Abstract
Meta-analyses of European populations has successfully identified genetic variants in over 150 loci associated with lipid levels, but results from additional ethnicities remain limited. Previously, we reported two novel lipid loci identified in a sample of 7,657 African Americans using a gene-centric array including 50,000 SNPs in 2,100 candidate genes. Initial discovery and follow-up of signals with P < 10−5 in additional African American samples confirmed CD36 and ICAM1. Using an additional 8,244 African American female samples from the Women’s Health Initiative SNP Health Association Resource genome-wide association study dataset, we further examined the previous meta-analyses results by attempting to replicate 20 additional putative lipid signals with P < 10−4. Replication confirmed rs868213, located in a splice donor region of exocyst complex component 3-like 1 (EXOC3L1) as a novel signal for HDL (additive allelic effect β = 0.02; P = 1.4 × 10−8; meta-analyses of discovery and replication). EXOC3L1 is strongly expressed in vascular endothelium and forms part of the exocyst complex, a key facilitator of the trafficking of lipid receptors. Increasing sample sizes for genetic studies in nonEuropean populations will continue to improve our understanding of lipid metabolism.
Keywords: high density lipoprotein, genetics, vascular biology, exocyst complex component 3-like 1
Greater than 150 lipid-associated loci have been discovered using genome-wide association studies (GWASs) mainly based on individuals of European ancestry (1, 2). The number of variants discovered and the power to detect variants of lower allele frequency and effect size is directly proportional to the number of participants in the study, demonstrated by the success of gradually larger meta-analyses (1). Genetic variants generally appear to exert similar effects across ethnicities if tested in a sufficiently powered cohort, but differences in allele frequency and regional linkage disequilibrium (LD) between ethnicities allows for the identification of novel contributors to lipid metabolism (3–5). While efforts to identify additional variants associated with blood lipids in African-American populations have been successful (3, 6–8), the identification of novel genetic loci in nonEuropean populations has been hampered by inadequate power due to limited sample size and genotyping platform designs made for optimal SNP coverage of European populations (3, 9).
The Institute for Translational Medicine and Therapeutics (ITMAT)-Broad-Candidate Gene Association Resource (IBC) array [also referred to as the CardioChip or HumanCVD Beadchip (Illumina)] was specifically designed to capture ∼50,000 SNPs across ∼2,100 loci, with pathways known or postulated to have roles in lipid traits captured at higher density than conventional GWAS arrays (9). SNPs were specifically selected from African HapMap samples to be included on the array to ensure coverage of African populations (9). Results of genetic associations with total cholesterol (TC), LDL cholesterol (LDL-C), HDL cholesterol (HDL-C), and TG concentrations in European-derived cohorts using the IBC array, as well as meta-analysis of multi-ethnic cohorts, have been published (2, 6). Follow-up replication of signals with P < 10−5 from the discovery cohort in African American samples confirmed two previously unreported lipid loci (CD36 and ICAM1) (6), which have since been validated independently (8).
We sought to improve the power to detect novel signals in African American populations by increasing sample size for meta-analyses using additional independent study samples. A meta-analysis of the original six African American populations (n = 7,657) and the replication population (n = 8,244) yielded a total sample size of 15,901. SNPs selected for replication in additional African Americans had 10−4 < P < 10−5 in our previous analysis (6) and independence from known lipid-related loci calculated by a minor allele frequency-specific r2 threshold (r2 < 0.3 if minor allele frequency > 1%, r2 < 0.6 if minor allele frequency < 1%).
METHODS
Sample collection
The six discovery cohorts [Atherosclerosis Risk in Communities (ARIC), Coronary Artery Risk Development in Young Adults (CARDIA), Cardiovascular Health Study (CHS), Multi-Ethnic Study of Atherosclerosis (MESA), Women’s Health Initiative (WHI), and Jackson Heart Study (JHS)] were included from our previous report (6), and all studies contributed individual-level IBC array genotypes and phenotypes (see Table 1). Summary level statistics for SNPs selected for replication were provided from a second independent cohort of African American females genotyped from the WHI SNP Health Association Resource (SHARe) GWAS dataset, termed here WHI-SHARe. There was no sample overlap between the WHI and WHI-SHARe genotyping initiatives. Further details of the populations, genotyping, and quality control analysis have been published elsewhere (6, 8). All participants provided informed written consent. Institutional Review Boards of each Candidate Gene Association Resource (CARe) cohort reviewed and approved the cohort’s interaction with CARe. The study described here was approved by the Committee on the Use of Humans as Experimental Subjects of the Massachusetts Institute of Technology.
