Abstract
The mammalian genome contains on the order of a million enhancer-like regions that are required to establish the identities and functions of specific cell types. Here, we review recent studies in immune cells that have provided insight into the mechanisms that selectively activate certain enhancers in response to cell lineage and environmental signals. We describe a working model wherein distinct classes of transcription factors define the repertoire of active enhancers in macrophages through collaborative and hierarchical interactions, and discuss important challenges to this model, specifically providing examples from T cells. We conclude by discussing the use of natural genetic variation as a powerful approach for decoding transcription factor combinations that play dominant roles in establishing the enhancer landscapes, and the potential that these insights have for advancing our understanding of the molecular causes of human disease.
Exploiting macrophages to understand enhancer biology and enhancer biology to understand macrophages
Macrophages are phagocytic cells of the innate immune system that reside in all tissues of the body and play key roles in responding to infection and injury through signaling downstream of pattern recognition receptors [1, 2] [3]. In addition to these general roles that operate throughout the body, each tissue-resident population of macrophages performs specific effector functions that contribute to the homeostasis of that tissue [2, 4]. Some of the diverse roles that macrophages have in vivo that are unique to their tissue environments include neuronal synaptic pruning by microglia in the brain [5], bone resorption and remodeling by osteoclasts [6], control of insulin sensitivity and adaptive thermogenesis in adipose tissue [7, 8] [7, 8] and surfactant recycling by lung alveolar macrophages [9]. Although the diverse functions of macrophages are normally adaptive, they can be co-opted to drive tissue pathology, particularly in the setting of chronic inflammatory diseases and cancer. For example, functions of macrophages that are important for pathogen recognition and initiation of inflammation play key roles the development and clinical complications of atherosclerosis [10, 11]. Conversely, functions of macrophages that are important for wound repair contribute to tumor growth and metastasis [12]. Understanding the mechanisms by which various macrophage populations achieve their tissue-specific functions and determining whether these functions can be modulated for therapeutic purposes remain largely unmet goals.
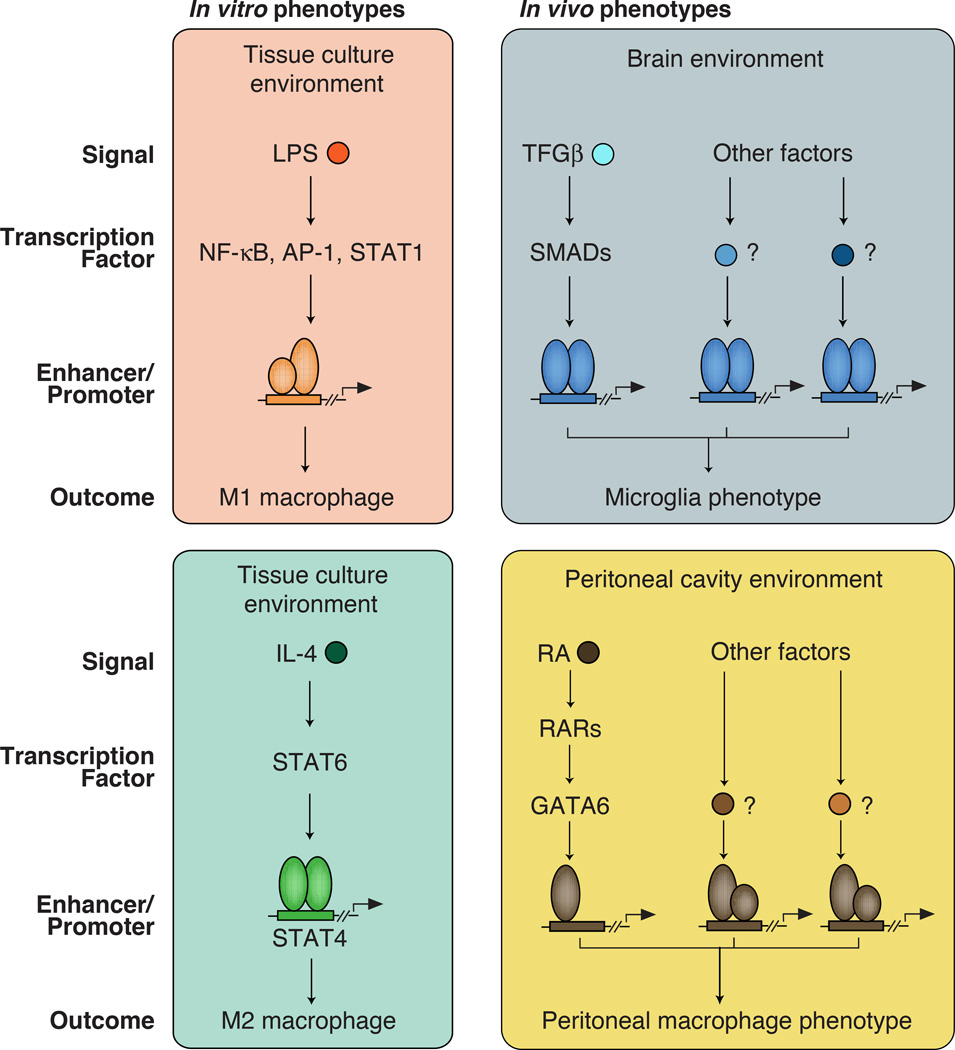
Distinct macrophage phenotypic polarization states have been characterized in vitro by studying responses to various ligands that result in alternative gene expression programs [13]. Two extensively characterized in vitro polarization programs are broadly categorized as classically activated/pro-inflammatory M1 macrophages or alternately activated/anti-inflammatory M2 macrophages [14, 15]. Treatment of macrophages with lipopolysaccharide (LPS), a component of gram-negative bacteria, drives M1 polarization through TLR4-dependent activation of members of the NF-κB, AP-1 and IRF families of transcription factors (Figure 1) [16, 17]. These factors induce the expression of hundreds of genes, many of which play key roles in innate immunity, inflammation, and initiation of adaptive immune responses. In contrast, treatment of macrophages with IL-4 drives M2 polarization through activation of STAT6, which induces a program of gene expression linked to immunity directed against parasitic infection (Figure 1)[14, 18].
