Abstract
LCAT is intimately involved in HDL maturation and is a key component of the reverse cholesterol transport (RCT) pathway which removes excess cholesterol molecules from the peripheral tissues to the liver for excretion. Patients with loss-of-function LCAT mutations exhibit low levels of HDL cholesterol and corneal opacity. Here we report the 2.65 Å crystal structure of the human LCAT protein. Crystallization required enzymatic removal of N-linked glycans and complex formation with a Fab fragment from a tool antibody. The crystal structure reveals that LCAT has an α/β hydrolase core with two additional subdomains that play important roles in LCAT function. Subdomain 1 contains the region of LCAT shown to be required for interfacial activation, while subdomain 2 contains the lid and amino acids that shape the substrate binding pocket. Mapping the naturally occurring mutations onto the structure provides insight into how they may affect LCAT enzymatic activity.
Keywords: lecithin:cholesterol acyltransferase, X-ray crystallography, high density lipoprotein, cholesterol, antibodies
LCAT (EC 2.3.1.43) is a plasma enzyme that catalyzes the conversion of cholesterol into long-chain cholesteryl esters (CEs). This reaction occurs on the surface of the HDL particle where, after activation by ApoA-I (the structural protein of HDL particles), LCAT hydrolyzes phosphatidylcholine (lecithin) at the sn-2 position and subsequently transfers the fatty acyl to cholesterol (1). The increased hydrophobicity of the resulting CE forces it into the interior core of the HDL particle and in the process transforms HDL from nascent discoidal particles into larger CE-enriched spherical particles. By reducing cholesterol on the surface of the HDL particle, LCAT maintains a chemical gradient that facilitates a unidirectional net flow of cholesterol from the cell surface of peripheral tissues to HDL particles in the blood (2). Mature HDL particles ultimately transport cholesterol (in the form of CE) back to the liver in pathways that involve cholesteryl ester transfer protein, LDL/VLDL, LDL receptor, and scavenger receptor class B type I. Hence, LCAT has been hypothesized to play a central role in driving HDL remodeling and reverse cholesterol transport (RCT) from the peripheral tissues to the liver.
Loss-of-function mutations in the LCAT protein, along with their corresponding phenotypes, have been described (3). Familial LCAT deficiency (FLD) is caused by a complete loss of LCAT activity, either through an absence of the LCAT protein itself or the presence of a mutant LCAT protein lacking any corresponding LCAT activity. In patients, FLD is characterized by HDL deficiency along with corneal opacity, anemia, and loss of renal function. Fish-eye disease (FED) is a partial LCAT deficiency and is characterized by the presence of LCAT protein with decreased LCAT activity. Patients with FED manifest with low HDL and corneal opacity later in life. Interestingly, a definitive correlation between loss-of-function LCAT mutations and coronary heart disease remains elusive (4).
Glomset (5, 6) first described LCAT enzymatic activity in 1962 and subsequently purified the protein from plasma. LCAT purified from plasma has a mass of ∼65 kDa as assessed by SDS-PAGE. Later analysis showed that LCAT is a 416-amino-acid protein with a calculated molecular mass of ∼47 kDa, and that the extra mass of plasma LCAT is a result of N-linked carbohydrates at Asn20, Asn84, Asn272, and Asn384. Glycosylation of LCAT has been shown to be important for its activity and secretion (7, 8). LCAT is predicted to have an α/β hydrolase fold with Ser181, Asp345, and His377 required for catalytic activity (9–11); however, the low sequence homology between LCAT and other proteins with available structures has made accurate modeling of the LCAT structure challenging. Here, we describe the crystallization of human LCAT and report the high-resolution crystal structure of this protein.
MATERIALS AND METHODS
LCAT expression and purification
Human LCAT 1-416 (Cys31Tyr) with a tobacco etch virus (TEV) cleavable C-terminal His tag was expressed using the BacMam expression system in HEK 2936e (12) cells. The Cys31Tyr mutation was found to increase the melting temperature of LCAT by 6.5°C using differential scanning calorimetry, and this protein expresses at higher levels than Cys31 wild-type protein. Kifunensine (5 mM) was added to the media in order to restrict the formation of complex glycans. The protein was captured on a HisTrap FF column and eluted with an imidazole gradient. LCAT-containing fractions were pooled, loaded on to a Source Q column, and eluted with 25 mM Tris pH 7.5, 500 mM NaCl. The protein was then purified by size exclusion chromatography using a Superdex 200 column in 25 mM Tris pH 7.5, 150 mM NaCl. Deglycosylation with endoglycosidase H (Endo H) was performed at 37°C for 4 h, followed by cleavage with TEV protease overnight on ice. Further purification was performed with Superdex 200 and MonoQ columns using the same buffer system. LCAT retains in vitro enzymatic activity throughout this purification procedure.
Antibody generation, complex formation, and crystallization
Mice were immunized with human LCAT, and antibodies against LCAT were isolated. Antibodies were selected for their ability to bind to conformational epitopes of LCAT, based on comparative binding to native and heat-denatured LCAT. Fab fragments of three antibodies were expressed in Escherichia coli and purified for complex formation with LCAT. The LCAT/Fab complexes were made by mixing a 1.5 M excess of Fab with purified LCAT. Size exclusion chromatography using a Superdex 200 column was used to purify the LCAT/Fab complex from excess Fab. The purified complexes were washed into 10 mM Tris pH 7.5, 25 mM NaCl and concentrated to 10–20 mg/ml for crystallization, and crystals were obtained from only one of the three complexes. Diffraction quality crystals were grown with the LCAT/Fab1 complex in 0.1 M sodium acetate pH 4.5, 0.2 M zinc acetate, 0.01 M cobalt chloride, 4–8% polyethylene glycol (PEG) 1500. Many data sets were collected, with the best crystals diffracting to ∼3.4 Å using synchrotron radiation. Various cryoprotectants and dehydration techniques were used in an attempt to increase the resolution without success. Although heavy atom screening did not produce a usable heavy atom derivative, it was discovered during this process that raising the pH by transferring crystals to a solution containing 0.1 M Tris pH 7, 0.2 M zinc acetate, 0.01 M cobalt chloride, 8% PEG 1500 immediately prior to cryoprotection in the same solution supplemented with 27% glycerol frequently increased the diffraction limit of the crystals.
