Abstract
Barth syndrome (BTHS), an X-linked disease associated with cardioskeletal myopathy, neutropenia, and organic aciduria, is characterized by abnormalities of cardiolipin (CL) species in mitochondria. Diagnosis of the disease is often compromised by lack of rapid and widely available diagnostic laboratory tests. The present study describes a new method for BTHS screening based on MALDI-TOF/MS analysis of leukocyte lipids. This generates a “CL fingerprint” and allows quick and simple assay of the relative levels of CL and monolysocardiolipin species in leukocyte total lipid profiles. To validate the method, we used vector algebra to analyze the difference in lipid composition between controls (24 healthy donors) and patients (8 boys affected by BTHS) in the high-mass phospholipid range. The method of lipid analysis described represents an important additional tool for the diagnosis of BTHS and potentially enables therapeutic monitoring of drug targets, which have been shown to ameliorate abnormal CL profiles in cells.
Keywords: cardiomyopathy, lysophospholipids, mass spectrometry, matrix-assisted laser desorption/ionization, mitochondria, phospholipids, phospholipids/metabolism, tafazzin, time-of-flight
Barth syndrome (BTHS) is a rare, life-threatening, X-linked recessive metabolic disease characterized by infantile or childhood onset of cardiomyopathy, skeletal myopathy, growth delay, neutropenia, 3-methylglutaconic aciduria (3-MGCA), abnormal mitochondrial structure, and variable mitochondrial respiratory chain dysfunction (1–4). It results from loss-of-function mutations of the tafazzin (TAZ) gene (5).
The major consequence of this loss-of-function is the deficient remodeling of the mitochondrial phospholipid cardiolipin (CL), a process that normally leads to the mature acyl composition (6, 7). CL is the signature lipid of mitochondria, where it is an important constituent of the inner membrane, essential for supercomplex formation, oxidative phosphorylation (i.e., mitochondrial energy metabolism), and protein import, and is capable of triggering mitophagy and mitochondria-mediated apoptosis (8–16). Because the remodeling of CL is deficient in BTHS, biochemical abnormalities in patients include a decreased level of mature CL (CLm), an increased level of monolysocardiolipin (MLCL), and altered CL acyl composition [i.e., the presence of immature CL (CLi) species].
Historically, the diagnosis of BTHS has relied on identification of the clinical symptoms accompanied by neutropenia and 3-MGCA and has then been confirmed by the finding of TAZ mutations. More recently, assays of CL alone or of the ratio of CL to MLCL have been used in Europe. However, each of these diagnostic approaches has problems. There are multiple reports of false-negative disease detection by urinary organic acid screening (4). TAZ sequencing is relatively slow and expensive, and CL analysis involves many steps including extraction of lipids and isolation of CL molecular species by complex chromatographic techniques. Consequently, although measurement of CL/MLCL ratio has 100% diagnostic sensitivity and specificity, it is only available in a few clinical laboratories worldwide (4). This forms a critical barrier to diagnosis in a disease that a major Australian study suggested was responsible for 4.8% of all cases of cardiomyopathy in male children (17). Consequently, in September 2014, only 175 males are known to be living with disease worldwide (data courtesy Barth Syndrome Foundation USA).
Up to now, CL and MLCL analyses in BTHS, with or without diagnostic intent, have been set up on lymphoblasts (18), lymphocytes (19), platelets (20–22), fibroblasts (23, 24), neutrophils (25), leukocytes (26), bloodspots (27), and tissues (19). All these reports, including the validated methods (26, 27), are based on lipid extraction followed by HPLC and ESI/MS analysis.
MALDI-TOF/MS currently represents a further valid tool in lipid analysis (28, 29). It has been shown that this emerging analytical technique can be used to directly acquire lipid profiles of biological samples, thus skipping extraction and separation steps (30–33). We successfully adopted this experimental approach to describe the lipid composition of bacterial and mitochondrial membranes (30, 31, 33) and to detect lipid markers in blood serum (32). Importantly, we found that the phosphatidylcholine-to-lysophosphatidylcholine ratio could be easily determined by MALDI-TOF/MS analyses of native blood serum and followed as a clinical parameter in various diseases (32). Relevantly, we also reported that CL could be easily detected in direct MALDI-TOF/MS total lipid profiles, by using 9-aminoacridine (9-AA) as matrix (31), even in the lipid fingerprint of whole cells.
On these bases, we developed a MALDI-TOF/MS method of lipid analysis of intact white blood cells for the clinical monitoring of the MLCL and CL relative levels in BTHS. Moreover, using the method described by Schlame et al. (34), we used vector algebra to evaluate the differences in CL and MLCL molecular composition of BTHS patients compared with controls.
In our method, both blood volume and time required for the analysis can be reduced because lipid extraction and separation can be skipped, simplifying the test. To guarantee the gaining of an informative lipid fingerprint even when using low-performance MALDI-TOF/MS instruments, we developed a simple mini-lipid-extraction protocol of minute amounts of white blood cell suspension; this additional minor step requires only 10–15 min more, significantly increases the signal-to-noise ratio, and increases the resolution of mass spectrometric peaks.
We believe that a rapid, less invasive, and novel screening test to monitor CL and its derivatives in blood could be of help in the diagnosis and has the potential for therapeutic monitoring of BTHS.
