SUMMARY
Oncogene-induced senescence (OIS) is a tumor suppressive mechanism typified by stable proliferative arrest, a persistent DNA damage response and the senescence-associated secretory phenotype (SASP), which helps to maintain the senescent state and triggers bystander senescence in a paracrine fashion. Here we demonstrate that the tumor suppressive histone variant macroH2A1 is a critical component of the positive feedback loop that maintains SASP gene expression and triggers the induction of paracrine senescence. MacroH2A1 undergoes dramatic genome-wide relocalization during OIS, including its removal from SASP gene chromatin. The removal of macroH2A1 from SASP genes results from a negative feedback loop activated by SASP-mediated endoplasmic reticulum stress. Endoplasmic reticulum stress leads to increased reactive oxygen species and persistent DNA damage response including activation of ATM, which mediates removal macroH2A1 from SASP genes. Together, our findings indicate that macroH2A1 is a critical control point for the regulation of SASP gene expression during senescence.
INTRODUCTION
In the absence of additional genomic insults, activation of an oncogene is often insufficient to transform a cell (Land et al., 1982). Instead, oncogene-induced senescence (OIS), an important tumor suppressive mechanism, is triggered leading to a stable proliferative arrest (Gorgoulis and Halazonetis, 2010; Serrano et al., 1997). OIS is marked by a variety of additional features including morphological changes, persistent DNA damage response (DDR) and senescence-associated secretory phenotype (SASP) (Campisi, 2005). The SASP is a complex pro-inflammatory transcriptional response made up of genes encoding cytokines, chemokines and metalloproteases that mediate a variety of effects (Coppé et al., 2010; Kuilman and Peeper, 2009). SASP factors help to maintain senescence in an autocrine fashion by participating in a positive feedback loop that supports their expression (Acosta et al., 2008a, 2008b). SASP can also function in a paracrine fashion to trigger senescence in bystander cells (Acosta et al., 2013). The SASP helps to recruit immune cells that promote the clearance of senescent cells from tissues (Xue et al., 2007). Not all of the effects of SASP are tumor suppressive, as SASP has also been shown to promote cancer cell proliferation and invasion (Coppé et al., 2008; Krtolica et al., 2001; Liu and Hornsby, 2007).
The SASP response is robustly regulated at the transcriptional level. Given the important roles of the SASP in mediating the senescent phenotype, relatively little is known about how the SASP transcriptional response is regulated. The transcription factors C/EBPβ and NF-κB both play positive roles in regulating SASP gene transcription (Acosta et al., 2008b; Kuilman et al., 2008). Alterations in chromatin modifications, such as H3K9me2, have been implicated in regulating SASP gene expression (Takahashi et al., 2012). Here, we demonstrate an additional feature of chromatin, the histone variant macroH2A1, is a critical control point in the transcriptional regulation of SASP genes.
Another feature of OIS and cellular senescence is persistent DDR (d’Adda di Fagagna, 2008; Schmitt et al., 2002). Several sources of DNA damage have been implicated in triggering the senescence-associated DDR including DNA hyper-replication, depletion of deoxyribonucleotide pools and increased production of reactive oxygen species (ROS) (Mannava et al., 2013; Di Micco et al., 2006; Weyemi et al., 2012). Senescence-associated DDR is mediated by activation of ataxia-telangiectasia, mutated (ATM), which catalyzes phosphorylation and activation of its downstream effectors CHK1, CHK2, p53 and H2AX contributing to proliferative arrest (Mallette et al., 2007). While H2AX phosphorylation (γH2AX) is downstream of ATM activation, we show that another histone variant, macroH2A1, is upstream of the activation of ATM and is critical for persistent DDR during OIS. Interestingly, DDR has also been implicated in the induction of senescence and SASP gene expression (Di Micco et al., 2006; Poele et al., 2002; Rodier et al., 2009; Takahashi et al., 2012).
The expression of the histone variant macroH2A1 increases during OIS (Sporn et al., 2009). MacroH2A1 participates in formation of senescence-associated heterochromatic foci (SAHF) (Zhang et al., 2005). MacroH2A1, which consists of an amino-terminal H2A-like region fused to a macrodomain by a flexible linker, can replace H2A in a subset of nucleosomes (Gamble and Kraus, 2010). MacroH2A1 plays important roles in either positively or negatively regulating transcription by recruiting transcriptional coregulators such as PARP-1, PELP1, CBP and HDAC1 (Chakravarthy et al., 2005; Chen et al., 2014; Gamble et al., 2010; Hussey et al., 2014). MacroH2A1 functions in a variety of physiological and pathological processes including senescence, tumor suppression, inhibiting proliferation and inhibiting stem cell reprogramming (Cantariño et al., 2013; Creppe et al., 2012; Gamble and Kraus, 2010; Kreiling et al., 2011; Novikov et al., 2011; Sporn et al., 2009).
MacroH2A1’s splice variants, macroH2A1.1 and macroH2A1.2, have distinct functions. MacroH2A1.1’s macrodomain can specifically interact with poly(ADP-ribose) (PAR) produced by PAR polymerases (PARPs) (Kustatscher et al., 2005; Timinszky et al., 2009). MacroH2A1.1 expression is lost in several cancer types probably due to its ability to inhibit cancer cell proliferation (Novikov et al., 2011; Sporn and Jung, 2012; Sporn et al., 2009). MacroH2A1.1 and macroH2A1.2 also have distinct roles in regulating transcriptional responses and are enriched in distinct chromatin environments (Chen et al., 2014). Additionally, macroH2A1.1 specifically regulates expression of its target genes by regulating H2B acetylation in a manner that requires its interaction with PAR.
Here, we examine how the genome-wide pattern of macroH2A1 deposition changes during OIS and how these alterations play a critical role in regulation of the transcriptional response underlying the SASP. We found that macroH2A1 undergoes a dramatic genomic relocalization during OIS, with SASP genes in particular undergoing a dramatic loss of macroH2A1. While macroH2A1 is not required for OIS-mediated proliferative arrest, it is a critical component of the positive feedback loop regulating SASP gene expression. MacroH2A1 is also required to render bystander cells susceptible to paracrine senescence. Intriguingly, macroH2A1 is required for OIS-associated persistent DDR triggered by endoplasmic reticulum (ER) stress. The activation of ATM in response to ER stress is at the heart of a negative feedback loop that leads to the removal of macroH2A1 from SASP genes, downregulating their expression. Overall, our results highlight the critical role macroH2A1 plays in coordinating SASP during OIS.
RESULTS
MacroH2A1 genome-wide occupancy is dramatically altered during OIS
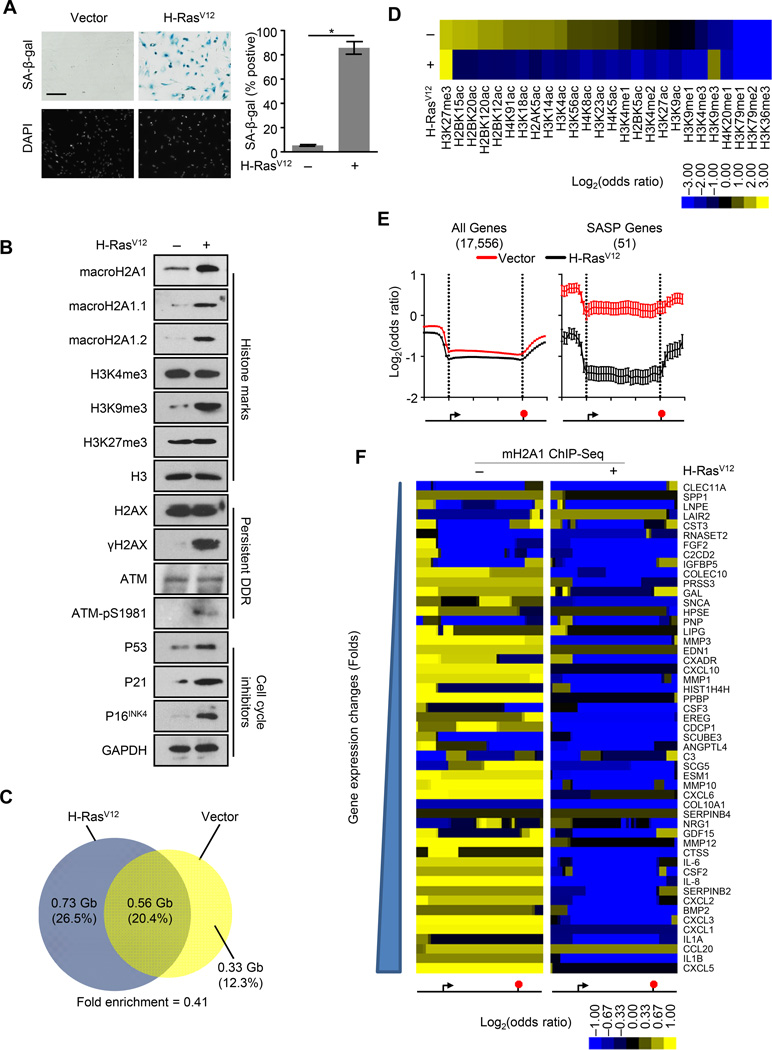
Both increased macroH2A1 expression and altered macroH2A1 sub-nuclear localization have been observed during OIS (Sporn et al., 2009; Zhang et al., 2005). However, the alterations in the location of macroH2A1 along the two-dimensional genome have not been studied in the context of senescence. Previously, we used ChIP-seq to determine the genome-wide localization of macroH2A1 in IMR90 human primary lung fibroblasts (Chen et al., 2014). To determine the genome-wide pattern of macroH2A1 incorporation upon OIS, we performed ChIP-seq for macroH2A1 from IMR90 cells two weeks after retroviral-mediated expression of H-RasV12. The IMR90 cells demonstrated the typical hallmarks of senescence including persistent growth arrest, increased senescence-associated β-galactosidase (SA-β-gal) activity (Figure 1A), increased levels of cell cycle inhibitors, persistent DDR, increased levels of H3K9me3 and macroH2A1 (Figure 1B).