Data collection and structure determination
Using this cryoprotection technique, a 2.65 Å data set was collected (Shamrock Structures LLC) at the Canadian Light Source (CMCF1, 08ID) and processed with iMOSFLM (13) and Scala (14) from the CCP4 Program Suite (15). The LCAT/Fab1 crystals grow in the R3:H space group with a = b = 168.59 and c = 93.57 Å with ∼50% solvent and one complex in the asymmetric unit. The crystal structure of Fab1 was determined previously at 2.3 Å resolution (data not shown) and used as a starting model for molecular replacement. Using Phaser (16), the variable and constant domains of Fab1 were placed in the asymmetric unit. Subsequent 2Fo-Fc electron density for LCAT was quite poor, but the α/β hydrolase core was discernable. This portion of LCAT was added to the model, but the 2Fo-Fc electron density remained too poor to allow further model building. Phases calculated after solvent flattening with DM (17) improved the electron density for the LCAT region of the asymmetric unit enough to continue model building. The initial model was built with multiple rounds of model building in Quanta (Accelrys), solvent flattening in DM and refinement with CNX (18), followed by removal of the solvent flattening step at the later stages of model building. Final refinement and model building was performed using PHENIX (19) and Coot (20), respectively. Validation with MolProbity (21) shows an overall score of 2.25 with 92% of the amino acids in Ramachandran favored regions. The final model has an R of 17.6% and an Rfree of 25.1% and includes 365 amino acid residues from LCAT, 432 amino acid residues from Fab1, 104 water molecules, and 16 zinc ions. Structure figures were made using PyMOL (22).
LCAT enzymatic assays
LCAT activity (in the form of LCAT alone or LCAT covalently linked to an antibody Fc domain) was determined by quantification of the conversion of cholesterol to CE on human ApoA-I lipoprotein substrate. The reaction substrate consisted of human ApoA-I proteoliposomes containing 67 μM l-α-Phosphatidylcholine Type XVI-E (Sigma P3556), 8 μM cholesterol (Sigma C8667), 4 μM [4-14C]cholesterol (Perkin Elmer NEC018250UC), and 12 μg/ml Apolipoprotein AI (Meridian Life Sciences). This assay also contained 1% human serum albumin (fatty acid free), 7 mM Tris pH 7.4, 4 mM EDTA, 100 mM NaCl, and 2 mM β-mercaptoethanol. Reactions were run in duplicate and incubated in a 37°C water bath for 1 h. Following the reaction, lipids were extracted with a 10-fold volume addition of 100% ethanol. Lipid-containing supernatants were obtained by centrifugation and then dried under a stream of nitrogen gas, resuspended in chloroform, and spotted on TLC plates (Silica gel 60A, Whatman 4865-821). Cholesterol and CE were separated by running plates in a TLC chamber with petroleum ether-ether-acetic acid (100:20:0.5 by volume). Cholesterol and CE bands were detected by exposing to image plates and subsequent reading on a Fujifilm FLA-5100 image reader. Ratios of cholesterol to CE were determined using ImageQuant software (Fujifilm).
To determine whether Fab1 would modulate the activity of the phospholipase step of the LCAT reaction on a soluble substrate, we used a commercially available LCAT activity assay kit from Roar Biomedical. The kit provides a soluble LCAT substrate that upon hydrolysis by LCAT fluoresces at 390 nm. Recombinant LCAT (Cys31Tyr) was incubated with Fab1 (1:1 molar ratio) and the soluble substrate at 37°C for 4 h. The hydrolysis reaction was assessed by measuring the emission at 390 nm. We performed the experiment at three concentrations of Fab1 and LCAT (0.1 nM, 1 nM, and 10 nM) and compared with parallel experiments with LCAT only (no Fab).
RESULTS
Protein preparation for crystallization
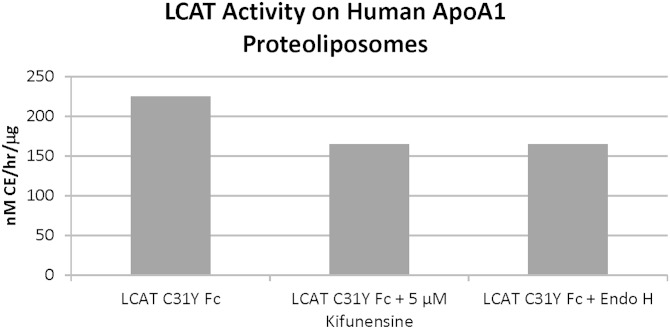
In order to reduce the heterogeneity and conformational flexibility inherent in highly glycosylated proteins, we pursued deglycosylation of LCAT for crystallization. Previous work has described the difficulty in expressing and purifying completely deglycosylated LCAT by incorporating mutations into the protein (23), so we decided to deglycosylate LCAT postexpression by treatment with glycosidases. Treatment with peptide-N-glycosidase F (PNGase F) reduces the size of LCAT from ∼65 kDa to 47 kDa as assessed by SDS-PAGE. Unfortunately, the resulting protein appeared aggregated by size exclusion chromatography and did not retain LCAT enzymatic activity (data not shown). The aggregation of proteins after PNGase F treatment has been previously described during the preparation of proteins for crystallography (24). We then followed a protocol similar to that described by Chang et al. (24) and expressed LCAT in the presence of kifunensine, followed by deglycosylation with Endo H. Endo H treatment of LCAT expressed in this manner results in an ∼47 kDa protein. The Endo H-treated protein is monomeric by size exclusion chromatography and retains similar in vitro activity to the fully glycosylated LCAT (Fig. 1).
Fig. 1.
LCAT retains activity following deglycosylation with Endo H. Activity assay showing that LCAT-Fc (Cys31Tyr) expressed in the presence of kifunensine and subsequently deglycosylated with Endo H retains activity comparable to untreated LCAT. Similar results were observed with wild type and LCAT (Cys31Tyr) alone (no Fc).
Although Endo H-deglycosylated LCAT was well behaved and could be concentrated to 15 mg/ml without forming irreversible aggregates as determined by dynamic light scattering, we had no success in crystallizing this protein. In order to facilitate crystallization, we isolated three monoclonal antibodies (mAbs) that bind to conformational epitopes of LCAT. The Fab fragments from these antibodies were prepared for use in crystallization experiments. Furthermore, we utilized a mutated version of LCAT (LCAT Cys31Tyr) that has higher thermostability than the wild-type protein. Crystals were only obtained with complexes from one of the Fab fragments (Fab1), and the LCAT/Fab1 complex crystals grew in a number of conditions with two observed crystal forms. Crystals from a primitive hexagonal crystal form diffracted poorly, and final space group determination was difficult. Crystals grown in a trigonal crystal form diffracted to 2.65 Å and were used to successfully solve the structure.
