SUMMARY
The product of the Brca1 tumor-suppressor gene is involved in multiple aspects of the cellular DNA damage response (DDR), including activation of cell cycle arrests and DNA double-stranded break (DSB) repair by homologous recombination (HR). Prior reports demonstrated that Brca1 recruitment to areas of DNA breakage depended on Rap80 and the RNF8/RNF168 E3 ubiquitin ligases. Here we extend these findings by showing that Rap80 is only required for the binding of Brca1 to regions flanking the DSB, while Brca1 binding directly to DNA breaks requires Nbs1. These differential recruitment mechanisms differentially affect Brca1 functions: 1) Rap80-dependent recruitment of Brca1 to chromatin flanking DNA breaks is required for Brca1 phosphorylation at serine 1387 and 1423 by ATM and, consequently, for the activation of S and G2 checkpoints; and 2) Brca1 interaction with Nbs1 upon DSB induction results in an Nbs1-dependent recruitment of Brca1 directly to the DNA break and is required for non-homologous end-joining repair. Together, these findings illustrate that spatially distinct fractions of Brca1 exist at the DSB site, which are recruited by different mechanisms and execute different functions in the DDR.
Keywords: Brca1, Rap80, DNA double-strand break, DNA repair, cell cycle arrest
INTRODUCTION
Both external exposures and natural metabolic processes provide constant challenges to the integrity of our cellular DNA. An intricate network of DNA damage response (DDR) pathways has evolved to enable cells to cope with various types of DNA lesions. One of the most significant components of this machinery is the product of the human cancer suppressor gene, Brca1, which plays a critical role in maintaining genomic integrity. Brca1 has multiple functions in the human DDR, including repair of DNA double-strand breaks (DSBs) and activation of cell cycle checkpoints upon DSB induction (1). Mutations in the Brca1 gene result in genomic instability, predisposing individuals to breast and ovarian cancer (1). On the other hand, loss of functional Brca1 sensitizes cancer cells to radiation and particular types of chemotherapy, highlighting the critical role of Brca1 in the DDR (2).
There are two major pathways that repair DSBs in mammalian cells, the error-prone non-homologous end joining (NHEJ) and the error-free homologous recombination (HR). NHEJ, which is the dominant DSB repair pathway in mammalian cells (3), can function throughout the cell cycle, whereas HR is limited to the S and G2 phases as it requires a homologous sequence on the sister chromatid. Among its multiple functions in the DDR, Brca1 plays a well-established role in repair of DSBs by HR. Brca1 interacts with CtIP at the break site (4) promoting DSB processing (5), an event that is required for initiation of HR. In line with these findings, Brca1 is required for the generation of the single stranded DNA regions that are formed as a consequence of the DSB processing (6). Consequently, Brca1 promotes accumulation of Rad51 at the DSB (7), which facilitates sister chromatid invasion during HR. However, in addition to the “classic” role of Brca1 in HR, some lines of evidence suggest that it may also be involved in DSB repair by NHEJ. Two studies have shown that the re-ligation of a linearized plasmid by NHEJ is impaired in the absence of Brca1 (8,9). However, while suggesting a potentially new and intriguing role of Brca1 in DNA repair, evidence gained from these studies is limited due to the assessment of the NHEJ activity using artificially introduced plasmid DNA, which differs significantly from chromatin. Therefore, the question remains whether Brca1 is involved in repair of genomic DSBs by NHEJ in mammalian cells.
Another important function of Brca1 is the activation of cell cycle checkpoints in response to DNA damage. Brca1 is a target of the ATM kinase, which is rapidly activated following DSB induction (10) and recruited to the DSB site in an MRN-dependent manner resulting in amplification of ATM signaling (11). ATM phosphorylates Brca1 at serine residues 1387 and 1423 in response to DSB induction and these phosphorylation events are required for induction of the S and G2/M checkpoints, respectively (12,13).
In light of the multiple functions of Brca1 in the DDR, identification of mechanisms involved in recruiting Brca1 to sites of DNA breakage has been of great interest. The E3 ubiquitin ligase, RNF8, is recruited to γH2AX domains at the DSB site, with MDC1 serving as a mediator between RNF8 and γH2AX (14,15). RNF8 initiates the formation of ubiquitin conjugates at histone proteins that are sustained by the E3 ubiquitin ligase RNF168 (16,17). Thereafter, a multi-protein complex containing Rap80 links Brca1 to the ubiquitin conjugates (18-20), thus recruiting it to the chromatin. However, a previously published study suggested that the RNF8/RNF168-Rap80 pathway might recruit only a portion of the Brca1 pool to the DSB (7). Using laser microirradiation to induce DSBs in BrdU-treated cells, Bekker-Jensen et al. investigated the recruitment of various DDR factors to the break site and observed that a down-regulation of MDC1, a DDR factor that was later shown to recruit RNF8 to the DSB (15), only partially reduced the presence of Brca1 in the laser track (7). In contrast, 53BP1 recruitment, which also depends on the RNF8 activity (15), was fully inhibited by the MDC1 knock-down (7). Together, these data suggested that an additional mechanism of Brca1 recruitment that is independent of the RNF8/RNF168-Rap80 pathway might exist.
