891 | 70.0% (0.005) |
| Pancreatic cancer | 2 (552/1795) | 0.93; 0.80–1.09; 0.359 | 0.0% (0.980) | 0.92; 0.68–1.24; 0.574 | 0.0% (0.768) | 0.83; 0.66–1.04; 0.103 | 0.0% (0.965) |
| Gastric cancer | 2 (636/908) | 1.20; 0.92–1.56; 0.182 | 69.3% (0.071) | 1.40; 0.75–2.63; 0.292 | 76.8% (0.038) | 1.39; 0.96–2.02; 0.080 | 62.5% (0.103) |
| Esophageal cancer | 1 (311/454) | 1.09; 0.89–1.33; 0.433 | NA | 1.18; 0.78–1.78; 0.438 | NA | 1.17; 0.84–1.63; 0.356 | NA |
| Gallbladder cancer | 1 (236/734) | 0.90; 0.73–1.11; 0.304 | NA | 0.79; 0.51–1.21; 0.276 | NA | 0.91; 0.66–1.24; 0.544 | NA |
| Hepatocellular cancer | 1 (497/500) | 1.50; 1.25–1.80; <0.001 | NA | 1.55; 1.05–2.29; 0.028 | NA | 1.22; 0.84–1.78; 0.302 | NA |
| Ethnicity (HWE=YES) | |||||||
| Caucasian | 6 (1916/2354) | 0.98; 0.82–1.18; 0.823 | 76.0% (0.001) | 0.96; 0.66–1.38; 0.809 | 75.0% (0.001) | 1.00; 0.79–1.28; 0.972 | 68.2% (0.008) |
| Asian | 5 (1707/3692) | 1.13; 0.92–1.38; 0.232 | 80.8% (<0.001) | 1.21; 0.89–1.64; 0.232 | 64.1% (0.025) | 1.11; 0.87–1.41; 0.410 | 64.1% (0.025) |
| Mixed | 2 (1106/1087) | 1.02; 0.90–1.15; 0.788 | 8.2% (0.297) | 1.03; 0.81–1.31; 0.789 | 0.0% (0.362) | 1.03; 0.70–1.52; 0.868 | 72.6% (0.056) |
| Study design (HWE=YES) | |||||||
| Hospital | 8 (2967/3272) | 1.09; 0.92–1.28; 0.331 | 80.8% (<0.001) | 1.15; 0.86–1.55; 0.342 | 72.9% (0.001) | 1.09; 0.88–1.35; 0.442 | 68.9% (0.002) |
| Population | 5 (1762/3861) | 0.99; 0.90–1.07; 0.724 | 0.0% (0.520) | 0.97; 0.81–1.15; 0.682 | 0.0% (0.654) | 1.00; 0.84–1.19; 0.975 | 42.5% (0.138) |
| Matched status (HWE=YES) | |||||||
| Yes | 10 (3615/5831) | 1.04; 0.91–1.20; 0.581 | 78.9% (<0.001) | 1.06; 0.84–1.36; 0.615 | 70.2% (<0.001) | 1.04; 0.87–1.24; 0.645 | 67.2% (0.001) |
| No | 1 (451/631) | 1.06; 0.89–1.26; 0.529 | NA | 1.11; 0.77–1.61; 0.565 | NA | 1.12; 0.85–1.46; 0.429 | NA |
| NA | 2 (663/671) | 1.03; 0.78–1.35; 0.836 | 68.6% (0.074) | 1.06; 0.64–1.76; 0.822 | 64.9% (0.091) | 1.05; 0.70–1.58; 0.823 | 67.0% (0.082) |
| Sample size (HWE=YES) | |||||||
| Total sample size ≥1000 | 4 (1906/3383) | 1.07; 0.98–1.16; 0.139 | 0.0% (0.476) | 1.15; 0.97–1.37; 0.119 | 0.0% (0.618) | 1.10; 0.93–1.29; 0.274 | 33.8% (0.209) |
| Total sample size <1000 | 9 (2823/3750) | 1.03; 0.87–1.22; 0.759 | 81.7% (<0.001) | 1.02; 0.76–1.37; 0.911 | 74.4% (<0.001) | 1.03; 0.84–1.27; 0.790 | 69.0% (0.001) |
| Genotyping (HWE=YES) | |||||||
| Non-RFLP | 9 (3296/5457) | 1.05; 0.94–1.18; 0.379 | 64.9% (0.004) | 1.06; 0.92–1.23; 0.417 | 17.4% (0.288) | 1.05; 0.92–1.18; 0.489 | 30.3% (0.176) |
| RFLP | 4 (1433/1676) | 1.24; 0.96–1.61; 0.100 | 83.9% (<0.001) | 0.97; 0.52–1.81; 0.925 | 86.3% (<0.001) | 1.04; 0.69–1.56; 0.869 | 83.8% (<0.001) |
HWE – Hardy-Weinberg equilibrium; OR – odds ratio; 95% CI – 95% confidence interval; NA – not available, RFLP – restriction fragment length polymorphism.