LCAT structure
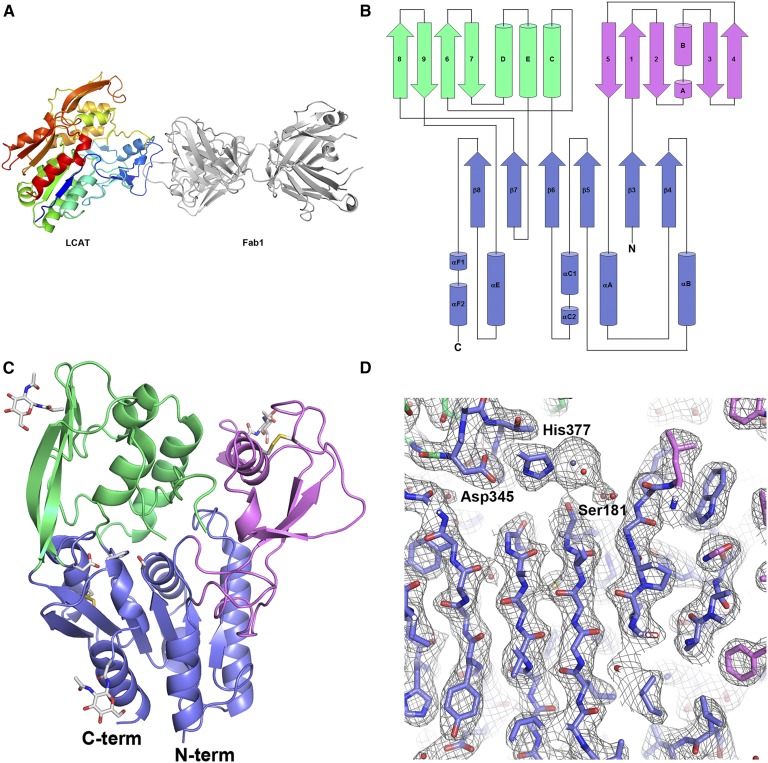
We solved the structure of LCAT in complex with the tool molecule Fab1 by molecular replacement, using a previously solved in house structure of Fab1 as the search molecule (Table 1). In the final structure, electron density is present for amino acids 21–230 and 238–396 of LCAT, along with the majority of amino acids from Fab1 (Fig. 2A, D). Three of the four asparagine amino acids with N-linked carbohydrates (Asn84, Asn272, and Asn384) are ordered within the structure, and electron density for the N-acetylglucosamine sugars left after Endo H cleavage is visible at each of these positions. Two disulfide bonds are present as predicted, Cys50-Cys74 and Cys313-Cys356 (9). Electron density for numerous poorly coordinated metal ions is present throughout the structure. These have all been modeled as zinc ions due to the relative concentrations of zinc and cobalt in the crystallization solution.
TABLE 1.
Data collection and refinement statistics
| Crystal Name | LCAT (Cys31Tyr) | LCAT (WT) |
| Data collection | ||
| Wavelength (Å) | 0.9795 | 1.2499 |
| Space group | R3:H | P32 |
| Cell dimensions | ||
| a, b, c (Å) | 168.59, 168.59, 93.57 | 166.42, 166.42, 97.66 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 40–2.65 (2.79–2.65)a | 83–3.60 (3.79–3.60) |
| Completeness | 100 (100) | 99.4 (99.0) |
| Redundancy | 4.1 (4.1) | 5.4 (4.9) |
| Rsym | 8.9 (64.4) | 16.4 (90.6) |
| I/σI | 10.3 (2.2) | 7.0 (2.1) |
| Refinement | ||
| Resolution (Å) | 34–2.65 | 72–3.60 |
| Molecules/ASU | 1 | 3 |
| Reflections | ||
| Total | 28,792 | 34,806 |
| Working set | 27,378 | 33,056 |
| Test set | 1,414 | 1,750 |
| Rwork/Rfree | 0.176/0.251 | 0.196/0.280 |
| Number of atoms | ||
| Protein | 6,236 | 17,112 |
| Carbohydrate | 42 | 126 |
| Ion | 16 | 0 |
| Water | 104 | 0 |
| RMSDs | ||
| Bond lengths (Å) | 0.14 | 0.01 |
| Bond angles (°) | 1.54 | 0.99 |
ASU, asymmetrical unit; RMSD, root-mean-square deviation.
Numbers in parentheses are highest resolution shell.
Fig. 2.
Crystal structure of LCAT. A: The overall structure of the LCAT/Fab1 complex. LCAT is shown in a rainbow multicolor depiction from the N to C terminus (blue to red). Fab1 (gray) binds to subdomain 1 at the N-terminal region of LCAT. B: Diagram of LCAT showing the secondary structure and chain trace of the protein. Secondary structure elements within the α/β hydrolase domain are numbered in canonical form. Secondary structure elements outside of the α/β hydrolase domain are labeled sequentially. The α/β hydrolase domain is colored blue, subdomain 1 is colored violet, and subdomain 2 is colored green. C: Structure of LCAT only with Fab1 removed for clarity. Regions of LCAT are colored as in B. The catalytic triad residues, N-linked carbohydrate sites, and disulfide bonds of LCAT are highlighted in stick representation. D: 2Fo-Fc electron density contoured at 1 σ focusing on the catalytic triad and β sheet of the α/β hydrolase core. Amino acids of the catalytic triad are labeled. Strong electron density for the Zn ion is present next to His377.
As expected, LCAT adopts an α/β hydrolase fold. This family of proteins is characterized by a predominately parallel β-sheet sandwiched on both sides by α-helices. The all parallel β-sheet in LCAT contains only six β-strands rather than the eight β-strands described in the canonical α/β hydrolase structure (25). The first β-strand of the LCAT α/β hydrolase core corresponds with canonical strand 3, and canonical numbering is used in the description of the LCAT structure (Fig. 2B). The active site containing the catalytic triad is found within this core region of the protein. Additional common characteristics of lipases with the α/β hydrolase fold are insertions or extra domains that help shape the substrate binding site and/or a flexible lid that can adopt multiple conformations to block or allow access to the substrate binding site (26). In LCAT, insertions between β3/αA (residues 32–119), β6/β7 and β7/αE (residues 214–303 and residues 319–344) form two subdomains at the “top” of the hydrolase core α/β sandwich (Fig. 2B, C). Using the PDBeFold service (27), no structurally related proteins were found in the Protein Data Bank for either of these subdomains.