TABLE 1.
Participating studies
| Study | Study Design | Participant Age (years) | N | Individual-Level Data | Genotyping Array |
| Discovery | |||||
| ARIC | Prospective cohort, asymptomatic randomly collected in four US centers | 45–64 | 2,687 | Yes | IBC array |
| CARDIA | Prospective cohort examining young adults collected in four US centers | 18–30 | 1,024 | Yes | IBC array |
| CHS | Prospective cohort of asymptomatic older adults collected in four US centers | 65–80 | 701 | Yes | IBC array |
| MESA | Prospective cohort of asymptomatic participants | 45–84 | 1,584 | Yes | IBC array |
| WHI | Prospective cohort of postmenopausal women from across US | 50–79 | 1,584 | Yes | IBC array |
| JHS | Prospective cohort of asymptomatic adults collected in Jackson, MS | 21–48 | 1,118 | Yes | IBC array |
| Replication | |||||
| WHI-SHARe | Prospective cohort of postmenopausal women from across US | 50–79 | 8,153 | No | Affymetrix 6.0 |
N, number of participants included in current study; ARIC, Atherosclerosis Risk in Communities; CARDIA, Coronary Artery Risk Development in Young Adults; CHS, Cardiovascular Health Study; MESA, Multi-Ethnic Study of Atherosclerosis; JHS, Jackson Heart Study; WHI-SHARe, second phase of Women’s Health Initiative genotyping.
All participants were greater than 21 years of age. Lipid phenotypes were taken from baseline or first measurements for all fasting individuals, as described in the original reports. All measurements were converted to millimoles per liter, with TC and HDL-C measurements converted from milligrams per deciliter by dividing by 38.67, and TG measurements converted from milligrams per deciliter by dividing by 88.57. TG and HDL values were log-transformed to satisfy normality. LDL-C was calculated according to the Friedewald formula: L ∼ TC − HDL − kTG, where k is 0.45 for millimoles per liter (or 0.20 if measured in milligrams per deciliter) (10). If TG values were >4.51 mmol/l (>400 mg/dl), then LDL-C was treated as a missing value.
Statistical methods
Gender stratified association analysis was performed in each participating study using an additive genetic model including age, sex, type 2 diabetes diagnosis, body mass index, and smoking history as covariates, as well as adjusting for 10 principal components of ancestry in PLINK (https://www.cog-genomics.org/plink2) (11). Principal components were calculated using Eigenstrat (12). Sensitivity analysis including and excluding covariates did not alter our findings. Meta-analyses were performed by two independent analysts using a fixed-effect inverse-variance approach in two different software packages, MANTEL (http://www.broadinstitute.org/~debakker/mantel.html) (13) and METAL (http://www.sph.umich.edu/csg/abecasis/metal/) (14). Results were highly concordant, reflecting a robust data analyses pipeline.
RESULTS
African American meta-analysis
Pruning of discovery cohort meta-analysis results for independence from previously reported loci and 10−4 < P <10−5 yielded three SNPs associated with plasma TC, three SNPs associated with LDL-C, six SNPs associated with HDL-C, and seven SNPs associated with TG (20 total lipid-SNP associations). Each of these SNPs was carried forward for replication in 8,244 WHI-SHARe African American females.
The results for all 20 SNPs in the discovery and WHI-SHARe replication samples are presented in Table 2. In the replication sample, six SNPs were associated with the same lipid trait and with the same direction of effect as the discovery report with P < 0.05 (Table 2). The strongest association observed was rs868213 near exocyst complex component 3-like 1 (EXOC3L1) (P = 4.4 × 10−4) with increased HDL concentration (β = 0.014). Meta-analysis of discovery and replication studies led to a genome-wide significant signal at the EXOC3L1 locus (P = 1.4 × 10−8) for association with HDL. Located on the long arm of chromosome 16, rs868213 is at position 67,220,457 (Hg19 build37). The minor allele of rs868213 is observed in the studied African American cohorts at 43%, while HapMap populations indicate allele frequencies of 2% in JPT (Japanese in Tokyo), 4% in CHB (Han Chinese in Beijing), 6% in CEPH (Utah residents with Northern and Western European ancestry), 8% in MXL (people with Mexican ancestry in Los Angeles, CA), and 52% of YRI (Yoruba in Ibadan, Nigeria).
