Abstract
With the first cancer-targeted microRNA drug, MRX34, a liposome-based miR-34 mimic, entering phase I clinical trial in patients with advanced hepatocellular carcinoma in April 2013, miRNA therapeutics are attracting special attention from both academia and biotechnology companies. Although to date the most studied non-coding RNAs (ncRNAs) are miRNAs, the importance of long non-coding RNAs (lncRNAs) is increasingly being recognized. Here we summarize the roles of miRNAs and lncRNAs in cancer, with a focus on the recently identified novel mechanisms of action, and discuss the current strategies in designing ncRNA-targeting therapeutics, as well as the associated challenges.
Keywords: drugs, microRNAs, long non-coding RNAs, therapy, cancer, diseases
Introduction
Non-coding RNAs (ncRNAs) comprise multiple classes of RNA transcripts that are not transcribed into proteins, but have been shown to regulate the transcription, stability or translation of protein-coding genes in the mammalian genome 1, 2. To date, the most studied ncRNAs are microRNAs (miRNAs, usually 19–24 nucleotide in length), although many other classes of experimentally identified ncRNAs with various lengths and characteristics have been reported in the literature (see Box 1).
BOX 1. Various types of non-coding RNAs.
miRNAs
MicroRNAs; they are 19–24 nucleotide-long, single-stranded RNAs, initially transcribed from the genome as primary miRNAs, and processed into precursor and mature forms through a biogenesis machinery including Drosha and Dicer. They exert biological functions by regulating transcription and/or translation of protein-coding genes.
endo-siRNAs
Endogenous small interfering RNAs; they are ~20 nucleotide-long, single-stranded RNAs processed from several sources of endogenous double stranded RNAs including transposable elements, cis-natural antisense transcripts, and trans-natural antisense transcripts and hairpin RNA transcripts. Endo-siRNAs load onto AGO2, and repress transposon transcripts or endogenous mRNAs 133.
piRNAs
PIWI-interacting RNAs; they are single-stranded RNAs with 21–35 nucleotides in length and play roles in germline transposon silencing and gametogenesis 134.
snoRNAs
Small nucleolar RNAs; they are 60 to 300 nucleotide-long RNAs playing roles in the modification of ribosomal RNAs 135.
sdRNAs
sno-derived RNAs; they are 20ey are RNAsNAs-long miRNA-like RNAs originating from H/ACA box snoRNAs, or ~17 NAsnt and >27 nt RNAs that derived from C/D box snoRNAs 136.
tiRNAs
Transcription initiation RNAs; they are nucleus-localized 18-nucleotide RNAs originated from sequences immediately downstream of RNA polymerase II transcription start site 137.
moRNAs
MicroRNA-offset-RNAs; they are produced from human microRNA precursors, but their expression levels are quite low and unrelated to those of associated microRNAs 138.
NATs
Natural antisense transcripts; they are endogenous RNA molecules with partial or full sequence complementarity to other transcripts. There are two types of NATs: cis-NATs are transcribed from the same genomic loci of their sense transcripts but from the opposite DNA strand, while trans-NATs are originated from distinct genomic regions from those encoding their targeted sense transcripts 105, 139.
CircRNAs
Circular RNAs; they are endogenous RNAs with covalently linked ends which can serve as miRNA sponges. One example is that a ~1500 nucleotide-long circRNA, predominately located in human and mouse brain, contains multiple binding sites for miR-7 and sequesters this miRNA 103, 140.
lncRNAs
Long non-coding RNAs; they are non-protein-coding RNA transcripts longer than 200 nucleotides with multiple functions and mechanisms of action 1, 12.
lincRNAs
Long intergenic non-coding RNAs; they are lncRNAs transcribed from non-coding DNA sequences in between protein-coding genes 141. They represent a category of lncRNAs.
long enhancer ncRNAs
lncRNAs with enhancer-like function regulating neighboring protein-coding genes 107, 142.
T-UCRs
Transcribed Ultraconserved Regions; they are lncRNAs with significant biological function that interact with microRNAs and overlap the genomic ultraconserved regions 106, 117.
Since the first demonstration of the involvement of miRNAs in the development of cancer a decade ago 3, the role of miRNAs in multiple human diseases has been intensively investigated 4. During the last decade, over 25,000 papers deposited on PubMed report on various aspects of miRNA genomics, biogenesis, mechanisms of action, pathway involvement, phenotypes in experimental models and disease abnormalities. About 40% of these publications are dedicated to the role of miRNAs in cancer. These studies unveiled a novel mechanism of posttranscriptional regulation that is profoundly altered in malignant cells 5. Using high throughput techniques such as expression microarrays or next generation sequencing it was shown that miRNAs are dysregulated in almost all human types of cancer 6, 7, and specific signatures of aberrantly expressed miRNAs harbor diagnostic, prognostic and theranostic implications 8–11. In addition, miRNA expression patterns allow an accurate discrimination between different types of cancer, and the identification of the tissue of origins of poorly differentiated tumors 6. More recently, other types of ncRNAs, such as lncRNAs, were found to play dynamic roles in transcriptional and translational regulation 1, and to be involved in a variety of human diseases including cancer 8, 12. These findings suggest the functional participation of multiple types of ncRNAs in normal physiological activities and disease phenotypes (Table 1). The fact that about 75% of the human genome is transcribed into RNA, while only 3% is transcribed into protein-coding mRNAs 13–15, indicates that the number of ncRNAs is potentially much higher than that of protein-coding genes. Therefore, ncRNAs could represent goldmines for basic research, biomarker discovery and therapeutic applications 10, 16–21.
Table 1.
Examples of non-coding RNA involvement in cancer*.
| lncRNA or miRNA | Type (with genomic location) | Cancer Involvement | Mechanism of action (examples of target genes) | Refs |
|---|---|---|---|---|
| miR-10b | miRNA (chr #2) | Breast cancer | Promotes breast cancer metastasis (HOXD10) | 158 |
| miR-15a/16-1 | miRNA cluster (chr #13) | Downregulated in chronic lymphocytic leukemia, diffuse large B-cell lymphoma, multiple myeloma, prostate and pancreatic cancers | Induce apoptosis in leukemia cells and regulate cell cycle (BCL2, CCND1, CDK6, DMTF1, MCL1, VEGF, P53) | 3, 54, 159 |
| miR-17-92 | miRNA cluster (chr #13) | Overexpression in lung and colon cancer, lymphoma, multiple myeloma, medulloblastoma | Increases tumor growth and tumor vascularization (BIM1, CDKN1A, E2F1, E2F2, E2F3, HIF-1A, PTEN, TGFBR2) | 7, 160, 161 |
| miR-21 | miRNA (chr #17) | Overexpression in glioblastoma, breast, lung, prostate, colon, stomach, esophageal and cervical carcinomas, and diffuse large B-cell lymphoma | Promotes invasion and metastasis in colorectal cancers; knockdown induces apoptosis in glioblastoma cells (BCL2, MASPIN, PDCD4, PTEN, TPM1, RECK) | 37, 38, 40, 162, 163 |
| miR-22 | miRNA (chr #17) | Breast cancer | Regulate breast Cancer Stemness and Metastasis (TET) | 45 |
| miR-31 | miRNA (chr #9) | Breast cancer | Inhibit breast cancer metastasis (RhoA) | 83 |
| miR-34a, b, c | miRNA family (chrs #1 and 11) | Downregulated in pancreatic cancer, Burkitt’s lymphoma without c-Myc translocation, CD44+ prostate cancer, and human primary breast tumors with lymph node metastases. | Transcriptionally activated by p53; miR-34a inhibits prostate cancer stemness and metastasis; miR-34a, b, and c suppress breast cancer cell invasion and metastasis (BCL2, CCND1, CCNE2, CDK4,MYC, MYCN, MET, HMGA2, SIRT1, CD44, Fra-1) | 164–168 |
| miR-155 | miRNA (chr #21) | Overexpression in pediatric Burkitt’s lymphoma, Hodgkin’s disease, diffuse large B- cell lymphoma and in breast, lung, colon, and pancreatic cancers. | Pre-B cell proliferation and lymphoblastic leukemia/high- grade lymphoma in miR-155 transgenic mice (AID, TP53INP1) | 42, 169 |
| miR-335 | miRNA (chr #7) | Breast cancers | Inhibit breast cancer metastasis (SOX4, Tenascin-C) | 170 |
| miR-373/520c | miRNA (chr #19) | Breast cancer | Promote migration and invasion of breast cancer cells in vitro and in vivo (CD44) | 171 |
| Let-7 family | miRNA family (multiple locations) | Suppressor - Downregulation in lung, breast, gastric, ovary, prostate and colon cancers, and chronic lymphocytic leukemia. Oncogenic - Overexpression in acute myeloid leukemia |
Suppressor - Represses cell proliferation/growth (CCND1, CDK6, HOXA9, MYC, RAS, TLR4) Oncogenic - Let-7a represses NF2 and decreases chemotherapy-induced apoptosis in vitro (CASP3) |
172–174 |
| Bc200 | lncRNA (chr #2) | Multiple cancers | Protein binding | 175, 176 |
| HOTAIR | lncRNA (chr #12) | Multiple cancers - Promote breast cancer metastasis | Reprogram chromatin state - Epigenetic regulation (HOXD10) | 108, 109 |
| MALAT-1 | lncRNA (chr #11) | Multiple cancers | RNA splicing, small RNA production, protein interaction | 115, 177, 178 |
| PCA3/DD3 | lncRNA (chr #9) | Upregulated in prostate cancer | Modulates androgen receptor signaling | 179 |
| PTENP1 | pseudogene (chr #9) | Downregulated in prostate cancer | Increase PTEN expression via miRNA decoy | 113 |
Here, we summarize recent insights into the physiological function of miRNAs and their involvement in disease, focusing on cancer, and discuss how these insights can be used for the development of new anti-cancer drugs. We also discuss emerging insights into the role of lncRNAs and their potential as targets for novel treatment paradigms.
miRNA generation and function
Mature miRNAs are evolutionally conserved single-stranded RNAs. The generation of mature miRNAs is a multi-step process, which starts with the initial transcription of their genes by RNA polymerase II. This results in long (varying from several hundreds to several thousands of nucleotides), capped and polyadenylated primary miRNAs. These primary transcripts are processed by the ribonuclease (RNase) III Drosha-DGCR8 nuclear complex into hairpin structure precursor miRNAs of 60–100 nucleotides, which are subsequently transported from the nucleus to the cytoplasm by exportin-5, and further cleaved by the RNase enzyme Dicer into double-stranded miRNAs. The two strands are separated by helicases, and the mature strand incorporates into the RNA-induced silencing complex (RISC). Typically, mature miRNAs regulate gene expression through sequence-specific binding to the 3′-untranslated region (3′-UTR) of a target messenger RNA (mRNA), however several lines of evidence indicate that miRNAs can also bind to other regions of a target mRNA 22,23. The miRNA:mRNA interaction usually causes translational repression and/or mRNA cleavage, and thus reduces the final protein output.