Figure 1. MacroH2A1 undergoes large-scale genome-wide rearrangement upon OIS.
(A) Left, representative SA-β-gal and DAPI counterstaining of IMR90 cells subjected to retroviral-mediated expression of H-RasV12 or empty vector as a control for 14 days. Scale bar represents 200 µm. Right, histogram depicting the percentage of SA-β-gal positive cells. Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
(B) Immunoblots for macroH2A1, various histone marks, DDR factors and cell cycle inhibitors from IMR90 cells treated as in (A).
(C) Two-way Venn diagrams depicting the overlap between ChIP-seq identified bound regions for macroH2A1 in IMR90 cells treated as in (A). Numbers indicate the DNA coverage (Gb) and percentage of DNA in each section.
(D) Heat map depicting the log2(odds ratio) for the overlap between macroH2A1-enriched domains identified in IMR90 cells treated as in (A) and 26 histone PTMs.
(E) Metagene analysis of ChIP-seq data for macroH2A1 from IMR90 cells treated as in (A). Left, all autosomal protein coding genes. Right, fifty-one SASP genes defined from gene expression microarray data. The data represent the average signal in ten 1-kb windows upstream of the TSS, 30 windows spanning the gene body and ten 1-kb windows downstream of the end of the gene. The location of the TSS and the end of the gene were depicted by the left and right vertical dotted line respectively. Error bars, s.e.m. (n = 17,556 and 51 for red and black curves respectively).
(F) Heat map of macroH2A1 occupancy in IMR90 cells treated as in (A) for the 51 individual SASP genes. The data are represented as ten 1 kb windows upstream of the TSS, 30 windows spanning the gene body, and ten 1 kb windows downstream of the end of the gene. See also Figure S1.
By comparing the pattern of macroH2A1 incorporation in normal and OIS cells, we found that macroH2A1 undergoes a dramatic relocalization during senescence. Nearly 33% of macroH2A1-containing genomic regions in normal cells are no longer enriched for macroH2A1 in senescent cells. Additionally, over 56% of macroH2A1-containing regions in OIS cells were devoid of macroH2A1 in normal cells (Figure 1C). This corresponds to a 45% increase in the proportion of the genome found in macroH2A1-enriched domains (from 33% to 46.9% of the genome). We confirmed the specificity of the macroH2A1 antibody and the ability of our ChIP-seq analysis to detect regions of chromatin where macroH2A1 association is gained or lost upon OIS, using loci-specific ChIP-qPCR (Figure S1).
We previously demonstrated that macroH2A1 participates in two physically and functionally distinct chromatin environments either marked by H3K27me3 or a set of histone acetylations including H2B acetylated at K12, K15, K20 and K120, H2A acetylated at K5, H3 acetylated at K4, K14 and K18, and H4 acetylated at K91 (Chen et al., 2014). Co-occupancy with these histone acetylations is a key determinant for gene regulation by macroH2A1. By comparing our macroH2A1 ChIP-seq data from normal and OIS IMR90 cells to publically available data for 26 histone marks in IMR90 cells, we observed a striking difference in the types of chromatin where macroH2A1 association is lost or gained (Figure 1D) Surprisingly, we found that macroH2A1 occupancy is globally reduced specifically at sites of co-occupancy with the set of histone acetylations described above. In addition, we observed an increased association of macroH2A1 in senescent cells with facultative and constitutive heterochromatin marks, H3K27me3 and H3K9me3 respectively, which is consistent with the involvement of macroH2A1 in the formation of SAHF (Zhang et al., 2005).
MacroH2A1 regulates SASP and persistent DDR during OIS
The genes composing the SASP are transcriptionally upregulated during senescence (Coppé et al., 2008; Kuilman and Peeper, 2009). Interestingly, we found that macroH2A1 was specifically enriched on SASP genes (Figures 1E and 1F). SASP genes are also enriched for H2B acetylations typically seen on genes regulated by macroH2A1 (Figure S1) (Chen et al., 2014). Additionally, we observed that macroH2A1 was specifically removed from SASP genes during OIS (Figures 1E and 1F). Together, these observations led us to hypothesize that macroH2A1 plays a role in the regulation of SASP gene expression.
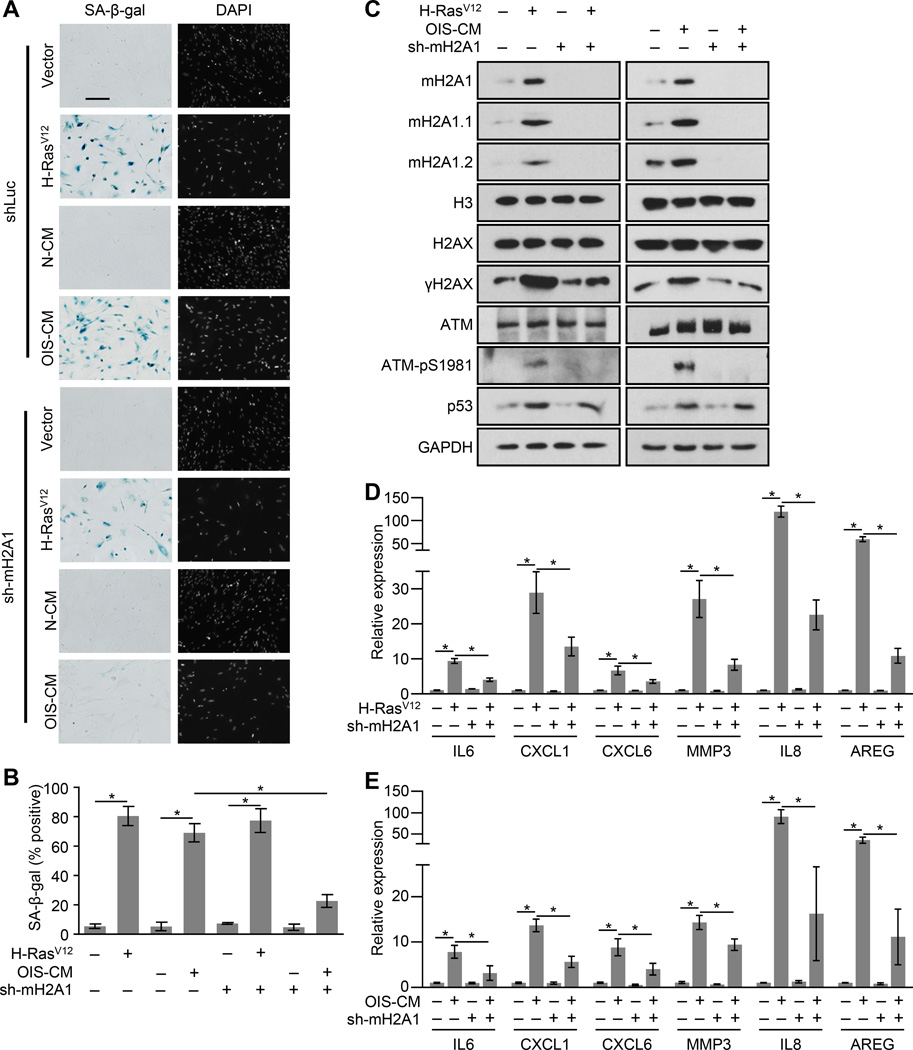
In order to determine the function of macroH2A1 during OIS we infected either macroH2A1-depleted or control cells with H-RasV12. Two weeks post-infection we found that both control and macroH2A1-depleted cells had a similar preponderance of senescent cells as monitored by SA-β-gal activity, increased expression of cell cycle inhibitors and reduced fraction of cells in S phase (Figures 2A—2C, S2). Interestingly, there were major differences in two key features of OIS. As assessed by S1981 phosphorylation, unlike controls cells, macroH2A1-depleted cells did not have increased levels of activated ATM (Figure 2C). Consistently, macroH2A1-depeleted senescent cells also did not accumulate γH2AX compared to control senescent cells. In addition, we examined the expression of several SASP genes in control and macroH2A1-depleted cells upon OIS by RT-qPCR. We observed that loss of macroH2A1 significantly reduced the induction of SASP gene transcription upon OIS (Figure 2D). Overall, this suggests that macroH2A1 is required for both the senescence-associated persistent DDR and the upregulation of SASP gene expression, two senescence hallmarks.