TABLE 2.
SNPs carried forward for replication and meta-analysis in 15,901 African American participants
| Trait | SNP | Nearby Gene | RAF | Discovery | Replication | Combined | |||
| β (SE) | P | β (SE) | P | β (SE) | P | ||||
| TC | rs1757214 | CACNB2 | 0.38 | 0.311 (0.071) | 1.26 × 10−5 | 1.348 (0.685) | 0.049 | 0.322 (0.071) | 5.12 × 10−6 |
| TC | rs2463117 | PRKCH | 0.76 | 0.087 (0.020) | 1.00 × 10−5 | 0.171 (0.744) | 0.82 | 0.087 (0.020) | 1.33 × 10−5 |
| TC | rs13087941 | CAV3 | 0.29 | 0.077 (0.018) | 3.70 × 10−5 | 0.028 (0.746) | 0.97 | 0.077 (0.018) | 1.89 × 10−5 |
| TC | rs10965224 | CDKN2B | 0.36 | 0.067 (0.017) | 5.47 × 10−5 | 1.02 (0.702) | 0.15 | 0.068 (0.017) | 7.03 × 10−5 |
| LDL | rs2463117 | PRKCH | 0.76 | 0.079 (0.020) | 5.76 × 10−5 | −0.132 (0.759) | 0.86 | 0.079 (0.02) | 8.01 × 10−5 |
| LDL | rs9382462 | HCRTR2 | 0.10 | 0.111 (0.028) | 2.89 × 10−5 | −2.566 (1.177) | 0.030 | 0.110 (0.028) | 9.18 × 10−5 |
| LDL | rs7885619 | GAB3 | 0.86 | 0.094 (0.026) | 8.48 × 10−5 | 2.016 (0.994) | 0.042 | 0.095 (0.026) | 2.50 × 10−4 |
| HDL | rs868213 | EXOC3L1 | 0.43 | 0.028 (0.007) | 4.94 × 10−5 | 0.014 (0.004) | 0.00044 | 0.020 (0.003) | 1.42 × 10−8 |
| HDL | rs9940665 | SLC9A5 | 0.42 | 0.027 (0.007) | 8.85 × 10−5 | 0.009 (0.004) | 0.019 | 0.015 (0.004) | 1.57 × 10−5 |
| HDL | rs3852700 | FHOD1 | 0.63 | 0.029 (0.007) | 3.15 × 10−5 | 0.008 (0.004) | 0.050 | 0.014 (0.004) | 1.01 × 10−4 |
| HDL | rs9441835 | MIA3 | 0.36 | 0.032 (0.008) | 9.53 × 10−5 | 0.007 (0.004) | 0.098 | 0.011 (0.004) | 1.64 × 10−3 |
| HDL | rs7136463 | VWF | 0.59 | 0.027 (0.007) | 6.95 × 10−5 | −0.005 (0.005) | 0.32 | 0.007 (0.004) | 0.078 |
| HDL | rs4304399 | COL5A1 | 0.35 | 0.027 (0.007) | 9.43 × 10−5 | −0.006 (0.006) | 0.33 | 0.007 (0.005) | 0.13 |
| TG | rs880263 | HRH4 | 0.95 | 0.078 (0.019) | 3.66 × 10−5 | 0.018 (0.016) | 0.2701 | 0.043 (0.012) | 4.56 × 10−4 |
| TG | rs9425309 | C1orf105 | 0.66 | 0.036 (0.009) | 6.69 × 10−5 | 0.008 (0.007) | 0.2639 | 0.019 (0.006) | 7.85 × 10−4 |
| TG | rs10220716 | GABRA5 | 0.27 | 0.036 (0.009) | 6.85 × 10−5 | −0.001 (0.008) | 0.8821 | 0.015 (0.006) | 0.010 |
| TG | rs1371427 | — | 0.52 | 0.124 (0.031) | 9.18 × 10−5 | 0.012 (0.007) | 0.07867 | 0.017 (0.006) | 0.010 |
| TG | rs7207419 | CLTC | 0.79 | 0.042 (0.010) | 6.02 × 10−5 | −0.008 (0.009) | 0.4173 | 0.014 (0.007) | 0.032 |
| TG | rs2411330 | KLK1 | 0.08 | 0.060 (0.015) | 7.07 × 10−5 | −0.01 (0.013) | 0.4255 | 0.02 (0.010) | 0.041 |
| TG | rs80051981 | — | 0.02 | 0.444 (0.110) | 6.28 × 10−5 | −0.008 (0.023) | 0.7432 | 0.011 (0.023) | 0.62 |
RAF, risk allele frequency; β, regression β coefficient. Genome-wide significant result is in boldface.