Furthermore, Brca1 has been shown to reside in distinct multi-protein complexes (1,21) with different functions of Brca1 in the DDR being attributed to specific protein macrocomplexes (1). Notably, several Brca1 macrocomplexes do not contain Rap80 (21), suggesting an existence of a Rap80-independent recruitment mechanism of Brca1 to the break site. Here, we demonstrate that different molecular mechanisms recruit Brca1 to distinct chromatin regions near the DSB. While the Rap80-dependent pathway recruits Brca1 to chromatin regions flanking the DSB, Brca1 is recruited directly to the DNA break through an interaction with Nbs1. Further, we show that Brca1 is involved in re-ligation of genomic DSBs by NHEJ and that the Brca1 fraction residing in the vicinity of the DSB, but not in the flanking chromatin, is required for DSB religation. On the other hand, Brca1 that is recruited to the flanking regions appears to control DSB processing and induction of S-phase and G2/M cell cycle arrests.
MATERIALS AND METHODS
Cell Culture, Chemicals, siRNA
MCF7 and HeLa cells were grown in DMEM supplemented with 10% FBS. 4-OHT (Sigma, St Louis, MO) was added to a final concentration of 1 μM. Shield-1 (Cheminpharma, New Haven, CT) was added to a final concentration of 1 μM for 3 h to stabilize the ddI-PpoI fusion protein. ON-TARGETplus SMARTpool siRNA towards Brca1, NBS1, Rap80, ATM (Dharmacon, Boulder, CO) or ON-TARGETplus Non-targeting siRNA (Dharmacon, Boulder, CO) was transfected into cells using Dharmafect-1 transfection reagent (Dharmacon, Boulder, CO) according to manufacturer's protocols to a final concentration of 100 nM. Olaparib (AZD2281) (Selleckchem, Houston, TX) and CP466722 (22) were added to the tissue culture medium at a final concentration of 5 μM 1 h prior to I-PpoI induction and remained in the medium for the duration of the experiment.
Antibodies
Antibodies used for immunoblotting were: beta-actin (C4, Chemicon), ATM (MAT3; gift from Y. Shiloh, Tel-Aviv, Israel), pKAP1 S824 (Bethyl laboratories), pCHK2 T68 (Cell Signalling), pATM S1981 (Epitomics), Brca1 (OP92 and OP93, Calbiochem, La Jolla, CA), Nbs1 (NB100-143, Novus, Littleton, CO), Rap80 (70822, Abcam, Cambridge, MA), pBrca1 S1387 (A300-007A, Bethyl, Montgomery, TX), pBrca1 S1423 (2838, Abcam, Abcam, Cambridge, MA).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed as previously described (23). Antibodies used for the ChIP assay were: Normal rabbit IgG (Cell Signaling, Beverly, Massachusetts), Brca1 (OP92 and OP93, Calbiochem, La Jolla, CA), Nbs1 (NB100-143, Novus, Littleton, CO), γH2AX (05-636, Millipore, Billerica, MA), Rap80 (ab70822, Abcam, Cambridge, MA), MDC1 (Abcam, Cambridge, MA), RNF8 (ab4183, Abcam, Cambridge, MA). Appropriate negative controls using normal rabbit IgG were performed for all ChIP experiments.
DNA Repair Assays
Assessments of DNA double strand break repair by PCR after I-PpoI induction were performed as previously described (23). Comet assays were performed using the Trevigen CometAssay Kit (Trevigen, Gaithersburg, MD) according to the manufacturer's protocol. Cell images were assessed by the TriTek CometScore 1.6.1.21 software. Olive tail moment (OTM) was determined as a product of DNA in the tail and the mean distance of migration in the tail.
Cell Survival Analysis
10 days following cytotoxic treatment cells were incubated with MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide). The product formazan was eluted with isopropanol containing 0.04 M HCl and quantified using a spectrophotometer. % survival was calculated in relation to untreated control for each sample.
Immunoblot and Protein Immunoprecipitation
Immunoblots were performed as previously described (24). For immunoprecipitations, cells were washed with PBS and lysed on ice in TNG buffer (50mM Tris (pH 7.5), 200mM NaCl, 50mM NaF, protease inhibitors) for 30 min. Samples were pre-cleared with Protein A / Protein G agarose beads (Calbiochem, La Jolla, CA) and incubated with indicated antibodies or normal rabbit IgG and Protein A / Protein G agarose beads overnight, washed with TGN buffer and analyzed by immunoblot. Antibodies used were Nbs1 (NB100-143, Novus, Littleton, CO), Brca1 (OP92 and OP93, Calbiochem, La Jolla, CA).
Cell Cycle Checkpoint Assays
PI staining
Cells were fixed in 70% ethanol and stored at – 20° C. Cells were washed with PBS, and incubated with 10 μg/ml PI (propidium iodide) and 250 μg/ml RNAse A and analyzed by FACSCalibur (BD, Franklin Lakes, NJ). Data was assessed by the WinMDI 2.9 software.