By digestive cancer sites, significance was only observed for hepatocellular cancer under allelic (OR=1.50; 95% CI: 1.25–1.80; P<0.001) and homozygous genotypic (OR=1.55; 95% CI: 1.02–2.29; P=0.028) models, although this finding was based on 1 eligible study. Moreover, considering the magnitude of risk estimates, albeit nonsignificant, for different sites of digestive cancer, it is suggestive of heterogeneous carcinogenic mechanisms.
Further stratifying studies according to ethnicity, study design, matched status, sample size (at a cutoff of 1000) and genotyping platform failed to identify any significance between Asp148Glu variant and digestive cancer risk. Given the limited sample sizes in some strata, it is, however, premature to negate the potential confounding effects of these characteristics in interpreting significant heterogeneity. For example, genetic susceptibility of Asp148Glu variant to digestive cancer was ethnicity-specific, as 148Glu/Glu genotype carriers were 1.21 times (OR=1.21; 95% CI: 0.89–1.64; P=0.232) more likely to develop digestive cancer when compared to those with 148Asp/Asp genotype in Asian populations, yet this genotype seemed to be a protective or neutral factor in Caucasians (OR=0.96; 95% CI: 0.66–1.38; P=0.809).
Meta-regression analysis
To further seek other sources of heterogeneity resulting from continuous covariates, a meta-regression model was constructed by incorporating age (P=0.338), gender (P=0.485), BMI (P=0.279), smoking (P=0.431), drinking (P=0.450) and family history of cancer (P=0.721), and still all regression coefficients did not differ significantly from zero.
Discussion
In this study, we aimed to summarize available data on the association between APE1 gene Asp148Glu variant and digestive cancer risk through a comprehensive meta-analysis involving 16 articles and 12664 subjects. Our findings suggested that APE1 gene Asp148Glu variant might not be a risk-conferring factor for digestive cancer. Moreover, conformity to Hardy-Weinberg equilibrium was identified as a potential source of significant overall heterogeneity.
Several possible limitations must be recognized prior to interpreting our findings. First, this meta-analysis is based on the summaries of retrospective case-control studies, which rarely establish causal relationship, and it is encouraging to incorporate the concept of Mendelian randomization into observational association studies [31]. Second, only 1 variant Asp148Glu in APE1 gene was covered in this study, which might not be sufficient to address the complex genetic architecture of digestive cancer. Third, only published articles written in English language were retrieved for inclusion and some unpublished small and/or negative articles might be missing, leading to the potential existence of publication bias. Fourth, it is essential to examine gene-environment and gene-gene interactions at the level of both individual studies and meta-analysis. To achieve this goal, one usually needs to perform a meta-analysis of individual participant data, which is not always practical for the majority of published meta-analyses. Five, although both subgroup and meta-regression analyses were undertaken to explore the potential sources of heterogeneity, it is still obsessing a majority of comparisons in this meta-analysis. Nevertheless, considering that residual confounding by incompletely considered physiologic covariates might exist in our findings, it seems unlikely that the effect estimates could be explained by confounding.