MATERIALS AND METHODS
Biological samples
Blood samples from 24 healthy donors (all males, average age 38.5 years) and 8 BTHS patients (all males, average age 10.8 years) were used in the study. In addition, a patient with a recently described ameliorated phenotype has been also analyzed (35). Samples of healthy donors were obtained with classical blood withdrawal according to institutional guidelines and anonymization, at the Blood Bank of the Policlinic Hospital of Bari. Samples of BTHS patients were obtained during routine venesection for clinical monitoring in patients seen for annual review by the National Health Service national United Kingdom BTHS clinic at Bristol Royal Hospital for Children. Written informed consent of healthy donors, parents, and patients (where appropriate) and approvals by the respective ethical committees were obtained.
Isolation of white blood cells
Red blood cells (RBCs) were sedimented with the dextran polymer (molecular mass >100 kDa), then hypotonic lysis was applied to eliminate residual RBCs. Dextran sedimentation was performed as follows: 0.111 ml of 20% dextran solution (molecular mass >100 kDa, in 0.9% NaCl) was added to 1 ml of whole blood in a 1.5 ml tube. The suspension was pipetted and dispersed gently 20 times with an automatic pipette avoiding air bubbles, which retain RBCs at the top of the tube. RBCs were sedimented for 1 hour at room temperature. Then the yellow supernatant was collected with a syringe, transferred to a 15 ml tube, and then centrifuged at 400 g for 15 min at room temperature (no brake, swing-bucket rotor).
Hypotonic lysis of residual RBCs was performed as follows: after the last centrifugation, the supernatant was discarded, and the pellet obtained was resuspended in 0.6 ml of iced bi-distilled water (ddH2O). After 10–15 s, 0.2 ml of 0.6 M KCl was added to the cell suspension to restore the correct osmolarity, and then the final volume was adjusted to 2.5 ml with PBS, containing 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM K2HPO4, pH 7.4. The suspension was then centrifuged at 400 g for 15 min at room temperature (no brake, swing-bucket rotor). The supernatant was sucked and discarded. The remaining leukocyte pellet was washed again with 2.5 ml PBS, recollected, resuspended in 200 μl of ddH2O, and immediately frozen at −80°C (see Fig. 1). Unless otherwise specified, lipid analyses performed on frozen samples are reported in the present study.
Fig. 1.
Work flow chart (left) and graphical abstract (right). From the blood withdrawal to the test results, only 2 h are necessary. MALDI-TOF/MS-based BTHS diagnosis in four steps: 1) native sample deposition, 2) matrix layering, 3) laser desorption and ionization, and 4) lipid fingerprint analysis.
In our experience, in these conditions, the total protein concentration was >0.9 and <1.8 μg/μl, measured by Bradford assay (Bio-Rad Protein Assay Kit; BioRad Laboratories, Germany).
Miniextraction of lipids from leukocytes
We set up a modified lipid extraction protocol (the “miniextraction” method) applicable to a very small quantity of white blood cell suspension. This quick procedure was performed as follows: the leukocyte suspension was aliquoted, and the equivalent volume for 20 μg of protein was transferred to a tube and spun at 16,000 g for 30 s. The supernatant was pipetted and discarded, and 10 μl of CHCl3 was added to the remaining pellet. The pellet was pipetted and dispersed repeatedly, favoring lipid extraction. Then, 10 μl of the matrix solution (10 mg/ml 9-AA in 2-propanol-acetonitrile, 60:40, v/v) was added to the pellet in CHCl3 and mixed. The pellet was again pipetted and dispersed repeatedly. After centrifugation at 16,000 g for 30 s, the supernatant was deposited in droplets of 0.35 μl on the MALDI target (sample plate) to be analyzed.
MALDI-TOF/MS
Analysis of intact leukocyte membranes.
After white blood cell isolation, the MALDI-TOF/MS lipid profiles of intact cells can be achieved directly. Intact samples, prepared as described in Isolation of White Blood Cells, were deposited on the MALDI target with a “double layer” deposition method as follows: a 1 μl droplet of the leukocyte suspension, diluted in distilled water to a concentration of 0.5 μg/μl, was deposited on the MALDI target and dried under a cold air stream (first layer). The resultant solid deposition was then covered by a thin second layer (0.4 μl droplet) of the 9-AA matrix solution (30 mg/ml in 2-propanol-acetonitrile, 60:40, v/v). After solvent evaporation, the sample could be analyzed.
Analysis of miniature lipid extracts.
The supernatant obtained at the end of the lipid-mini-extraction procedure, containing lipids and 9-AA as matrix, was deposited on the MALDI target (0.35 μl, “dried droplet” deposition method).
The main analytical steps of our method are schematically reported in the flow chart of method in Fig. 1.
MS settings.