BrdU / PI staining
Cells were either irradiated with 10 Gy IR or left untreated. 45 min after irradiation 20 μM BrdU (bromdeoxyuridine) was added to the culture medium for 15 min. Thereafter, cells were fixed in 70% ethanol and stored at – 20° C. Cells were washed with PBS, treated with 2N HCl and subsequently incubated with anti-BrdU (M0744, DaKo, Carpinteria, CA) and anti-mouse IgG (F0257, Sigma, St. Louise, MO) in PBS + 0.5% BSA + 0.1% Tween 20, washed and incubated with 10 μg/ml PI (propidium iodide) and 250 μg/ml RNAse A and analyzed by FACSCalibur (BD, Franklin Lakes, NJ). Data was assessed by the WinMDI 2.9 software.
pH3 / PI staining
Cells were either irradiated with 10 Gy IR or left untreated. Thereafter, cells were fixed in 70% ethanol and stored at – 20° C. Cells were washed with PBS, incubated in PBS/0.25 % Triton X-100 on ice for 15 min followed by a subsequent incubation with anti-phospho-H3 (Ser 10) (06-570, Millipore, Billerica, MA) antibody and goat anti-rabbit IgG FITC conjugated antibody (111-096-144, Jackson ImmunoResearch Labs, West Grove, PA) in PBS + 1% BSA. Cells were washed and incubated with 10 μg/ml PI (propidium iodide) and 250 μg/ml RNAse A and analyzed by FACSCalibur (BD, Franklin Lakes, NJ). Data was assessed by the WinMDI 2.9 software.
RESULTS
Spatiotemporal recruitment kinetics of Brca1 and members of the RNF8/Rap80-pathway to the DSB site
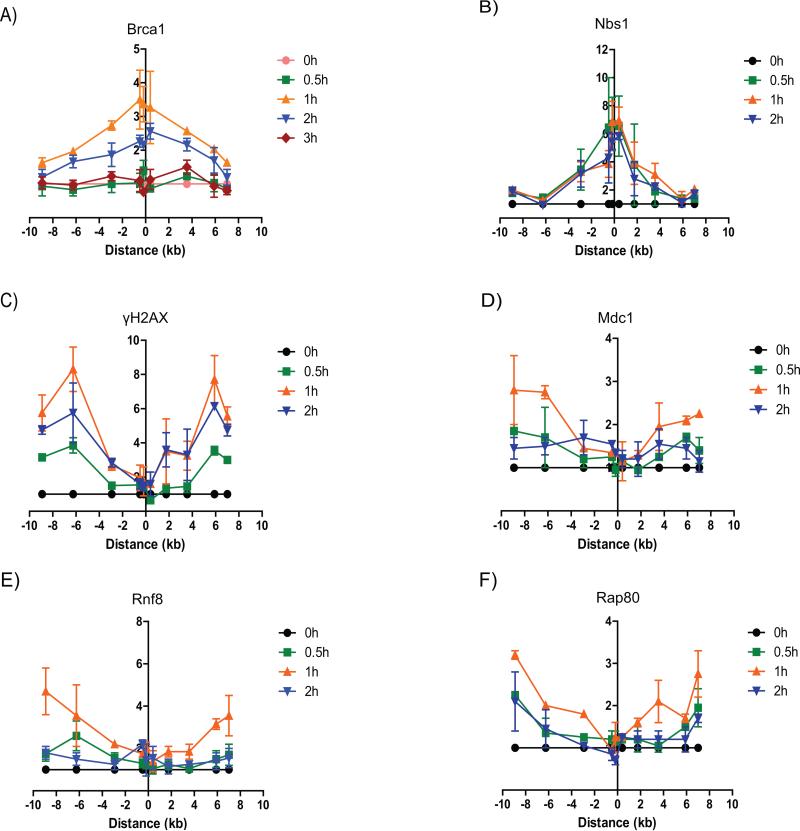
While immunofluorescent focus formation and “laser microirradiation” assays have been quite informative in studying protein dynamics and modifications associated with DNA damage and repair, they are limited in their ability to provide detailed spatial resolution for protein binding to chromatin. We recently reported a system in which DNA breaks could be introduced by the homing endonuclease, I-PpoI, at defined sites in the human genome in a highly regulated manner and in which assessments of DSB repair and protein movements could be done with significantly improved resolution (23). Using this approach, we found that Brca1 is recruited both directly to the DSB site and to the regions surrounding the DSB, with moderately higher levels being detected in the immediate vicinity of the DSB (Figure 1A). Consistent with our previous findings (23,24), Nbs1 is detectably recruited only in the vicinity of the break, while γH2AX is conversely present in the flanking chromatin, but not directly at the DSB (Figure 1B, C). Notably, γH2AX serves as a platform for recruitment of MDC1 (25,26). Consistent with this, MDC1 and the downstream factors RNF8 and Rap80, showed a similar recruitment pattern accumulating in the DSB flanking regions, but not directly at the break (Figure 1D-F). In terms of kinetics, Nbs1 recruitment preceded the other proteins, plateauing at 30 minutes after I-PpoI induction, compared to peak binding at 60 minutes for Brca1 and the members of the γH2AX-MDC1-RNF8/RNF168-Rap80 pathway (Figure 1). Further, levels of Nbs1 and γH2AX remained at the plateau level for the duration of the experiment in contrast to decreasing chromatin binding of Brca1, MDC1, RNF8 and Rap80 by 120 minutes (Figure 1). Thus, Brca1 is recruited both directly to the break and to the DSB surrounding regions, while Nbs1 is present only in the vicinity of the break and the γH2AX-MDC1-RNF8/RNF168-Rap80 pathway members accumulate only in the DSB flanking chromatin.
Figure 1. Spatiotemporal recruitment kinetics of Brca1 and other DDR factors to the DSB.