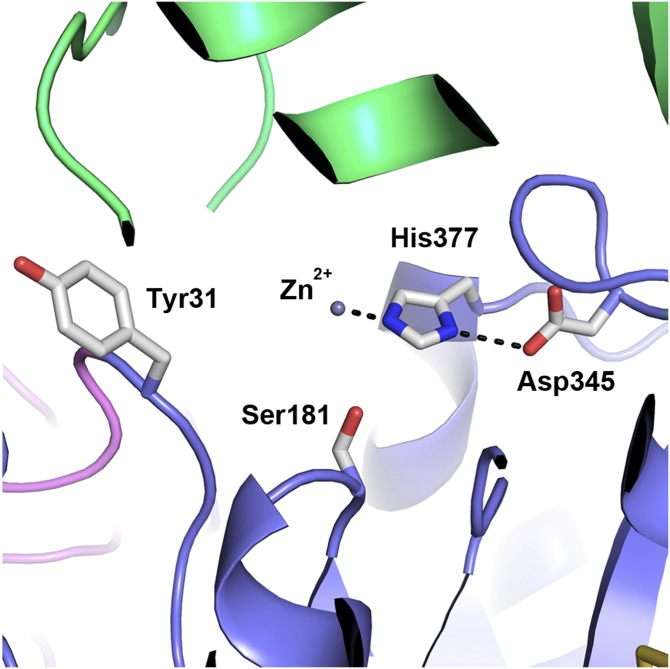
Sequence and mutational analyses have previously identified the catalytic triad of LCAT as Ser181, Asp345, and His377. Indeed, these amino acids are positioned in the structurally conserved location of catalytic residues in α/β hydrolases (Fig. 3) (25). Ser181 is located at the “nucleophilic elbow” between β5 and αC. The catalytic aspartic acid is frequently found in the loop that links β7 and αE, whereas in LCAT Asp345 is located between the 319–344 insertion and αE. His377 is located in the loop between β8 and αF. In this structure His377 does not form a hydrogen bond with Ser181, possibly pulled out of position due to the presence of the metal ion bound in the catalytic site. The oxyanion hole, required to stabilize the tetrahedral intermediate during the catalytic reaction, was initially described as Phe103 and Leu182 in LCAT (11), although more recently Holleboom et al. (28) named Cys31 and Leu182 as the possible oxyanion hole amino acid residues. Indeed, the oxyanion hole of LCAT is formed by backbone amide nitrogen atoms from Cys31 (tyrosine in this structure) and Leu182.
Fig. 3.
Close-up view of the LCAT active site. Amino acids of the LCAT catalytic triad (Ser181, Asp345, and His377) are shown in stick representation. His377 forms hydrogen bonds with Asp345 and a Zn ion at the catalytic site. The Cys31Tyr mutation (one of the amino acid residues that make up the oxyanion hole) is also shown. The regions of LCAT are colored the same as in Fig. 2B.
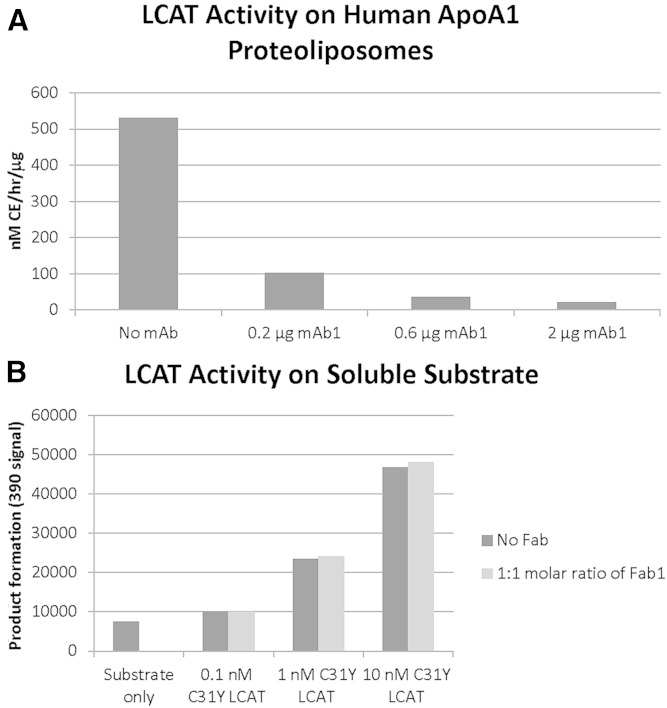
The first subdomain (residues 32–119) consists of a large number of short α-helices and β-strands, but with the majority of amino acids in this subdomain (>60%) lacking any secondary structure. This subdomain of LCAT contains a disulfide bond (Cys50-Cys74), and the amino acids between this disulfide bond have been predicted to belong to the lid (11). In lipases, the lid frequently plays dual roles; blocking/allowing access to the active site through conformational changes and being involved in “interfacial activation” in the presence of a lipid-water interface. Although not a “lid” in the sense that this subdomain does not cover the active site (Fig. 2C), it has been previously shown that this region in LCAT is required for interfacial activation. Adimoolam and Jonas showed that LCATΔ53-71 expresses at similar levels as wild-type LCAT, retains activity on water soluble substrates but is inactive on interfacial substrates (29). Jin et al. (30) used surface plasmon resonance to show that LCATΔ53-71 is incapable of binding to HDL surfaces. Finally, Peelman et al. (31) found that Trp61 was critical for LCAT activity on HDL and that an LCAT peptide (LCAT 56-68) had membrane destabilizing properties. Interestingly, this is also the region of LCAT involved in the Fab1 interaction. Fab1 binds to LCAT by inserting complementarity determining region 3 from the heavy chain into a deep pocket formed by the amino acid stretch connecting Cys50-Cys74. The low amount of secondary structure in subdomain 1 indicates that this region may be highly flexible, and Fab1 binding may assist in the crystallization of LCAT by reducing conformational variability. Additionally, activity assays indicate that while mAb1 (the parent antibody of Fab1) or Fab1 inhibit LCAT activity on ApoA-I-containing proteoliposomes, Fab1 does not affect LCAT activity on soluble substrates (Fig. 4A, B), supporting the hypothesis that this region of LCAT is involved in interfacial activation.
Fig. 4.
Fab1 inhibits LCAT activity on lipid-associated but not soluble substrates. A: Increasing concentrations of mAb1 (the parent antibody of Fab1) inhibit the full LCAT reaction and prevent the formation of CEs on ApoA-I-containing proteoliposome particles. Activity shown is per 0.05 μg of LCAT enzyme. Similar inhibition was seen with Fab1 (data not shown). B: LCAT and LCAT/Fab1 complex have similar phospholipase activity on soluble substrates.