Figure 1. Different signaling pathways in macrophages lead to diverse phenotypic outcomes.
Left panels: Macrophages can be polarized toward M1, M2 phenotypes in in vitro by exposure to LPS or IL-4 respectively.
Right panels: The brain and peritoneal cavity contain high levels of TGFβ or retinoic acid (RA) respectively, that are important determinants of the distinct phenotypes of microglia and resident peritoneal macrophages. However, these are only a subset of what are as yet mostly unknown signals that must be integrated to establish tissue-specific gene expression signatures.
Recent studies indicate that tissue macrophages exhibit distinct gene expression programs that underlie their tissue-specific functions[19–21]. Furthermore, tissue environment has been shown to be a significant determinant of the gene expression patterns and the underlying transcriptional regulatory elements that are characteristic of a particular macrophage subtype [20, 21]. The specific signals that dictate tissue-specific programs of macrophage gene expression are for the most part unknown. Some recently identified examples include TGFβ, which is essential for maintenance of microglia phenotypes [22] (Figure 1), retinoic acid, which is required for development/maintenance of large peritoneal macrophages [23] (Figure 1), IL-4, which is required for maintenance of homeostatic beige adipose tissue macrophages (Figure 1)[8], and RANKL, which is required for the development of osteoclasts [24]. Importantly, while these molecules are established to be important for the phenotypic characteristics of particular macrophage subsets in vivo, they represent only a part of the total spectrum of signals sensed by the macrophage within each tissue environment (Figure 1). Furthermore, as discussed below, signal-dependent transcription factors such as NF-κB primarily regulate gene expression by acting on pre-existing enhancers, which have recently been shown to differ among tissue macrophage subsets [20, 21]. The implication of these findings is that the quantitative and qualitative responses of different tissue macrophages to the same signal, such as LPS, are likely to vary in a tissue-specific manner. Therefore, while in vitro studies of M1 and M2 macrophage activation provide powerful models to investigate mechanisms of signal-dependent gene expression, studies of the enhancer and promoter landscapes of macrophages in vivo provide important insights into how complex environmental signals regulate their development and function in distinct tissues.
In this review, we briefly introduce the current state of enhancer biology along with advances in the field of macrophage gene regulation. We begin with the characteristics of enhancers and underscore their dynamic behavior in cell lineage specification and environmental signaling contexts. We describe a working model in which distinct classes of sequence-specific transcription factors, referred to as lineage-determining and signal-dependent transcription factors (LDTFs and SDTFs), define the repertoire of active enhancers in macrophages through collaborative and hierarchical interactions. We note some important challenges to this model, specifically providing examples from T cells. Next, we highlight how natural genetic variation can be leveraged as a powerful tool to identify sets of collaborating transcription factors that establish enhancers in different cells. To this end, studies utilizing genetic variation in tissue-resident subsets of macrophages are discussed to exemplify the importance of tissue environment on enhancer selection. We conclude with a discussion of how these findings, which combine genetic variation and enhancer function, are highly informative for interrogating the molecular causes of human disease.
The million enhancer question
All cells in the body contain essentially the same genome. The mechanisms that govern how different cell types uniquely interpret the same set of instructions, and thereby achieve specialized functional roles, are incompletely understood. In recent years, it has become clear that on the genome scale, DNA sequences called enhancers, more so than promoters, orchestrate the majority of cell type-specific patterns of gene expression [25–29]. Although the distinction between enhancers and promoters are becoming increasingly blurred [30], as discussed further below, Text Box 1 outlines key properties of each.