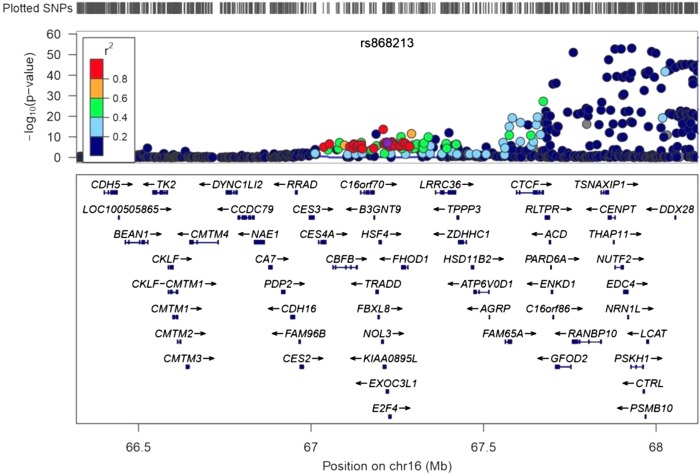
We then assessed whether the lower minor allele frequency of rs868213 in European populations could account for the lack of HDL-rs868213 association from accessible large-scale public lipid GWAS datasets. The IBC lipid genetics meta-analysis found a nonstatistically significant association of the minor allele of rs868213 with increased HDL in 66,240 Europeans (β = 0.0071; P = 0.10) (2). The larger Global Lipid Genetics Consortium (http://www.sph.umich.edu/csg/abecasis/public/lipids2013/) found that in 92,817 individuals of European ancestry, the minor allele of rs868213 was associated with an increase of HDL (β = 0.054; P = 1.01 × 10−7) (1). Other nearby SNPs were genome wide significantly associated with HDL (rs2233455, β = 0.056; P = 2.27 × 10−14, 12 kb away), but the locus was not identified as distinct from the associations seen with SNPs in LCAT (Fig. 1), likely due to higher levels of LD observed in European populations. The lead LCAT SNP, rs255052, is >800 kb upstream and in minimal LD with rs868213 in African Americans (r2 = 0.158). An additional SNP, rs3729639, found 5 kb away and in strong LD with rs868213 in African Americans, was found associated with HDL (β = 0.09; P = 1.9 × 10−11) in a CARe African American GWAS, but was not identified as independent of LCAT (15).
Fig. 1.
Global Lipids Genetics Consortium European ancestry association results for chromosome 16 demonstrating significant association of rs868213 with HDL-C but LD with rs255052 in LCAT. Figure created in LocusZoom (http://csg.sph.umich.edu/locuszoom/) with public data (http://www.sph.umich.edu/csg/abecasis/public/lipids2013/).
To further assess the independence of the rs868213-HDL association from upstream LCAT in an African American population, a secondary analysis was performed with the minor allele dosage of rs255052 included as a covariate in the model. The lead SNP in the LCAT locus, rs255052, is prevalent in both European and African American populations and has been consistently associated with HDL concentrations. The rs868213-HDL association remained significant despite correction for rs255052 (discovery P = 0.048; replication P = 0.086; meta-analysis P = 0.0080).