MALDI-TOF mass spectra were acquired on a Bruker Microflex RLF mass spectrometer (Bruker, Germany) or alternatively on a Bruker Autoflex II, referred to as either Microflex or Autoflex, respectively. Both systems utilize a pulsed nitrogen laser, emitting at 337 nm; the extraction voltage is 20 kV, and gated matrix suppression is applied to prevent detector saturation (up to 400 Th on the Microflex and up to 900 Th on the Autoflex instrument). The laser fluence is kept ∼5% above threshold (of CL and MLCL) to have a good signal-to-noise ratio. All spectra were acquired in the reflector mode using delayed pulsed extraction; only spectra acquired in negative ion mode are shown in this study. Baseline correction, peaks areas, intensities, spectral mass resolutions, and signal-to-noise ratios were determined by the instrument software “Flex Analysis 3.0” (Bruker Daltonics, Bremen, Germany). Smoothing was obtained by applying the SavitskyGofay algorithm; baseline subtraction was achieved by selecting the TopHat algorithm.
The Bruker Autoflex has a stronger laser fluence and a longer TOF than the Microflex, resulting in a higher resolution and sensitivity. Unless otherwise specified, most of the data included in this study were obtained with the Microflex instrument.
(MLCL + CLi)/CLm ratio as diagnostic parameter
Defects in the TAZ gene typically determine the appearance of MLCL and CLi forms together with the reduction of CLm forms in the CL fingerprint. In the lipid profile of fibroblasts of healthy donors, two CLm species are typically present.
We considered the (MLCL + CLi)/CLm defined as the sum of the values of the mass spectrometric peak area of MLCL species and of CLi species on the value of the mass spectrometric peaks area of CLm species detected in the same negative MALDI-TOF mass spectrum of a given sample. In the practice to calculate the ratio, only the first isotopologue of the following species were considered: MLCL 52:2 at m/z 1,165.8 and MLCL 54:1 at m/z 1,191.8; CLi 68:2 at m/z 1,404.0 and CLi 70:3 at m/z 1,430.0; CLm 72:8 at m/z 1,447.9 and CLm 72:7 at m/z 1,449.9. In addition, a correction for the overlapping between the M + 2 isotopologue of the CLm 72:8 and the monoisotopic peak of CLm 72:7 has been introduced. In brief, the X + 2 factor of carbon relates to the probability of the ion having two atoms of 13C. This probability is much smaller than the probability that an ion will have a single atom of 13C. Consider the binomial expression (a + b)n, where a is the percent abundance of the most abundant isotope, b is the abundance of the second isotope of an X + 1 element, and n is the number of atoms of the X + 1 element. Solving this expansion results in an X + 2 factor for the probability of having two atoms of carbon being 0.0060 times the square of the number of carbon atoms (0.006 × nC2). Carbon is usually the most ubiquitous element in ions encountered in organic mass spectrometry; therefore, based on the large numbers of carbon atoms that may be present and the abundance of 13C, the X + 2 factor for carbon can be significant. For a CL having 72 carbons, the X + 2 factor is 31.1%. In our calculations, we therefore subtracted the 31.1% of the peak area of the CL (18:2)4 from the peak area of the CL (18:2)3(18:1)1.
For a detailed list of assigned CL and MLCL species present in the lipid fingerprints in this study, see Table 1. Sodium adducts were not taken in consideration because their presence was negligible in the lipid profiles considered in the present study. In measurements on intact cells, their presence was minimized by the double wash applied to the final pellet. In measurements on lipid miniextracts, when samples contained high amounts of sodium adducts, their abundance was reduced by washing the dried sample/matrix spot with cold water. For this purpose, 2 μl of ice-cold water was placed on top of the spot. After a few seconds, the water was sucked up with a small piece of filter paper. The wash was repeated once.
TABLE 1.
List of detected peaks of interest in the MALDI-TOF mass spectra assigned to CL and MLCL species
| m/z [M-H]− | Assignment |
| 1,165.8 | MLCL 52:2 |
| 1,191.8 | MLCL 54:1 |
| 1,404.0 | CLi 68:2 |
| 1,430.0 | CLi 70:3 |
| 1,448.0 | CLm 72:8 |
| 1,450.0 | CLm 72:7 |
| 1,456.0 | CLi 72:4 |
To speed up data analysis, we set up an algorithm in MATLAB 7.0 (The MathWorks Inc.) that automatically elaborates MS spectra exported as (x,y) coordinates in a ASCII file (*.txt). The algorithm can read the exported spectra relative to each subject and calculate the (MLCL + CLi)/CLm ratio as defined. The algorithm is described in detail in supplementary Note 1.
Compositional distance
According to Schlame et al. (34), we used vector algebra to compare CL and MLCL molecular compositions of leukocytes from patients and healthy controls considered in our study, as described in the following. By using the statistical analysis of compositional data, we compared the different molecular composition, measuring not only the quality of the differences but also their magnitude. To make this comparison, CL and MLCL molecular composition of each subject (MA, MB, etc.) is considered as a point vector (M) in an n-dimensional space, where n is the number of the molecular species considered. We considered two MLCL species, two CLm species, and two CLi species, listed in Table 1. Each point vector M, therefore, has six components in the different dimensions M = (m1, m2, m3, m4, m5, m6). The module of these components is given by the signal area of its relative CL or MLCL species in the lipid profile of the considered subject (average value in at least three spectra). To normalize data, we imposed the condition that the sum of the different components mi must be equal to 1:
The difference between two molecular compositions MA and MB, can therefore be evaluated as the distance between their respective point vectors (i.e., compositional distance), which is given by the Pythagorean theorem in multiple dimensions:
Because mi components are normalized fractional intensities, in our conditions the compositional distance can vary from 0 to .