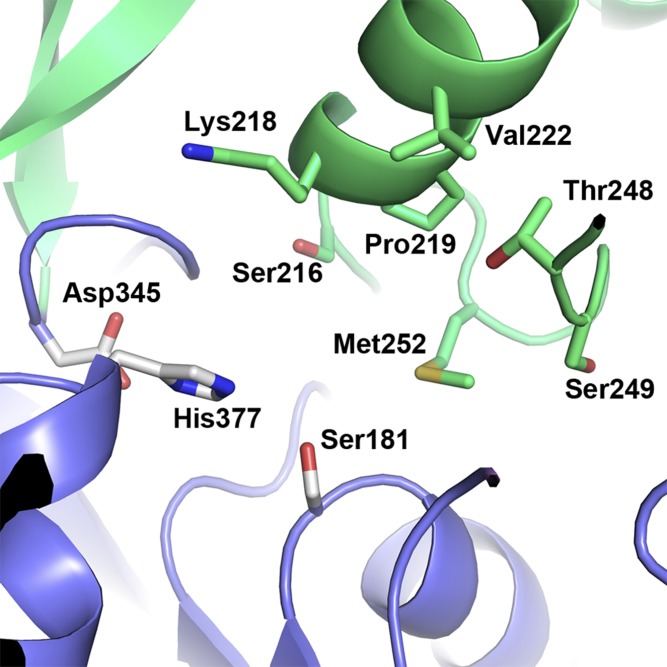
The second subdomain, formed by the insertions between β6/β7 and β7/αE (residues 214–303 and residues 319–344), is in much closer proximity to the catalytic site. This subdomain is composed of a four-stranded β-sheet that packs against a three-helix bundle and forms a cap that folds over the top of the catalytic site. Within this second subdomain, Ser216, Lys218, Pro219, Val222, Thr248, Ser249, and Met252 help shape the back wall of the substrate binding pocket (Fig. 5). Amino acids 225–248 form a flexible loop (as indicated by increased B-factor and missing electron density for amino acids 231–237) that partially blocks access to the catalytic site and is structurally positioned in a location more analogous with lids observed in other lipase structures (32–35).
Fig. 5.
Amino acids in subdomain 2 help shape the substrate binding pocket. Amino acids from subdomain 2 that line the back of the substrate binding pocket are shown in green stick representation. Amino acids from the catalytic triad are shown as white sticks.
DISCUSSION
The crystal structure of LCAT reveals a core α/β hydrolase domain that contains the catalytic triad, along with two additional subdomains. Subdomain 1 contains the region of LCAT shown to be required for interfacial activation while subdomain 2 contains the lid and amino acids that form the back wall of the substrate binding pocket. The possibility remains that in the structure observed here, LCAT is in a “closed” conformation present as soluble LCAT circulates in the plasma and conformational changes could occur upon the interaction between LCAT and the HDL particle. In addition to the more frequently described rearrangement of the α/β hydrolase lid region that allows increased access to the active site and contributes substrate specificity (35), a conformational change may also occur in subdomain 1. Taken together, the experiments that pinpoint the requirement of amino acids between the Cys50-Cys74 disulfide bond for interfacial activation, the likely high flexibility of subdomain 1, and the inhibitory characteristics of Fab1 indicate that upon interaction with the HDL particle the peptide between the Cys50-Cys74 disulfide bond may flip out and interpolate into the particle membrane. A conformational change in this region of the protein could result in both the stabilization of LCAT on the particle surface and the opening of an additional channel to provide substrate greater access to the catalytic site.
When the LCAT protein comes in contact with the HDL particle, the large pocket bound by helices A, B, αA, C, and αF1 and strands 9 and 10 likely faces the particle membrane where it exposes the active site to the lipid surface containing the phosphatidylcholine and cholesterol substrates. Phosphatidylcholine from the HDL particle surface is able to react with Ser181, resulting in the acylation of the catalytic serine amino acid. By remaining on the surface of the particle the acyl enzyme can then transfer the acyl chain preferentially to cholesterol (rather than other acceptors), forming the CE product, which subsequently segregates to the internal core of the HDL particle [comprehensively described by Jonas (1)]. Although we attempted to crystallize LCAT in complex with multiple covalent substrate mimetics and cholesterol, we were unable to identify electron density for any of these small molecules in the structure. A more complete description of the LCAT reaction will need to wait for further studies that successfully show LCAT in complex with various substrate molecules.
A large number of naturally occurring mutations have been observed in the LCAT protein that result in an overall reduction of LCAT enzymatic activity and cause two main disease conditions, FLD and FED (3, 36–39). By analyzing the locations of these mutations within the crystal structure, we are able to predict how these mutations may contribute to decreased LCAT activity. Although many of the amino acids involved in the FED phenotype tend to be scattered across the surface of LCAT (with a small cluster made up by Thr123, Asn131, and Asn391) and an explanation for their effect remains difficult, many of the mutations involved in the FLD phenotype cluster in specific regions and a functional explanation is possible (Table 2 and Fig. 6). In addition, a subset of the LCAT FLD mutations has previously been shown to dramatically reduce the levels of secreted LCAT protein (40), and the location of these mutations intimately within the LCAT α/β hydrolase core indicates a likely structural significance and requirement for proper protein folding.
TABLE 2.
LCAT FLD mutations and their predicted mechanism of inhibition
| Mutation | Location within Structure | Mechanism of Inhibition |
| Gly30Ser | α/β Hydrolase core | Disrupt oxyanion hole |
| Leu32Pro | Subdomain 1 | Disrupt oxyanion hole |
| Gly33Arg | Subdomain 1 | Disrupt oxyanion hole |
| Gly71Arg | Subdomain 1 | Affect interaction with lipid surface |
| Arg135Trp/Gln | α/β Hydrolase core | Disrupt α/β hydrolase core |
| Arg140His/Cys | α/β Hydrolase core | Disrupt α/β hydrolase core |
| Arg147Trp | α/β Hydrolase core | Disrupt oxyanion hole |
| Tyr156Asn | α/β Hydrolase core | Disrupt α/β hydrolase core |
| Gly183Ser | α/β Hydrolase core | Disrupt catalytic Ser181 |
| Leu209Pro | α/β Hydrolase core | Disrupt catalytic Ser181 |
| Lys218Asn | Subdomain 2 | Affect substrate binding |
| Asn228Lys | Subdomain 2 | Destabilize lid |
| Arg244Gly | Subdomain 2 | Destabilize lid |
| Met252Lys | Subdomain 2 | Affect substrate binding |
| Thr274Ala/Ile | Subdomain 2 | Disrupt subdomain 2 |
| Cys313Tyr | α/β Hydrolase core | Disrupt catalytic Asp345 |
| Thr321Met | Subdomain 2 | Disrupt catalytic Asp345 |
| Gly344Ser | Subdomain 2 | Disrupt catalytic Asp345 |
| Thr347Met | α/β Hydrolase core | Affect substrate binding |
| Leu372Arg | α/β Hydrolase core | Disrupt catalytic His377 |
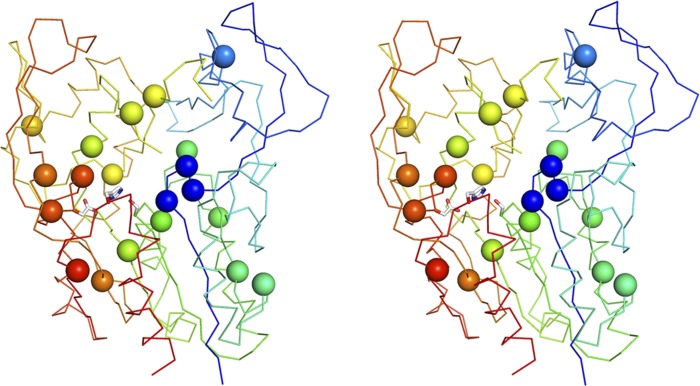
Fig. 6.