Overall, this “traditional” mechanism of action of miRNAs as negative regulators of gene expression has recently been challenged by the discovery of new and unexpected mechanisms of action (discussed in detail in Box 2). This includes evidence that miRNAs can also increase the translation of a target mRNA by recruiting protein complexes to the Au-rich elements of the mRNA 24, or indirectly increase the target protein output by de-repressing the mRNA translation via interaction with proteins that block the target gene translation 25, cause global protein synthesis by enhancing ribosome biogenesis 26, or switch the regulation from repression to activation of target gene translation in cell cycle arrest conditions 24.
BOX 2. Novel insights into miRNA properties.
The discovery that miRNAs target the 3′-UTR of genes and downregulate the expression of protein-coding genes in the cytoplasm has been significantly expanded in the last six years with the following additional discoveries that demonstrate unexpected complexity for their mechanism of action:
MicroRNAs can be localized in the nucleus – for example, human miR-29b has been shown to predominantly localize to the nucleus 143. It will be more challenging to target nuclear miRNAs with anti-miRNA strategies.
In addition to 3′-UTRs, miRNAs target other genetic regions at the DNA (promoter regions) or RNA level (5′-UTR, coding regions) 22, 23, 144, 145, other ncRNAs such as T-UCRs 106 and even proteins 25. For instance, miR-122 facilitates HCV RNA replication by binding directly to two adjacent sites close to the 5′ end of HCV RNA 76. Thus miRNAs can have much larger than anticipated effects on whole transcriptome, as the spectrum of targets is much wider than that of coding genes.
MiRNAs can upregulate (and not only downregulate) translation by diverse mechanisms. For example, miR-369-3 was shown to interact with AU-rich elements in the tumor necrosis factor alpha (TNF-alpha) mRNA and recruit the protein complex composing Argonaute 2 (AGO2) and fragile X mental retardation-related protein 1 (FXR1) to the TNF-alpha mRNA, leading to increased protein translation during cell cycle arrest 24. In the same study, the condition of cell cycle arrest switched the regulation of miRNA let-7 on targeted genes from translational repression to translational activation. It was also shown that miR-10a interacts with the 5′-UTR of ribosome protein-encoding mRNAs to enhance ribosomal biogenesis, which induces global protein synthesis and causes oncogenic transformation of murine NIH3T3 cells 26. In another study, miR-328 increases the translation of the myeloid-specific transcription factor CCAAT/enhancer binding protein alpha (CEBPA) in chronic myelogenous leukemia cells, not by directly binding to CEBPA mRNA, but by directly binding to PCBP2, a poly(rC)-binding protein that interacts with a C-rich element located in the 5′-UTR of CEBPA mRNA and inhibits its translation 25. However, whether this activation of protein translation represents a general phenomenon or just exceptions of miRNA regulatory mechanisms remains to be determined.
MiRNAs interact with other ncRNAs and various types of mRNA transcripts in a “competing endogenous RNA” (ceRNA) network 112. Two co-expressed transcripts that are targeted by the same collection of miRNAs are functionally coupled to one another as a result of the finite amount of available miRNA: a transient change in the amount of one transcript will impact on the apparent abundance of the other transcript as a result of the concomitant change in the amount of miRNA that is available.
MiRNAs can also be packaged into multivesicular bodies (MVB’s) and released into the extracellular environment as exosomes. This allows them to act as hormones, defined as secreted molecules that trigger a receptor-mediated response in a different cell or tissue 28, 33, 146. It has been shown that macrophages influenced breast cancer cell invasion through exosome-mediated delivery of oncogenic miR-223 147, and pre-treatment of mice with tumor-derived exosomes accelerates lung metastasis formation 148. Therefore, targeting miRNAs secreted by a specific cell could impact on a different cell type.
MiRNAs can act as agonists of Toll-like receptors through interaction with Tlr7 and TLR8, triggering downstream pathway activation 27, 92. Therefore, modulation of miRNAs (e.g. miR-29a) might lead not only to variations in target mRNA expression (e.g. DNMTs) 149, but also to changes in TLR-mediated signaling (e.g. NF-κB pathway).
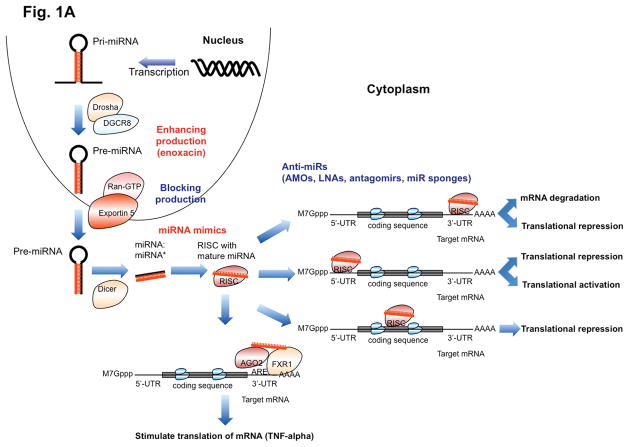
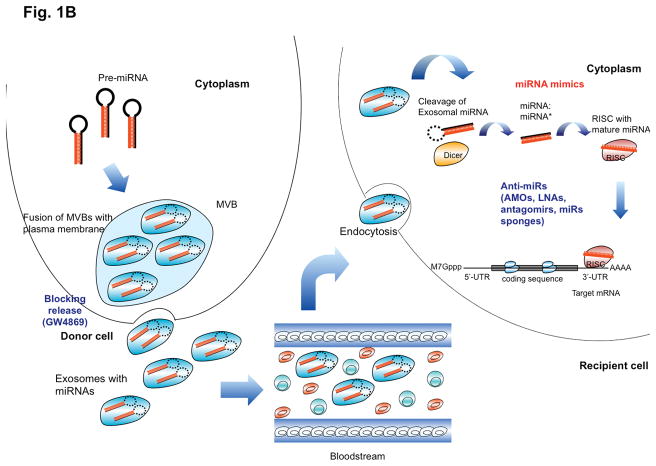
MiRNAs have been found to function not only within cells but they are also abundant in the bloodstream and can act at neighboring cells and at more distant sites within the body in a hormone-like fashion, indicating that they can mediate both short- and long-range cell-cell communication 27, 28. MiRNAs, together with RNA-binding proteins (such as Nucleophosmin 1 and AGO2), can be packaged and transported extracellularly by exosomes or microvesicles 29–32. Likewise, precursor miRNAs inside the donor cell can be stably exported in conjunction with RNA-binding proteins or by binding to high-density lipoprotein 31. Additionally, passive leakage from cells due to injury, chronic inflammation, apoptosis or necrosis, or from cells with short half-lives such as platelets, is thought to be another way of release. Circulating miRNAs enter the bloodstream and are taken up by the recipient cells by endocytosis and further bind to intracellular proteins such as Toll-like receptors (TLRs) 27. It is hypothesized that miRNAs bind to specific as-yet unidentified membrane receptors present on the recipient cells 33. Each step of miRNA generation and function, both intracellularly (Figure 1A) and in its endocrine function (Figure 1B) can potentially be therapeutically targeted.
Figure 1. Mechanisms of action of miRNAs and the use of therapeutic agents to block or activate their function.
(A) The diverse mechanisms of miRNA activity are presented together with the related miRNA-targeting strategies. In blue are the agents that reduce the miRNA activity, while in red are the agents that increase miRNA activity. Initially transcribed as primary miRNA (pri-miRNA), processed into precursor miRNA (pre-miRNA) by a microprocessor complex composed of Drosha and DGCR8, transported from nucleus to the cytoplasm by Exportin 5 in the presence of the Rna-GTP cofactor, and further processed into mature form by Dicer, miRNA recruits on the RISC and regulates the output of protein-coding genes through diverse mechanisms. The interaction of miRNA with the 3′-untranslated region (3′-UTR) of protein-coding genes is considered as the main mechanism, which usually leads to a decrease in protein output by either mRNA degradation or translational repression. Recent studies also suggest that miRNA can interact with the 5′-untranslated region (5′-UTR) of protein-coding genes via complementarity, and cause translational repression 23 or activation of the targeted proteins 26. Similarly, miRNA can also target the coding sequence, and repress the translation of targeted genes 22. Moreover, miRNAs can interact with regulatory protein complexes such as AGO2 and FXR1, and indirectly upregulate translation of a target gene 24. Whether the “non-canonical” mechanisms represent a general mechanism or just exceptions to the canonical one remains to be determined. Various means can be used to enhance (such as by Enoxacin) or block (by any defects in the biogenesis machinery) general miRNA production, however this approach is not specific. More specific regulation of miRNA activity can be achieved by using miRNA mimics or anti-miRs such as LNAs, antagomirs and miR sponges, which bind and thereby functionally block specific miRNAs. While most of the miRNA therapeutics are still in preclinical development, one LNA anti-miR (miravirsen) and one miRNA mimic (MRX34) have reached clinical trials. (B) miRNAs can also be packaged into multivesicular bodies (MVBs), released into the extracellular environment as exosomes, carried through the circulation system and act on recipient cells. This has been found to play a role in cancer development (see Box 2). Blocking of such secreted miRNAs can be achieved by interfering with their secretion from the cells of origin (e.g. cancer cells), for example with inhibitors of neutral sphingomyelinase such as GW4869 27. Alternatively anti-miR strategies can be employed to interfere with the function of secreted miRNAs in the recipient cell. Conversely, miRNA mimics with suitable formulation, for example with lipid encapsulation, of secreted miRNAs can be employed to enhance their function.
miRNAs and their roles in cancer
MiRNAs play a variety of roles in cancer development and progression (see Table 1 and Box 3). The first evidence of miRNA involvement in cancer was discovered in a quest to identify tumor suppressors in the frequently deleted 13q14 region in chronic lymphocytic leukemia (CLL): the miRNA cluster miR15a/16-1 was found to be deleted or downregulated via epigenetic silencing in 69% of CLL patients analyzed 3. Subsequent studies confirmed the tumor suppressor function of miR-15a/16-1, and identified several other suppressor miRNAs such as the let-7 family and miR-34 family 5. These tumor suppressor miRNAs exert function by targeting oncogenic protein-coding genes for degradation 34. For instance, miR-15a/16-1 targets the anti-apoptotic proteins BLC2 and MCL1; let-7 negatively regulates the oncogenes KRAS and MYC; miR-34 mediates p53 signaling by targeting CDK4, MYC and MET 35 (Table 1).
BOX 3. Principles of ncRNA involvement in cancer and their therapeutic consequences.
NcRNAs such as miRNAs and T-UCRs are frequently located within cancer associated genomic regions and can act as tumor suppressors or oncogenes 150. Several mouse models strongly suggest that alterations in miRNA expression alone cause a cell to become neoplastic: miR-155 transgenic and miR-21 transgenic mice develop acute lymphoblastic/high-grade lymphoma 37, 42 while mice in which the tumor-suppressor cluster miR-15/16 is functionally deleted develop chronic lymphocytic leukemia 151.
Each ncRNA (including miRNA and lncRNA) can regulate the expression of numerous target genes, and conversely multiple types of ncRNAs can regulate the same target gene. This interplay is involved in regulation of various physiological processes and the pathophysiology of many diseases, including all types of human cancer analyzed so far 4, 8.