We set up another MATLAB algorithm that can automatically elaborate exported MS spectra and calculate the compositional distance of a suspected patient from the control group, leading to a fast diagnosis. The algorithm is described in detail in supplementary Note 2.
RESULTS
CL fingerprint of healthy donors and BTHS patients
Here we report lipid profiles from control leukocytes obtained by the blood bank of the main hospital in Bari (Italy) and of BTHS patients from Bristol Royal Hospital for Children (United Kingdom). Both groups of the samples were prepared by the method described in Materials and Methods. Analyses were performed on frozen samples kept at −80°C for up to 6 weeks. In preliminary experiments, we found no differences in the lipid profiles of samples stored for 2, 4, or 6 weeks (not shown).
Figure 2 shows the comparison of representative MALDI-TOF/MS lipid fingerprint of intact leukocytes, obtained from control subjects and BTHS-affected boys, in the CL and MLCL mass (m/z) range (i.e., CL fingerprint). CL and MLCL species detected in these mass spectra are listed in Table 1.
Fig. 2.
BTHS diagnostic leukocyte CL fingerprints. Lipid profiles of intact leukocytes of a healthy control (CTRL, upper) and a representative BTHS patient (BTHS, lower) are shown. Peaks of CL and MLCL species considered in this study are indicated (in red). The CLm form is highlighted in turquoise. Peaks labeled with an asterisk are assigned to gangliosides and are not of interest for the present study. Spectra were acquired with the Bruker Microflex RLF mass spectrometer.
In the upper panel of Fig. 2, a representative CL fingerprint of a healthy donor is shown. The only cluster of peaks of interest present in this spectrum belongs to the CLm species, tetralinoleoyl CL (18:2)4 at m/z 1,448.0 and CL (18:2)3(18:1)1 at m/z 1,450.0. Therefore, it is evident that two species of CLm are typically present in the lipid profile of leukocytes of healthy donors: the tetralinoleoyl CL plus the CL carrying three linoleoyl fatty acid chains and one oleoyl fatty acid chain; in our method of analyses, the two species are easily recognized and together considered in the healthy phenotype.
In the lower panel of Fig. 2, the representative CL fingerprint of a boy affected by BTHS shows a different pattern of peaks compared with controls. The CLm species are reduced and present together with a number of different CLi species, listed and assigned in Table 1. Peaks of MLCL species at m/z 1,165.8 and 1,191.8 appear in parallel. Please note that peaks labeled with an asterisk are assigned to gangliosides, not relevant to the aims of our study. The structures of the molecules corresponding to the peaks at m/z 1,230.5 and 1,263.9 have been elucidated by post-source decay mass spectrometry analyses (not shown).
In summary, data in Fig. 2 show that in BTHS patients TAZ gene mutation causes reduction of CLm level and consequent accumulation of both MLCL and CLi species (see also Fig. 3). It is therefore evident that CL and MLCL species can be revealed by direct MALDI-TOF/MS analyses of intact leukocytes, thus avoiding cell fractionation and lipid separation.
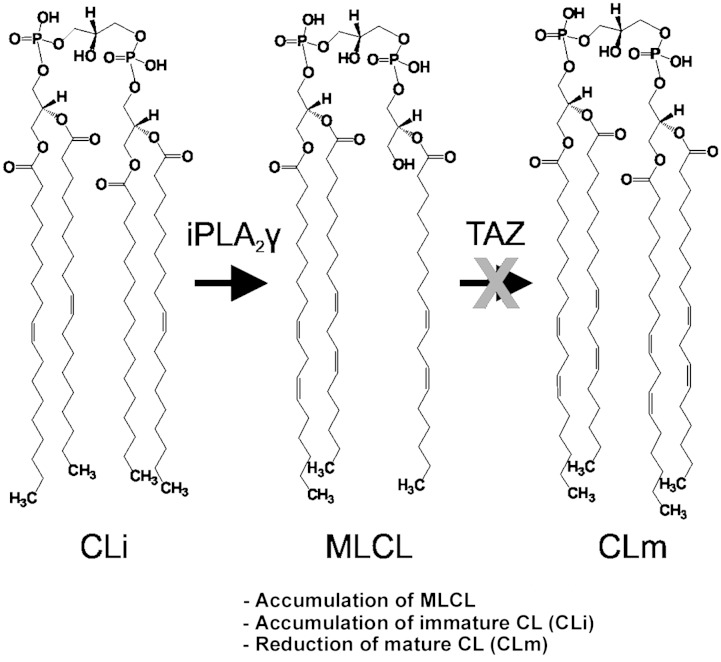
Fig. 3.
Remodeling enzymatic pathway of CL. iPLA2γ, calcium-independent phospholipase A2 γ.