ChIP assay showing the recruitment of (A) Brca1, (B) Nbs1, (C) γH2AX, (D) MDC1, (E) RNF8 and (F) RAP80 to the DSB induced by I-PpoI on chromosome 1 in MCF7 cells expressing ddI-PpoI that were cultivated in medium containing 0.1% fetal bovine serum for 24 h prior to DSB induction. Time indicated is hours following addition of 4-OHT. The I-PpoI cleavage site on chromosome 1 is located at distance 0. Data of two independent experiments are shown as mean +/− SEM. The y-axis displays the fold change in relative occupancy normalized to the control.
Differential roles for the RNF8/Rap80-pathway and Nbs1 in Brca1 recruitment to chromatin
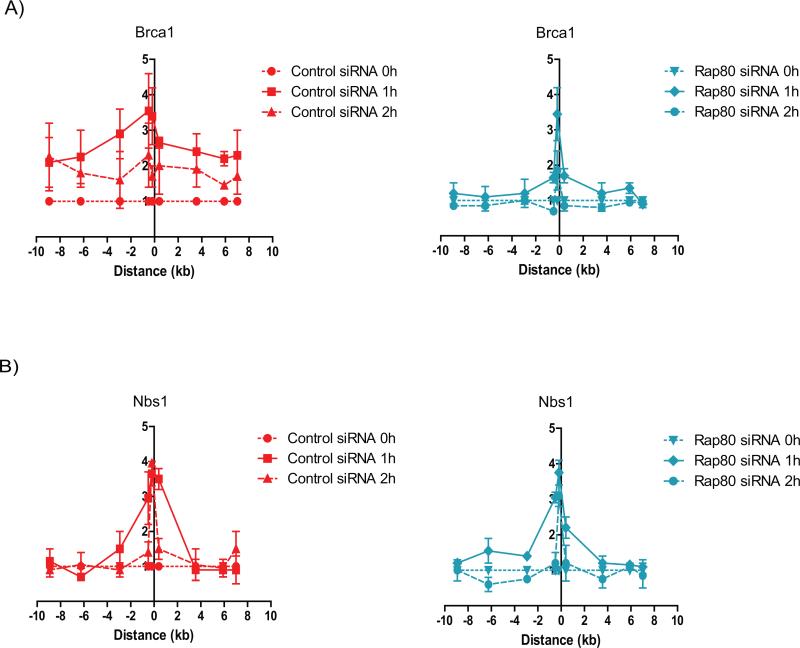
RNF8 and RNF168 activities affect multiple DDR processes, including the recruitment of 53BP1 (15,17,27). Thus, to investigate mechanisms involved in Brca1 recruitment to the DSB and flanking region, we knocked-down Rap80 instead of RNF8 in order to minimize the effect on DDR proteins other than Brca1. Rap80 knock-down abrogated the recruitment of Brca1 to the DSB flanking regions without affecting the levels of Brca1 in the vicinity of the break (Figure 2A), thus indicating that the RNF8/RNF168-Rap80 pathway is responsible for recruiting Brca1 to the flanking chromatin, but not directly to the DSB. Importantly, Rap80 depletion did not disrupt Nbs1 recruitment (Figure 2B), demonstrating a localized and specific effect. Since a recent study demonstrated that the interaction between Brca1 and Rap80 is dependent on Brca1 PARYlation by PARP1 (28), we examined the effect of PARP1 inhibition on Brca1 recruitment to the DSB. Indeed, PARP1 inhibition reduced Brca1 recruitment to the DSB surrounding regions without affecting Brca1 in the vicinity of the DSB (Figure S1A), thus, having a similar effect on the Brca1 recruitment pattern as Rap80 depletion. Additionally, we observed that ATM inhibition affected Brca1 recruitment to the DSB surrounding regions (Figure S1B), consistent with its role in facilitating the formation of γH2AX domains surrounding the DSB that serve as platform for Mdc1 recruitment (25,26).
Figure 2. Rap80 recruits Brca1 to the DSB surrounding regions.
ChIP of (A) Brca1 and (B) Nbs1 in MCF7 cells expressing ddI-PpoI as described in Figure 1. Cells were transfected with either non-targeting control siRNA (left panels) or RAP80 targeting siRNA (right panels).
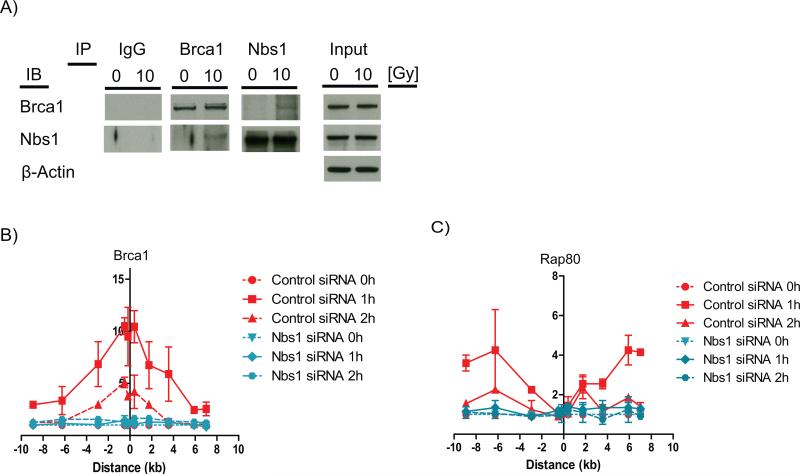
These results raised the question of the mechanism(s) responsible for recruitment of Brca1 directly to the DSB. Since the MRN complex accumulates mainly in the vicinity of the DSB, but not the surrounding regions (e.g. Figure 1B), it was a logical candidate for recruiting Brca1 directly to the break. In support of this hypothesis, Brca1 was reported to be present in a multi-protein complex with MRN (21). Therefore, we asked whether DSB induction by IR affects the interaction between Brca1 and Nbs1 and whether Brca1 recruitment to the DSB depended on Nbs1. Reciprocal co-immunoprecipitation demonstrated an increased interaction between Brca1 and Nbs1 after 10 Gy IR (Figure 3A) and Nbs1 knock-down abolished Brca1 accumulation both at the DSB and around the break site (Figure 3B). Since the MRN complex is essential for initiation of DDR signaling, including ATM recruitment to the DSB, which is required for γH2AX domain formation and MDC1 recruitment (25), we suspected that the abrogation of Brca1 recruitment to the surrounding chromatin after Nbs1 knock-down was likely due to the disruption of the RNF8/RNF168-Rap80 pathway. Consistent with this, we found that Nbs1 knock-down abolishes Rap80 accumulation in the DSB surrounding regions (Figure 3C). These results indicate that two spatially distinct Brca1 fractions exist at the DSB site. One Brca1 fraction is recruited directly to the DSB through an interaction with the MRN complex, whereas another Brca1 fraction is recruited to the DSB flanking chromatin by the RNF8/RNF168-Rap80-pathway.