Despite these limitations, our stratified findings suggest that APE1 gene Asp148Glu variant might be a susceptible locus for the development of hepatocellular cancer, suggesting that digestive cancer is characterized by marked genetic heterogeneity. This genetic heterogeneity is not surprising in light of the heterogeneous pathogenesis for different sites of cancer [32], necessitating the construction of a database of candidate genes and variants responsible for different sites of cancer. As stated by Burrell et al., there is extensive genetic diversity both between and within cancer, which poses a significant challenge to personalized cancer medicine [33]. Moreover, the effect of Asp148Glu variant on cancer susceptibility has strong biological plausibility since this variant resides in the carboxy-terminus region of APE1 gene, the region containing the endonuclease activity required for DNA repair [34]. Functional investigations showed that individuals carrying APE1 gene 148Glu allele had higher levels of APE1 mRNA expression when compared with those with the 148Asp/Asp genotype [35]. At present, the mechanism linking APE1 gene Asp148Glu variant and hepatocellular cancer is not clear, and thus if involved, this variant might, by affecting DNA repair activity or gene function via altering the stability of mRNA, be implicated in the pathogenesis of hepatocellular cancer. In addition, we cannot rule out the possible involvement of APE1 gene Asp148Glu variant or others in strong linkage disequilibrium in other sites of cancer, considering the sample size involved in this meta-analysis. Nevertheless, considering the limited studies with inadequate sample sizes for most subgroups, our stratified findings should be considered preliminary and be viewed as hypothesis-generating for future large and well-designed studies.
Deviation from Hardy-Weinberg equilibrium was identified as a potential source of heterogeneity in our subgroup analyses. In reality, conformity to Hardy-Weinberg equilibrium weakened the association between Asp148Glu variant and digestive cancer risk. In the evaluation of case-control studies, assessment of Hardy-Weinberg equilibrium for a given genetic locus among controls is considered an important criterion [36]. Generally, deviation from Hardy-Weinberg equilibrium should imply some potential biases in the selection of controls or genotyping misclassifications, which tend to inflate the change of a false-positive association [37]. In view of this fact, all following subgroup and meta-regression analyses that sought to explore the potentially sources of heterogeneity were undertaken in studies with Asp148Glu genotypes in Hardy-Weinberg equilibrium. Unfortunately, none of the other confounding factors can explain significant heterogeneity of Asp148Glu in susceptibility to digestive cancer. Meta-regression per se is analogous to simple regression where an outcome variable is predicted according to the values of 1 or more explanatory variables. However, it is of importance to acknowledge that meta-regression, albeit enabling coverage of various continuous covariates, does not have the methodological rigor of a properly designed study that is intended to test the effect of these covariates formally [38]. We therefore must regard our findings as preliminary, which should be viewed as hypothesis-generating and call for validation in future large and well-design studies.
Conclusions
The results of this meta-analysis collectively suggest that APE1 gene Asp148Glu variant is not a risk-conferring factor for digestive cancer. For practical reasons, we hope that this study will not remain just another endpoint of research, but instead serve as a beginning to establish background data to unravel the contributory role of APE1 gene and its genetic alterations in the development of digestive cancer and other solid tumors.
Supplementary Data
Filled funnel plots of APX1 gene Asp148Glu variant with digestive cancer risk under 3 genetic models.
Footnotes
Source of support: This work was financially supported by Taishan Scholars Construction Engineering; National Natural Science Foundation of China (81400771 and 81171303), Shandong Provincial Natural Science Foundation (ZR2014HL028 and ZR2010HM091), A Project of Shandong Province Higher Educational Science and Technology Program (J14LE01) and Binzhou Medical University Scientific Research Funds (BY2013KYQD17 and BY2013KYQD18)
Conflicts of interest statement
None of the authors have any conflict of interest to disclose.