An ameliorated form of BTHS has been characterized for the first time by Bowron et al. (35), who described seven BTHS patients from three families with TAZ mutations. One of these patients has been also examined with our method. The MALDI-TOF/MS CL fingerprint of the patient without CLm deficiency is reported in supplementary Fig. 1. Compared with the CL fingerprints shown in Fig. 2, it can be seen that in the variant form of BTHS peaks belonging to CLm species (see Table 1) are lower than in the control fingerprint (Fig. 2, upper spectrum) and have about the same intensities of the CLi species. Moreover, peaks of MLCLs are present at very low concentrations in the fingerprint of this variant form of BTHS. Indeed the diagnosis of the ameliorated phenotypes is based mainly on the appearance of CLi species.
The difference in the lipid profile may be associated with the different clinical features seen in this group of patients. They appear to have a milder form of BTHS: most do not have skeletal myopathy, and none has neutropenia, although in one family three boys died in infancy indicating the possibility of both ameliorated and severe disease within the same family (35). Two subjects with genetically confirmed BTHS from this group had their diagnosis missed in error due to measurement of only CL, emphasizing the importance of including either CLi or MLCL species when setting up a diagnostic test for BTHS. It is of interest that this group of BTHS patients with TAZ mutations does not have the severe CLm deficiency characteristic of BTHS, but it does have some CLi and MLCL, which distinguishes these patients from controls and may indicate a role for these abnormal forms of CL in the pathogenesis of the disorder (35).
(MLCL + CLi)/CLm ratio as diagnostic parameter
To validate the new diagnostic method, we determined the relative levels of CL and MLCL species in leukocytes from each subject in the study. Here we show that CL (and MLCL) MALDI-TOF/MS fingerprint can be used to recognize BTHS-affected individuals. Lipid profiles of intact leukocytes and corresponding lipid miniextracts have been analyzed in parallel to validate the method; the CL fingerprints of either intact leukocytes and their respective lipid extracts of all BTHS patients are compared with those of some healthy controls and reported in supplementary Fig. 2A, B, respectively.
It should be considered that not only the presence of MLCL peaks is diagnostic for BTHS but also the presence of CLi species is typical of this disease. Therefore, in the present study we introduced the (MLCL + CLi)/CLm ratio as diagnostic parameter to identify BTHS pathological conditions. Calculated MLCL + CLi/CLm ratios in patients with BTHS and controls are shown in Fig. 4. The ratio has been calculated by considering the area of CL and MLCL peaks in lipid profiles of leukocytes from 24 healthy donors and 8 BTHS-affected boys; MALDI-TOF/MS lipid profiles of either intact leukocytes or their respective lipid extracts have been considered (Fig. 4). Fig. 4A shows that using this novel parameter it is possible to distinguish BTHS patients from controls. Groups of controls and patients are better separated in Fig. 4B, which reports lipid ratios, calculated from areas of peaks belonging to the lipid species of interest, in the miniextracts of control and BTHS leukocytes. Therefore, it is evident that the addition of the miniextraction step can be critical to gain diagnostic power with a low-performance instrument such as the Bruker Microflex RLF mass spectrometer. On the other hand, Fig. 4C shows that acquisition of MALDI-TOF/MS lipid fingerprint of intact leukocytes can ensure the distinction between all BTHS patients and controls when a high-performance mass spectrometer such as the Bruker Autoflex II is used. A representative example of CL fingerprints of control and BTHS patient acquired with Autoflex mass spectrometer is shown in supplementary Fig. 3. Characteristics of these instruments are described in Materials and Methods. Reference cutoff values have been calculated and are indicated in the legend of Fig. 4.
Fig. 4.
(MLCL + CLi)/CLm ratios in controls and BTHS patients. Error bars indicate SDs obtained in measurements of three spectra for each subject [24 healthy donors as controls (CTRL) and 8 patients (BTHS)]. Ratios have been calculated from CL fingerprints of either intact leukocytes (intact) or after the addition of organic solvents to quickly solubilize lipids (extract), as described in Materials and Methods. A and B: Results obtained by using the low-performance Bruker Microflex RLF mass spectrometer. C: Results obtained by Bruker Autoflex II, with the same number of patients’ samples but with a reduced number of healthy controls. High-p. MS, high-performance mass spectrometry. The y-axis shows a logarithmic scale. Upper cutoff values of controls, calculated as in Bowron et al. (26), are 4.21 for A, 0.64 for B, and 1.16 for C.
In conclusion (MLCL + CLi)/CLm ratio calculated from MALDI-TOF/MS lipid fingerprint of whole leukocytes is a valid diagnostic parameter to distinguish between controls and BTHS patients. We have also shown that our analytical approach can be used even with a low-performance mass spectrometer simply by adding the minor miniextraction step.
Compositional distance as diagnostic parameter
We also used vector algebra to evaluate the effect of the different TAZ mutations on CL and MLCL molecular composition. The statistical approach used is described in Materials and Methods. Briefly, each different molecular composition is considered as a vector M, and distances between the different vectors (i.e., the compositional distance) are used to evaluate differences in CL and MLCL molecular composition of the different subjects in the study. In Fig. 5, the compositional distances have been plotted. In the chart, healthy controls have been considered as a unique group and represented as a single point (± SD). For this control group, the compositional distance is the average value of the distance between the molecular components of each control subject from all the others. It should not be far from zero because healthy donors have very similar lipid fingerprints, which can be easily verified in the chart. BTHS patients have very similar lipid fingerprints and are therefore considered as a unique group and represented as a single point (± SD). We calculated the distances between the vectors representing molecular compositions of patients and the vectors representing control compositions and then plotted the average value. This statistical approach has been applied on the same group of spectra considered in Fig. 4.