Figure 3. Nbs1 is required for Brca1 recruitment to the DSB site.
(A) Reciprocal co-immunoprecipitation of Brca1 and NBS1 in either untreated MCF7 cells or in MCF7 cells 30 min following 10 Gy IR. Normal rabbit IgG was used as control. ChIP of (B) Brca1 and (C) RAP80 in MCF7 cells expressing ddI-PpoI as described in Figure 1. Cells were transfected with either non-targeting control siRNA or NBS1 targeting siRNA.
Brca1 at the DSB is involved in DNA repair
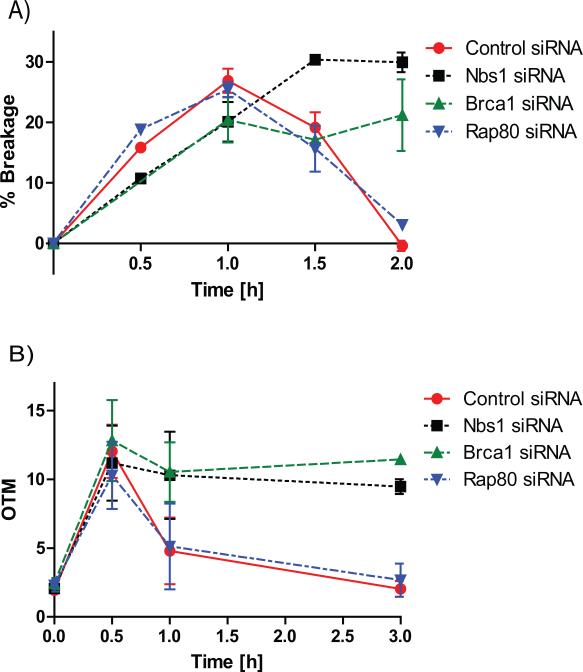
Differential mechanisms involved in recruitment of spatially distinct Brca1 fractions raised the possibility of different functions for these two sites of Brca1 binding in the DDR. Though several studies reported that Brca1 promotes re-ligation of linearized plasmids by NHEJ (8,9), plasmid DNA lacks many structural and functional properties of the DNA packaged into chromatin. Thus, the role of Brca1 in NHEJ repair of genomic DNA has been somewhat unclear. By arresting cells in the G1 phase of the cell cycle, the I-PpoI system can specifically assess re-ligation of DNA by NHEJ repair (23). Down-regulation of Brca1 significantly inhibited NHEJ repair of DSBs induced either by I-PpoI (Figure 4A, Figure S2-3) or by ionizing irradiation (comet assay; Figure 4B, Figure S2-3). This raised the question of which population(s) of chromatin-bound Brca1 were affecting DNA repair. While Nbs1 knock-down, which affects Brca1 recruitment to both direct and flanking sites, had the expected negative impact on NHEJ repair (Figure 4A, B), Rap80 knock-down, which abrogates the binding of Brca1 only to the flanking sites, did not affect NHEJ repair (Figure 4A, B). Thus, the Brca1 fraction bound to chromatin flanking the DSB is dispensable for NHEJ repair. For completeness, we also tested the effects of Brca1, Nbs1 and Rap80 knock-downs on DSB repair in cycling cells, where both NHEJ and HR would contribute to repair of DSBs in cells containing S and G2 populations. Similar to the observations in G1-arrested cells, depletion of Rap80 did not affect DSB repair in cycling cells, while depletion of either Brca1 or Nbs1 significantly reduced DSB repair (Figure S4).
Figure 4. DSB re-ligation by NHEJ depends on Brca1 and NBS1 but not RAP80.
(A) DNA repair measured by quantitative real-time PCR spanning the unique I-PpoI cleavage site at chromosome 1 in MCF7 cells expressing ddI-PpoI that were cultivated in medium containing 0.1% fetal bovine serum for 24 h prior to DSB induction. Time indicated is hours following addition of 4-OHT. Data of three independent experiments are shown as mean +/− SEM. (B) Comet assay performed in MCF7 cells that were untreated (time point 0 h) or treated with 10 Gy IR and harvested at indicated time points after irradiation. Cells were cultivated in medium containing 0.1% fetal bovine serum for 24 h prior to DSB induction. OTM, olive tail moment. (A-B) Cells were transfected with either non-targeting control siRNA, Brca1, NBS1 or RAP80 targeting siRNA.