In silico analysis of rs868213 and EXOC3L1
EXOC3L1 is highly expressed in developing human umbilical vein endothelial cells, and siRNA studies have shown that the gene plays an important role in tubular network formation (16). In silico bioinformatic evaluation of rs868213 identified that it alters a putative splice site consensus site of EXOC3L1. Significant changes in splicing regulatory protein binding are expected with the nucleotide change produced by rs868213, according to information theory analysis (supplementary Fig. 1). Thus, we reviewed ENCODE RNA-seq data for evidence of alternative splicing and possible exon skipping within EXOC3L1. In four out of thirteen cell lines evaluated, including HeLa (cervical carcinoma), MCF7 (breast tumor), HCT6 (HCT116; colon tumor), and NHLF (lung fibroblasts) cell lines, alternative splicing of the exon immediately following rs868213 was observed (supplementary Fig. 2). Exclusion of the exon would remove residues 429 to 461 of the EXOC3L1 protein. UniProt annotation indicates that amino acids 1-370 are required for interaction with other components of the exocyst complex and should be maintained despite the loss of residues 429 to 461 (17), though functional consequences of the deletion remain unknown.
Other SNPs in LD with rs868213 may explain the observed association, so we reviewed evidence for expression quantitative trait loci (eQTL) mediated by rs868213 and SNPs showing LD (Yoruba r2 > 0.2) across a range of tissues using three available eQTL browsers (Genevar: https://www.sanger.ac.uk/resources/software/genevar/; GTEx: http://www.gtexportal.org/; seeQTL: http://gbrowse.csbio.unc.edu/). Suggestive evidence for an eQTL was found in a monocyte dataset in four genes: ELMO3 (logP = 7.78); NOL3 (logP = 4.35); DUS2L (logP = 7.49); and LRRC29 (logP = 5.92) (supplementary Fig. 3) (18). However, these genes were all associated below the study-wide significance threshold (logP = 11.24), and although monocytes are a disease relevant tissue, no other data were available to further support the eQTL candidates in lipid biology. High-throughput genome confirmation capture analysis (4Dgenome: http://4dgenome.int-med.uiowa.edu/) (19) of the region extending around rs868213 indicated no evidence for physical interaction between rs868213 and the LCAT locus, further supporting independence of the association from LCAT. There was evidence of potential interaction between the rs868213 locus and the CES2-CES3-CES4A cluster (supplementary Fig. 3).
DISCUSSION
Building upon previous genetic association studies, we attempted to replicate nominally associated variants in a larger meta-analysis including 15,901 African Americans. The most interesting finding was the genome-wide significant association of rs868213 with HDL-C. Found in a gene-rich region 800,000 bp upstream of LCAT, SNPs surrounding rs868213 were found to be associated with HDL in meta-analyses of European populations, but were overlooked due to LD with LCAT SNPs. Higher rs868213 allele frequency and lower LD between rs868213 and LCAT SNPs in African Americans increased power to detect the novel association with HDL in the current study. Maintenance of the rs868213-HDL association after correction for the effect of lead LCAT SNP rs255052 further supports an independent mechanism.
Bioinformatic analysis identified that rs868213 is located within an EXOC3L1 putative splice site consensus site, with variable exon skipping observed within RNA sequencing data, supporting a potential functional consequence. Extensive analysis revealed little evidence for an eQTL explaining the observed association. High-throughput genome confirmation capture analysis of the LD region extending around rs868213 indicated no evidence for physical interaction between rs868213 and the LCAT locus, further supporting independence of the association from LCAT.
EXOC3L1 is required for the exocytosis of insulin granules from pancreatic β cells, and is differentially methylated and expressed in patients with type 2 diabetes (20). Barkefors et al. (21) reported that the close homolog EXOC3L2 is also expressed extensively in the endothelium, and siRNA studies demonstrate that EXOC3L2 silencing inhibits VEGF receptor 2 phosphorylation and VEGFA-directed migration of cultured endothelial cells. Although EXOC3L1 and other members of the exocyst complex are unprecedented as drug targets, druggable binding pockets were located in EXOC3L1 using structure-based evaluation via the DoGSiteScorer tool (22), indicating the potential for investigational pharmacological modification of EXOC3L1 activity.