Fig. 5.
Compositional distances of CL fingerprints of controls and BTHS patients. The same samples considered in Fig. 4 have been used to estimate differences in molecular composition of CL and its lysoderivative species. Average values ± SDs are provided for controls (CTRL) and typical BTHS patients (BTHS). A and B: Results obtained by using the low-performance Bruker Microflex RLF mass spectrometer. C: Results obtained by Bruker Autoflex II. High-p. MS, high-performance mass spectrometry.
Analyzing intact leukocytes by the Bruker Microflex RLF mass spectrometer, BTHS patients appeared sufficiently “distant” from controls, meaning that the CL and MLCL molecular composition of patients is sufficiently different from controls to achieve their diagnosis (Fig. 5A). Nevertheless, with the same instrument, BTHS patients can be neatly distinguished from controls when considering MALDI-TOF mass spectra of the corresponding lipid extracts (Fig. 5B).
Finally, a good separation between BTHS patients and controls was obtained by using spectra of intact leukocytes acquired by the Bruker Autoflex II mass spectrometer (Fig. 5C).
DISCUSSION
Diagnosis of BTHS is difficult. Unambiguous diagnostic testing for BTHS can be performed by determination of the relative amounts and distribution of (monolyso-)CL species and confirmed by TAZ gene sequencing or vice versa. Unfortunately, molecular analysis of the TAZ gene can lead to false-negative results when mutations are present in regulating or relevant noncoding sequences; in addition, more than 120 distinct TAZ mutations have been described (4, 5, 36).
Furthermore, current lipidomic-based diagnostic tests require relevant experience in HPLC-ESI/MS analysis. Nowadays, MALDI-TOF mass spectrometers are widely distributed among clinical laboratories worldwide and do not require high analytical expertise. We have described a simple and fast novel method for measurement of CL in intact leukocytes by MALDI-TOF/MS that requires only 1 ml of blood. As lipid analyses of intact leukocyte membranes may result in poorly resolved spectra, as in the case of BTHS 7 and 8 patients in supplementary Fig. 2A, we have developed a fast method to solubilize lipids and gain a better resolving power. In general, when the signal-to-noise ratio of the mass spectrum of intact leukocytes is ≤3, the introduction of the additional step of the miniextract is highly recommended.
The main advantage of the present method over existing approaches is that it can simultaneously detect diagnostic CL and MLCL species in the total lipid profile by a single run of mass spectrometric analysis immediately after isolation of leukocytes.
Although shotgun lipidomic ESI/MS analysis and quantitation of cellular lipidomes can be directly performed on crude extracts of various biological samples (37), to the best of our knowledge, this approach has not yet been used to study CL profiles of whole white blood cells. A direct infusion-based ESI/MS approach for gaining a CL fingerprint has been previously used to study lipids of the heart tissue of a BTHS animal model (38).
The diagnosis for BTHS patients here described not only relies on the decrease of CLm, but also considers the presence of CLi species, which together with the increase of MLCL suggest a defect in the CL metabolism, determined by TAZ mutations. Even in the case in which low levels of MLCL are found in patients, still the appearance of the immature species should alert the biologist or physician to the possibility of the presence of a serious disease. Indeed MALDI-TOF/MS allows the direct analysis of native samples, skipping time-consuming steps of standard lipid extraction and separation procedures. The success of our experimental approach is due to the use of 9-AA as matrix, which is particularly suitable for CLs detection (31), even when these species are only minor components in complex lipid profiles.
In conclusion, the method described here is straightforward to perform and can be easily integrated into the routine work of a clinical biochemistry laboratory. We hope that this novel simplified test, using a different analytical approach, could increase the number of laboratories capable of diagnosing BTHS with the consequent identification of new hidden cases.
Supplementary Material
Acknowledgments
The authors thank the personnel of the Blood Bank of the Policlinic of Bari (Italy) for their kind collaboration, Jürgen Schiller (University of Leipzig, Germany) for the use of the Bruker Autoflex II mass spectrometer and for many inspiring discussions, Leonardo Angelini for help in developing the MATLAB algorithm, Jörg Flemmig in leukocyte isolation, and Roberto Letizia for graphical advice.