Figure 2. MacroH2A1 is required for SASP gene expression and persistent DDR during OIS and paracrine senescence.
(A) Representative SA-β-gal and DAPI counterstaining of IMR90 cells expressing an shRNA against luciferase (Luc KD, as a control) or macroH2A1 (mH2A1 KD) subjected to retroviral-mediated expression of H-RasV12 for OIS or empty vector as a control for 14 days or subjected to paracrine senescence with cultured media from H-RasV12–mediated senescent IMR90 cells (OIS-CM) or cultured media from IMR90 cells expressing empty vector (as a control, N-CM) for 14 days, as indicated. Scale bar represents 200 µm.
(B) Histogram depicting the percentage of SA-β-gal positive-staining of cells described in (A). Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
(C) Immunoblots for macroH2A1, DDR factors and p53 from cells as described in (A).
(D, E) Reverse transcription coupled to qPCR (RT-qPCR) of IMR90 cells described in (A) for the indicated genes during OIS (D) or paracrine senescence (E). Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests. See also Figure S2.
MacroH2A1 is required for paracrine senescence
The SASP factors induced during senescence have been implicated in both pro-oncogenic and tumor suppressive functions (Gorgoulis and Halazonetis, 2010). The SASP reinforces the senescent phenotype in an autocrine fashion and can also stimulate paracrine senescence in bystander cells by participating in a positive feedback loop that activates the transcription of SASP genes (Acosta et al., 2008b, 2013; Coppé et al., 2008). Treatment of IMR90 cells with conditioned media from OIS cells leads to a senescent phenotype typified by growth arrest, SA-β-gal expression, increased macroH2A1 expression, upregulation of cell cycle inhibitors, and a persistent DDR (Figure S2). However, unlike control cells, macroH2A1-depleted cells do not senesce in response to SASP factor-containing conditioned media from senescent cells (Figures 2A—2C). In addition, macroH2A1-depleted cells do not robustly activate transcription of SASP genes in response to SASP factor-containing conditioned media compared to controls (Figure 2E).
Several of the SASP factors can independently trigger senescence in a paracrine fashion (Acosta et al., 2008b; Kortlever et al., 2006; Kuilman et al., 2008; Wajapeyee et al., 2008). The ability of IL8, one of these SASP factors, to induce senescence is also abrogated in when macroH2A1 is depleted (Figure S2). Consistently, the ability of IL8, like SASP-containing conditioned media, to cause senescence, activate a persistent DDR and activate SASP gene expression is significantly abrogated in cells depleted of macroH2A1 (Figure S2). Overall, this suggests that macroH2A1 is a critical component of the SASP positive feedback loop that helps to maintain senescence and render cells susceptible to paracrine senescence.
MacroH2A1.1 together with PARP is sufficient to trigger SASP and senescence
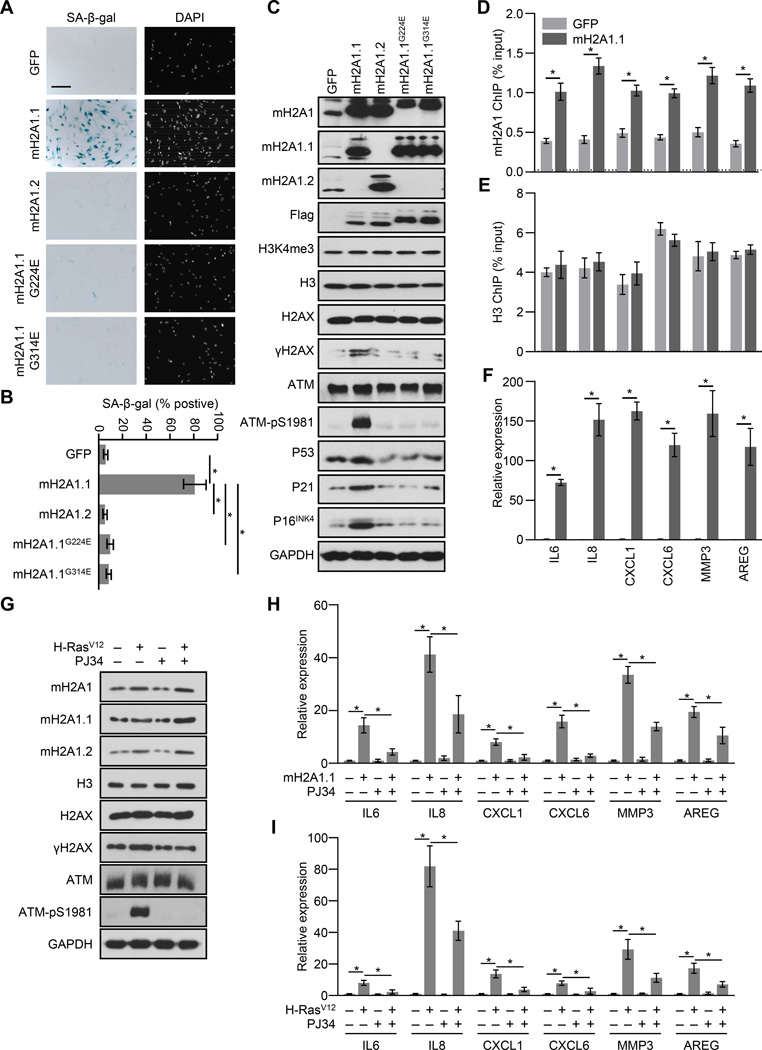
MacroH2A1 exists as either of two splice variants, macroH2A1.1 or macroH2A1.2, the former of which can interact with poly(ADP-ribose)ylated factors and is often suppressed in many types of cancer. MacroH2A1.1 plays a dominant role in regulating the expression of genes found to harbor H2B acetylations on K12, K15, K20 and K120 (Chen et al., 2014). While both macroH2A1.1 and macroH2A1.2 levels are elevated during senescence, many SASP genes exist in H2B acetylated chromatin suggesting that macroH2A1.1 may play a specific role in regulating these genes (Figure S1). When we ectopically expressed macroH2A1.1, macroH2A1.2 or either of two macroH2A1.1 point mutants that can no longer interact with poly(ADP-ribose) (Kustatscher et al., 2005), we found that only ectopic expression of macroH2A1.1 could stimulate senescence (Figure 3). The senescence triggered by macroH2A1.1 exhibited SA-β-gal activity, increased levels of cell cycle inhibitors and persistent DDR marked by activated ATM and γH2AX (Figures 3A—C). In addition, ectopic expression of macroH2A1.1 led to increased macroH2A1 occupancy at SASP genes, while not affecting overall nucleosome occupancy as assayed by H3 ChIP (Figures 3D and 3E). Consistent with a positive role for macroH2A1 in SASP gene expression, ectopic expression of macroH2A1.1 led to a robust SASP transcriptional response (Figure 3F).
Figure 3. Ectopic expression of macroH2A1.1 specifically triggers senescence, SASP expression and persistent DDR.
(A) Representative SA-β-gal and DAPI counterstaining of IMR90 cells after retroviral transduction with expression constructs for GFP (as a control), macroH2A1.1, macroH2A1.2 or two macroH2A1.1 point mutants (G224E and G314E) that are incapable of interacting with PAR isoforms, for 14 days.