NcRNAs such as miRNAs and lncRNAs are also involved in tumor metastasis as activators or suppressors. This can be exemplified by the miR-10b 81, which promoted distant metastasis of breast cancer to the lung by negatively regulating homeobox D10 (HOXD10), and thus increasing expression of pro-metastatic gene, RHOC, and the miR-31 83, which suppressed breast cancer cell invasion and metastasis by targeting ITGA5, RDX and RHOA. LncRNAs have also been linked to tumor metastasis, as exemplified by the involvement of HOTAIR in breast cancer metastasis 108.
Genetic variations in miRNA (and potentially also lncRNA) genes and their precursors, target sites, and genes encoding components of the miRNA processing machinery (such as the exportin-5 or TARBP2 for miRNA processing) can affect cell phenotype and disease susceptibility 152, 153.
A subclass of miRNAs, the epi-miRNAs, can directly control the epigenetic machinery (e.g. by directly targeting DNA methyltransferases such as in the case of miR-29 family 149). Conversely, miRNA expression can be downregulated via promoter hypermethylation 154.
Aberrant ncRNA (miRNAs, T-UCRs and lincRNAs) gene expression signatures characterize cancer cells and ncRNA profiling can be used for the diagnosis, for establishing a prognosis, and to indicate treatment-responsiveness of cancer patients. Since ncRNA profiles allow to accurately and specifically differentiate tissue and disease type, ncRNAs are of interest for the development of biomarkers for early diagnosis and prognosis 9, 12.
Circulating miRNAs are potentially non-invasive cancer biomarkers 28.
Messenger RNA inhibition using miRNAs could be used therapeutically for the treatment of specific types of cancer 17.
The interplay between miRNAs, lncRNAs such as lincRNAs, T-UCRs and pseudogenes, and protein-coding genes forms a complex network of interactions in normal tissues and this is dys-regulated in diseased tissues 12.
Three years after the first identification of miRNAs as tumors suppressors, several miRNAs including miR-21, miR-17-92 cluster and miR-155 were revealed to exhibit oncogenic activity in carcinogenesis. One of the most well characterized oncogenic miRNAs is miR-21, which has been found to be upregulated in all types of cancer so far analyzed. 36. Multiple studies demonstrate the oncogenic functions of miR-21; for example it has been shown that miR-21 overexpression induces pre-B-cell lymphoma in mice 37, promotes KRAS-dependent lung carcinogenesis by activating the Ras/MEK/ERK pathway 38, and enhances metastasis of colorectal cancers by targeting PDCD4 39. Similarly, the miR-17-92 cluster located at 13q22 is frequently upregulated in a wide range of cancer types including lymphoma, lung, breast, stomach, colon and pancreatic cancer, through amplification or transcriptional activation 36. MiRNAs from this cluster are direct downstream targets of the oncogene MYC, and they attenuate MYC-induced apoptosis to promote the formation of B-cell lymphoma in mice 40, 41. The concept of miRNAs as oncogenic driving force of cancer was demonstrated by showing that overexpression of miR-155 alone is sufficient to cause lymphoblastic leukemia/high-grade lymphoma in transgenic mice 42.
It has further been found that some miRNAs can act as specific activators or suppressors of tumor metastasis 43. Functional studies in animal models showed that miR-9, miR-10b, miR-103/miR-107, miR-373 and miR-520c are drivers or promoters of tumors metastases, whereas miR-31, miR-34a, miR-126, miR-206 and miR-335 suppress tumor metastases through diverse mechanisms 44. A recent study revealed an interesting interaction between miRNAs by showing that miR-22 increases mammary stem cell population and promotes breast cancer metastasis in transgenic mouse by reducing miR-200, which has antimetastatic function, via direct targeting of the TET, a methylcytosine dioxygenases responsible for demethylation of the miR-200 promoter 45. Although metastasis-related miRNAs generally act by regulating migration and invasion of cancer cells, miR-126 has been shown to impair the ability of breast cancer cells to recruit endothelial cells in the tumor microenvironment into the metastatic niche, thus inhibiting metastatic colonization 46.
Complicating the issue of miRNA involvement in cancer and therapeutic targeting, it has been found that some miRNAs can behave like an oncogene in one cell type, and a tumor suppressor in others. For example, miR-221, when overexpressed in liver cancer, exerts an oncogenic function by downregulating the tumor suppressor PTEN 47 but in erythroblastic leukemia it acts as tumor suppressor by reducing the expression of the KIT oncogene 48. The same bivalent effects were also identified for the let-7 family (Table 1). One paradigm that emerged is the existence of miRNA/transcription factor feedback circuitries composed of multiple miRNAs and protein-coding genes, which can be involved in the pathogenesis of cancer. An example is the microRNA/TP53 circuitry in CLL composed of five miRNAs (miR-15a/16-1, miR-34a, -34b, and -34c) and four coding genes including the transcription factor TP53, the anti-apoptotic oncogenes BCL2, MCL1 and ZAP70 (the 70-kDa-zeta-associated protein). The expression levels of the gene members of this circuitry were demonstrated to be a powerful predictor of CLL patient survival 49. In acute T-cell lymphoblastic leukemia, a small set of miRNAs composed of miR-19b, miR-20a, miR-26a, miR-92 and miR-223 were found to cooperatively downregulate tumor suppressor genes including IKARO, PTEN, BIM, PHF6, NF1 and FBXW7, and promote leukemia development in a NOTCH1-driven leukemia mouse model 50. Similarly, the miR-17-92 cluster miRNAs and the protein-coding genes MYC and E2F1 form a network involved in lymphoma development 51. The complex interaction of miRNAs and protein-coding genes in cancer indicates that treatments targeting only protein-coding genes may not be sufficient in controlling cancer progression.
Considerations for miRNA therapeutics
As cancer is a multigenic disease, the main advantage of miRNA therapeutics (defined as strategies restoring or blocking miRNA function) is that miRNAs target multiple coding genes or non-coding genes that can be involved in a specific pathway or in redundant pathways involved in cancer development. In other words, the ability of miRNAs to target genes implicated in the same pathway and/or in interacting pathways provides the rationale for the use of a small number of miRNAs to achieve a broad silencing of pro-tumoral pathways and, for this reason, miRNAs are superior to a mixture of siRNAs specifically designed to reduce the expression of a given number of target genes. For instance, miR-124 has been shown to target multiple protein-coding genes such as p38 MAP Kinase, STAT3 and AKT2 in the epidermal growth factor receptor (EGFR) signaling 52. Since constitutive activation of EGFR signaling has been linked with multiple cancer types including lung, colon and breast cancers 53, it can be hypothesized that miR-124 replacement by mimics or vector-encoded miR-124 may simultaneously downregulate the expression of these three protein-coding genes, and thereby reverse cancer progression by inactivation of the EGFR signaling. In another scenario, miR-15a and miR-16-1, both with reduced expression in CLL, target the anti-apoptotic proteins BCL2 and MCL1 54, and therefore a personalized miRNA therapy with agents that either boost the expression of or mimic miR-15a and miR-16-1, or with vector-encoded miR-15a and miR-16-1 (see section below), can be envisioned for CLL patients that have both reduced miR-15a/16-1 cluster expression and overexpression of BCL2 and MCL1 protein in their malignant cells. The use of these two miRs could not only cause reduction for BCL2 and MCL1 protein expression, but produce an inhibitory effect on other anti-apoptotic targets such as RAD51C, PDCD6IP, GRP78 and PDIA2 54, and consequently this multiple targeting causes an additive effect on blocking the antiapoptotic signaling. Therefore, when targeting particular cancer-related pathways, the combination of miRNAs (e.g. miR-15a and miR-16) could have more potent effects than the combination of the specific siRNAs. Mutations in miRNAs, although previously reported 55, are rare due to the small size of miRNA, and resistance development to miRNA therapeutics would likely require multiple mutations in multiple genes. Circulating miRNAs could be used as biomarkers to identify the patients for whom this approach would be suitable (see Box 4 for discussion of miRNAs as biomarkers).
BOX 4. Circulating miRNAs as non-invasive biomarkers.
The biomarker potential of body fluid miRNAs relies mainly on their high stability and resistance to storage and handling. Expression patterns of serum miRNAs that are specific for lung cancer, colorectal cancer and diabetes have been identified, indicating that serum miRNAs provide fingerprints for various diseases 155. Correlations between circulating miRNA levels and the response to a given anticancer agent were also observed and may be useful in predicting patterns of resistance and sensitivity to particular drugs. For example, circulating miR-21 levels were higher in hormone-refractory prostate cancer patients whose disease was resistant to docetaxel-based chemotherapy, compared to patients with chemo-sensitive disease 156. High levels of miR-141 expression in the plasma have been associated with poor prognosis in patients with colorectal cancers 157.
However, there are also substantial challenges associated with miRNA-targeted approaches. As discussed above, miRNA activity can be dependent on the cellular environment, and the same miRNA can have different targets (due to different sets of coding and non-coding gene expression) in the same organism (but in different cell types) and consequently opposite effects. Therefore, the modulation of the specific miRNA with miRNA therapeutics might have beneficial effects in one cell type and harmful effects in another.
Further complicating the issue is the finding (reported for various miRNAs) that the mature products generated from each strand of the same hairpin RNA structure, termed 5p and 3p, can bind to different mRNAs and display bivalent behavior. For example, miR-28-5p and miR-28-3p are transcribed from the same RNA hairpin and are downregulated in colorectal cancer cells 56. In contrast, in some BCR-ABL negative myeloproliferative neoplasms, miR-28 is upregulated 57. In vitro experiments with colorectal cancer cells showed that the overexpression of either miR-28-5p or miR-28-3p had different effects due to the fact that the two miRNAs interacted with different mRNAs: miR-28-5p altered the expression of CCND1 and HOXB3 proteins, whereas miR-28-3p regulated the expression of the NM23-H1 metastasis-suppressor protein. Overexpression of miR-28-5p reduced colorectal cancer cell proliferation, migration, and invasion in vitro, whereas miR-28-3p increased colorectal cancer cell migration and invasion in vitro 56. Similarly, it was reported that miR-125a-5p and miR-125a-3p, which are downregulated in non-small cell lung cancer, exhibit distinct effects in vitro on the migration and invasion of lung cancer cells 58. Such information has a direct implication for the design of miRNA gene therapy trials: if a precursor miRNA inserted in a viral vector is to be used and both strands are produced, the identification of the specific roles of each strand in the same cell type is mandatory. If opposite effects are observed (such as for miR-28 or miR-125), then such precursors should be excluded from use and the respective active mature miRNA should be either delivered as miR-mimics (e.g. miR-28-5p, miR-125-3p) or targeted (e.g. miR-28-3p, miR-125-5p) (Table 2).
TABLE 2.