Footnotes
Abbreviations:
- 9-AA
- 9-aminoacridine
- BTHS
- Barth syndrome
- CL
- cardiolipin
- CLi
- immature cardiolipin
- CLm
- mature cardiolipin
- MLCL
- monolysocardiolipin
- RBC
- red blood cell
- TAZ
- tafazzin
This work was supported by a USA Barth Syndrome Foundation grant (A.C.). The authors are indebted to the patients and parents attending the National Health Service National Barth Syndrome Clinic at Bristol Royal Hospital for Children who gave permission for taking of the blood samples used in the development of this assay and to the Barth Syndrome Trust (United Kingdom) and Association Barth France for providing support.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Steward C. G., Martin R. P., Hayes A. M., Salmon A. P., Williams M. M., Tyfield L. A., Davies S. J., Newbury-Ecob R. A. 2004. Barth syndrome (X linked cardiac and skeletal myopathy, neutropenia, and organic aciduria): rarely recognised, frequently fatal [abstract]. Arch. Dis. Child. 89: A48. Abstract G140. [Google Scholar]
- 2.Barth P. G., Scholte H. R., Berden J. A., Van der Klei-Van Moorsel J. M., Luyt-Houwen I. E., Van ’t Veer-Korthof E. T., Van der Harten J. J., Sobotka-Plojhar M. A. 1983. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J. Neurol. Sci. 62: 327–355. [DOI] [PubMed] [Google Scholar]
- 3.Barth P. G., Van den Bogert C., Bolhuis P. A., Scholte H. R., van Gennip A. H., Schutgens R. B., Ketel A. G. 1996. X-linked cardioskeletal myopathy and neutropenia (Barth syndrome): respiratory-chain abnormalities in cultured fibroblasts. J. Inherit. Metab. Dis. 19: 157–160. [DOI] [PubMed] [Google Scholar]
- 4.Clarke S. L., Bowron A., Gonzalez I. L., Groves S. J., Newbury-Ecob R., Clayton N., Martin R. P., Tsai-Goodman B., Garratt V., Ashworth M., et al. 2013. Barth syndrome. Orphanet J. Rare Dis. 8: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whited K., Baile M. G., Currier P., Claypool S. M. 2013. Seven functional classes of Barth Syndrome mutation. Hum. Mol. Genet. 22: 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vreken P., Valianpour F., Nijtmans L. G., Grivell L. A., Plecko B., Wanders R. J., Barth P. G. 2000. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem. Biophys. Res. Commun. 279: 378–382. [DOI] [PubMed] [Google Scholar]
- 7.Schlame M., Ren M. 2006. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 580: 5450–5455. [DOI] [PubMed] [Google Scholar]
- 8.Mileykovskaya E., Dowhan W. 2009. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim. Biophys. Acta. 1788: 2084–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claypool S. M., Koehler C. M. 2012. The complexity of cardiolipin in health and disease. Trends Biochem. Sci. 37: 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlame M., Rua D., Greenberg M. L. 2000. The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 39: 257–288. [DOI] [PubMed] [Google Scholar]
- 11.Gohil V. M., Hayes P., Matsuyama S., Schägger H., Schlame M., Greenberg M. L. 2004. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. J. Biol. Chem. 279: 42612–42618. [DOI] [PubMed] [Google Scholar]
- 12.Haines T. H., Dencher N. A. 2002. Cardiolipin: a proton trap for oxidative phosphorylation. FEBS Lett. 528: 35–39. [DOI] [PubMed] [Google Scholar]
- 13.Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., Schagger H. 2003. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278: 52873–52880. [DOI] [PubMed] [Google Scholar]
- 14.Claypool S. M., Boontheung P., McCaffery J. M., Loo J. A., Koehler C. M. 2008. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Mol. Biol. Cell. 19: 5143–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu F. Y., McBride H., Acehan D., Vaz F. M., Houtkooper R. H., Lee R. M., Mowat M. A., Hatch G. M. 2010. The dynamics of cardiolipin synthesis post mitochondrial fusion. Biochim. Biophys. Acta. 1798: 1577–1585. [DOI] [PubMed] [Google Scholar]
- 16.Kagan V. E., Chu C. T., Tyurina Y. Y., Cheikhi A., Bayir H. 2014. Cardiolipin asymmetry, oxidation and signaling. Chem. Phys. Lipids. 179: 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nugent A. W., Daubeney P. E., Chondros P., Carlin J. B., Cheung M., Wilkinson L. C., Davis A. M., Kahler S. G., Chow C. W., Wilkinson J. L., et al. 2003. The epidemiology of childhood cardiomyopathy in Australia. N. Engl. J. Med. 348: 1639–1646. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y., Sutachan J. J., Plesken H., Kelley R. I., Schlame M. 2005. Characterization of lymphoblast mitochondria from patients with Barth syndrome. Lab. Invest. 85: 823–830. [DOI] [PubMed] [Google Scholar]
- 19.Houtkooper R. H., Rodenburg R. J., Thiels C., van Lenthe H., Stet F., Poll-The B. T., Stone J. E., Steward C. G., Wanders R. J., Smeitink J., et al. 2009. Cardiolipin and monolysocardiolipin analysis in fibroblasts, lymphocytes, and tissues using high-performance liquid chromatography-mass spectrometry as a diagnostic test for Barth syndrome. Anal. Biochem. 387: 230–237. [DOI] [PubMed] [Google Scholar]