After evaluation of all data across the rs868213 locus, we found EXOC3L1 to be the most likely candidate on the basis of both functional impact and biological plausibility. EXOC3L1 is a component of the exocyst complex, an evolutionarily conserved multisubunit protein complex implicated in molecular trafficking and tethering secretory vesicles to the plasma membrane (23). The exocyst complex has an important role in polarized vesicular trafficking of transmembrane proteins, specifically including the LDL receptor (24) and the HDL scavenger receptor class B member I (SR-BI) (25). Considering all the available genetic and bioinformatic evidence, we propose that variation in EXOC3L1 function, particularly in the vascular endothelium where this gene is abundantly expressed, could lead to altered exocyst function impacting HDL homeostasis.
The current study raises three important points for interpretation of future genetic association studies in lipid traits: 1) large GWAS meta-analysis results are publicly available and should be used as a reference and in silico replication when examining putative regions; 2) as power to detect variants of modest effect size has increased, researchers should be exhaustive in their attempts to explain differences observed between studies; and perhaps most importantly 3) due to smaller haplotype blocks and additional areas of recombination, African populations can be very helpful for identifying additional signals within large GWAS signals from Europeans.
Should we make efforts to continue to grow population datasets from additional ethnicities? Despite greater than 150 lipid loci identified in meta-analyses of Europeans, the concordance of direction of effect between cohorts and ethnicities of additional non-genome-wide significant variants suggests many variants and genes are yet to be discovered (1). Each associated locus is also likely to include multiple common, uncommon, and rare variants which independently contribute to the variance of the trait. Moreover, multiple different genes within a gene-rich locus may be contributing to trait variation. Collection of the broad swath of variants and incorporation into suitable models will undoubtedly improve our predictive abilities and explain “missing heritability.” As technological development reduces the cost and increases the resolution of genetic information acquisition, efforts to obtain and biobank samples will need to focus upon improving the representation of nonEuropean populations.
In conclusion, the current meta-analysis of 15,901 African Americans identified an association between rs868213 in EXOC3L1 and HDL-C.
Supplementary Material
Acknowledgments
The Candidate Gene Association Resource (CARe) Consortium wishes to acknowledge the support of the National Heart, Lung, and Blood Institute and the contributions of the research institutions, study investigators, field staff, and study participants. The authors thank the contributors and patients of the WHI-SHARe project, a long-term national health study. The WHI-SHARe GWAS had data cleaning and harmonization performed at the Fred Hutchinson Cancer Research Center in Seattle, WA.
Footnotes
Abbreviations:
- CARe
- Candidate Gene Association Resource
- eQTL
- expression quantitative trait loci
- EXOC3L1
- exocyst complex component 3-like 1
- GWAS
- genome-wide association study
- HDL-C
- HDL cholesterol
- IBC
- Institute for Translational Medicine and Therapeutics (ITMAT)-Broad-Candidate Gene Association Resource
- LD
- linkage disequilibrium
- LDL-C
- LDL cholesterol
- SHARe
- SNP Health Association Resource
- TC
- total cholesterol
- WHI
- Women’s Health Initiative
C.C.E. was supported by a Rubicon grant from The Netherlands Organization for Scientific Research (NWO). Y.L., G.Z., and Q.D. were supported by National Institutes of Health Grants R01HG006292 and R01HG006703. M.R.B. was funded by the portfolio of translational research of the National Institutes for Health Research Biomedical Research Unit at Barts and The London School of Medicine and Dentistry. The WHI SNP Health Association Resource GWAS was funded by the National Heart, Lung, and Blood Institute.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Willer C. J., Schmidt E. M., Sengupta S., Peloso G. M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M. L., Mora S., et al. ; Global Lipids Genetics Consortium. 2013. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45: 1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asselbergs F. W., Guo Y., van Iperen E. P., Sivapalaratnam S., Tragante V., Lanktree M. B., Lange L. A., Almoguera B., Appelman Y. E., Barnard J., et al. 2012. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am. J. Hum. Genet. 