(B) Histogram depicting the percentage of SA-β-gal positive-staining of cells described in (A). Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
(C) Immunoblots for macroH2A1, various histone marks, DDR factors and cell cycle inhibitors from IMR90 cells lines described in (A).
(D, E) ChIP-qPCR for macroH2A1 (D) and H3 (E) from IMR90 cells expressing GFP or macroH21.1 at the indicated SASP genes. The horizontal dotted line indicates upper limit of the 95% confidence interval of the signal from no-antibody control ChIPs. Error bars, s.e.m. (n= 3 independent cell passages and retroviral infections). * p < 0.05 from a two-tailed Student’s t-test.
(F) RT–qPCR for the indicated genes from IMR90 cells described in (D) for the indicated SASP genes. Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
(G) Immunoblots for macroH2A1 and DDR factors from IMR90 cells subjected to retroviral-mediated expression of H-RasV12 (+) for OIS or empty vector (−) as a control and 10 µM PJ34 (+) or DMSO (−) as a control as indicated for 3 days.
(H) RT-qPCR for the indicated genes from IMR90 cells after retroviral transduction with expression constructs for macroH2A1.1 (+) or GFP (−) as a control and 10 µM PJ34 (+) or DMSO (−) as a control as indicated for 3 days.
(I) RT-qPCR for the indicated genes from IMR90 cells treated as in (G).
A specific role for macroH2A1.1 in the regulation of SASP gene expression suggests that PARPs may also play a role in this process. Indeed, PARP-1 has previously been shown to regulate SASP gene expression (Ohanna et al., 2011). Using the PARP inhibitor PJ34, we determined that macroH2A1.1-mediated SASP expression requires PARP activity (Figure 3H). In addition PARP inhibition also blocks H-RasV12-induced persistent DDR and SASP gene expression (Figures 3G and 3I). Overall, these results demonstrate a required role for the macroH2A1.1—PARP-1 axis in the regulation of the SASP.
Senescence-associated ER stress and increased ROS requires macroH2A1
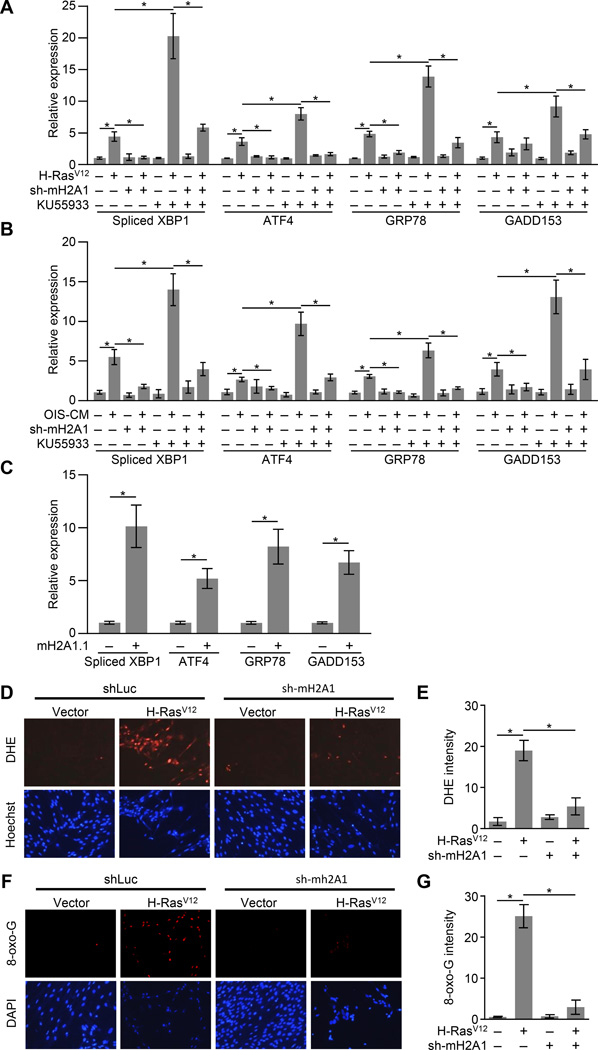
The secretory state of senescent cells has been shown to trigger an ER stress response (Denoyelle et al., 2006; Dörr et al., 2013; Matos et al., 2014; Zhu et al., 2014). ER stress triggers the activation of the unfolded protein response (UPR), which can be monitored by RT-qPCR for spliced XBP1, which is spliced at the ER membrane upon ER stress, and downstream UPR target genes, such as ATF4, GRP78 and GADD153, which are transcriptionally upregulated upon ER stress (Hetz, 2012). We detected ER stress in response to H-RasV12-mediated OIS, SASP-mediated paracrine senescence, or macroH2A1.1-induced senescence (Figures 4A—4C). ER stress in response to either H-RasV12 or SASP-containing conditioned media was abrogated in the absence of macroH2A1 (Figures 4A and 4B).
Figure 4. Senescence-associated ER stress, ROS and oxidative DNA damage requires macroH2A1.
(A) RT-qPCR for the indicated UPR genes from IMR90 cells expressing shRNA against luciferase as a control (−) or macroH2A1 (+) subjected to retroviral-mediated expression of H-RasV12 (+) for OIS or empty vector (−) as a control. Where indicated cells were treated with 10 µM of the ATM inhibitor KU55933 (+) or DMSO (−) as a control for 3 days. Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
(B) As for (A) except cells were treated with SASP-containing conditioned media from H-RasV12-mediated senescent IMR90 cells (OIS-CM, +) or conditioned media from IMR90 cells transduced with empty vector (−) as a control.
(C) RT-qPCR for the indicated UPR genes from IMR90 cells after retroviral transduction with expression constructs for GFP (−) as a control or macroH2A1.1 (+) for 14 days.
(D) Representative fluorescence microscopy images for ROS detection in IMR90 cells expressing shRNA against luciferase as a control (−) or macroH2A1 (+) subjected to retroviral-mediated expression of H-RasV12 (+) for OIS or empty vector (−) as a control. Cells were treated with 10 µM DHE for 30 min prior to collection and counterstained with Hoechst. Scale bar represents 200 µm.
(E) Histogram depicting the average fluorescence of cells described in (D). Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
(F) Representative immunofluorescence microscopy for 8-oxo-dG and counterstained with DAPI in IMR90 cells treated as described in (D). Scale bar represents 200 µm.
(G) Histogram depicting the average fluorescence of cells described in (F). Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
See also Figure S3.
ER stress can lead to increased ROS which can in turn lead to DNA damage (Chen et al., 2012; Higa and Chevet, 2012). Consistently, when macroH2A1 is depleted and senescence-associated ER stress is ameliorated, ROS levels and oxidative DNA damage are also reduced (Figures 4D—4G). Overall, this data is consistent with a model in which macroH2A1 promotes SASP gene expression which leads to enhanced ER stress and consequently more ROS and oxidative DNA damage.