Box 1. Characteristics of Promoters and Enhancers.
| Promoters | Enhancers |
|---|---|
|
|
By cataloging enhancers using epigenetic chromatin marks across hundreds of tissues, cell types, and activation states, the total number of enhancers in the human genome is estimated to be on the order of one million [27–29]. From this vast palette, a given cell type typically selects about 30–50 thousand enhancerlike regions that determine its identity and functional potential [25–29]. A fundamental question is therefore to elucidate the molecular determinants regulating enhancer activity.
General features of enhancers
Enhancers are discrete regions of the genome that function to increase transcription from nearby promoters [31] (reviewed in [32, 33]). In the pre-genomics era, enhancers were first identified as stretches of DNA that, when inserted up- or down-stream of transgenes, were able to augment the genes’ expression irrespective of orientation [31].
In eukaryotes, DNA is wrapped around nucleosomes into chromatin, which serves as a regulatory barrier to transcription factors. Enhancer elements are bound by sequence-specific transcription factors that are able to compete with nucleosomes to generate a nucleosome-free region of DNA. These binding events can be measured by assays of increased DNA accessibility such as DNase I hypersensitivity or the assay for transposase-accessible chromatin, ATAC-seq [34, 35]. It is important to note that not all transcription factors are able to recognize their DNA binding motifs in the context of compact, or closed chromatin. The post-genomics era has led to the observation that enhancers exhibit distinctive patterns of modifications on adjacent histone tails, and that chromatin immunoprecipitation sequencing (ChIP-seq) is an effective technique to identify these elements [36]. Rigorous proof that a specific genomic region performs enhancer function requires evidence that mutation or deletion results in reduced activity of the associated gene promoter. Enhancer function can be tested in vivo with transgenic mice [37] or by other, massively parallel reporter assays [38–41]. However, it is important to keep in mind that the vast majority of enhancer elements discovered by genomics methods remain annotated based on indicative chromatin features rather than on in vivo mutation.
In a given cell, enhancer elements can be broadly categorized as inactive, primed, poised, or active [25, 36, 42]. An inactive enhancer is defined as DNA that is either sequestered as heterochromatin, is actively repressed by DNA methylation, or generally lacks the marks of an alternate enhancer state. A primed enhancer is defined by mono- or di-methyl modifications on histone H3 lysine 4 (H3K4me1/2) [43] but lacks additional active marks (see below). Particularly during early embryogenesis, poised enhancers can additionally be marked with tri-methylation of histone H3 on lysine 27 (H3K27me3), which is a marker of active repression and is mutually exclusive with acetylation on the same residue [44]. Finally, active enhancers generally exhibit acetylation of histone H3 lysine 4 (H3K27ac) [44, 45]. Interestingly, active enhancers are also actively transcribed by RNA Polymerase II, giving rise to enhancer RNA, or eRNA [28, 46–48]. Some studies have demonstrated that chromatin looping is facilitated by eRNAs [49–51]. Consistent with this, changes in eRNA levels correlate with changes in target gene expression [51–55], making eRNA an accurate marker of enhancer activity.
Enhancer selection by lineage-determining transcription factors
Enhancer selection is defined here as the process by which an enhancer element in the genome is converted from an inactive to a primed, poised, or active state. Important classes of transcription factors, called pioneer or lineage-determining transcription factors (LDTFs), are able to initiate enhancer selection by competing with nucleosomes to bind their DNA recognition motifs and establish a nucleosome-free region. This process is accompanied by concurrent or subsequent recruitment of chromatin-modifying enzymes that read, write, and erase histone marks [56, 57].