The principal types of RNA therapeutic drugs.
| Agent | Target RNA | What are these agents? | Mechanism of action | Preclinical or clinical applications | |
|---|---|---|---|---|---|
| Block | Antisense Oligonucleotid es (ASOs) | all RNAs | Single stranded, chemically modified DNA-like molecule that is 13 to 25 nt in length and designed to be complementary to a selected RNA. | Formation of an RNA-ASO duplex through Watson-Crick binding, leading to RNase-H mediated cleavage of the target RNA 180 | Preclinical studies |
| Ribozymes or Dnazymes | lncRNAs | A ribozyme, or RNA enzyme, is an RNA molecule that can catalyze a chemical reaction. A Dnazyme, or deoxyribozyme, is a catalytic DNA that cleaves target RNA in a site-specific manner. | Three steps, cyclically repeated: Watson-Crick base pairing with a complementary target sequence, then site-specific cleavage of the substrate and finally release of the cleavage products. | Preclinical studies | |
| siRNAs | all RNAs | A siRNA is a double strand (ds) RNA that is homologous to the target RNAs including mRNAs and ncRNAs, both targeted based on perfect sequence complementarity. | The siRNAs are incorporated into a multiprotein RNA-induced silencing complex, leaving the antisense strand to guide the complex to its homologous mRNA or ncRNA target for endonucleolytic cleavage of RNA 181. | Preclinical studies | |
| AMOs | miRNAs | ASOs targeted at miRNA | Same mechanism as the ASO. | Preclinical studies | |
| LNA anti-miRs | miRNAs | The LNAs anti miRNAs represent LNA modified ASOs. LNAs are bicyclic RNA analogues where the ribose is locked in a C3′-endo conformation by the introduction of a 2′-O,4′-C methylene bridge 79 | Same mechanism as the ASO/AMO. | Phase I and 2a (for HCV) 74, 75 | |
| Antagomirs | miRNAs | Single-stranded 23 nt RNA molecules complementary to the targeted miRNA that have been modified to increase the stability of the RNA and protect it from degradation. The modifications included a partial phosphorothioate backbone in addition to 2′-Omethoxyethyl 80. | The antagomirs silence miRNA in a way still not completely known; the miRNA/antagomir – duplexes induce degradation of the miRNA and recycling of the antagomir 80. | Preclinical studies | |
| miR sponges | miRNAs | RNAs containing multiple tandem binding sites to a miRNA of interest and are transcribed from expression vectors 82. | By competing with the native targets of miRNAs, these highly expressed transcripts reduce miRNA effects, and thus result in increased expression of the miRNA’s native targets 82. | Preclinical studies | |
| SMIRs | miRNAs | Small molecule chemical compounds | Block activities of specific miRNAs by structurebased docking onto the precursor or mature from of miRNA structure. | Preclinical studies | |
| Restore | Small molecules | miRNAs | Hypomethylating agents (Decitabine or 5- azacytidine) and enoxacin | Non-specific induction of miRNA expression | Preclinical studies |
| miRNA mimics | miRNAs | Double stranded synthetic RNAs that mimic endogenous miRNAs | Restore the expression and function of a specific miRNA | Phase I | |
| miRNA expression vectors | miRNAs | Vectors expressing a specific type of miRNA | Restore the expression and function of a specific miRNA | Preclinical studies |
Note: nt – nucleotide; AMO - anti-microRNA antisense oligodeoxyribonucleotide; LNA – locked nucleic acids; SMIRs: small-molecule drugs that target specific miRNAs (SMIRs)
The development of miRNA therapeutics
miRNA therapeutics are being devised that either down-regulate or block the function of oncogenic miRNAs, or upregulate the expression of miRNAs with tumour suppressive function (Figure 1). The main causes of reduced expression of tumor suppressor miRNAs in human cancers are genetic deletion of the miRNA loci or epigenetic silencing via CpG island hypermethylation in promoter of the miRNA genes 11. Molecular approaches are pursued that reverse epigenetic silencing, or enhance the biogenesis of miRNAs, and levels of silenced or deleted miRNAs can be restored by the direct administration of miRNA formulations, either naked, coupled to a carrier, or delivered via a viral vector. Likewise, for strategies that block miRNA functions, both oligonucleotide-based and small molecules approaches are being explored. As discussed above, miRNAs can be modulated either intracellularly in the cells that produce them, or they can be targeted in their endocrine function (see Figure 1B). The different approaches are discussed below.
Restoring miRNA levels with small molecules
Epigenetic silencing of miRNA can be reversed by hypomethylating agents such as decitabine or 5-azacytidine. Both agents have been approved for the treatment of myelodisplastic syndromes 59, and have been shown to re-induce the expression of multiple messenger RNAs as well as ncRNAs including miRNAs 60. However, this is a “non-specific” miRNA effect and the spectrum of induced miRNAs is different from cell to cell 61, 62. Another example is the small molecule enoxacin (Penetrex), a fluoroquinolone used as an antibacterial compound, which has been shown to enhance the production of a subset of miRNAs by binding to the miRNA biosynthesis protein TARBP2 63. Treatment of RKO and HCT116 colon cancer cells with enoxacin caused a general upregulation of miRNA expression in vitro. Enoxacin treatment increased expression of 24 miRNAs in mice and reduced tumor growth in xenograft, orthotopic and metastatic mouse models 63. Interestingly, the enoxacin’s growth-inhibitory effect was substantially compromised both in a colon cancer cell line with inactivating mutation in theTARBP2 gene, and in in vivo studies with TARBP2-deficient mice 63, indicating that miRNA regulation by enoxacin is the main mechanism for its anti-cancer effect. These examples highlight the key role of disrupted miRNA expression patterns in cancer, and demonstrate the effectiveness of restoring the distorted full spectrum of miRNAs that are downregulated in cancer cells.
Restoring miRNA levels with oligonucleotide-based approaches
A more targeted approach to boosting the levels of particular miRNAs than the strategies discussed above is the restoration of the expression and function of one or a limited number of miRNAs, usually located in a cluster (such as miR-15a and miR-16-1 at 13q14.3), either with miRNA mimics or with miRNAs encoded in expression vectors.
MiRNA-mimic molecules are double stranded synthetic miRNA oligonucleotides, and when transfected into cells, they are processed into single strand form and regulate protein-coding genes through miRNA-like function; however, an effective delivery system is necessary to improve stability and uptake, and thus the efficacy of miRNA mimics 64. One strategy is to couple miRNA mimics to nanoparticles coated with tumor-specific antibodies, as exemplified by a study showing that targeted delivery of miR-34a using nanoparticles coated with a neuroblastoma-specific anti-disialoganglioside GD2 antibody inhibits neuroblastoma tumor growth in a murine orthotopic model 65. It was also shown that systemically delivered neutral lipid emulsions of miR-34a and let-7 mimics induced a significant reduction in tumor burden in a KRAS-activated mouse model of non-small-cell lung cancer (NSCLC) 66. Moreover, formulations with atelocollagen lead to the efficient delivery of miR-34a into tumors and inhibited colon cancer progression in a subcutaneous xenograft mouse model 67.
miRNA expression vectors are engineered with promoters that allow for the expression of the miRNA of interest in a tissue- or tumor-specific fashion. This approach has been demonstrated in hepatocellular carcinoma (HCC) both in vitro and in a xenograft mouse model. MiR-26a is reduced in human primary HCC (with respect to the normal liver tissue counterpart) and its re-expression in hepatic malignant cells leads to antitumor activity 68. Systemic administration of miR-26a in a mouse model of HCC using an adeno-associated virus (AAV) was shown to inhibit cancer cell proliferation, induce tumor specific apoptosis and slow disease progression with minimal toxicity 69. Interestingly, a recombinant miR-26a expression vector driven by a dual promoter for alpha-fetoprotein and human telomerase reverse transcriptase was shown to be specifically expressed in liver tumor cells and caused a reduction in HCC formation, at least in part by direct downregulation of estrogen receptor-alpha 70.
The strategies of restoring miRNA function have yielded the first miRNA replacement therapeutic in the clinical pipeline – MRX34, a liposome-formulated miR-34 mimic with a diameter of ~120 nm in clinical trial for patients with advanced or metastatic liver cancer by intravenous injection. Preclinical study has shown that tail vein injection of MRX34 remarkably reduced tumor growth and significantly enhanced mouse survival with a favorable safety profile in orthotopic mouse models of hepatocellular carcinoma 71.
Blocking miRNA function with oligonucleotide-based approaches
Current strategies for inhibitory miRNA-targeting (Table 2) are mainly based on antisense oligonucleotides (so called anti-miRs), and comprise locked nucleic acids (LNA) and tiny LNA anti-miR constructs, antagomirs and miRNA sponges.
Amongst these drugs, LNAs hold the highest affinity to the targeted miRNA since the “locked” modification of the ribose ring, engineered through the addition of a methylene bridge connecting the 2′-O atom and the 4′-C atom, renders in an ideal conformation for Watson-Crick binding to the targeted miRNA. This allows a short sequence of ~22 nucleotides, equaling the targeted miRNA size, to achieve effective blocking 72. Several LNA-based approaches are currently undergoing preclinical investigation for cancer. For instance, high expression of miR-380-5p was associated with a poor outcome in neuroblastoma with MYCN amplification and in vivo delivery of a LNA-anti-miR-380-5p efficiently reduced the tumor size in an orthotopic mouse model of neuroblastoma, likely through reversing the suppression of p53 by miR-380-5p 73. However, the agent furthest in development is not an anti-cancer compound but an antiviral; Miravirsen (also known as SPC3649), a LNA-based antisense molecule against miR-122 developed by Santaris Pharma A/S (Horsholm, Denmark) was recently evaluated in Phase 1 and Phase 2a clinical trials for the treatment of hepatitis C (HCV) 74, 75. The rationale for these studies was based on the observation that liver specific miR-122 binds to two miR-122 target sites in the 5′ non-coding region of the HCV genome, leading to an upregulation of viral RNA levels 76.
Tiny LNAs, specifically designed to target the 5′-seed region (the 2–8 nucleotides) of miRNAs with short sequences of 8 nucleotides, were shown to concurrently bind and sequester miRNAs sharing the same seed sequence, and thereby increase the expression of miRNA-suppressed protein-coding genes 77. Intravenous injection of unconjugated tiny LNAs achieved long-term silencing of the targeted miRNAs in a mouse orthotopic breast tumour model with high specificity and efficacy 77. A concern with tiny LNAs could be off-target effects on mRNAs with perfect complementarity, however comprehensive RNA and protein profiling showed that tiny LNAs did not significantly alter the output of such protein-coding genes 77. In another study, a tiny LNA targeting miR-155 inhibited Waldenstrom Macroglobulinemia and CLL cell proliferation in vitro, and significantly decreased the number of leukemic cells in vivo when administered in unconjugated form by tail vain injection 78. However, it should be pointed out that although tiny LNAs have the advantage of targeting multiple miRNAs from the same family, longer oligonucleotides are necessary if a more specific effect towards an individual miRNA is preferred. The high efficacy of LNAs, their resistance to degradation and efficient uptake by many tissues eliminate the need for sophisticated formulation and delivery associated with most other oligonucleotide treatments. Moreover, the levels of the plasma transaminases and bilirubin are normal and no acute or subchronic toxicities were observed in primates treated with LNA anti-miRs 79.