ER stress triggers a negative feedback loop regulating SASP gene expression
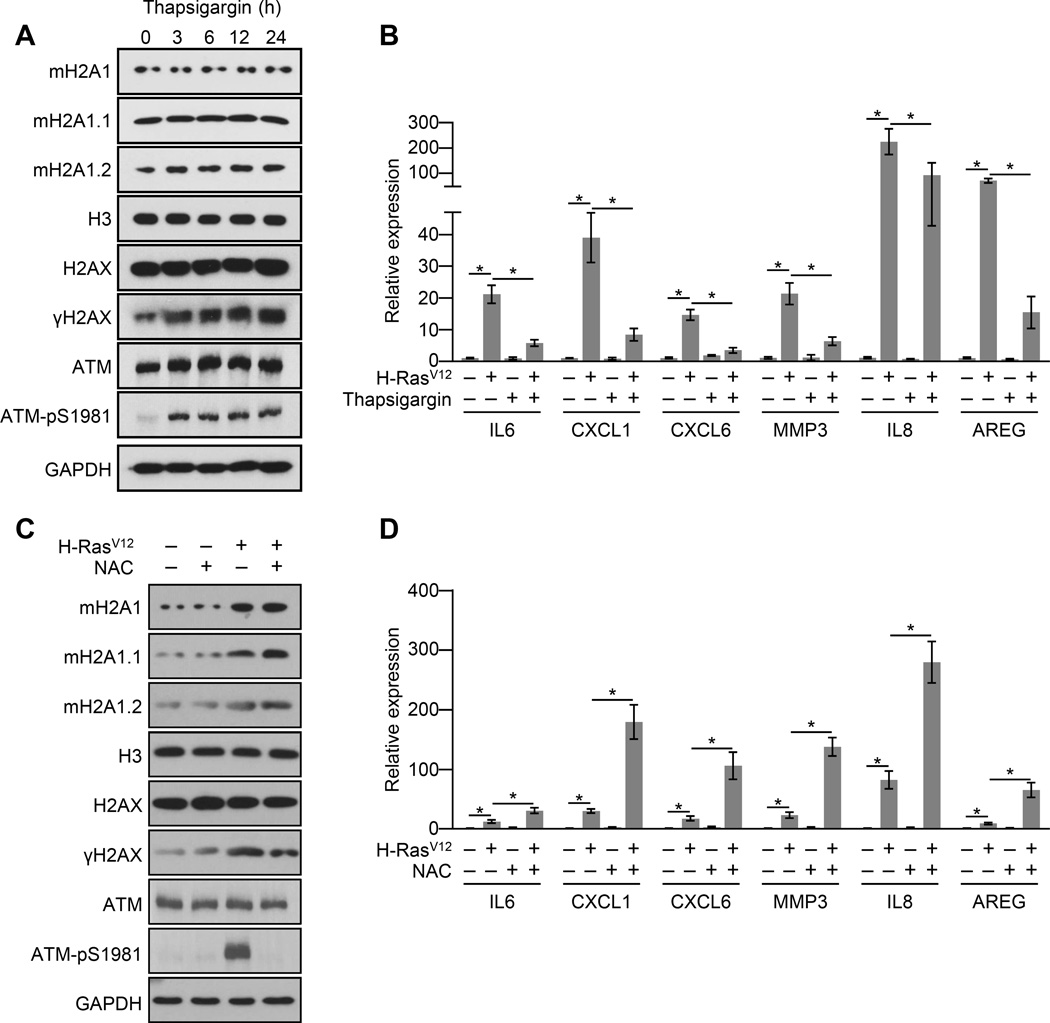
The lack of a persistent DDR in macroH2A1-depleted IMR90 cells undergoing OIS or treatment with SASP factor-containing conditioned media is consistent with the conclusion that the DDR is, at least in part, a consequence of ER stress caused by SASP gene expression. Consistently, drug-induced ER stress with thapsigargin led to an increase in γH2AX and activated ATM in IMR90 cells (Figure 5A). But, if macroH2A1 is a positive regulator of the SASP, why is it removed from SASP genes during OIS? We hypothesized that the ER stress may be triggering a negative feedback loop attenuating SASP gene transcription. To test this we treated cells undergoing OIS with thapsigargin to cause additional and persistent ER stress unrelated to that caused by the SASP to determine the effect on SASP gene transcription. We found that chemical induction of ER stress led to a dramatic reduction in SASP gene transcription (Figure 5B).
Figure 5. ER stress represses SASP gene expression in a ROS dependent manner.
(A) Immunoblots for macroH2A1 and the indicated DDR factors from IMR90 cells treated with 0.25 µM of the ER stress inducing agent, thapsigargin, for the indicated times.
(B) RT-qPCR for the indicated SASP genes from IMR90 cells subjected to retroviral-mediated expression of H-RasV12 (+) for OIS or empty vector (−) as a control and, where indicated, treated with 0.25 µM of the ER stress inducer thapsigargin (+) or DMSO (−) for 3 days. Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
(C) Immunoblots for macroH2A1 and the indicated DDR factors from IMR90 cells subjected to retroviral-mediated expression of H-RasV12 (+) for OIS or empty vector (−) as a control and, where indicated, treated with 10 mM of the antioxidant N-acetyl-cysteine (NAC, +) or DMSO (−) for 3 days.
(D) RT-qPCR for the indicated SASP genes from IMR90 cells treated as in (C). Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
See also Figure S4.
We next focused on how the ER stress negative feedback signal might be transduced to the nucleus. The connection between macroH2A1 and ER stress, increased ROS levels and persistent DDR during OIS combined with the fact that ER stress leads to the generation of ROS (Higa and Chevet, 2012; Jaronen et al., 2014), led us to test the role of ROS in mediating the negative feedback signal from the ER to the nucleus. Treatment of cells undergoing H-RasV12-mediated senescence with the antioxidant N-acetyl-cysteine (NAC) simultaneously reduces the OIS-associated persistent DDR and augments SASP gene transcription during OIS and paracrine senescence (Figures 5C, 5D and S4). These results demonstrate that an ER stress pathway transduced by elevated ROS levels exists to negatively regulate SASP gene transcription and thereby attenuate ER stress.
ATM mediates ER stress-induced SASP gene repression by mobilizing macroH2A1
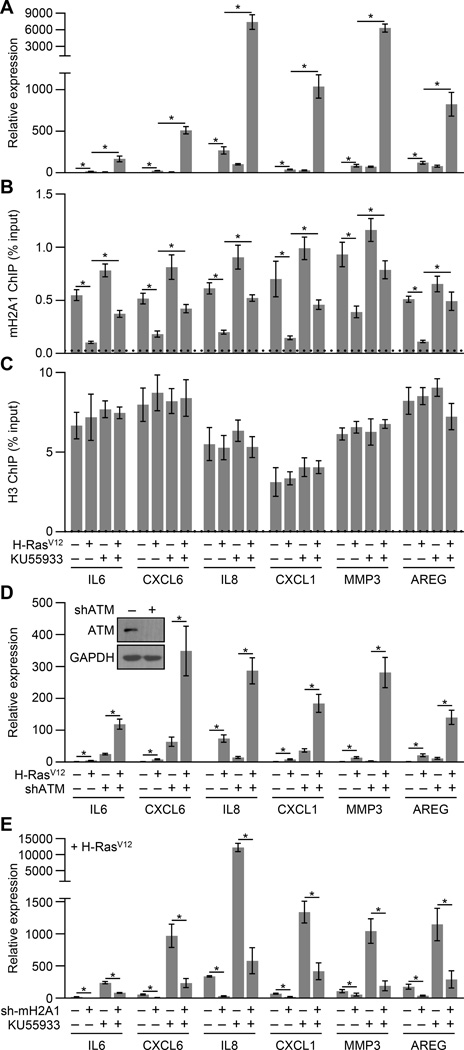
We hypothesized that ER-generated ROS-mediated repression of SASP genes might require aspects of the persistent DDR seen during OIS. When we treated cells with ATM inhibitors, caffeine or KU55933, concurrently with H-RasV12 ectopic expression, we found that inhibition of ATM dramatically increased (hundreds of fold in some cases) the expression of SASP genes during OIS (Figures 6A). Consistently, inhibition of ATM activity or depletion of ATM during H-RasV12 or SASP-mediated senescence led to increased levels of ER stress (Figures 4A, 4B and S3). We further confirmed the role of ATM in repressing SASP gene transcription during OIS by either stably or transiently depleting ATM with three independent shRNAs (Figures 6D and S5).
Figure 6. ATM represses SASP expression during OIS by triggering removal of macroH2A1 from SASP genes.
(A) RT-qPCR for the indicated SASP genes from IMR90 cells subjected to retroviral-mediated expression of H-RasV12 (+) for OIS or empty vector (−) as a control and where indicated cells were treated with 10 µM of the ATM inhibitor KU55933 (+) or DMSO (−) as a control for 3 days. Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
(B,C) ChIP-qPCR for macroH2A1 (B) and H3 (C) from IMR90 cells treated as described in (A). Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
(D) RT-qPCR for the indicated SASP genes from IMR90 cells expressing an shRNA against ATM (+, shATM) or luciferase (−, shLuc) as a control subjected to retroviral-mediated expression of H-RasV12 (+) for OIS or empty vector (−) as a control for 3 days. Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests. Inset, immunoblots for ATM and GAPDH from shLuc (−) and shATM (+) IMR90 cells.
(E) RT-qPCR for the indicated SASP genes from IMR90 cells expressing an shRNA against macroH2A1 (+) or luciferase (−) as a control subjected to retroviral-mediated expression of H-RasV12 for OIS for 3 days. Error bars, s.e.m. (n = 3 independent cell passages and retroviral infections). *p < 0.05 from two-tailed Student’s t-tests.
See also Figure S5.