Chromatin-bound Brca1 in the DSB flanking regions is required for cell cycle checkpoints and regulates DSB processing
Since Brca1 residing in the DSB flanking regions was dispensable for DSB repair, we sought to identify the role of this Brca1 fraction in the DDR. A previously published report suggested that the Brca1-Rap80 protein complex suppresses excessive DNA end resection at the DSB (29,30). Consistent with this finding, we observed that a knock-down of Rap80 in proliferating cells resulted in elevated levels of RPA32 at the break site (Figure S5), which reflects an increase in the amount of DNA resection. Importantly, an increase in RPA32 levels due to Rap80 depletion was only observed in the DSB surrounding regions (Figure S5), where Brca1 recruitment is mediated by Rap80, but not directly at the break.
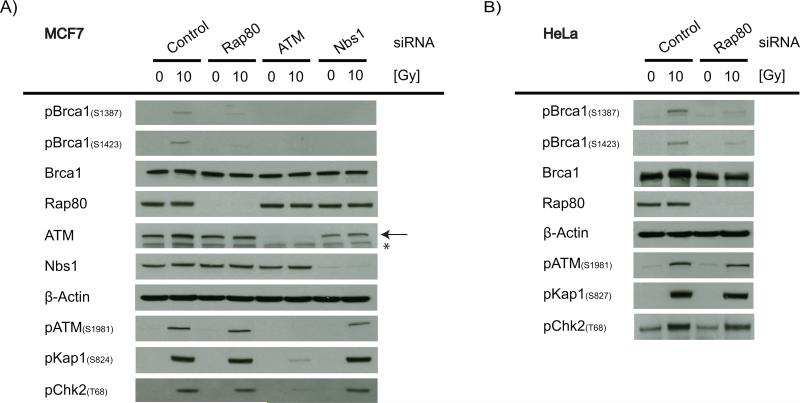
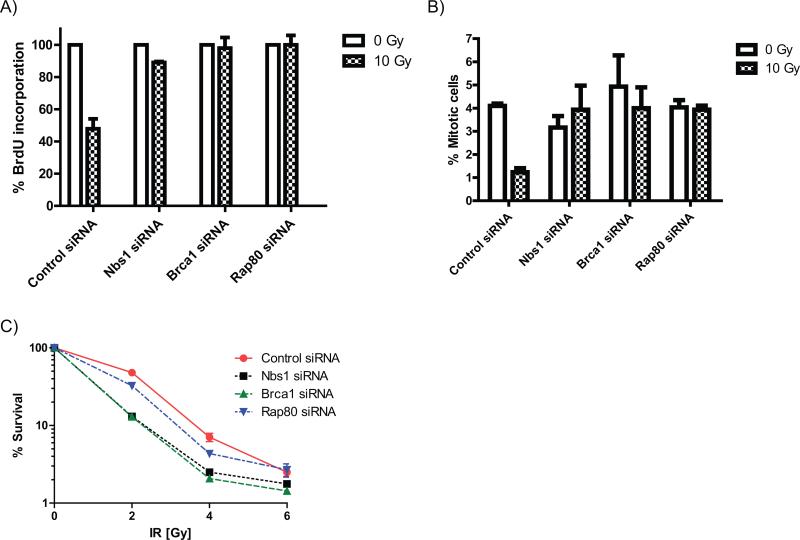
DSB flanking chromatin is also a region where ATM-dependent signaling is amplified in a γH2AX dependent manner (25). Following DSB induction, activated ATM phosphorylates Brca1 at serine residues 1387 and 1423, modifications required for activation of the S and G2/M checkpoints, respectively (12,13). Therefore, we asked whether Brca1 recruitment to the DSB surrounding regions by Rap80 facilitates the phosphorylation of Brca1 by ATM, thus triggering the DNA damage-induced cell cycle checkpoints. Rap80 depletion in two different cell lines significantly reduced the phosphorylation of Brca1 at serine residues 1387 and 1423 without affecting the phosphorylation of other ATM targets, such as Kap1 and Chk2 (Figure 5 A, B). A knock-down of Nbs1, which would abrogate Brca1 recruitment to both the DSB and the flanking regions, resulted in a similar degree of reduction of the ATM-dependent Brca1 phosphorylation (Figure 5A), suggesting that Brca1 phosphorylation occurs predominantly in the DSB flanking chromatin. Consistent with the abrogation of Brca1 phosphorylation, knock-down of Brca1, Nbs1 or Rap80 all abrogated the IR-induced S (Figure 6A) and G2/M (Figure 6B) checkpoints to a similar extent. Interestingly, the effects of the Rap80 depletion on cell survival after IR was less pronounced than that seen with the knock-down of Brca1 or Nbs1 (Figure 6C), consistent with the lack of requirement for Rap80 in DSB repair.
Figure 5. RAP80-dependent recruitment of Brca1 to the DSB surrounding regions is required for Brca1 phosphorylation by ATM.