References
- 1.Buschman MD, Rahajeng J, Field SJ. GOLPH3 links the golgi, DNA damage, and cancer. Cancer Res. 2015;75:624–27. doi: 10.1158/0008-5472.CAN-14-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roszkowski K, Jozwicki W, Blaszczyk P, et al. Oxidative damage DNA: 8-oxoGua and 8-oxodG as molecular markers of cancer. Med Sci Monit. 2011;17(6):CR329–33. doi: 10.12659/MSM.881805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loilome W, Kadsanit S, Namwat N, et al. Impaired antioxidant enzyme activity and increased DNA repair enzyme expression in hamster liver tissues related to cholangiocarcinoma development. Asian Pac J Cancer Prev. 2012;13(Suppl):59–64. [PubMed] [Google Scholar]
- 4.Kim MH, Kim HB, Yoon SP, et al. Colon cancer progression is driven by APEX1-mediated upregulation of Jagged. J Clin Invest. 2013 doi: 10.1172/JCI65521. pii: 65521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim MH, Kim HB, Acharya S, et al. Ape1/Ref-1 induces glial cell-derived neurotropic factor (GDNF) responsiveness by upregulating GDNF receptor alpha1 expression. Mol Cell Biol. 2009;29:2264–77. doi: 10.1128/MCB.01484-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fishel ML, Jiang Y, Rajeshkumar NV, et al. Impact of APE1/Ref-1 redox inhibition on pancreatic tumor growth. Mol Cancer Ther. 2011;10:1698–708. doi: 10.1158/1535-7163.MCT-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fung H, Demple B. A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell. 2005;17:463–70. doi: 10.1016/j.molcel.2004.12.029. [DOI] [PubMed] [Google Scholar]
- 8.Raffoul JJ, Heydari AR, Hillman GG. DNA repair and cancer therapy: targeting APE1/Ref-1 using dietary agents. J Oncol. 2012;2012:370481. doi: 10.1155/2012/370481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin CH, Chen PM, Cheng YW, et al. The APE1 Asp/Asp genotype and the combination of APE1 Asp/Asp and hOGG1-Cys variants are associated with increased p53 mutation in non-small cell lung cancer. J Epidemiol. 2012;22:537–42. doi: 10.2188/jea.JE20120048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canbay E, Cakmakoglu B, Zeybek U, et al. Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Curr Med Res Opin. 2011;27:1295–302. doi: 10.1185/03007995.2011.573544. [DOI] [PubMed] [Google Scholar]
- 11.Wang M, Qin C, Zhu J, et al. Genetic variants of XRCC1, APE1, and ADPRT genes and risk of bladder cancer. DNA Cell Biol. 2010;29:303–11. doi: 10.1089/dna.2009.0969. [DOI] [PubMed] [Google Scholar]
- 12.Gu M, Dong X, Zhang X, et al. Strong association between two polymorphisms on 15q25.1 and lung cancer risk: a meta-analysis. PLoS One. 2012;7:e37970. doi: 10.1371/journal.pone.0037970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yri OE, Ekstrom PO, Hilden V, et al. Polymorphisms in genes encoding interleukin-10 and drug metabolizing enzymes GSTP1, GSTT1, GSTA1 and UGT1A1 influence risk and outcome in Hodgkin lymphoma. Leuk Lymphoma. 2012;53:1934–44. doi: 10.3109/10428194.2012.682307. [DOI] [PubMed] [Google Scholar]
- 14.Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowden J, Tierney JF, Copas AJ, et al. Quantifying, displaying and accounting for heterogeneity in the meta-analysis of RCTs using standard and generalised Q statistics. BMC Med Res Methodol. 2011;11:41. doi: 10.1186/1471-2288-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berndt SI, Huang WY, Fallin MD, et al. Genetic variation in base excision repair genes and the prevalence of advanced colorectal adenoma. Cancer Res. 2007;67:1395–404. doi: 10.1158/0008-5472.CAN-06-1390. [DOI] [PubMed] [Google Scholar]
- 17.Brevik A, Joshi AD, Corral R, et al. Polymorphisms in base excision repair genes as colorectal cancer risk factors and modifiers of the effect of diets high in red meat. Cancer Epidemiol Biomarkers Prev. 2010;19:3167–73. doi: 10.1158/1055-9965.EPI-10-0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canbay E, Agachan B, Gulluoglu M, et al. Possible associations of APE1 polymorphism with susceptibility and HOGG1 polymorphism with prognosis in gastric cancer. Anticancer Res. 2010;30:1359–64. [PubMed] [Google Scholar]