But, how does ATM activation suppresses SASP? Given the central role macroH2A1 plays in the positive feedback loop maintaining SASP gene expression during senescence, we hypothesized that ATM might repress SASP gene expression by antagonizing macroH2A1 function. Inhibition of ATM did not have an effect on macroH2A1 expression or nuclear localization (Figure S5B). We monitored macroH2A1 occupancy at SASP genes during H-RasV12-mediated OIS induction with or without inhibition of ATM activity. We found that the typical reduction of macroH2A1 occupancy that occurs during senescence is abrogated upon ATM inhibition (Figures 6B, 6C and S5). Additionally, experiments in macroH2A1-depleted cells demonstrate that the increase in SASP gene expression seen when ATM is inhibited during OIS requires macroH2A1 (Figures 6E and S5). Interestingly, ATM activity is not only responsible for the removal of macroH2A1 from SASP genes but is also required for its expanded incorporation into H3K27me3-marked chromatin (Figure S5). Overall, these results indicate that ATM antagonizes macroH2A1-mediated expression of SASP genes during senescence by triggering the removal of macroH2A1 from SASP genes and establishes an ER stress pathway that functions to limit ER stress by repressing the transcription of SASP genes through ROS-mediated DNA damage signaling and ATM.
DISCUSSION
Here we show that the genome-wide distribution of macroH2A1 is dramatically altered during OIS. The chromatin of senescent cells undergoes three-dimensional relocalization during senescence where regions of heterochromatin containing macroH2A1, HMGA1, and H3K9me3 or H3K27me3 form into dense SAHF (Narita et al., 2003, 2006; Zhang et al., 2005). However, during OIS neither H3K27me3 nor H3K9me3 undergo large-scale changes in their pattern of deposition along the linear genome (Chandra et al., 2012). In addition to the three-dimensional alterations in macroH2A1 nuclear localization during OIS (Zhang et al., 2005), macroH2A1 undergoes a large-scale rearrangement across the linear genome. This suggests macroH2A1 deposition and removal from chromatin are dynamically regulated. Little is known about the machinery that regulates macroH2A1 chromatin incorporation and dissociation. The ATP-dependent chromatin remodeling enzyme ATRX plays a role in removing macroH2A1 from subtelomeric chromatin (Ratnakumar et al., 2012). Here we demonstrate that ATM activation triggers a process that also leads to the removal of macroH2A1. Future studies are required to determine the mechanism of ATM-dependent removal of macroH2A1 from chromatin and if ATRX is required for this process.
The SASP factors maintain their own expression through a positive feedback loop. When SASP gene transcription is induced, it leads to secretion of SASP factors, which can function in an autocrine fashion. Both IL8 and CXCL1 can activate their receptor CXCR2 which not only leads to a proliferative arrest, but also to the transcriptional activation of SASP genes (Acosta et al., 2008b). Our data demonstrate that macroH2A1 is a critical component of the positive feedback loop supporting SASP gene expression during both OIS and paracrine senescence (Figure 7). However, while macroH2A1 is required for the growth arrest observed in SASP-mediated paracrine senescence, the histone variant is not required for proliferative arrest during OIS. This is likely because H-RasV12 alone can mediate stabilization of p53 in a manner that requires activation of PRAK kinase (Sun et al., 2007). Alternatively, SASP autocrine and paracrine effects through CXCR2 have been suggested to lead to proliferative arrest through generation of ROS, which leads to DDR and p53 stabilization (Guo et al., 2013). Our experiments demonstrate that macroH2A1 is required for persistent DDR during OIS or SASP-mediated paracrine senescence and together explains why cells lacking macroH2A1 are not susceptible to paracrine senescence. Expression of macroH2A1.1, the macroH2A1 variant sufficient to cause senescence and SASP gene expression, is often downregulated in cancer (Novikov et al., 2011; Sporn and Jung, 2012; Sporn et al., 2009). We hypothesize that loss of macroH2A1.1 may render cancer cells resistant to SASP-mediated senescence and may explain their ability to bypass cellular senescence, an important barrier to oncogenesis.
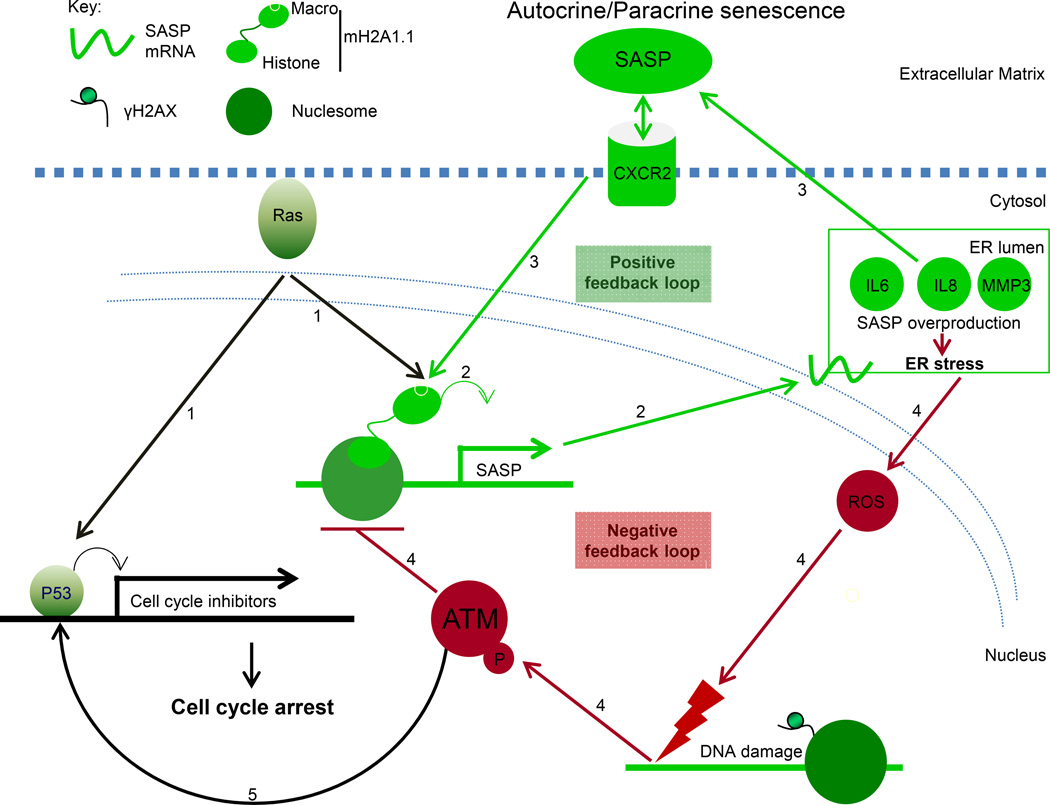
Figure 7. Model depicting the role of macroH2A1 as a critical control point in the regulation of SASP gene expression.
(1) Activated Ras both mediates a proliferative arrest and upregulates the expression of macroH2A1.
(2) Increased macroH2A1.1 levels promote the expression of SASP genes.
(3) Secreted SASP factors can act in an autocrine or paracrine fashion by engaging cell surface receptors, such as CXCR2, which can upregulate the expression of macroH2A1, forming a positive feedback loop which supports SASP gene expression.
(4) The SASP leads to ER stress which triggers ROAMM (reactive oxygen and ATM-mediated macroH2A1 mobilization). The ROS-mediated DNA damage activates ATM which triggers the removal of macroH2A1 from SASP gene chromatin, providing negative feedback on the expression of SASP gene expression.
(5) The activation of ATM can also contribute to the senescent proliferative arrest in a manner similar to activated Ras.
In addition to the macroH2A1-mediated positive feedback loop supporting SASP gene expression, we demonstrate there is a negative feedback loop that limits ER stress during senescence by suppressing SASP gene expression. ER stress is a feature of senescence triggered by induced expression of the secreted factors involved in the SASP (Denoyelle et al., 2006; Dörr et al., 2013; Matos et al., 2014; Zhu et al., 2014). The formation of correct disulfide bonds is critical for many of the components of the SASP. For example CXC type cytokines such as IL8 require the formation of two disulfide bonds for folding and function (Fernandez and Lolis, 2002). The rearrangement of disulfide bonds is catalyzed in the ER by protein disulfide isomerases (PDIs), which are critical for the folding of disulfide-containing secreted factors (Higa and Chevet, 2012). Increased PDI activity due to unfolded protein can lead to increased ROS though both its ability to regulate the NADPH oxidase NOX4 and futile rounds of thiol-disulfide isomerization in coordination with the flavoreductase Ero1 (Higa and Chevet, 2012; Jaronen et al., 2014). Consistently, NOX4 upregulation can trigger senescence (Kodama et al., 2013; Weyemi et al., 2012).