Immunoblot showing the ATM-specific phosphorylation of Brca1 at serine 1387 and 1423 and the activation of the ATM pathway in (A) MCF7 and (B) HeLa cells 30 min after treatment with 10 Gy IR. Cells were transfected with either non-targeting control siRNA, RAP80, NBS1 or ATM targeting siRNA. (←) ATM band, (*) Non-specific band.
Figure 6. Brca1 recruitment to the DSB surrounding regions promotes induction of the cell cycle arrest in S and G2.
(A) S-phase checkpoint analysis in MCF7 cells by quantification of BrdU incorporation into DNA following 10 Gy IR. BrdU incorporation is shown normalized to non-irradiated control. (B) G2/M checkpoint analysis in MCF7 cells by phospho-H3 staining following 10 Gy IR. (C) MTT assay in MCF7 cells showing cell survival following IR. (A-C) MCF7 cells were transfected with either non-targeting control siRNA, NBS1, Brca1 or RAP80 targeting siRNA. Data of three independent experiments are shown as mean +/− SEM. In 4C the SEM is too small to be shown for several time points.
DISCUSSION
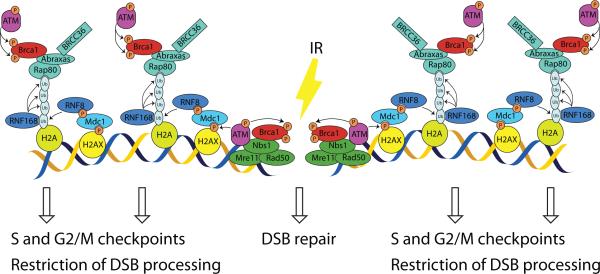
By taking advantage of the spatial resolution afforded by introduction of I-PpoI-induced DSBs at defined sites in the human genome, we found that Brca1 recruitment to DSBs actually represents two separate processes (Figure 7). In response to DNA breakage, Brca1 binds to Nbs1 protein and is recruited directly to the sites of DNA breaks, where its recruitment is required for efficient DNA repair. The results definitively implicated Brca1 residing in the direct vicinity of the DSB in NHEJ repair of endogenous DNA. Given that the initiation of DSB processing that is required for HR is dependent on the MRN complex (25), while the Rap80/Brca1 complex represses DNA resection as discussed below, we speculate that Brca1 bound directly to the break may also facilitate repair by HR. In addition, Brca1 is recruited to chromatin flanking sites of DNA breakage in a manner dependent on Rap80. In this DSB flanking region, ATM-mediated phosphorylation of Brca1 occurs and is required for both S-phase and G2/M checkpoints. Notably, our model is consistent with a previous report showing that a simultaneous knock-down of Nbs1 and H2AX is required to achieve a complete abrogation of Brca1 foci formation following irradiation (31).
Figure 7. Model of spatially differential Brca1 recruitment to the DSB.
The data presented herein support the following steps involved in Brca1 recruitment to chromatin and function after DNA breakage: 1) Following DSB induction, Brca1 interacts with Nbs1, resulting in recruitment of a small fraction of Brca1 directly to the DNA break site where it facilitates DSB repair; 2) ATM phosphorylates histone protein H2AX, resulting in formation of γH2AX domains in the DSB flanking regions that serve as a platform for MDC1 and RNF8 recruitment; 3) RNF8 and RNF186 E3 ubiquitin ligases facilitate the conjugation of ubiquitin chains to the histone H2A; 4) Rap80 links Brca1 to these poly-ubiquitin chains, thus recruiting a larger Brca1 fraction to the DSB flanking chromatin; 5) Brca1 recruitment to the flanking chromatin increases the amount of Brca1 molecules interacting with active ATM, resulting in an efficient phosphorylation of Brca1 at serine residues 1387 and 1423, which is required for the activation of S-phase and G2/M checkpoints, respectively; 6) The Rap80/Brca1 complex residing in the DSB flanking regions also represses an excessive DSB processing, thus tuning HR.
Consistent with previous reports implicating the Rap80/Brca1 complex in suppression of excessive DSB processing (28-30), we observed an increase in DNA resection in the DSB flanking regions in the absence of Rap80. These findings suggest an additional role for the Brca1 fraction that is recruited by Rap80 to the DSB flanking chromatin in tuning HR by preventing excessive DSB processing. We suspect that a much larger fraction of Brca1 protein is bound to the DSB flanking regions than directly to the DSB site, thus making it difficult to assess the relative amount and importance of the directly-bound fraction of Brca1 by other techniques. The approaches described here were sufficient to clarify that this fraction is both recruited by a different mechanism and functionally distinct. However, some questions remain with regard to the MRN-dependent Brca1 recruitment in the vicinity of the break, including whether the Brca1-Nbs1 interaction is direct or whether it requires other proteins as mediators. In addition, it remains to be clarified which domains of Nbs1 and Brca1 are responsible for the interaction and whether it is regulated by the post-translational modifications of Brca1 and Nbs1 that occur following DNA damage induction.