Location of FLD mutations in the LCAT structure. Stereo image highlighting LCAT FLD mutations as Cα spheres. The FLD mutations scatter throughout the LCAT structure but are mainly concentrated around the catalytic machinery. LCAT is shown in a rainbow multicolor depiction from the N to C terminus (blue to red). The catalytic triad is shown in white stick depiction.
A large number of the FLD-causing mutations cluster around the catalytic machinery or substrate binding pocket of the LCAT protein and likely disrupt the catalytic reaction of LCAT or its ability to efficiently bind substrate. A first group of four FLD mutations is found in close proximity to the oxyanion hole amino acid 31. The Gly30Ser, Leu32Pro, and Gly33Arg mutations all likely affect the peptide backbone conformation in the region of amino acid 31. Introducing side chain-containing amino acids at position 30 or 33 possibly adds strain to the peptide backbone, while Leu32 is located in a region of the Ramachandran plot that disfavors proline. Arg147 is located on a loop directly behind amino acid 31 and forms hydrogen bonds with Thr79, Asn92, and Asp145. The FLD mutation at this site (Arg147Trp) removes the ability of this amino acid to form these interactions and adds a bulky aromatic residue that may destabilize the position of this loop and cause a knock-on effect for the proper orientation of amino acid 31. Furthermore, a side chain-containing amino acid at position 30 (rather than glycine) may sterically hinder access to the oxyanion hole and/or the catalytic serine.
A second group, Gly183Ser and Leu209Pro, likely disrupts the position of catalytic Ser181. Gly183 is in the GXSXG motif, conserved in α/β hydrolases and required for the constrained strand-turn-helix motif that presents the catalytic serine at the top of the nucleophilic elbow. Leu209 is located at the top of β6, and a proline at this position would place the side chain within the β-sheet, breaking hydrogen bonding to neighboring β5 and possibly affecting Ser181 that directly follows β5.
A third group, made up of Cys313Tyr, Thr321Met, and Gly344Ser, possibly affects the catalytic Asp345. Cys313 is involved in a disulfide bond with Cys356, ∼11 Å from Asp345. Cys356 is located within αE, the helix directly following Asp345. Breaking this disulfide bond could possibly increase conformational flexibility of αE, decreasing stability of Asp345. Both Thr321 and Gly344 are in quite close proximity to Asp345, and mutating these amino acids would disrupt the loop that contains Asp345.
Fourth, the Leu372Arg mutation is located at the top of β8, 5 amino acids upstream from catalytic His377. Replacing this hydrophobic amino acid that packs within the protein with a large charged arginine likely disrupts the fold of LCAT in this region and the position of His377.
Finally, Lys218Asn, Met252Lys, and Thr347Met are all found on the back wall of the pocket above Ser181. It is likely that these mutations affect substrate binding. Additionally, Thr347Met is in very close proximity to Asp345 and His377, and this mutation may also affect the positioning of those catalytic amino acids.
Additional FLD mutations can be found at regions that have possible functional or structural significance. The Gly71Arg mutation is found between the Cys50-Cys74 disulfide bond, within the fusogenic peptide required for activity on interfacial substrates. Both Asn228 and Arg244 are found on the lid above the active site, form a hydrogen bond with each other and possibly help anchor the lid. Mutating either of these residues would break this hydrogen bond. Arg135, Arg140, and Tyr156 all form hydrogen bonds within the α/β hydrolase core. Additionally, Tyr156 sits on the hydrophobic face of aliphatic helix αB. Mutation of any of these 3 amino acids may affect the general stability of the α/β scaffold. Finally, Thr274 forms two hydrogen bonds with surrounding residues and may be required for proper folding of the subdomain formed by the insertions between β6/β7 and β7/αE.