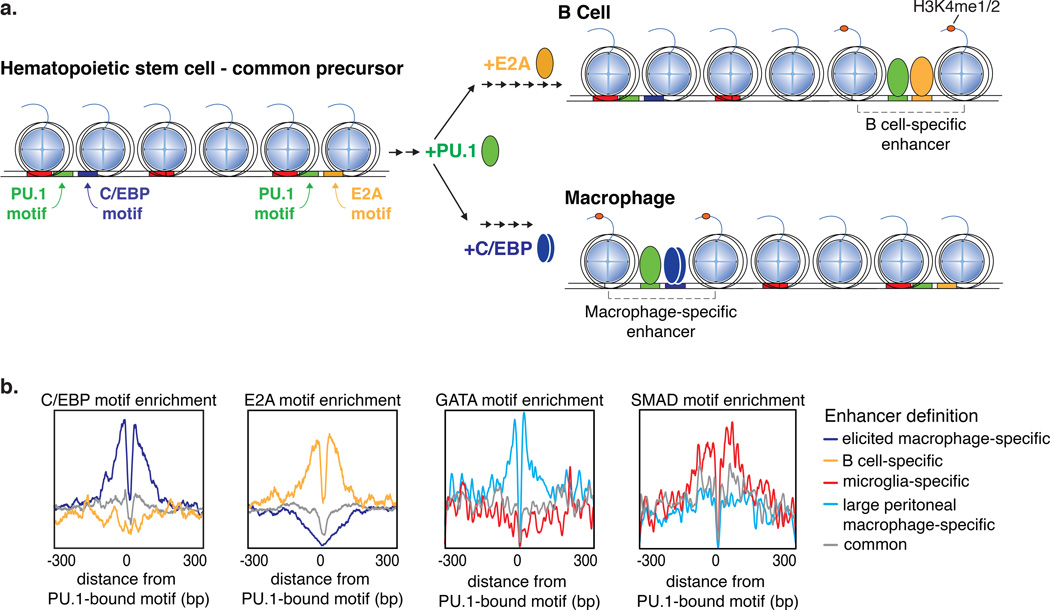
Studies of macrophages and B cells illustrate enhancer selection by collaborative interactions among lineage-determining transcription factors [58]. Differentiation from the common precursor, the haematopoietic stem cell, to either the lymphoid lineage, and mature B cells, or the myeloid lineage, and macrophages requires expression of the transcription factor PU.1 [59, 60] (Figure 2a). Despite a common requirement for PU.1, roughly half of PU.1 enhancer binding is cell type-specific between B cells and macrophages [58, 61]. Cell type-specific PU.1 binding is explained by the local distribution of DNA motifs and the expression of additional lineage-determining transcription factors that collaborate with PU.1 to select enhancers (Figure 2). Specifically, DNA sequence motifs recognized by B cell lineage transcription factors E2A, EBF1, NF-κB and OCT2 are enriched near (<100 base pairs) the B cell-specific PU.1-bound enhancer cistromes [58]. Conversely, these regions are depleted of macrophage lineage factor recognition motifs. In contrast, in the macrophage, PU.1 co-localizes with macrophage lineage-determining factors C/EBPα/β or AP-1 (Figure 2b)
Figure 2. Combinations of closely spaced transcription factor motifs are bound by lineage-determining transcription factors during enhancer selection.
a. Upon differentiation from the hematopoietic stem cell to mature macrophage or B cells requires collaborative enhancer selection involving the transcription factor PU.1 and additional lineage-determining factors E2A for B cells and C/EBPα/β for macrophages.
b. The enrichment profile of motifs for collaborating transcription factors around PU.1-bound PU.1 motifs are distributed rather than showing a fixed spacing relationship, suggesting that enhancer selection occurs independent of protein-protein interactions. Motifs for the collaborating lineage-determining factors C/EBP, E2A, GATA and SMAD are enriched in enhancers specific to thioglycolate-elicited peritoneal macrophages, B cells, large peritoneal macrophages, and microglia, respectively.
Importantly, gain and loss of function experiments indicated that the binding of PU.1 and alternate lineage determining factors is mutually dependent, a property that we refer to as collaborative binding. For example, binding of C/EBP factors to many of their genomic locations in macrophages was dependent on PU.1, while many PU.1 binding sites in B cells were dependent on E2A [58]. The distance distribution between PU.1 and the other DNA motifs suggest that the transcription factors compete with nucleosomes to bind DNA independent of direct protein-protein interactions between the lineage transcription factors [20, 58, 62] (Figure 2b). While protein-protein interactions between such factors likely augment enhancer selection in some situations, the ability to select enhancers independent of spacing requirements allows for evolution to act on many combinations of different sets of transcription factors and DNA motif configurations. Consistent with this flexibility, the precise genomic location of cell-specific enhancers relative to target genes are largely not conserved between mouse and man [63]. Nonetheless, the cell type-specific combinations of lineage-determining transcription factor motifs, and corresponding binding, that establish functional transcriptional networks do appear to be conserved between species [64]. This suggests that while the units of information have become shuffled since mice and man diverged, the meaningful combinations of transcription factors that drive specific functions have largely remained the same. The functional evidence for this flexibility is exemplified by the correct expression of transgenes between species. For example, when the human globin locus is inserted in mouse it is expressed with the same fetal-to-adult switch as it is in humans [65, 66].
Recent studies indicate that all cells contain ~300–500 regions of the genome that are characterized by a particularly high density of features of active enhancers, referred to as super-enhancers or stretch enhancers [67–70]. Although the vast majority of these regions remain to be functionally validated, they can most likely be considered analogous to locus control regions (LCRs) [71], which were initially discovered as crucial regions controlling globin gene expression [72–74]. Notably, each cell type contains a different repertoire of super-/stretch enhancers that co-localize with genes that are particularly important for that cell type’s identity and function. For example, super-/stretch enhancers are typically associated with genes encoding LDTFs, key receptors and proteins with major cell-specific functions. Furthermore, super-/stretch enhancers are occupied by combinations of lineage determining factors, which suggests a mechanism for reinforcement of their own expression. Thus, knowledge of a cell’s super-/stretch enhancer repertoire can provide insights into the identities of genes that the cell has prioritized for expression. Interestingly, many of the super-/stretch enhancers identified in large peritoneal macrophages were environment-dependent [20].