Canonical UPR pathways seek to ameliorate ER stress through several mechanisms including increased synthesis of ER chaperones, inhibition of translation and regulated IRE1-dependent decay (RIDD) of mRNA (Hollien et al., 2009; Ron and Walter, 2007). The increased level of ROS caused by ER stress leads to DNA damage, activating a DDR involving ATM (Chen et al., 2012). Our data demonstrate that active ATM triggers a process leading to removal of macroH2A1 from SASP genes, which keeps ER stress in check in a process we have termed the ROAMM (reactive oxygen and ATM-mediated macroH2A1 mobilization) pathway (Figure 7). The ROAMM pathway explains why ATM inhibition leads to dramatic increases in SASP gene expression and ER stress. In the absence of ATM-mediated negative feedback from ER stress leading to the removal of macroH2A1 from SASP genes, the SASP positive feedback loop runs unopposed triggering a dramatic increase in both SASP expression and ER stress.
Overall, our data demonstrate the central importance of macroH2A1 as a control point for both positive and negative regulation of SASP gene expression. The SASP is an important therapeutic target in cancer treatment and aging. In healthy individuals, the SASP aids in recruitment of immune cells to clear senescent cells from tissues (Kang et al., 2011; Krizhanovsky et al., 2008; Xue et al., 2007). In aging or immunocompromised individuals SASP fails to lead to the clearance of senescent cells which may contribute to a persistent inflammatory state (Baker et al., 2011; Schmitt, 2007). Treatment of cancer patients with various chemotherapeutics can lead to senescence of a subset of cancer cells and induce SASP which has been implicated in promoting oncogenesis, chemoresistance and metastasis (Dörr et al., 2013; Fumagalli and d’Adda di Fagagna, 2009). Further understanding of the mechanism by which macroH2A1 regulates SASP gene expression and how it is antagonized by ER stress and ATM will lead to improved methods for treating post-therapy “senescent” cancer cells and preventing relapse.
EXPERIMENTAL PROCEDURES
Cell culture
All cells used in this study were derived from IMR90 primary human fetal lung fibroblasts (ATCC). The details of cell lines produced for and used in this study are presented in Supplemental Experimental Procedures.
SA-β-Gal staining and fluorescence microscopy
Details of the SA-β-Gal staining, brdU incorporation, immunofluorescence and ROS detection protocols are presented in Supplemental Experimental Procedures. Cells were quantified over four separate fields containing at least 80 cells.
Immunoblots and acid-extraction of histones
Immunoblots and acid-extraction of histones were performed as previously described (Chen et al., 2014). A list of antibodies is available in Table S1. Detailed methods are available in Supplemental Experimental Procedures.
Chromatin Immunoprecipitation (ChIP)
Chromatin Immunoprecipitation (ChIP) was performed as previously described (Chen et al., 2014). Primers used for ChIP-qPCR are listed in Table S2. Detailed ChIP protocol is presented in Supplemental Experimental Procedures.
ChIP-Seq
IMR90 cells were subjected to retroviral-mediated expression of H-RasV12 or empty vector as a control. Following 14 days selection in 1 µg/ml puromycin, a portion of the cells were stained for SA-β-Gal activity to confirm senescence. The cells were processed for ChIP-seq as previously described (Chen et al., 2014). Reads mapped to the hg19 draft of the human genome and ISOR processed data were uploaded to GEO (GSE64601). Our previous macroH2A1 ChIP-seq from normally growing IMR90 cells is available from GEO (GSE54847). ChIP-seq data for the 26 additional histone marks in IMR90 cells were downloaded from the GEO website (GSE16256).
Expression microarray analysis
SASP genes were identified from previously published publically available expression microarray data (GSE40349) according to the details in Supplemental Experimental Procedures.
RT-qPCR
RNA purification and RT-qPCR were performed as described previously (Chen et al., 2014). Expression primers used in this study are listed in Table S3. The detailed RT-qPCR method is presented in Supplemental Experimental Procedures.
Statistical analysis
All SA-β-Gal staining, ROS detection, immunofluorescence, ChIP and RT-qPCR experiments were repeated at least 3 times with independent biological samples. All immunoblots were performed at least twice using independent biological samples. Results are presented as means ± s.e.m. Two-tailed Student’s t-tests were used to determine the significance of differences between samples indicated in figures. For ChIP-Seq analysis, Fisher exact tests have been applied to determine the significance of enrichment in count data.
Supplementary Material
ACKNOWLEDGMENTS
We thank S.B. Horwitz, H.M. McDaid and C.S. Rubin from Albert Einstein College of Medicine for helpful discussions and sharing equipment and reagents. The high-throughput sequencing was performed by the Einstein Epigenomics Core Facility. Microscopy was performed at the Einstein Analytical Imaging Facility supported by the US National Institutes of Health (NIH) Cancer Center Support Grant (P30 CA013330). This work was supported by the NIH training grant T32GM007491-38 (P.D.R) and grant R01CA155232 (M.J.G).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
ChIP-seq data have been deposited in the Gene Expression Omnibus database under accession code GSE64601.
SUPPLEMENTAL INFORMATION
Supplemental information includes five figures, three tables and supplemental experimental procedures.
The authors declare no competing financial interests.
REFERENCES
- Acosta J, O’Loghlen A, Banito A, Raguz S, Gil J. Control of senescence by CXCR2 and its ligands. Cell Cycle. 2008a;7:2956–2959. doi: 10.4161/cc.7.19.6780. [DOI] [PubMed] [Google Scholar]
- Acosta JC, O’Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008b;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- Acosta JC, Banito A, Wuestefeld T, Georgilis A, Janich P, Morton JP, Athineos D, Kang T-W, Lasitschka F, Andrulis M, et al. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat. Cell Biol. 2013;15:978–990. doi: 10.1038/ncb2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Cantariño N, Douet J, Buschbeck M. MacroH2A - An epigenetic regulator of cancer. Cancer Lett. 2013;336:247–252. doi: 10.1016/j.canlet.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Gundimella SKY, Caron C, Perche P, Pehrson JR, Khochbin S, Luger K. Structural characterization of the histone variant macroH2A. Mol. Cell. Biol. 2005;25:7616–7624. doi: 10.1128/MCB.25.17.7616-7624.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T, Kirschner K, Thuret J-Y, Pope BD, Ryba T, Newman S, Ahmed K, Samarajiwa Sa, Salama R, Carroll T, et al. Independence of repressive histone marks and chromatin compaction during senescent heterochromatic layer formation. Mol. Cell. 2012;47:203–214. doi: 10.1016/j.molcel.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BPC, Li M, Asaithamby A. New insights into the roles of ATM and DNA-PKcs in the cellular response to oxidative stress. Cancer Lett. 2012;327:103–110. doi: 10.1016/j.canlet.2011.12.004. [DOI] [PubMed] [Google Scholar]
- Chen H, Ruiz PD, Novikov L, Casill AD, Park JW, Gamble MJ. MacroH2A1.1 and PARP-1 cooperate to regulate transcription by promoting CBP-mediated H2B acetylation. Nat. Struct. Mol. Biol. 2014;21:981–989. doi: 10.1038/nsmb.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé J-P, Patil CK, Rodier F, Sun Y, Muñoz DP, Goldstein J, Nelson PS, Desprez P-Y, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé J-P, Desprez P-Y, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creppe C, Posavec M, Douet J, Buschbeck M. MacroH2A in stem cells: a story beyond gene repression. Epigenomics. 2012;4:221–227. doi: 10.2217/epi.12.8. [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- Denoyelle C, Abou-Rjaily G, Bezrookove V, Verhaegen M, Johnson TM, Fullen DR, Pointer JN, Gruber SB, Su LD, Nikiforov Ma, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat. Cell Biol. 2006;8:1053–1063. doi: 10.1038/ncb1471. [DOI] [PubMed] [Google Scholar]
- Dörr JR, Yu Y, Milanovic M, Beuster G, Zasada C, Däbritz JHM, Lisec J, Lenze D, Gerhardt A, Schleicher K, et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature. 2013;501:421–425. doi: 10.1038/nature12437. [DOI] [PubMed] [Google Scholar]
- Fernandez E, Lolis E. Structure, function, and inhibition of chemokines. Annu. Rev. Pharmacol. Toxicol. 2002;42:469–499. doi: 10.1146/annurev.pharmtox.42.091901.115838. [DOI] [PubMed] [Google Scholar]