Antagomirs are cholesterol-conjugated synthetic RNAs with a 2′-O-methyl linkage and phosphorothioate modification. They function as anti-miRs by being complementary to the full sequence of the targeted miRNA and blocking miRNA function 80 (Table 2). While chemical modification with cholesterol aims to increase the cellular uptake, the 2′-O-methyl modification and the phosphorothioate modification are designed to improve the binding affinity and prevent the nuclease degradation of oligonucleotides, respectively. In a study investigating the role of miR-10b in metastasis using an orthotopic xenograft murine model of metastatic breast cancer, the researchers silenced miR-10b with a specific antagomir via intravenous delivery 81. They observed that silencing miR-10b did not reduce the growth of the primary tumor, but there was an impressive reduction in the formation of lung metastases. Interestingly, the effects of the antagomir could be fully replicated by using anti-miR-10b sponges (see below). However, when the lung nodules developed from disseminated cells directly injected into the tail vein, antagomir-10b had no effect on lung metastasis formation. These findings indicate that miR-10b is not involved in the late stage of metastasis formation. The toxicity of the antagomir was limited to a slight reduction (just below the normal range) in white blood cell count, a 8%–9% increase in liver and spleen size, a 2-fold increase of serum bilirubin (which was still in the normal range) and slight increase of transaminases 81. It should be noted that although antagomirs are routinely being used as experimental tools, most miRNA therapeutic agents in the developmental stage employed other modifications such as 2′-fluoro or LNA, largely due to the fact that antagomirs require high dosages for effective miRNA blocking 72.
MiRNA sponges are RNAs that are designed to contain multiple tandem binding sites that are complementary to a heptamer in the seed sequence of the miRNA of interest, and thus a single type of sponge can be used to block an entire miRNA seed family. Since the efficacy of miRNA sponges depends also on the concentrations of sponge RNAs, they are usually encoded in either plasmid or viral expression vectors driven by a strong promoter such as the CMV promoter, which can be stably transfected to allow for long-term delivery of the miRNA sponge after a single application 82. MiRNA sponges have been used to study tumor metastasis-related miRNAs, and using orthotopic mouse models, it was found that sponges targeting miR-31 83, miR-9 84, or miR-10b 81 dramatically reduced the expression of the respective miRNAs and effectively blocked breast cancer metastasis. In another study, sponges targeted at the seed regions of the miR-17~92 cluster simultaneously silenced each miRNA member, i.e., miR-17, miR-18a, miR-19 and miR-92a 85. Compared with miRNA sponges targeting individual miRNAs (for example, miR-92a alone), the combined targeting of the miR-17-92 cluster showed a stronger inhibitory effect on proliferation of WEHI-231 B-cell lymphoma cells in vitro 85. With regards to the stable transfection of vectors encoding miRNA sponges, it has been reported that recombinant AAV vectors encoding an anti-miR-122 sponge depleted the corresponding miRNA in the liver of treated mice and reduced serum cholesterol by >30% for 25 weeks compared with the control group 86. Although this is not a cancer-related study, the long-lasting effect of miRNA sponges has an immediate application in cancer treatment since anticancer strategies often require persistent target regulation. However, it should be noted that, since sponge uses competitive RNAs without chemical modification, the binding affinity is relatively low and the concentration needed for effective miRNA blocking is likely to be much higher compared with LNA or antagomirs. It remains to be determined whether an excess of sponge transcripts which are not sequestered by the target miRNAs can have undesired functions.
The challenges associated with anti-miR treatment include off-target effects, which can lead to unwanted responses in tissues other than the intended ones. Although combined modifications with antagomir and LNA substitutions may improve the in vivo bioavailability, stability and specificity of the anti-miR towards a specific miRNA, a specific delivery system that targets only tumor cells in the intended tissues is lacking. Another obstacle for anti-miR therapeutics is the complexity of the assessment of anti-miR efficacy. This is because anti-miR treatment may not always reduce miRNA expression levels. For instance, miravirsen was shown to sequester miR-122 by forming a highly stable heteroduplex, rather than degrade the mature miR-122 74. It is still inconclusive at which conditions the anti-miR treatment will cause degradation of the targeted miRNAs, partially complicated by the possible interference of anti-miR:miRNA duplex on detection methods such as PCR and northern blots 87. In order to evaluate an anti-miR efficacy, it is necessary to profile not only the expression level of miRNAs in cancer cells, but also the extent of target de-repression. Since miRNAs have multiple mRNA targets, and as suggested by a recent study, the miRNA modulation of protein-coding gene targets is a fine-tuning effect rather than an on-off effect 88. High throughput profiling of global mRNA and protein changes in samples could provide more comprehensive information regarding the specificity and effectiveness of a particular anti-miR treatment.
Small molecules targeting miRNAs (SMIRs)
While the bulk of research focuses on oligonucleotide-based approaches, there are several reports on efforts to identify small-molecule drugs that target specific miRNAs (SMIRs) and modulate their activities. For example, a high throughput screening of chemical compounds and structure-activity relationship analyses identified diazobenzene and its derivatives as effective inhibitors of pri-miR-21 formation, however the sequence specificity was not comprehensively tested 89. Small molecule inhibitors such as 2,4-dichloro-N-naphthalen-2-ylbenzamide, 6-[(4ar,8as)-octahydroquinolin-1(2h)-ylsulfonyl]-1,2,3,4-tetrahydroquinoline of the liver-specific miRNA miR-122 have also been identified in a reporter plasmid assay and their specificity towards miR-122 was demonstrated 90. Despite these findings, the underlying mechanisms of action of SMIRs are not clear. The structural features of miRNAs such as the stem loops in pre-miRNAs may partially uncover the internal bases, scattering the local electronegative distribution, and thus facilitate SMIR binding to the grooves and pockets on the surfaces of a specific miRNA 91. The advantages of SMIRs are that they are chemical compounds, and thus conventional drug development can be applied for such compounds. The limitations of SMIRs are their intrinsic poor specificity, possible unwanted miRNA-independent effects and their more complicated design compared with oligonucleotide-based therapeutics.
Blocking extracellular miRNAs in exosomes
As described above, it was recently discovered that miRNAs (such as miR-21, miR-29a and miR-16) can be released by cancer cells within exosomes. When engulfed by macrophages in the tumor microenvironment the miRNAs co-localize with TLR8 in the endosome of the recipient cells, and activate the single strand RNA-specific TLR8 in humans (or its murine homologue Tlr7) receptor. This leads to an increased secretion of IL-6 and TNF-alpha by the immune cells, which can increase the proliferation and metastatic potential of cancer cells 27. Whether the endogenous miRNAs can have similar effects on TLR8, and whether this miRNA effect is sequence-specific, remains to be determined. It was also shown that extracellular let-7 (a highly abundant regulator of gene expression in the central nervous system) can activate Tlr7 in mice and induce neurodegeneration through neuronal Tlr792. Intriguingly, let-7b levels are higher in the cerebrospinal fluid of patients with Alzheimer’s disease, indicating that miRNA-mediated activation of TLRs could have implications beyond cancer 91. This novel mechanism of action and its potential central role in cancer dissemination and neurodegenerative diseases harbors important translational implications. It was demonstrated that ceramide-dependent secretion is one of the mechanisms leading to the release of exosome miRNAs, and the small molecule GW4869, an inhibitor of neutral sphingomyelinase that plays a key role in ceramide biosynthesis, effectively blocked the miRNA-mediated aberrant cross-talk between cancer cells and surrounding immune cells within the tumor microenvironment in NSCLC 27.
Combination approaches with miRNA therapeutics
The concept of combinations of various miRNA agents in “cocktails”, together with chemotherapeutic agents or molecularly targeted agents, might benefit patients by resulting in synergistic effects. Encouraged by positive results from the Phase II clinical trial of miravirsen in controlling HCV infection, a phase II clinical trial on combination of this anti-miR with clinically used antiviral drugs such as telaprevir and ribavirin was initiated and is now recruiting participants (clinical trial identifier: NCT01872936). For a cancer such as CLL, two strategies for the inhibition of RNA expression could be envisioned. The “sandwich RNA inhibition strategy” would focus on a major molecular alteration clearly linked with CLL pathogenesis through the use of multiple agents. For example, clinical studies in relapsed or refractory CLL showed a variable efficacy (in some studies a significant increase in the overall survival while in others no advantage) of oblimersen sodium (Genasense), an antisense oligonucleotide designed to specifically bind to human BCL2 mRNA 93. The fluctuation of oblimersen efficacy has been suggested to be related to delivery difficulties in the unfavorable tumor environment 94. The physiological presence of miRNAs in most of the tissues and the findings that miRNAs are very stable in the body fluids 33 indicate a possiblity that strategies using miRNA mimics may bypass the lower efficiency due to delivery obstacles. Thus regimens using a cocktail of oligonucleotides antisense or siRNA against BCL2 and miRNAs targeting BCL2 (that is specifically overexpressed in malignant B cells and not in surrounding non-malignant blood, lymph node or bone marrow cells), such as miR-15a and miR-16, might conceivably achieve a better therapeutic outcome for indolent CLL. Two recent in vitro studies showed that combined treatment with BCL2 siRNA and miR-15a synergistically enhanced methotrexate-induced apoptosis in vitro in Raji cells 95, 96. Another strategy, the “multiplex RNA-inhibition strategy”, could target various molecular defects in the same pathway. For instance, strategies restoring the function of miR-15a and miR-34a may achieve a better effect in inducing apoptotic response by reducing anti-apoptotic proteins such as MCL1 (by miR-15a), SIRT1 (by miR-34a) and BCL2 (by miR-15a and miR-34a).
Sensitization of tumors to therapy by combined use of miRNA and chemotherapeutic agents
Glucocorticoids are effective agents in the treatment of lymphoid malignancies and resistance to glucocorticoid-induced apoptosis is associated with poor prognosis of MLL-AF4 ALL 97. In MLL-AF4 acute lymphocytic leukemia cells, downregulation of miR-128b and miR-221 is implicated in glucocorticoid resistance 98. It was demonstrated that miR-128b negatively regulates oncogenes such as MLL, AF4, and both MLL-AF4 and AF4-MLL fusion genes, and a mutation in the miR-128b gene was shown to lead to glucocorticoid resistance because of reduced production of mature miR-128b 99. Thus the restoration of the levels of miR-128b could potentially revert the glucocorticoid resistance and combined treatment with anti-miR-128b and glucocorticoid could be envisioned. In another study, ectopic expression of miR-30c, a good prognostic marker for human breast cancer, was shown to sensitize cancer cells to doxorubicin treatment in a xenograft mouse model of triple negative human breast tumors. This sensitization was achieved by negative regulation of TWF1 and IL-11, two protein-coding genes that regulate drug sensitivity 100. Similarly, the combination of 5-fluorouracil with an adenoviral vector expressing the tumor suppressor miR-145 was shown to exert a stronger anti-proliferative effect compared to 5-fluorouracil treatment alone, both in in vitro and in in vivo models of breast cancer 101. In another study, LNA-anti-miR-21 was shown to increase the treatment efficacy of a secreted variant of the cytotoxic agent tumor necrosis factor-related apoptosis inducing ligand (S-TRAIL) on glioma. LNA-anti-miR-21 enhanced S-TRAIL-induced caspase activation and thereby the apoptotic response, and the combination leads to complete eradication of gliomas in the murine brain xenografted with human gliomas U87 cells 102.