- 19.Gu D, Wang M, Wang S, et al. The DNA repair gene APE1 T1349G polymorphism and risk of gastric cancer in a Chinese population. PLoS One. 2011;6:e28971. doi: 10.1371/journal.pone.0028971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang WY, Gao YT, Rashid A, et al. Selected base excision repair gene polymorphisms and susceptibility to biliary tract cancer and biliary stones: a population-based case-control study in China. Carcinogenesis. 2008;29:100–5. doi: 10.1093/carcin/bgm247. [DOI] [PubMed] [Google Scholar]
- 21.Jelonek K, Gdowicz-Klosok A, Pietrowska M, et al. Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J Appl Genet. 2010;51:343–52. doi: 10.1007/BF03208865. [DOI] [PubMed] [Google Scholar]
- 22.Jiao L, Bondy ML, Hassan MM, et al. Selected polymorphisms of DNA repair genes and risk of pancreatic cancer. Cancer Detect Prev. 2006;30:284–91. doi: 10.1016/j.cdp.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasahara M, Osawa K, Yoshida K, et al. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J Exp Clin Cancer Res. 2008;27:49. doi: 10.1186/1756-9966-27-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Li S, Wu Z, et al. Polymorphisms in genes of APE1, PARP1, and XRCC1: risk and prognosis of colorectal cancer in a northeast Chinese population. Med Oncol. 2013;30:505. doi: 10.1007/s12032-013-0505-z. [DOI] [PubMed] [Google Scholar]
- 25.Moreno V, Gemignani F, Landi S, et al. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12:2101–8. doi: 10.1158/1078-0432.CCR-05-1363. [DOI] [PubMed] [Google Scholar]
- 26.Nakao M, Hosono S, Ito H, et al. Selected polymorphisms of base excision repair genes and pancreatic cancer risk in Japanese. J Epidemiol. 2012;22:477–83. doi: 10.2188/jea.JE20120010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palli D, Polidoro S, D’Errico M, et al. Polymorphic DNA repair and metabolic genes: a multigenic study on gastric cancer. Mutagenesis. 2010;25:569–75. doi: 10.1093/mutage/geq042. [DOI] [PubMed] [Google Scholar]
- 28.Pardini B, Naccarati A, Novotny J, et al. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res. 2008;638:146–53. doi: 10.1016/j.mrfmmm.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Tse D, Zhai R, Zhou W, et al. Polymorphisms of the NER pathway genes, ERCC1 and XPD are associated with esophageal adenocarcinoma risk. Cancer Causes Control. 2008;19:1077–83. doi: 10.1007/s10552-008-9171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng X, Liu S, Yu H, et al. DNA repair capacity, DNA-strand break repair gene polymorphisms, and the incidence of hepatocellular carcinoma in southwestern Guangxi of China. DNA Cell Biol. 2012;31:1384–91. doi: 10.1089/dna.2012.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sleiman PM, Grant SF. Mendelian randomization in the era of genomewide association studies. Clin Chem. 2010;56:723–28. doi: 10.1373/clinchem.2009.141564. [DOI] [PubMed] [Google Scholar]
- 32.Diaz-Cano SJ. Tumor heterogeneity: mechanisms and bases for a reliable application of molecular marker design. Int J Mol Sci. 2012;13:1951–2011. doi: 10.3390/ijms13021951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burrell RA, McGranahan N, Bartek J, et al. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–45. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 34.Hsieh MM, Hegde V, Kelley MR, et al. Activation of APE/Ref-1 redox activity is mediated by reactive oxygen species and PKC phosphorylation. Nucleic Acids Res. 2001;29:3116–22. doi: 10.1093/nar/29.14.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu H, Zhao H, Wang LE, et al. Correlation between base-excision repair gene polymorphisms and levels of in-vitro BPDE-induced DNA adducts in cultured peripheral blood lymphocytes. PLoS One. 2012;7:e40131. doi: 10.1371/journal.pone.0040131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Little J, Bradley L, Bray MS, et al. Reporting, appraising, and integrating data on genotype prevalence and gene-disease associations. Am J Epidemiol. 2002;156:300–10. doi: 10.1093/oxfordjournals.aje.a000179. [DOI] [PubMed] [Google Scholar]
- 37.Dennis J, Hawken S, Krewski D, et al. Bias in the case-only design applied to studies of gene-environment and gene-gene interaction: a systematic review and meta-analysis. Int J Epidemiol. 2011;40:1329–41. doi: 10.1093/ije/dyr088. [DOI] [PubMed] [Google Scholar]
- 38.Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439–44. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Filled funnel plots of APX1 gene Asp148Glu variant with digestive cancer risk under 3 genetic models.