Chromatin dynamics
Chromatin dynamics in hematopoietic development has proven to be a powerful system to study enhancer state transitions during lineage specification [21, 61, 75]. Hematopoiesis initiates with the self-renewing multipotent hematopoietic stem cell (HSC) that differentiates into either the common lymphoid progenitor (CLP) or common myeloid progenitor (CMP) [76]. CMPs further differentiate into lineage-committed progenitors called megakaryocyte-erythroid progenitors (MEPs) or granulocyte-macrophage progenitors (GMPs). From CLPs, MEPs and GMPs arise all terminally differentiated cell types in the blood, including erythrocytes, monocytes, macrophages, dendritic cells, B cells, T cells, and NK cells.
For the roughly fifty thousand enhancers identified using histone modifications across all stages of hematopoiesis, 90% changed enhancer state during differentiation [61]. For these dynamic enhancers, 60% transitioned from being primed in the HSC (H3K4me1-positive) to inactive (H3K4me1-negative) in subsequent lineages that failed to maintain the primed status. For example, the Gata2 locus is primed in HSCs, and remains so in MEPs, but is lost in B and T cells. The reciprocal 40% of dynamic enhancers transitioned from an inactive state in the HSC to a primed or active state in subsequent stages. Examples of these de novo enhancers include the apparent priming at loci for myeloid genes IL-1β, CD14 and S100a8, B cell gene loci Ebf1 and Cr2 and T cell gene loci Bcl11b and CD3g. Notably, the loci for genes that are ultimately expressed in lineage-specific patterns were found to be primed with H3K4me1 at developmental stages prior to exhibiting H3K27ac or RNA expression. In fact, 32% of activated enhancers (H3K27ac-positive) in terminally differentiated cells were primed at a previous stage by H3K4me1 alone. Results of other studies utilizing different model systems have come to the same conclusion that the genetic loci of lineage-specific genes are often primed prior to transcriptional activation [77–82].
Studies in the erythroid lineage of hematopoiesis have shown that Gata2 serves as a multipotency factor and binds important regulatory sequences in the erythroid progenitor, such as near the Gata2 and globin genes [83–85]. Upon erythrocyte commitment, Gata2 is exchanged for Gata1 with coincident changes in chromatin modifications and gene expression patterns. These studies highlight another important mechanism of chromatin priming whereby two members of the same transcription factor family are exchanged to fine-tune chromatin state.
Lineage-determining transcription factors direct signal responsiveness
Enhancer selection by lineage-determining transcription factors results in primed enhancers, but may not result in active enhancers (as measured by acetylation on histone H3 tails at lysine 27, or H3K27ac [45] and enhancer transcription [46, 47]).
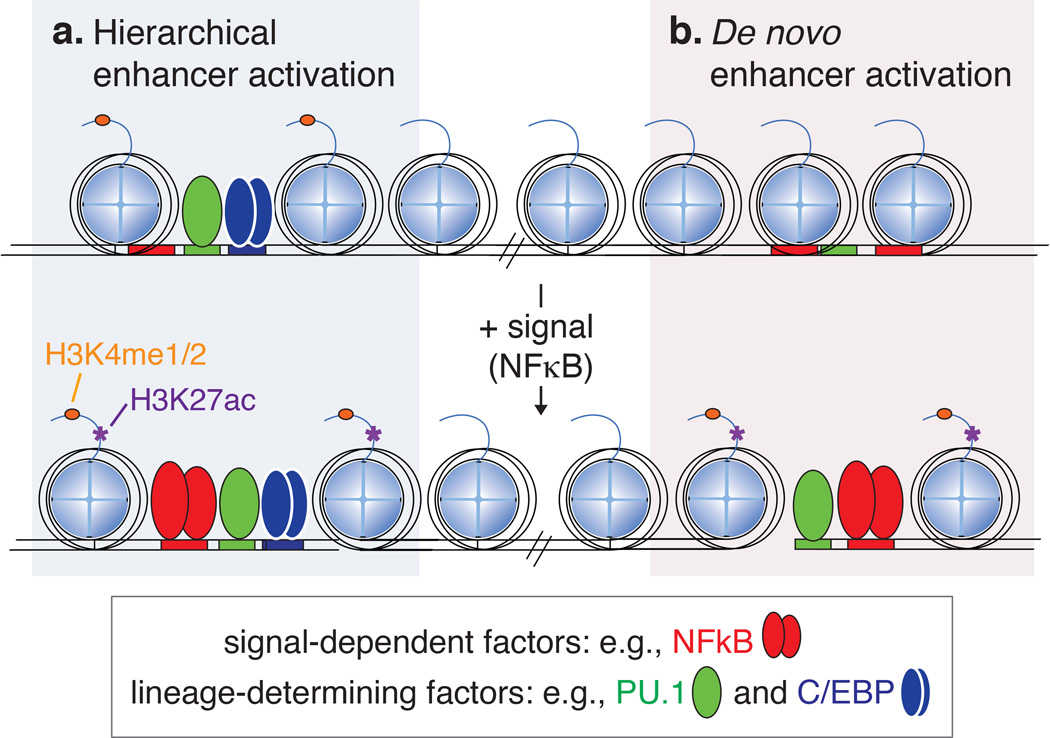
The transition to an active enhancer state can either be initiated from a primed state, whereby lineage factors have already established a nucleosome-free region, or from an inactive or closed state [32, 61, 86] (Figure 3). Both mechanisms of enhancer activation involve collaborative interactions between lineage-determining transcription factors and signal-dependent transcription factors (SDTF). Examples illustrating these mechanisms are discussed here for two transcription factors important in macrophage biology: liver X receptor (LXR) and NF-κB.