Involvement of lncRNAs in cancer and therapeutic implications
ncRNAs other than miRNAs might also hold potential for new anticancer approaches 21. Long non-coding RNAs (lncRNAs) are excellent candidates in this respect. LncRNAs are defined as RNA molecules that are longer than 200 nucleotides (see Box 1 and Box 3) that are not translated into proteins. Based on their structural or functional characteristics, they can be further separated into multiple subgroups such as circular RNAs (circRNAs) 103, natural antisense transcripts (NATs) 104, 105, transcribed ultraconserved regions (T-UCRs) 106, long enhancer ncRNAs 107, and long intergenic ncRNAs (lincRNAs). Recent studies suggest that lncRNAs are predominately located in the nucleus, however the functional mechanisms of lncRNAs have not yet well elucidated, and members of the same subfamilies of lncRNAs may display distinct mechanisms of action.
LincRNAs, which are transcribed from DNA sequences in between protein-coding genes were identified using histone marker signatures associated with RNA polymerase II, specifically trimethylation of lysine 4 and lysine 36 of histone 3 (K4K36) 108, 109. About 20% of lincRNAs regulate transcriptional activity of protein-coding genes by guiding the polycomb repressive complex 2 (PRC2) to specific genomic loci 110, 111. As a typical example, HOTAIR, a 2.2-kilobase lincRNA residing in the HOXC locus, was demonstrated to redirect PRC2 to specific genomic loci via direct interaction with PRC2 and thus cause downregulation of a specific set of genes, thereby regulating cancer invasion and metastasis 108, 109.
According to a newly proposed “competing endogenous RNAs (ceRNAs) hypothesis”, RNA transcripts of different types can communicate with each other using microRNA response elements, which have been symbolically referred to as letters of a new language 112. The tumor suppressor gene PTEN and the PTEN pseudogene PTENP1, a lncRNA sharing a high degree of sequence homology, are targeted by and thus can compete for the same set of miRNAs (namely, the miR-17, -21, -214, -9, and -26 families) 113. Accordingly, changes in PTENP1 expression indirectly affect PTEN levels by sequestering PTEN-targeted miRNAs. If PTENP1 expression levels decrease, more miRNAs will be available to target PTEN and ultimately downregulate PTEN expression levels. As a recent addition to the ceRNA mechanism, circRNAs, naturally occurring RNAs with a circular structure, have attracted much attention because they bind and sequester miRNAs, and thus de-repress the mRNA genes normally regulated by the miRNAs sequestered 103.
Another large group of lncRNAs are NATs, which are RNA transcripts encoded in the genome that have sequence complementarity to protein-coding RNA transcripts and regulate the transcription of protein-coding genes (for details see Box 1) 21,105. It was estimated that about one third of protein-coding genes process NATs 21. Interestingly, studies showed that the NAT regulatory mechanism is more likely a consequence of epigenetic modulation such as DNA methylation induced by NAT interaction with DNA methyltransferases and modification of chromatin structure by recruiting histone-modifying enzymes to the genomic locus, rather than the previously presumed direct degradation of the sense transcript through siRNA-like degradation mechanism for complementary sequences 21, 105, 114.
Multiple lncRNAs are abnormally expressed in human cancers and participate in cancer pathogenesis. To date, one of the best studied lincRNAs is HOTAIR, which controls HOXD locus gene expression, and was found to be highly expressed in breast cancer samples 108. Another lincRNA (MALAT1) was found to predict metastasis and survival in early stage NSCLC 115. Many p53-regulated lincRNAs were identified to be induced in response to DNA damage 116, and thus could be involved in the development of resistance to therapy (Table 1).
The expression levels of T-UCRs, which can be regulated by miRNAs via a direct interaction, are often altered in leukemias and in solid tumors106. This has significant implications for miRNA therapy, since some therapeutic effects mediated by miRNAs could be attributed to their downstream lncRNA targeting (e.g. miR-155 that was shown to directly target T-UCR 160 106). Recently, CCAT2, a lncRNA that is transcribed from the 8q24 cancer risk locus and is upregulated in microsatellite-stable colorectal cancers, was shown to promote oncogenic activity and induce chromosomal instability in colorectal cancers 117. In addition, CCAT2 regulates expression of c-MYC, located downstream of its own genomic locus, and is involved in the Wnt signaling network 117. These findings suggest that targeting lncRNAs such as CCAT2 will likely affect broad cancer-associated pathways.
LncRNA therapeutics
Several features of lncRNAs support the use of lncRNAs as therapeutic targets. First, expression levels of lncRNAs are usually lower than that of protein-coding genes, and this may be caused by selected expression of lncRNAs only in a subpopulation of cells 15. The specific expression pattern of lncRNAs in certain types of tissues or cells provides a unique opportunity for delicate regulation by lncRNA-targeting therapeutics (see Table 2). Second, chromatin modification represents one of the main mechanisms of action of lncRNAs, thus a rationale for targeting the interaction of lncRNAs with epigenetic factors such as PRC2 can be envisioned. Third, since many lncRNAs are located in the nucleus 15 and cis-regulate neighboring gene expression, specific gene-locus regulation can be achieved by lncRNA manipulation. A variety of lncRNA therapeutics are being investigated, and several companies are also actively developing lncRNA-targeting therapeutics for treatment of human diseases 21.
One of the strategies (Table 2 and Figure 2) to regulate lncRNA function is by applying specifically designed siRNAs against lncRNAs. Although many lncRNAs have been found to be predominantly located in the nucleus, and may thus be less accessible than messenger RNAs to siRNA targeting, several studies have demonstrated the successful knockdown of lncRNAs by siRNAs irrespective of their subcellular localization. For instance, an in vitro study showed that siRNA pools can reduce the expression of multiple lincRNAs to less than half of their original levels in human cells 111. In another study, siRNAs designed against PANDA, a lncRNA involved in DNA damage response, substantially reduced PANDA expression and consequently sensitized human fibroblasts to doxorubicin-induced apoptosis 118. A more recent publication showed that nuclear RNAs are susceptible to knockdown by siRNA in myotonic dystrophy both in vitro and in vivo 119. These studies suggest the feasibility of conventional siRNA treatment for negative regulation of lncRNAs with oncogenic functions. However, to achieve a general high in vivo efficacy, chemical modifications that improve stability, binding affinity, interference capacity and cellular uptake are necessary. In cases where an extensive secondary structure or the nucleotide sequence of the lncRNAs is unfavorable for an optimal siRNA design, other strategies can be developed to directly target lncRNAs. These include antisense oligonucleotides, as well as Ribozymes or Dnazymes that specifically target the regions involved in the tertiary structures of the lncRNA that are important for the interaction with downstream targets (Figure 2 and Table 2). The advantage of antisense oligonucleotides over siRNAs includes the independence on the RISC machinery, higher specificity and fewer off-target effects 120. Recent studies have shown that antisense oligonucleotides inhibited MALAT1 function and blocked metastatic spread of lung cancer cells in a mouse model 121. Hammerhead ribozyme, which binds to target sequence based on complementary and catalyzes the cleavage of the flanked RNA region, may also be utilized for the targeting of lncRNAs that are unfavorable for optimal siRNA design because of their short length and extensive secondary structures 120.
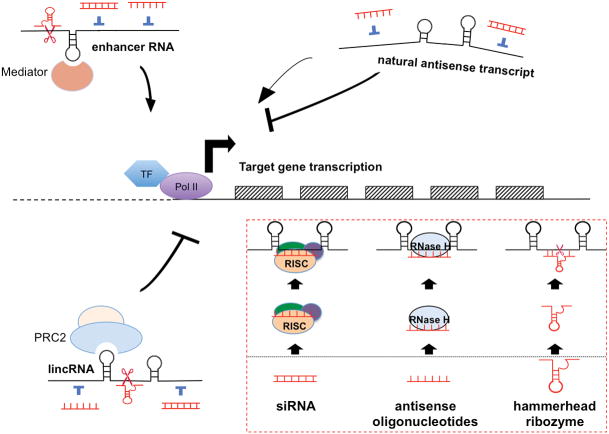
Figure 2. Mechanisms of action of lncRNAs and use of therapeutic agents to regulate their function.
The mechanisms of action of lncRNAs are more diversified than those of miRNAs, and several representative examples are shown. About 20% of long intergenic RNAs (lincRNAs) are bound to polycomb repressive complex 2 (PRC2) and inhibit transcriptional activity by trans regulation. Enhancer RNAs (eRNAs) associate with mediator proteins to modify chromatin structure and activate gene transcription in cis. Natural antisense transcripts (NATs) are another type of lncRNAs that are transcribed from either the same genomic site or distant from the gene locus where the sense transcript counterpart is produced. NATs repress, and in some cases can also activate, transcription of the targeted protein-coding genes via mechanisms such as DNA methylation and chromatin modification at the genomic loci of the targeted genes. Several methods including small interfering RNAs (siRNAs), antisense oligonucleotides and ribozymes can be used to block the lncRNA function. The double stranded siRNA duplex can be stably produced by vectors encoding short hairpin RNA (shRNA) or transiently transfected with synthetic double stranded short RNA. The antisense strand of the siRNA duplex loads on to the RNA-induced silencing complex (RISC) and degrades the targeted lncRNA. Antisense oligonucleotides are single stranded, chemically modified DNA-like molecules (13 to 25 nt in length) that are designed to be complementary to a targeted RNA. Antisense oligonucleotide forms a heteroduplex with the RNA, and RNase H recognizes the RNA-DNA heteroduplex and cleaves the RNA strand. Most of the current studies to target lncRNAs use siRNAs and antisense oligonucleotides as the main tools. However, unique features of hammerhead ribozymes, naturally occurring or artificially synthesized, including activity independence (not rely on the RISC, which mediates siRNA-induced degradation or RNase H, which is essential for activity of antisense oligonucleotides) and specificity in target recognition make it an ideal candidate for lncRNA therapeutics.
Another approach to lncRNA targeting uses synthetic RNA molecules that form hairpin structures which mimick lncRNAs. For example, GAS5 is a lncRNA that can bind to glucocorticoid receptor (GR) and inhibit the interaction of this transcription factor with DNA promoters, functioning as a decoy to block the transcription of target genes 122. Mutant GAS5 mimic lacking the GR binding site failed to inhibit GR-induced transcription 122. Based on this observation, the use of synthetic lncRNA mimics to restore the function of endogenous lncRNAs can be envisioned. Conversely, synthetic mutated lncRNAs may be used to competitively block the function of overexpressed cancer-related lncRNAs. However, the length of lncRNAs may be an obstacle for such applications.