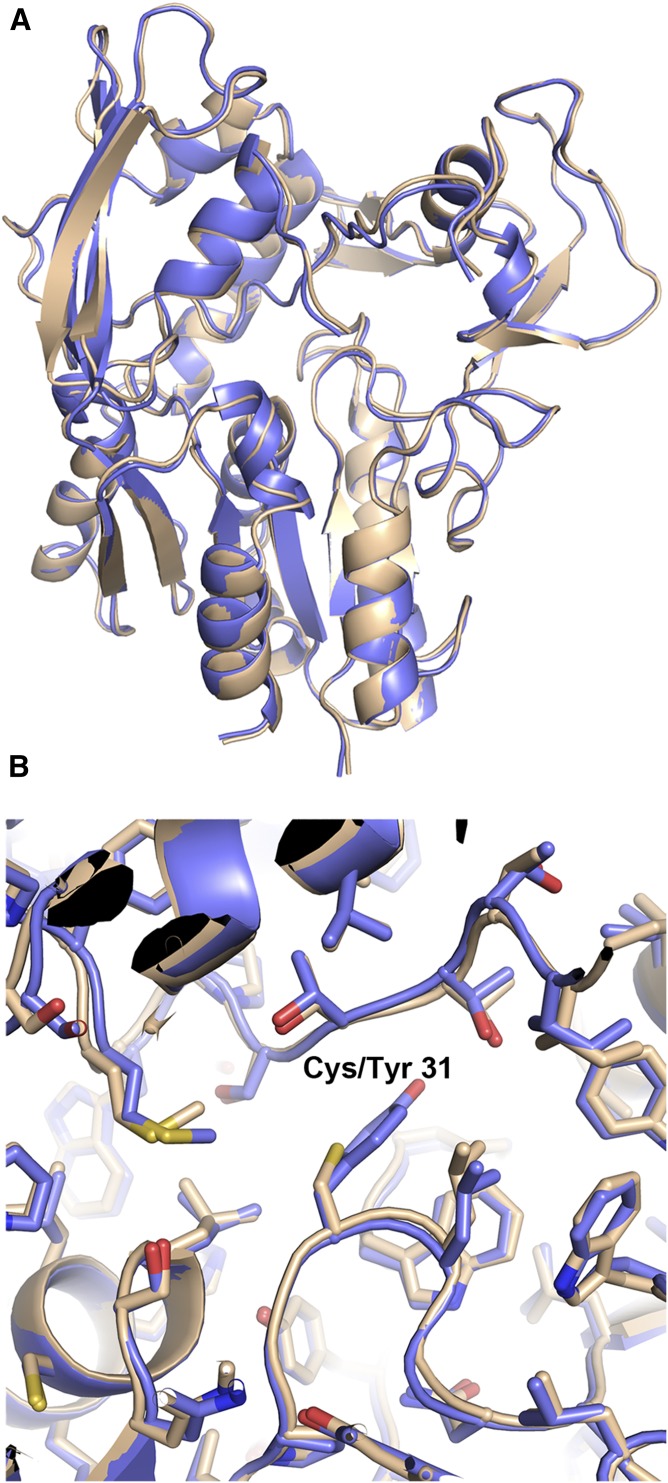
We provide here the first high-resolution description of the structure of the LCAT protein. This structure confirms the catalytic residues Ser181, Asp345, and His377 and consistent with the prediction of Holleboom et al. (28) identifies Cys31 and Leu182 as the amino acids that make up the oxyanion hole. Although a mutant version of LCAT (LCAT Cys31Tyr) was used for the solution and description of this structure, we also solved a lower-resolution structure of wild-type LCAT at 3.6 Å. Wild-type LCAT superposes quite well with LCAT Cys31Tyr (Cα RMSD = 0.388) and confirms the relevance of the observations reported here (Fig. 7). In addition, after this article was written, Glukhova et al. (41) published a very low-resolution structure of the LCAT protein (8.69 Å) and described structural characteristics similar to what we observe in our structure. Although this report helps to unravel one of the key components of the HDL maturation pathway and RCT, many aspects surrounding this protein remain unanswered. Further work investigating questions such as how LCAT interacts with HDL/ApoA-I, whether LCAT undergoes a conformational change upon interaction with HDL/ApoA-I, and how substrate binds in the LCAT active site will continue to advance our understanding of this important protein.
Fig. 7.
The structures of wild-type LCAT and LCAT Cys31Tyr. A: Overall superposition of wild-type (wheat) and Cys31Tyr (blue) LCAT. B: Close-up of amino acid 31 and surrounding region. The mutation introduced at amino acid 31 does not cause significant changes to this structure.
Acknowledgments
The authors thank Timothy Yu, Hila Roshanravan, Nik Sharkov, Ping Cao, David Shen, Tom Boone, Margaret Karow, John Delaney, and the entire LCAT project team for technical and managerial support.
Footnotes
Abbreviations:
- CE
- cholesteryl ester
- Endo H
- endoglycosidase H
- FED
- fish-eye disease
- FLD
- familial LCAT deficiency
- mAb
- monoclonal antibody
- RCT
- reverse cholesterol transport
- RMSD
- root-mean-square deviation
All authors are or have been employees and shareholders of Amgen. X-ray diffraction data were collected at beamlines 08ID at the Canadian Light Source (LCAT Cys31Tyr) and 22ID at the Advanced Photon Source (LCAT WT).
REFERENCES
- 1.Jonas A. 2000. Lecithin cholesterol acyltransferase. Biochim. Biophys. Acta. 1529: 245–256. [DOI] [PubMed] [Google Scholar]
- 2.Ohashi R., Mu H., Wang X., Yao Q., Chen C. 2005. Reverse cholesterol transport and cholesterol efflux in atherosclerosis. QJM. 98: 845–856. [DOI] [PubMed] [Google Scholar]
- 3.Kuivenhoven J. A., Pritchard H., Hill J., Frohlich J., Assmann G., Kastelein J. 1997. The molecular pathology of lecithin:cholesterol acyltransferase (LCAT) deficiency syndromes. J. Lipid Res. 38: 191–205. [PubMed] [Google Scholar]
- 4.Kunnen S., Van Eck M. 2012. Lecithin:cholesterol acyltransferase: old friend or foe in atherosclerosis? J. Lipid Res. 53: 1783–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glomset J. A. 1962. The mechanism of the plasma cholesterol esterification reaction: plasma fatty acid transferase. Biochim. Biophys. Acta. 65: 128–135. [DOI] [PubMed] [Google Scholar]
- 6.Glomset J. A. 1968. The plasma lecithins:cholesterol acyltransferase reaction. J. Lipid Res. 9: 155–167. [PubMed] [Google Scholar]
- 7.Qu S. J., Fan H. Z., Blanco-Vaca F., Pownall H. J. 1993. Effects of site-directed mutagenesis on the N-glycosylation sites of human lecithin:cholesterol acyltransferase. Biochemistry. 32: 8732–8736. [DOI] [PubMed] [Google Scholar]
- 8.O., K., Hill J. S., Wang X., McLeod R., Pritchard P. H. 1993. Lecithin:cholesterol acyltransferase: role of N-linked glycosylation in enzyme function. Biochem. J. 294: 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang C. Y., Manoogian D., Pao Q., Lee F. S., Knapp R. D., Gotto A. M., Jr., Pownall H. J. 1987. Lecithin:cholesterol acyltransferase. Functional regions and a structural model of the enzyme. J. Biol. Chem. 262: 3086–3091. [PubMed] [Google Scholar]
- 10.Francone O. L., Fielding C. J. 1991. Structure-function relationships in human lecithin:cholesterol acyltransferase. Site-directed mutagenesis at serine residues 181 and 216. Biochemistry. 30: 10074–10077. [DOI] [PubMed] [Google Scholar]
- 11.Peelman F., Vinaimont N., Verhee A., Vanloo B., Verschelde J. L., Labeur C., Seguret-Mace S., Duverger N., Hutchinson G., Vandekerckhove J., et al. 1998. A proposed architecture for lecithin cholesterol acyl transferase (LCAT): identification of the catalytic triad and molecular modeling. Protein Sci. 7: 587–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durocher Y., Perret S., Kamen A. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30: E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67: 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62: 72–82. [DOI] [PubMed] [Google Scholar]
- 15.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cowtan K. D. 1994. dm: an automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsl. Protein Crystallogr. 31: 34–38.