- 20.Valianpour F., Wanders R. J., Barth P. G., Overmars H., van Gennip A. H. 2002. Quantitative and compositional study of cardiolipin in platelets by electrospray ionization mass spectrometry: application for the identification of Barth syndrome patients. Clin. Chem. 48: 1390–1397. [PubMed] [Google Scholar]
- 21.Schlame M., Towbin J. A., Heerdt P. M., Jehle R., Di Mauro S., Blanck T. J. 2002. Deficiency of tetralinoleoyl-cardiolipin in Barth syndrome. Ann. Neurol. 51: 634–637. [DOI] [PubMed] [Google Scholar]
- 22.Schlame M., Kelley R. I., Feigenbaum A., Towbin J. A., Heerdt P. M., Schieble T., Wanders R. J., DiMauro S., Blanck T. J. 2003. Phospholipid abnormalities in children with Barth syndrome. J. Am. Coll. Cardiol. 42: 1994–1999. [DOI] [PubMed] [Google Scholar]
- 23.Valianpour F., Wanders R. J., Overmars H., Vreken P., Van Gennip A. H., Baas F., Plecko B., Santer R., Becker K., Barth P. G. 2002. Cardiolipin deficiency in X-linked cardioskeletal myopathy and neutropenia (Barth syndrome, MIM 302060): a study in cultured skin fibroblasts. J. Pediatr. 141: 729–733. [DOI] [PubMed] [Google Scholar]
- 24.van Werkhoven M. A., Thorburn D. R., Gedeon A. K., Pitt J. J. 2006. Monolysocardiolipin in cultured fibroblasts is a sensitive and specific marker for Barth Syndrome. J. Lipid Res. 47: 2346–2351. [DOI] [PubMed] [Google Scholar]
- 25.Kuijpers T. W., Maianski N. A., Tool A. T., Becker K., Plecko B., Valianpour F., Wanders R. J., Pereira R., Van Hove J., Verhoeven A. J., et al. 2004. Neutrophils in Barth syndrome (BTHS) avidly bind annexin-V in the absence of apoptosis. Blood. 103: 3915–3923. [DOI] [PubMed] [Google Scholar]
- 26.Bowron A., Frost R., Powers V. E. C., Thomas P. H., Heales S. J. R., Steward C. G. 2013. Diagnosis of Barth syndrome using a novel LC-MS/MS method for leukocyte cardiolipin analysis. J. Inherit. Metab. Dis. 36: 741–746. [DOI] [PubMed] [Google Scholar]
- 27.Kulik W., van Lenthe H., Stet F. S., Houtkooper R. H., Kemp H., Stone J. E., Steward C. G., Wanders R. J., Vaz F. M. 2008. Bloodspot assay using HPLC-tandem mass spectrometry for detection of Barth syndrome. Clin. Chem. 54: 371–378. [DOI] [PubMed] [Google Scholar]
- 28.Schiller J., Suss R., Fuchs B., Muller M., Zschornig O., Arnold K. 2007. MALDI-TOF MS in lipidomics. Front. Biosci. 12: 2568–2579. [DOI] [PubMed] [Google Scholar]
- 29.Sun G., Yang Z., Zhao S., Guan X., Han R., Gross W. 2008. Matrix assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte-matrix interactions. Anal. Chem. 80: 7576–7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angelini R., Babudri F., Lobasso S., Corcelli A. 2010. MALDI-TOF/MS analysis of archaebacterial lipids in lyophilized membranes dry-mixed with 9-aminoacridine. J. Lipid Res. 51: 2818–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angelini R., Vitale R., Patil V. A., Cocco T., Ludwig B., Greenberg M. L., Corcelli A. 2012. Lipidomics of intact mitochondria by MALDI-TOF MS. J. Lipid Res. 53: 1417–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angelini R., Vormieter G., Corcelli A., Fuchs B. 2014. A fast method for the determination of PC/LPC ratio in intact horse serum by MALDI-TOF-MS: an easy-to-follow lipid biomarker of inflammation. Chem. Phys. Lipids. 183: 169–175. [DOI] [PubMed] [Google Scholar]
- 33.Vitale R., Angelini R., Lobasso S., Capitanio G., Ludwig B., Corcelli A. 2015. MALDI-TOF MS lipid profiles of cytochrome c oxidases: cardiolipin is not an essential component of the Paracoccus denitrificans oxidase. Biochemistry. 54: 1144–1150. [DOI] [PubMed] [Google Scholar]
- 34.Schlame M., Blais S., Edelman-Novemsky I., Xu Y., Montecillo F., Phoon C. K., Ren M., Neubert T. A. 2012. Comparison of cardiolipins from Drosophila strains with mutations in putative remodeling enzymes. Chem. Phys. Lipids. 165: 512–519. [DOI] [PubMed] [Google Scholar]
- 35.Bowron A., Honeychurch J., Williams M., Tsai-Goodman B., Clayton N., Jones L., Shortland G. J., Qureshi S. A., Heales S. J., Steward C. G. 2015. Barth syndrome without tetralinoleoyl cardiolipin deficiency: a possible ameliorated phenotype. J. Inherit. Metab. Dis. 38: 279–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferri L., Donati M. A., Funghini S., Malvagia S., Catarzi S., Lugli L., Ragni L., Bertini E., Vaz F. M., Cooper D. N., et al. 2013. New clinical and molecular insights on Barth syndrome. Orphanet J. Rare Dis. 8: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han X., Gross R. W. 2005. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 24: 367–412. [DOI] [PubMed] [Google Scholar]
- 38.Kiebish M. A., Yang K., Liu X., Mancuso D. J., Guan S., Zhao Z., Sims H. F., Cerqua R., Cade T. W., Han X., et al. 2013. Dysfunctional cardiac mitochondrial bioenergetic, lipidomic, and signaling in a murine model of Barth syndrome. J. Lipid Res. 54: 1312–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.