It is important to note that the DSB-flanking chromatin is considered to be a region where ATM signaling is amplified in a manner that depends on the formation of the γH2AX domains that spread around the DSB (25,26). Here, we identified an additional role of the DSB-flanking regions in promoting ATM-dependent signaling by showing that these regions serve as a platform for Brca1 phosphorylation by ATM, which is required for an efficient checkpoint activation. Interestingly, although activation of the Brca1-dependent cell cycle checkpoints relies on Rap80, we found that a knock-down of Rap80 only moderately affects cell survival after exposure to IR. In contrast, a depletion of Nbs1 and Brca1 strongly reduces the surviving fraction. These observations are consistent with Rap80 lacking a role in DSB re-ligation. Further, our findings are in line with previously published reports showing different phenotypes of mice lacking Nbs1 or Brca1 versus Rap80-deficient mice. While a knockout of Brca1 or Nbs1 in mice is lethal (32,33), Rap80−/− mice are viable (34). Consistent with our results, MEFs derived from Rap80−/− mice are only moderately hypersensitive to IR compared to wild-type mice (34). Thus, efficient DNA repair appears to be the major determinant of cell survival after exposure to agents that induce DSBs, whereas activation of cell cycle arrests plays a minor role in cell survival, consistent with numerous studies in the literature. Taken together, these results provide new insights into the regulation of the functional diversity of Brca1 in the DDR and demonstrate the functional importance and role of the spatial arrangement of Brca1 at the DSB site.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to members of the Kastan lab for advice and technical support throughout the course of this study, especially Donald Fleenor and John Crutchley. This work was supported by grants from the NIH (R01CA071387, R01CA159826, and P30CA014236) and DFG (German Research Foundation; GO1894/1-1).
Footnotes
Conflict of Interest
None of the authors have any potential conflicts of interest in regards to this submission.
REFERENCES
- 1.Huen MS, Sy SM, Chen J. BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol. 2010;11(2):138–48. doi: 10.1038/nrm2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldstein M, Kastan MB. The DNA Damage Response: Implications for Tumor Responses to Radiation and Chemotherapy. Annual review of medicine. 2014 doi: 10.1146/annurev-med-081313-121208. [DOI] [PubMed] [Google Scholar]
- 3.Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Molecular cell. 2001;8(6):1163–74. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 4.Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. The Journal of biological chemistry. 1998;273(39):25388–92. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 5.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450(7169):509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlegel BP, Jodelka FM, Nunez R. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer research. 2006;66(10):5181–9. doi: 10.1158/0008-5472.CAN-05-3209. [DOI] [PubMed] [Google Scholar]
- 7.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173(2):195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang HC, Chou WC, Shieh SY, Shen CY. Ataxia telangiectasia mutated and checkpoint kinase 2 regulate BRCA1 to promote the fidelity of DNA end-joining. Cancer research. 2006;66(3):1391–400. doi: 10.1158/0008-5472.CAN-05-3270. [DOI] [PubMed] [Google Scholar]
- 9.Zhuang J, Zhang J, Willers H, Wang H, Chung JH, van Gent DC, et al. Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. Cancer research. 2006;66(3):1401–8. doi: 10.1158/0008-5472.CAN-05-3278. [DOI] [PubMed] [Google Scholar]
- 10.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 11.Kitagawa R, Bakkenist CJ, McKinnon PJ, Kastan MB. Phosphorylation of SMC1 is a critical downstream event in the ATM-NBS1-BRCA1 pathway. Genes & development. 2004;18(12):1423–38. doi: 10.1101/gad.1200304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu B, Kim S, Kastan MB. Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol Cell Biol. 2001;21(10):3445–50. doi: 10.1128/MCB.21.10.3445-3450.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu B, O'Donnell AH, Kim ST, Kastan MB. Phosphorylation of serine 1387 in Brca1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer research. 2002;62(16):4588–91. [PubMed] [Google Scholar]
- 14.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131(5):901–14. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131(5):887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 16.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136(3):420–34. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 17.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–46. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 18.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316(5828):1194–8. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316(5828):1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316(5828):1202–5. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 21.Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes & development. 2006;20(1):34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rainey MD, Charlton ME, Stanton RV, Kastan MB. Transient inhibition of ATM kinase is sufficient to enhance cellular sensitivity to ionizing radiation. Cancer research. 2008;68(18):7466–74. doi: 10.1158/0008-5472.CAN-08-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldstein M, Derheimer FA, Tait-Mulder J, Kastan MB. Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proc Natl Acad Sci U S A. 2013;110(42):16874–9. doi: 10.1073/pnas.1306160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berkovich E, Monnat RJ, Jr., Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nature cell biology. 2007;9(6):683–90. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 25.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends in cell biology. 2009;19(5):207–17. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 27.Fradet-Turcotte A, Canny MD, Escribano-Diaz C, Orthwein A, Leung CC, Huang H, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499(7456):50–4. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Y, Petit SA, Ficarro SB, Toomire KJ, Xie A, Lim E, et al. PARP1-Driven Poly-ADP-Ribosylation Regulates BRCA1 Function in Homologous Recombination-Mediated DNA Repair. Cancer discovery. 2014;4(12):1430–47. doi: 10.1158/2159-8290.CD-13-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes & development. 2011;25(7):685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coleman KA, Greenberg RA. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. The Journal of biological chemistry. 2011;286(15):13669–80. doi: 10.1074/jbc.M110.213728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan J, Chen J. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. The Journal of biological chemistry. 2010;285(2):1097–104. doi: 10.1074/jbc.M109.078436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van Laethem F, et al. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nature cell biology. 2005;7(7):675–85. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- 33.Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes & development. 1997;11(10):1226–41. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 34.Yin Z, Menendez D, Resnick MA, French JE, Janardhan KS, Jetten AM. RAP80 is critical in maintaining genomic stability and suppressing tumor development. Cancer research. 2012;72(19):5080–90. doi: 10.1158/0008-5472.CAN-12-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.