- 18.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- 19.Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emsley P., Lohkamp B., Scott W. G., Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen V. B., Arendall W. B., III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLano W. L. 2002. DeLano Scientific, Palo Alto, CA. [Google Scholar]
- 23.Lima V. L. M., Coelho L. C. B. B., Kennedy J. F., Owen J. S., Dolphin P. J. 2004. Lecithin-cholesterol acyltransferase (LCAT) as a plasma glycoprotein: an overview. Carbohydr. Polym. 55: 179–191. [Google Scholar]
- 24.Chang V. T., Crispin M., Aricescu A. R., Harvey D. J., Nettleship J. E., Fennelly J. A., Yu C., Boles K. S., Evans E. J., Stuart D. I., et al. 2007. Glycoprotein structural genomics: solving the glycosylation problem. Structure. 15: 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nardini M., Dijkstra B. W. 1999. Alpha/beta hydrolase fold enzymes: the family keeps growing. Curr. Opin. Struct. Biol. 9: 732–737. [DOI] [PubMed] [Google Scholar]
- 26.Jaeger K. E., Dijkstra B. W., Reetz M. T. 1999. Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 53: 315–351. [DOI] [PubMed] [Google Scholar]
- 27.Krissinel E., Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60: 2256–2268. [DOI] [PubMed] [Google Scholar]
- 28.Holleboom A. G., Kuivenhoven J. A., Peelman F., Schimmel A. W., Peter J., Defesche J. C., Kastelein J. J., Hovingh G. K., Stroes E. S., Motazacker M. M. 2011. High prevalence of mutations in LCAT in patients with low HDL cholesterol levels in The Netherlands: identification and characterization of eight novel mutations. Hum. Mutat. 32: 1290–1298. [DOI] [PubMed] [Google Scholar]
- 29.Adimoolam S., Jonas A. 1997. Identification of a domain of lecithin-cholesterol acyltransferase that is involved in interfacial recognition. Biochem. Biophys. Res. Commun. 232: 783–787. [DOI] [PubMed] [Google Scholar]
- 30.Jin L., Shieh J. J., Grabbe E., Adimoolam S., Durbin D., Jonas A. 1999. Surface plasmon resonance biosensor studies of human wild-type and mutant lecithin cholesterol acyltransferase interactions with lipoproteins. Biochemistry. 38: 15659–15665. [DOI] [PubMed] [Google Scholar]
- 31.Peelman F., Vanloo B., Perez-Mendez O., Decout A., Verschelde J. L., Labeur C., Vinaimont N., Verhee A., Duverger N., Brasseur R., et al. 1999. Characterization of functional residues in the interfacial recognition domain of lecithin cholesterol acyltransferase (LCAT). Protein Eng. 12: 71–78. [DOI] [PubMed] [Google Scholar]
- 32.Schrag J. D., Li Y., Cygler M., Lang D., Burgdorf T., Hecht H. J., Schmid R., Schomburg D., Rydel T. J., Oliver J. D., et al. 1997. The open conformation of a Pseudomonas lipase. Structure. 5: 187–202. [DOI] [PubMed] [Google Scholar]
- 33.Noble M. E., Cleasby A., Johnson L. N., Egmond M. R., Frenken L. G. 1993. The crystal structure of triacylglycerol lipase from Pseudomonas glumae reveals a partially redundant catalytic aspartate. FEBS Lett. 331: 123–128. [DOI] [PubMed] [Google Scholar]
- 34.Roussel A., Canaan S., Egloff M. P., Riviere M., Dupuis L., Verger R., Cambillau C. 1999. Crystal structure of human gastric lipase and model of lysosomal acid lipase, two lipolytic enzymes of medical interest. J. Biol. Chem. 274: 16995–17002. [DOI] [PubMed] [Google Scholar]
- 35.Labar G., Bauvois C., Borel F., Ferrer J. L., Wouters J., Lambert D. M. 2010. Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. ChemBioChem. 11: 218–227. [DOI] [PubMed] [Google Scholar]
- 36.Calabresi L., Pisciotta L., Costantin A., Frigerio I., Eberini I., Alessandrini P., Arca M., Bon G. B., Boscutti G., Busnach G., et al. 2005. The molecular basis of lecithin:cholesterol acyltransferase deficiency syndromes: a comprehensive study of molecular and biochemical findings in 13 unrelated Italian families. Arterioscler. Thromb. Vasc. Biol. 25: 1972–1978. [DOI] [PubMed] [Google Scholar]
- 37.Holleboom A. G., Kuivenhoven J. A., van Olden C. C., Peter J., Schimmel A. W., Levels J. H., Valentijn R. M., Vos P., Defesche J. C., Kastelein J. J., et al. 2011. Proteinuria in early childhood due to familial LCAT deficiency caused by loss of a disulfide bond in lecithin:cholesterol acyl transferase. Atherosclerosis. 216: 161–165. [DOI] [PubMed] [Google Scholar]
- 38.Peelman F., Verschelde J. L., Vanloo B., Ampe C., Labeur C., Tavernier J., Vandekerckhove J., Rosseneu M. 1999. Effects of natural mutations in lecithin:cholesterol acyltransferase on the enzyme structure and activity. J. Lipid Res. 40: 59–69. [PubMed] [Google Scholar]
- 39.Hörl G., Kroisel P. M., Wagner E., Tiran B., Petek E., Steyrer E. 2006. Compound heterozygosity (G71R/R140H) in the lecithin:cholesterol acyltransferase (LCAT) gene results in an intermediate phenotype between LCAT-deficiency and fish-eye disease. Atherosclerosis. 187: 101–109. [DOI] [PubMed] [Google Scholar]
- 40.Qu S. J., Fan H. Z., Blanco-Vaca F., Pownall H. J. 1995. In vitro expression of natural mutants of human lecithin:cholesterol acyltransferase. J. Lipid Res. 36: 967–974. [PubMed] [Google Scholar]
- 41.Glukhova A., Hinkovska-Galcheva V., Kelly R., Abe A., Shayman J. A., Tesmer J. J. 2015. Structure and function of lysosomal phospholipase A2 and lecithin:cholesterol acyltransferase. Nat. Commun. 6: 6250. [DOI] [PMC free article] [PubMed] [Google Scholar]