The targeting of NATs represent a unique opportunity for therapeutic regulation of specific genes 105. The inhibition of NATs by single-stranded oligonucleotides, termed antagoNATs, has been shown to disrupt the interaction of NAT with its complementary mRNA, and thus increase the expression of the protein-coding gene 123. To improve stability and cellular uptake, the chemical modifications used for miRNAs have also been applied to antagoNATs. In a recent study, a 14-nucleotide antagoNAT with a mixture of 2′-O-methyl RNA and LNA modifications targeting BDNF-AS efficiently inhibited its function and thus increased transcription of the sense BDNF mRNA both in vitro and in vivo 124. One advantage of antagoNATs is their ability to increase protein expression, and this upregulation of protein output is difficult to achieve by other conventional drug designs.
Challenges and Outlook
The development of miRNA and lncRNA targeted strategies is challenged by several obstacles. One is the successful delivery of the therapeutic agent to the target tissues. Therapeutics must overcome problems associated with oligonucleotides such as degradation by nucleases, renal clearance, failure to cross the capillary endothelium, ineffective endocytosis by target cells, or ineffective endosome release 125, 126. An additional challenge is represented by the release of RNA-based therapeutics formulated in complexes larger than 5 nm diameter from the blood to the target tissue through the capillary endothelium 127. Also, while local delivery into eye or skin has been shown to improve bioavailability in targeted sites, systemically delivered miRNA formulations and RNA-based miRNA targeting agents might be negatively impacted by the host immune system, since macrophages and monocytes can remove complexed RNAs from extracellular spaces 128. For instance, 21 base pair or longer dsRNAs can lead to a sequence-independent interferon response 129. Moreover, the limitations for use of siRNA therapeutics (such as carrier toxicity including haemolysis, thrombogenicity and complement activation induced by nanoparticles, mutagenesis potential with the viral vectors) 126 equally apply to miRNA therapeutic methods. Notwithstanding these limitations, in 2012 the gene therapy drug alipogene tiparvovec (Glybera, UniQure), an AAV-packed protein-coding gene for treatment of lipoprotein lipase deficiency, has been approved by the European Union 130, 131, indicating the plausibility of application of vector-based miRNA replacement or miRNA blocking with miRNA sponges in clinical practice.
Another challenge is the safety evaluation of miRNA-based therapeutics, such as the potential immune response of the delivery system, toxicity caused by the chemical modification or unexpected off-target effects that are likely to occur, given that each miRNA can affect hundreds of target genes. The findings that exosome miRNAs can activate TLRs and promote secretion of IL-6 and TNF-alpha 27 indicate a possibility that tumor suppressor miRNA mimics may also induce such activities and thus it is necessary to monitor this reaction when applying miRNA mimics for cancer therapeutics. This is further complicated by the recently identified novel mechanisms of action of miRNAs such as upregulation of protein expression, or regulation of protein-coding genes via complementarity to the coding region or 3′-UTR of the targeted genes. The recent finding that deletion of the oncogenic miR-17-92 cluster causes congenital syndromic developmental defects in humans suggest that it is necessary to consider non-cancer related effects of miRNAs in designing miRNA therapeutics for cancer treatment 132. This is particularly important when long-term delivery is required.
Due to the complexity and challenges associated with miRNA research, the only miRNA targeted therapeutic that has been tested in clinical trial so far is miravirsen. However, several pharmaceutical companies have miRNA therapeutics in their developmental pipelines. For instance, miRagen Therapeutics (Boulder, Colorado, USA) is currently focusing on the treatment of cardiovascular and muscle diseases by means of miRNA inhibition and replacement. Regulus Therapeutics (San Diego, California, USA) is actively exploring the value of anti-miRs in the treatment of diseases such as fibrosis, HCV infection, atherosclerosis and cancer. MRX34 (developed by Mirna Therapeutics, Austin, Texas, USA), a liposome-formulated mimic of the tumor suppressor miR-34, produced complete tumor regression in two separate orthotopic mouse models of liver cancer, with no observed immunostimulatory activity or toxicity to normal tissues (http://mirnadx.net/___Pipeline/Pipeline_MRX34.aspx). These results have prompted a clinical phase 1 trial (Clinical trial identifier: NCT01829971), which is currently recruiting patients with advanced or metastatic liver cancer.
What are the expectations for the future? We predict that new miRNA-targeting anti-cancer drugs with improved specificity and efficacy will enter the clinical stage and finally be used, in combination with chemotherapy and radiotherapy, for the treatment of cancer patients. The main obstacle of applying miRNA-targeting strategies for clinical use is how to precisely deliver the therapeutic agents into the targeted cells, without inducing unwanted responses in cells other than the intended ones. Nanoparticles specifically engineered for delivery to specific cells will further help with this goal. MiRNA dys-regulation has been involved also in other diseases beyond cancer. Therefore, it can be expected that clinical trials for diseases such as sepsis, which require prompt treatment but only for a short duration, would be very informative with regards to miRNA therapeutic efficacy. However, the development of ncRNA therapeutics for cancer will need to take the possible development of chronic toxicities into consideration.
The novel mechanisms of miRNA action (see Box 2) also offer opportunities for miRNA-targeting strategies. Indeed, the development of the first clinically evaluated miRNA-targeting agent is not based on the canonical mechanism of miRNA action, but initiated on the unusual finding that miR-122 binds to 5′ non-coding region of the HCV genome, and upregulates HCV RNA expression. Similarly, the miRNAs that upregulate protein translation can be targeted for desired control of protein production of oncogenes or tumor suppressors. It can also be envisioned that blocking production, transportation and release of exosome miRNAs may have beneficial effects in controlling cancer development, and this may be achieved by targeting other non-cancerous cell such as the inflammatory cells in the cancer microenvironment.
Acknowledgments
Dr. Calin is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. He is supported also as a Fellow at The University of Texas MD Anderson Research Trust, as a University of Texas System Regents Research Scholar and by the CLL Global Research Foundation. Work in Dr. Calin’s laboratory is supported in part by the NIH/NCI (CA135444), a Department of Defense Breast Cancer Idea Award, Developmental Research Awards in Breast Cancer, Ovarian Cancer, Brain Cancer, Prostate Cancer, Multiple Myeloma, Leukemia (P50 CA100632) and Head and Neck (P50 CA097007) SPOREs, a SINF MDACC_DKFZ grant in CLL, a SINF grant in colorectal cancer, the Laura and John Arnold Foundation, the RGK Foundation and the Estate of C. G. Johnson, Jr. Dr. Ling is an Odyssey Fellow, and his work is supported in part by the Odyssey Program and The Estate of C. G. Johnson. Dr. Fabbri’s lab is supported by the Jean Perkins Foundation, by Nautica Malibu Triathlon Funds, by the Pablove Foundation, by the St. Baldrick’s Foundation, by the Southern California Clinical and Translational Science Institute, by Funds from the Saban Research Institute, by award number P30CA014089 from the National Cancer Institute, by the Hugh and Audy Lou Colvin Foundation, and by the T.J. Martell Foundation. We apologize to all colleagues whose work was not cited because of space restrictions.
Glossary of Terms
- ASO
An antisense oligonucleotide is a single-stranded, chemically modified DNA-like molecule that is 17 to 22 nucleotides in length and designed to be complementary to a selected messenger RNA and thereby specifically inhibits the expression of that gene
- microRNA cluster
A group of at least two microRNAs located close on the genome (usually several hundreds of bases apart) that are generally transcribed in a unique transcript and also commonly regulated
- Noncoding RNAs (ncRNAs)
Any RNA molecule that is not translated into a protein
- Open Reading Frame (ORF)
A section of a sequence piece of DNA that begins with an initiation codon and ends with a nonsense codon. ORFs all have the potential to encode a protein or polypeptide
Biographies
Hui Ling received his M.D. degree from First Military Medical University, China in 2000 and his Ph.D. degree from National University of Singapore, Singapore in 2010. After graduation from medical school, he worked in China as a physician from 2000 to 2005. The training in clinical medicine and working experience in the hospital nurtured him a strong interest in biomedical research, especially in the field of cancer research. During his Ph.D. study from January 2006 to July 2010 in National University of Singapore, he investigated the anticancer potential of triterpenoids contained in Poria cocos, a herb widely used in Traditional Chinese Medicine. He also explored the beneficial activities of naturally occurring or semi-synthetic shogaols, the pungent components of dry ginger, against breast cancer initiation, progression and metastasis. In October 2010, he joined Dr. Calin’s laboratory in MD Anderson Cancer Center as a postdoctoral fellow studying the role of long non-coding RNAs in the initiation and progression of colorectal cancer. His research also involves the study of miRNA localization and its significance in cancer development.
Muller Fabbri is Assistant Professor of Pediatrics and Molecular Microbiology & Immunology at the Children’s Center for Cancer and Blood Diseases and Norris Comprehensive Cancer Center of Children’s Hospital Los Angeles and the University of Southern California, Los Angeles, California, USA. He received his M.D. degree in 1997 from the University of Pisa, Italy. He trained at Thomas Jefferson University, Philadelphia, Pennsylvania, USA first and then at the Ohio State University, Columbus, Ohio, USA where he started his research on the role of microRNAs and other non-coding RNAs in cancer. In particular, he was the first to identify a family of miRNAs (the miR-29 family) able to affect DNA methyltransferases and induce re-expression of epigenetically silenced tumor suppressor genes. In 2009 he was awarded a Sidney Kimmel Foundation Fellowship. He has recently identified a completely new mechanism of action of miRNAs: as ligands and agonists of Toll-like receptors, and modulators of tumor growth and dissemination through this aberrant cross-talk between cancer and immune cells within the tumor microenvironment.