Figure 3. Chromatin transitions to active enhancers involve interactions between cell lineage-determining transcription factors and signal-dependent factors.
a. Enhancers primed by lineage determining factors frequently require signal-dependent transcription factor binding to gain H3K27ac and become active.
b. Active enhancers can also be selected by interactions between signal-dependent factors and lineage determining factors.
LXR is a nuclear receptor and sequence-specific transcription factor that becomes available to bind DNA upon changes in cellular cholesterol levels in diverse cell types including macrophages [87]. NF-κB, like LXR, is also activated in a variety of cell types, including macrophages. NF-κB is usually sequestered in the cytoplasm, but upon signaling downstream of pattern recognition receptors (e.g., TLR4 ligation by LPS), NF-κB is free to enter the nucleus and find its recognition motif [88]. In macrophages, a large fraction of LXR binding is dependent on enhancer priming by PU.1, whereas PU.1 binding is not altered by deletion of LXRs [58]. These studies established a hierarchical relationship between the PU.1 and LXR. A similar picture was observed for NF-κB, in which ~ninety percent of TLR4-induced NF-κB binding events occured at primed enhancers [52, 89–91] (Figure 3a). Notably, a small fraction of NF-κB binding in macrophages was observed at sites of previously closed chromatin and binding at these loci occurred in collaboration with macrophage lineage-determining transcription factors PU.1 and AP-1 [52, 90] (Figure 3b). Similar observations have been made for other signal-dependent transcription factors [90]. The transition from a closed to an active state at these enhancers, called de novo or latent enhancers, play a role in sustained activation of NF-κB target genes. At de novo enhancers, NF-κB exhibits properties of both a lineage-determining and a signal-dependent transcription factor, in that it is required for the initial process of enhancer selection as a collaborative binding partner with PU.1 and/or C/EBPs, and this activity is signal-dependent. Latent enhancers are likely to provide mechanistic insights into the substantial remodeling of enhancer landscapes that occurs during developmental transitions, exemplified by intermediates in hematopoiesis [61].
Similar observations have been made in other cell types [58, 92–98]. For example, in erythroid cells, the lineage-determining GATA1 transcription factor directs binding of the respective Wnt and BMP signal-dependent factors TCF7L2 and SMAD [99]. In this system, ectopic expression of the myeloid lineage factor C/EBPα redirects TCF7L2 and SMAD to occupy myeloid enhancers. Given that many signal-dependent transcription factors, such as nuclear receptors, NF-κB, TCFs and SMADs, are induced across many cell types by common signaling pathways, cell type-specific enhancer priming by lineage factors provides a mechanistic explanation for how different cells integrate the same signal to output different patterns of gene response.
While the aforementioned studies, primarily from macrophages, suggest a straightforward definition of transcription factors as LDTFs or SDTFs, in T cells the picture seems to be less clear-cut. For example, the transcription factors Foxp3, Rorγt, and Tbet are indispensible for specialized subsets of T cells, and are thus accepted as master regulators of Treg, Th17, and Th1 cells, respectively [100–103]. Nonetheless, these factors do not open up chromatin [95, 98, 104]. For example, Foxp3 binding in Treg cells occurs at enhancers that are already accessible upon T cell receptor activation prior to Foxp3 expression [95]. Similarly, in Th17 cells, Rorγt binds to preformed regulatory elements that are pioneered by a cooperative complex involving Irf4 and Batf [104, 105]. In this setting, Rorγt functions to modulate the expression of a small set of lineage-specific genes. With respect to SDTF function in T cells, the signal-dependent binding of STAT proteins to enhancers in Th1 and Th2 cells was found to determine subset-specific enhancer activation. Enhancer activation specific to Th1 cells as measured by coactivator (p300) recruitment, however, was predominately independent of the Th1 master regulator Tbet [98].