- Fumagalli M, d’Adda di Fagagna F. SASPense and DDRama in cancer and ageing. Nat. Cell Biol. 2009;11:921–923. doi: 10.1038/ncb0809-921. [DOI] [PubMed] [Google Scholar]
- Gamble M, Kraus WL. Multiple facets of the unique histone variant macroH2A: From genomics to cell biology. Cell Cycle. 2010;9:70–69. doi: 10.4161/cc.9.13.12144. [DOI] [PubMed] [Google Scholar]
- Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev. 2010;24:21–32. doi: 10.1101/gad.1876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Halazonetis TD. Oncogene-induced senescence: the bright and dark side of the response. Curr. Opin. Cell Biol. 2010;22:816–827. doi: 10.1016/j.ceb.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Guo H, Liu Z, Xu B, Hu H, Wei Z, Liu Q, Zhang X, Ding X, Wang Y, Zhao M, et al. Chemokine receptor CXCR2 is transactivated by p53 and induces p38-mediated cellular senescence in response to DNA damage. Aging Cell. 2013;12:1110–1121. doi: 10.1111/acel.12138. [DOI] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13:89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Higa A, Chevet E. Redox signaling loops in the unfolded protein response. Cell. Signal. 2012;24:1548–1555. doi: 10.1016/j.cellsig.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey KM, Chen H, Yang C, Park E, Hah N, Erdjument-Bromage H, Tempst P, Gamble MJ, Kraus WL. The histone variant MacroH2A1 regulates target gene expression in part by recruiting the transcriptional coregulator PELP1. Mol. Cell. Biol. 2014;34:2437–2449. doi: 10.1128/MCB.01315-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaronen M, Goldsteins G, Koistinaho J. ER stress and unfolded protein response in amyotrophic lateral sclerosis-a controversial role of protein disulphide isomerase. Front. Cell. Neurosci. 2014;8:402. doi: 10.3389/fncel.2014.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T-W, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, Hohmeyer A, Gereke M, Rudalska R, Potapova A, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- Kodama R, Kato M, Furuta S, Ueno S, Zhang Y, Matsuno K, Yabe-Nishimura C, Tanaka E, Kamata T. ROS-generating oxidases Nox1 and Nox4 contribute to oncogenic Ras-induced premature senescence. Genes Cells. 2013;18:32–41. doi: 10.1111/gtc.12015. [DOI] [PubMed] [Google Scholar]
- Kortlever RM, Higgins PJ, Bernards R. Plasminogen activator inhibitor-1 is a critical downstream target of p53 in the induction of replicative senescence. Nat. Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiling Ja, Tamamori-Adachi M, Sexton AN, Jeyapalan JC, Munoz-Najar U, Peterson AL, Manivannan J, Rogers ES, Pchelintsev Na, Adams PD, et al. Age-associated increase in heterochromatic marks in murine and primate tissues. Aging Cell. 2011;10:292–304. doi: 10.1111/j.1474-9726.2010.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Yon M, Dickins Ra, Hearn S, Simon J, Miething C, Yee H, Zender L, Lowe SW. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Peeper DS. Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer. 2009;9:81–94. doi: 10.1038/nrc2560. [DOI] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LCW, Douma S, van Doorn R, Desmet CJ, Aarden La, Mooi WJ, Peeper DS. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG. Splicing regulates NAD metabolite binding to histone macroH2A. Nat. Struct. Mol. Biol. 2005;12:624–625. doi: 10.1038/nsmb956. [DOI] [PubMed] [Google Scholar]
- Land H, Parada L, Weinberg R. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1982;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- Liu D, Hornsby PJ. Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 2007;67:3117–3126. doi: 10.1158/0008-5472.CAN-06-3452. [DOI] [PubMed] [Google Scholar]
- Mallette Fa, Gaumont-Leclerc M-F, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannava S, Moparthy KC, Wheeler LJ, Natarajan V, Zucker SN, Fink EE, Im M, Flanagan S, Burhans WC, Zeitouni NC, et al. Depletion of deoxyribonucleotide pools is an endogenous source of DNA damage in cells undergoing oncogene-induced senescence. Am. J. Pathol. 2013;182:142–151. doi: 10.1016/j.ajpath.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos L, Gouveia AM, Almeida H. ER Stress Response in Human Cellular Models of Senescence. J. Gerontol. A. Biol. Sci. Med. Sci. 2014:1–13. doi: 10.1093/gerona/glu129. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre’ M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Narita M, Nuñez S, Heard E. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- Narita M, Narita M, Krizhanovsky V, Nuñez S, Chicas A, Hearn Sa, Myers MP, Lowe SW. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- Novikov L, Park JW, Chen H, Klerman H, Jalloh AS, Gamble MJ. QKI-mediated alternative splicing of the histone variant MacroH2A1 regulates cancer cell proliferation. Mol. Cell. Biol. 2011;31:4244–4255. doi: 10.1128/MCB.05244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohanna M, Giuliano S, Bonet C, Imbert V, Hofman V, Zangari J, Bille K, Robert C, Bressac-de Paillerets B, Hofman P, et al. Senescent cells develop a PARP-1 and nuclear factor-{kappa}B-associated secretome (PNAS) Genes Dev. 2011;25:1245–1261. doi: 10.1101/gad.625811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poele Rte, Okorokov A, Jardine L. DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 2002;62:1876–1883. [PubMed] [Google Scholar]
- Ratnakumar K, Duarte LF, LeRoy G, Hasson D, Smeets D, Vardabasso C, Bönisch C, Zeng T, Xiang B, Zhang DY, et al. ATRX-mediated chromatin association of histone variant macroH2A1 regulates α-globin expression. Genes Dev. 2012;26:433–438. doi: 10.1101/gad.179416.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Coppé J-P, Patil CK, Hoeijmakers WaM, Muñoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Schmitt Ca. Cellular senescence and cancer treatment. Biochim. Biophys. Acta. 2007;1775:5–20. doi: 10.1016/j.bbcan.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Schmitt C, Fridman J, Yang M, Lee S. A Senescence Program Controlled by p53 and p16 INK4a Contributes to the Outcome of Cancer Therapy. Cell. 2002;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin A, McCurrach M, Beach D, Lowe S. Oncogenic ras Provokes Premature Cell Senescence Associated with Accumulation of p53 and p16 INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Sporn JC, Jung B. Differential regulation and predictive potential of MacroH2A1 isoforms in colon cancer. Am. J. Pathol. 2012;180:2516–2526. doi: 10.1016/j.ajpath.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn JC, Kustatscher G, Hothorn T, Collado M, Serrano M, Muley T, Schnabel P, Ladurner aG. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene. 2009;28:3423–3428. doi: 10.1038/onc.2009.26. [DOI] [PubMed] [Google Scholar]
- Sun P, Yoshizuka N, New L, Moser Ba, Li Y, Liao R, Xie C, Chen J, Deng Q, Yamout M, et al. PRAK is essential for ras-induced senescence and tumor suppression. Cell. 2007;128:295–308. doi: 10.1016/j.cell.2006.11.050. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Imai Y, Yamakoshi K, Kuninaka S, Ohtani N, Yoshimoto S, Hori S, Tachibana M, Anderton E, Takeuchi T, et al. DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Mol. Cell. 2012;45:123–131. doi: 10.1016/j.molcel.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EHK, Scheffzek K, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- Wajapeyee N, Serra RW, Zhu X, Mahalingam M, Green MR. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell. 2008;132:363–374. doi: 10.1016/j.cell.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyemi U, Lagente-Chevallier O, Boufraqech M, Prenois F, Courtin F, Caillou B, Talbot M, Dardalhon M, Al Ghuzlan a, Bidart J-M, et al. ROS-generating NADPH oxidase NOX4 is a critical mediator in oncogenic H-Ras-induced DNA damage and subsequent senescence. Oncogene. 2012;31:1117–1129. doi: 10.1038/onc.2011.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins Ra, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Poustovoitov MV, Ye X, Santos Ha, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu Aa, Dunbrack RL, et al. Formation of MacroH2Acontaining senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Zhu B, Ferry CH, Markell LK, Blazanin N, Glick AB, Gonzalez FJ, Peters JM. The nuclear receptor peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) promotes oncogene-induced cellular senescence through repression of endoplasmic reticulum stress. J. Biol. Chem. 2014;289:20102–20119. doi: 10.1074/jbc.M114.551069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.