91: 823–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buyske S., Wu Y., Carty C. L., Cheng I., Assimes T. L., Dumitrescu L., Hindorff L. A., Mitchell S., Ambite J. L., Boerwinkle E., et al. 2012. Evaluation of the metabochip genotyping array in African Americans and implications for fine mapping of GWAS-identified loci: the PAGE study. PLoS One. 7: e35651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teo Y. Y., Small K. S., Kwiatkowski D. P. 2010. Methodological challenges of genome-wide association analysis in Africa. Nat. Rev. Genet. 11: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg N. A., Huang L., Jewett E. M., Szpiech Z. A., Jankovic I., Boehnke M. 2010. Genome-wide association studies in diverse populations. Nat. Rev. Genet. 11: 356–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbers C. C., Guo Y., Tragante V., van Iperen E. P., Lanktree M. B., Castillo B. A., Chen F., Yanek L. R., Wojczynski M. K., Li Y. R., et al. 2012. Gene-centric meta-analysis of lipid traits in African, East Asian and Hispanic populations. PLoS One. 7: e50198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musunuru K., Romaine S. P., Lettre G., Wilson J. G., Volcik K. A., Tsai M. Y., Taylor H. A., Schreiner P. J., Rotter J. I., Rich S. S., et al. 2012. Multi-ethnic analysis of lipid-associated loci: the NHLBI CARe project. PLoS One. 7: e36473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coram M. A., Duan Q., Hoffmann T. J., Thornton T., Knowles J. W., Johnson N. A., Ochs-Balcom H. M., Donlon T. A., Martin L. W., Eaton C. B., et al. 2013. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am. J. Hum. Genet. 92: 904–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keating B. J., Tischfield S., Murray S. S., Bhangale T., Price T. S., Glessner J. T., Galver L., Barrett J. C., Grant S. F., Farlow D. N., et al. 2008. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One. 3: e3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedewald W. T., Levy R. I., Fredrickson D. S. 1972. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 18: 499–502. [PubMed] [Google Scholar]
- 11.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 13.de Bakker P. I., Ferreira M. A., Jia X., Neale B. M., Raychaudhuri S., Voight B. F. 2008. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum. Mol. Genet. 17: R122–R128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willer C. J., Li Y., Abecasis G. R. 2010. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 26: 2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lettre G., Palmer C. D., Young T., Ejebe K. G., Allayee H., Benjamin E. J., Bennett F., Bowden D. W., Chakravarti A., Dreisbach A., et al. 2011. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genet. 7: e1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takase H., Matsumoto K., Yamadera R., Kubota Y., Otsu A., Suzuki R., Ishitobi H., Mochizuki H., Kojima T., Takano S., et al. 2012. Genome-wide identification of endothelial cell-enriched genes in the mouse embryo. Blood. 120: 914–923. [DOI] [PubMed] [Google Scholar]
- 17.UniProt Consortium. 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43: D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeller T., Wild P., Szymczak S., Rotival M., Schillert A., Castagne R., Maouche S., Germain M., Lackner K., Rossmann H., et al. 2010. Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS One. 5: e10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin F., Li Y., Dixon J. R., Selvaraj S., Ye Z., Lee A. Y., Yen C. A., Schmitt A. D., Espinoza C. A., Ren B. 2013. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 503: 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dayeh T., Volkov P., Salo S., Hall E., Nilsson E., Olsson A. H., Kirkpatrick C. L., Wollheim C. B., Eliasson L., Ãon T. R., et al. 2014. Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion. PLoS Genet. 10: e1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barkefors I., Fuchs P. F., Heldin J., Bergstrom T., Forsberg-Nilsson K., Kreuger J. 2011. Exocyst complex component 3-like 2 (EXOC3L2) associates with the exocyst complex and mediates directional migration of endothelial cells. J. Biol. Chem. 286: 24189–24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volkamer A., Kuhn D., Grombacher T., Rippmann F., Rarey M. 2012. Combining global and local measures for structure-based druggability predictions. J. Chem. Inf. Model. 52: 360–372. [DOI] [PubMed] [Google Scholar]
- 23.Heider M. R., Munson M. 2012. Exorcising the exocyst complex. Traffic. 13: 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grindstaff K. K., Yeaman C., Anandasabapathy N., Hsu S. C., Rodriguez-Boulan E., Scheller R. H., Nelson W. J. 1998. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell. 93: 731–740. [DOI] [PubMed] [Google Scholar]
- 25.Pagler T. A., Rhode S., Neuhofer A., Laggner H., Strobl W., Hinterndorfer C., Volf I., Pavelka M., Eckhardt E. R., van der Westhuyzen D. R., et al. 2006. SR-BI-mediated high density lipoprotein (HDL) endocytosis leads to HDL resecretion facilitating cholesterol efflux. J. Biol. Chem. 281: 11193–11204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.