George Adrian Calin received both his M.D. and Ph.D. degrees at Carol Davila University of Medicine in Bucharest, Romania. After working cytogenetic as undergraduate student with Dr. Dragos Stefanescu in Bucharest, he completed a cancer genomics training in Dr. Massimo Negrini’s laboratory at University of Ferrara, Italy. In 2000 he became a postdoctoral fellow at Kimmel Cancer Center in Philadelphia, PA, and while working with Dr. Carlo Croce’s in Philadelphia, Dr. Calin discovered in 2002 the link between human cancers and microRNAs. He is presently an Associate Professor in Experimental Therapeutics at MDACC and his main research interests are: 1) the involvement of non-coding RNAs in human diseases in general and of microRNAs in human cancers in particular, 2) the study of familial predisposition to human cancers, 3) the identification of ncRNA biomarkers in body fluids, and 4) the development of new RNA-based therapeutic options for cancer patients.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin GA, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–87. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 6.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 7.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 11.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–87. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337:1159–1161. doi: 10.1126/science.337.6099.1159. [DOI] [PubMed] [Google Scholar]
- 14.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 17.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thorsen SB, Obad S, Jensen NF, Stenvang J, Kauppinen S. The therapeutic potential of microRNAs in cancer. Cancer J. 2012;18:275–84. doi: 10.1097/PPO.0b013e318258b5d6. [DOI] [PubMed] [Google Scholar]
- 19.van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11:644–56. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 20.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. 2012;11:860–72. doi: 10.1038/nrd3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–46. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 22.Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–8. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 23.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104:9667–72. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 25.Eiring AM, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–65. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–71. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 27.Fabbri M, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–6. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 31.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pigati L, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5:e13515. doi: 10.1371/journal.pone.0013515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cortez MA, et al. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–77. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–64. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spizzo R, Nicoloso MS, Croce CM, Calin GA. SnapShot: MicroRNAs in Cancer. Cell. 2009;137:586–586 e1. doi: 10.1016/j.cell.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 36.Volinia S, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–99. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 38.Hatley ME, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frankel LB, et al. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 40.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mu P, et al. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–11. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 44.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–54. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song SJ, et al. MicroRNA-Antagonism Regulates Breast Cancer Stemness and Metastasis via TET-Family-Dependent Chromatin Remodeling. Cell. 2013;154:311–24. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–4. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 47.Pineau P, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–9. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Felli N, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci U S A. 2005;102:18081–6. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fabbri M, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA. 2011;305:59–67. doi: 10.1001/jama.2010.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mavrakis KJ, et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL) Nat Genet. 2011;43:673–8. doi: 10.1038/ng.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 52.Uhlmann S, et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol. 2012;8:570. doi: 10.1038/msb.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calin GA, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci U S A. 2008;105:5166–71. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calin GA, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 56.Almeida MI, et al. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142:886–896 e9. doi: 10.1053/j.gastro.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Girardot M, et al. miR-28 is a thrombopoietin receptor targeting microRNA detected in a fraction of myeloproliferative neoplasm patient platelets. Blood. 2010;116:437–45. doi: 10.1182/blood-2008-06-165985. [DOI] [PubMed] [Google Scholar]
- 58.Jiang L, et al. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer. 2010;10:318. doi: 10.1186/1471-2407-10-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Galm O, Herman JG, Baylin SB. The fundamental role of epigenetics in hematopoietic malignancies. Blood Rev. 2006;20:1–13. doi: 10.1016/j.blre.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 60.Lujambio A, et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–9. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 61.Bandres E, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–43. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 62.Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220–2. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- 63.Melo S, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci U S A. 2011;108:4394–9. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18:1121–6. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tivnan A, et al. Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticles. PLoS One. 2012;7:e38129. doi: 10.1371/journal.pone.0038129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trang P, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116–22. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci U S A. 2007;104:15472–7. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ji J, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–47. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kota J, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen L, et al. Tumor-specific expression of microRNA-26a suppresses human hepatocellular carcinoma growth via cyclin-dependent and -independent pathways. Mol Ther. 2011;19:1521–8. doi: 10.1038/mt.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lennox KA, Behlke MA. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011;18:1111–20. doi: 10.1038/gt.2011.100. [DOI] [PubMed] [Google Scholar]
- 73.Swarbrick A, et al. miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat Med. 2010;16:1134–40. doi: 10.1038/nm.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Janssen HL, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 75.Lieberman J, Sarnow P. Micromanaging hepatitis C virus. N Engl J Med. 2013;368:1741–3. doi: 10.1056/NEJMe1301348. [DOI] [PubMed] [Google Scholar]
- 76.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 77.Obad S, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371–8. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y, et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120:1678–86. doi: 10.1182/blood-2012-02-410647. [DOI] [PubMed] [Google Scholar]
- 79.Elmen J, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 80.Krutzfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 81.Ma L, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–7. doi: 10.1038/nbt.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valastyan S, et al. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137:1032–46. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84.Ma L, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–56. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kluiver J, et al. Rapid generation of microRNA sponges for microRNA inhibition. PLoS One. 2012;7:e29275. doi: 10.1371/journal.pone.0029275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xie J, et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat Methods. 2012;9:403–9. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Davis S, et al. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009;37:70–7. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dvinge H, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–82. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 89.Gumireddy K, et al. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47:7482–4. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Young DD, Connelly CM, Grohmann C, Deiters A. Small molecule modifiers of microRNA miR-122 function for the treatment of hepatitis C virus infection and hepatocellular carcinoma. J Am Chem Soc. 2010;132:7976–81. doi: 10.1021/ja910275u. [DOI] [PubMed] [Google Scholar]
- 91.Zhang S, Chen L, Jung EJ, Calin GA. Targeting microRNAs with small molecules: from dream to reality. Clin Pharmacol Ther. 2010;87:754–8. doi: 10.1038/clpt.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lehmann SM, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–35. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 93.O’Brien S, et al. Randomized phase III trial of fludarabine plus cyclophosphamide with or without oblimersen sodium (Bcl-2 antisense) in patients with relapsed or refractory chronic lymphocytic leukemia. J Clin Oncol. 2007;25:1114–20. doi: 10.1200/JCO.2006.07.1191. [DOI] [PubMed] [Google Scholar]
- 94.Jiang K. Biotech comes to its ‘antisenses’ after hard-won drug approval. Nat Med. 2013;19:252. doi: 10.1038/nm0313-252. [DOI] [PubMed] [Google Scholar]
- 95.Ding L, et al. Combined transfection of Bcl-2 siRNA and miR-15a oligonucleotides enhanced methotrexate-induced apoptosis in Raji cells. Cancer Biol Med. 2013;10:16–21. doi: 10.7497/j.issn.2095-3941.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hu X, et al. The effect of Bcl-2 siRNA combined with miR-15a oligonucleotides on the growth of Raji cells. Med Oncol. 2013;30:430. doi: 10.1007/s12032-012-0430-6. [DOI] [PubMed] [Google Scholar]
- 97.Pieters R, et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 2007;370:240–50. doi: 10.1016/S0140-6736(07)61126-X. [DOI] [PubMed] [Google Scholar]
- 98.Kotani A, et al. miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood. 2009;114:4169–78. doi: 10.1182/blood-2008-12-191619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kotani A, et al. A novel mutation in the miR-128b gene reduces miRNA processing and leads to glucocorticoid resistance of MLL-AF4 acute lymphocytic leukemia cells. Cell Cycle. 2010;9:1037–42. doi: 10.4161/cc.9.6.11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bockhorn J, et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun. 2013;4:1393. doi: 10.1038/ncomms2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim SJ, et al. Development of microRNA-145 for therapeutic application in breast cancer. J Control Release. 2011;155:427–34. doi: 10.1016/j.jconrel.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 102.Corsten MF, et al. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 103.Memczak S, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 104.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 105.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–43. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Calin GA, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–29. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 107.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Faghihi MA, Wahlestedt C. RNA interference is not involved in natural antisense mediated regulation of gene expression in mammals. Genome Biol. 2006;7:R38. doi: 10.1186/gb-2006-7-5-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ji P, et al. MALAT-1, a novel noncoding RNA, and thyroxin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 116.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ling H, et al. CCAT2, a novel non-coding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013 doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wheeler TM, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–5. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: Progress and prospects. Int J Biochem Cell Biol. 2013;45:1895–910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 121.Gutschner T, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rusk N. AntagoNATs boost gene expression. Nat Methods. 2012;9:437. doi: 10.1038/nmeth.2007. [DOI] [PubMed] [Google Scholar]
- 124.Modarresi F, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–9. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–84. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 126.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer. 2011;11:59–67. doi: 10.1038/nrc2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–38. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Broderick JA, Zamore PD. MicroRNA therapeutics. Gene Ther. 2011;18:1104–10. doi: 10.1038/gt.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pai SI, et al. Prospects of RNA interference therapy for cancer. Gene Ther. 2006;13:464–77. doi: 10.1038/sj.gt.3302694. [DOI] [PubMed] [Google Scholar]
- 130.Flemming A. Regulatory watch: pioneering gene therapy on brink of approval. Nat Rev Drug Discov. 2012;11:664. doi: 10.1038/nrd3835. [DOI] [PubMed] [Google Scholar]
- 131.Gruber K. Europe gives gene therapy the green light. Lancet. 2012;380:e10. doi: 10.1016/s0140-6736(12)61992-8. [DOI] [PubMed] [Google Scholar]
- 132.de Pontual L, et al. Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans. Nat Genet. 2011;43:1026–30. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–8. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim VN. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes Dev. 2006;20:1993–7. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- 135.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–8. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 136.Taft RJ, et al. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–40. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Taft RJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–8. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 138.Langenberger D, et al. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics. 2009;25:2298–301. doi: 10.1093/bioinformatics/btp419. [DOI] [PubMed] [Google Scholar]
- 139.Wang XJ, Gaasterland T, Chua NH. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol. 2005;6:R30. doi: 10.1186/gb-2005-6-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 141.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lai F, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 144.Forman JJ, Legesse-Miller A, Coller HA. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A. 2008;105:14879–84. doi: 10.1073/pnas.0803230105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Redis RS, Calin S, Yang Y, You MJ, Calin GA. Cell-to-cell miRNA transfer: From body homeostasis to therapy. Pharmacol Ther. 2012;136:169–74. doi: 10.1016/j.pharmthera.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yang M, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu Y, et al. Contribution of MyD88 to the tumor exosome-mediated induction of myeloid derived suppressor cells. Am J Pathol. 2010;176:2490–9. doi: 10.2353/ajpath.2010.090777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Klein U, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 152.Wojcik SE, et al. Non-codingRNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis. 2010;31:208–15. doi: 10.1093/carcin/bgp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fabbri M, Valeri N, Calin GA. MicroRNAs and genomic variations: from Proteus tricks to Prometheus gift. Carcinogenesis. 2009;30:912–7. doi: 10.1093/carcin/bgp063. [DOI] [PubMed] [Google Scholar]
- 154.Fabbri M. MicroRNAs and cancer epigenetics. Curr Opin Investig Drugs. 2008;9:583–90. [PubMed] [Google Scholar]
- 155.Chen X, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 156.Zhang HL, et al. Serum miRNA-21: elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate. 2011;71:326–31. doi: 10.1002/pros.21246. [DOI] [PubMed] [Google Scholar]
- 157.Cheng H, et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2011;6:e17745. doi: 10.1371/journal.pone.0017745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–8. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 159.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dews M, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–5. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 163.Meng F, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–43. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 167.Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–5. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yang S, et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene. 2012 doi: 10.1038/onc.2012.432. [DOI] [PubMed] [Google Scholar]
- 169.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tavazoie SF, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Huang Q, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–10. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 172.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 173.Sampson VB, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 174.Tsang WP, Kwok TT. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis. 2008;13:1215–22. doi: 10.1007/s10495-008-0256-z. [DOI] [PubMed] [Google Scholar]
- 175.Zalfa F, et al. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J Biol Chem. 2005;280:33403–10. doi: 10.1074/jbc.M504286200. [DOI] [PubMed] [Google Scholar]
- 176.Muddashetty R, et al. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J Mol Biol. 2002;321:433–45. doi: 10.1016/s0022-2836(02)00655-1. [DOI] [PubMed] [Google Scholar]
- 177.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–38. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–32. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Crawford ED, et al. Diagnostic Performance of PCA3 to Detect Prostate Cancer in Men with Increased Prostate Specific Antigen: A Prospective Study of 1,962 Cases. J Urol. 2012;188:1726–31. doi: 10.1016/j.juro.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 180.Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther. 2002;1:347–55. [PubMed] [Google Scholar]
- 181.Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–4. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]