Taken together, these studies in T cells challenge the working definitions of LDTFs and SDTFs, and what we mean by cell ‘fate’ and cell ‘state’. Because T cell subsets largely share the same set of enhancers, but adopt distinct functional roles based on responses to alternative signals that activate factors such as FoxP3 and RORγt, they could be considered analogous to M1 and M2 macrophages, which are generally considered to represent changes in cell ‘state’ rather than ‘fate’. In this view, Foxp3, Rorγt and Tbet would not be defined as LDTFs analogous to PU.1 and C/EBPs, as they do not pioneer chromatin accessibility to enable SDTF binding and function. However, if distinct cell lineages are defined on the basis of distinct functions, then NF-κB, STAT6, Foxp3, Rorγt and Tbet would all meet the criteria of being LDTFs.
Testing enhancer selection models using natural genetic variation
A collaborative and hierarchical model for selection and activation of cell-specific enhancers provides a framework for understanding how genetic variation perturbs enhancer function and target gene expression with cell specificity. The concept that enhancers are major determinants of cell-specific gene expression is central to the interpretation of certain types of non-coding variants associated with disease risk. Conversely, natural genetic variation can be used as a genome-wide ‘mutagenesis screen’ that enables testing specific hypothesis related to how enhancers are selected and activated. In mice this strategy, applied to C57BL/6J and BALB/cJ mouse macrophages, illuminated several principles of cis-regulatory elements [89]. At the very basic level, this approach confirmed on the genome-wide scale that genetic variation that mutates lineage-determining transcription factor motifs reduces binding of the respective transcription factor relative to the alternate strain’s loci with wild type alleles. The effect size of motif mutations was dependent on the position within a given motif. Comparison of genome-wide mutation with and without consequence enabled the empirical definition of functional binding motifs for PU.1 and C/EBPα and furthermore identified the core nucleotides most consequential to binding. Notably, when analyzing the surrounding sequence of ‘mutated’ motifs, the presence of an additional motif within ~20 base pairs buffered the effect of mutations on binding, thereby highlighting the importance of local sequence context.
Consistent with the collaborative model of enhancer selection by sets of lineage-determining transcription factors, motif mutations that directly reduce binding of the respective factor also significantly reduce binding of the collaborating factor, even if its motif is not mutated [89]. Interestingly, upon TLR4 ligation and signal-dependent activation of NF-κB, motif mutations in lineage factor motifs were three times as likely to reduce NF-κB binding compared to mutations in the κB motif itself. These findings are consistent with individual-specific binding of NF-κB in human lymphoblastoid cell lines [106, 107]. Thus, natural genetic variation can be used to validate the importance of specific transcription factor combinations predicted by genomic studies.
Using natural genetic variation to discover regulatory networks
Macrophages are important effector cells that reside in every tissue of the body [4]. Their diverse functions in different tissue environments as well as their essential roles in health and disease make them an important experimental system to study chromatin priming, signal integration, and cooperative interactions at enhancers. To this end, transcriptomes and primed and active enhancers were compared between macrophages resident in diverse tissues in the mouse [19–21]. Different macrophage populations exhibited both a common program of ‘core’ macrophage gene expression as well as highly divergent patterns of gene expression that were specific to different tissue environments. In parallel, each population of tissue macrophages exhibited both common and distinct sets of active enhancers (Figure 4a). Intriguingly, experiments in which specific populations of tissue macrophages were either placed in culture or were transplanted to another anatomic location demonstrated that marked changes occurred in both transcriptomes and enhancer landscapes [21]. These results indicate that macrophage phenotypes are under constant environmental regulation and that local signals specify the active enhancer repertoire that controls context-dependent gene expression.
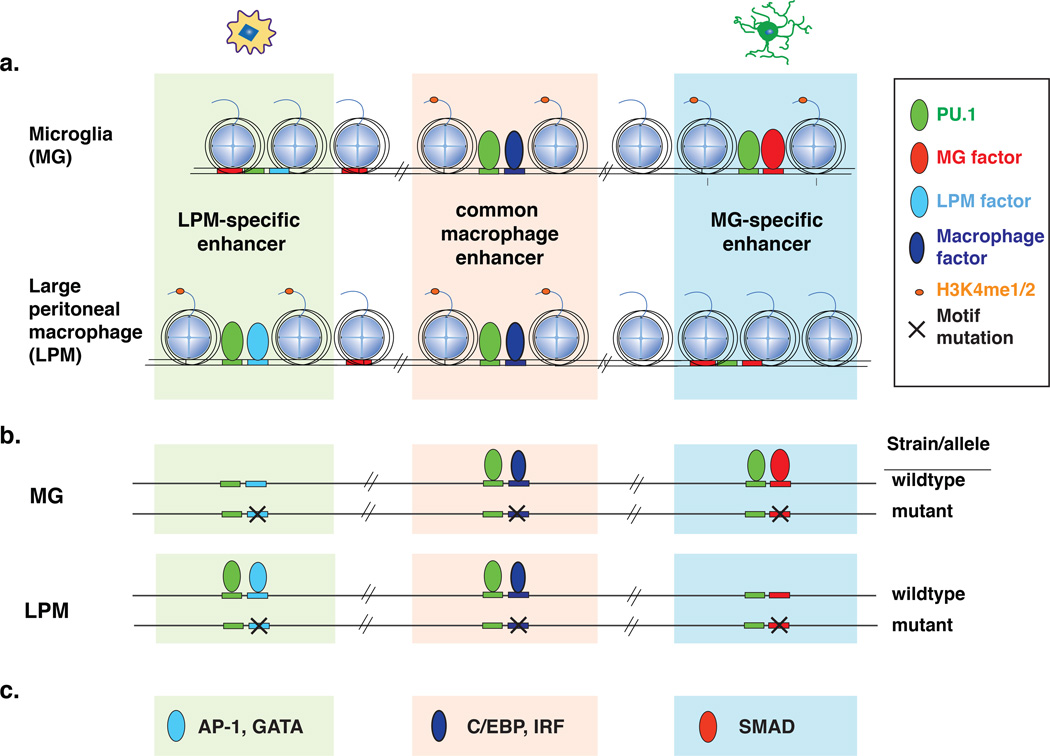
Figure 4. Natural genetic variation identifies transcription factor motifs that collaborate with PU.1 in distinct classes of enhancers.
a. Different transcription factors collaborative with PU.1 to select distinct classes of enhancers between microglia (MG) and large peritoneal macrophages (LPMs).
b. PU.1 binding is differently affected for distinct sets of collaborating motif mutations.
c. Motifs suggested to collaborate with PU.1 in each macrophage enhancer set include GATA in LPMs and SMAD in MG.
Motif enrichment analysis for cell type-specific enhancers suggested distinct sets of transcription factors that bind to each macrophage enhancer subset. This approach, however, does not establish that the implicated factors participate in enhancer selection and/or cooperative binding. Enhancers of all macrophage subsets are highly enriched for the ETS motif to which PU.1 binds, consistent with a requirement for PU.1 for the development of all tissue macrophages [2]. The observation that mutations in binding sites for C/EBP or AP-1 transcription factors could result in loss of PU.1 binding without mutations in the PU.1 binding site itself [89] suggested that genetic variation between diverse inbred mouse strains could be used to discover collaborative interactions between PU.1 and unknown lineage determining factors for each macrophage type (Figure 4b). Specifically, loss of PU.1 binding at a particular genomic location in one strain compared to another, without a mutation in the PU.1 recognition motif itself, could occur because of a mutation in the recognition motif for a collaborative transcription factor. Thus, characterization of the frequencies of mutations in all potential transcription factor recognition motifs in the vicinity of strain-similar versus strain-specific PU.1 binding could provide the basis for identifying collaborative factors important for PU.1 binding.
This approach was taken by performing ChIP-Seq for PU.1 in large peritoneal macrophages (LPMs) and microglia (MG) from three genetically diverse mouse strains (C57BL/6J, NOD/ShiLtJ, and SPRET/EiJ) that provide more than 40 million single nucleotide variants [20]. A sufficiently large number of strain-specific PU.1 binding sites was observed to allow the identification of dozens of motifs predicted to bind transcription factors that collaborate with PU.1 either in both LPMs and MG or specifically in one macrophage type (Figure 4). This approach yielded both known and unknown transcription factor candidates. For example, a GATA motif was selectively recovered in LPMs, which is consistent with the recently identified role of GATA6 in survival and proliferation of peritoneal macrophages downstream of retinoic acid signaling [19, 23, 108]. In line with its important function in LPMs, the Gata6 locus is associated with a super-enhancer in these cells, which is typical for cell fate-determining genes [69]. Conversely, SMAD motif mutations affected PU.1 specifically in MG, consistent with an essential role of TGFβ signaling in specifying microglia phenotypes [22, 109].
Remarkably, many of the putative collaborative transcription factors for PU.1 identified in LPMs were significantly down-regulated following transfer of LPMs to a tissue culture environment, in concert with loss of a large fraction of LPM-specific enhancers. Down-regulation of a subset of the collaborative factors could be prevented by treatment with retinoic acid, which was associated with maintenance of a corresponding subset of LPM-specific enhancers [20]. These results are consistent with the recent discovery of retinoic acid as an important peritoneal cavity-specific environmental factor [23].
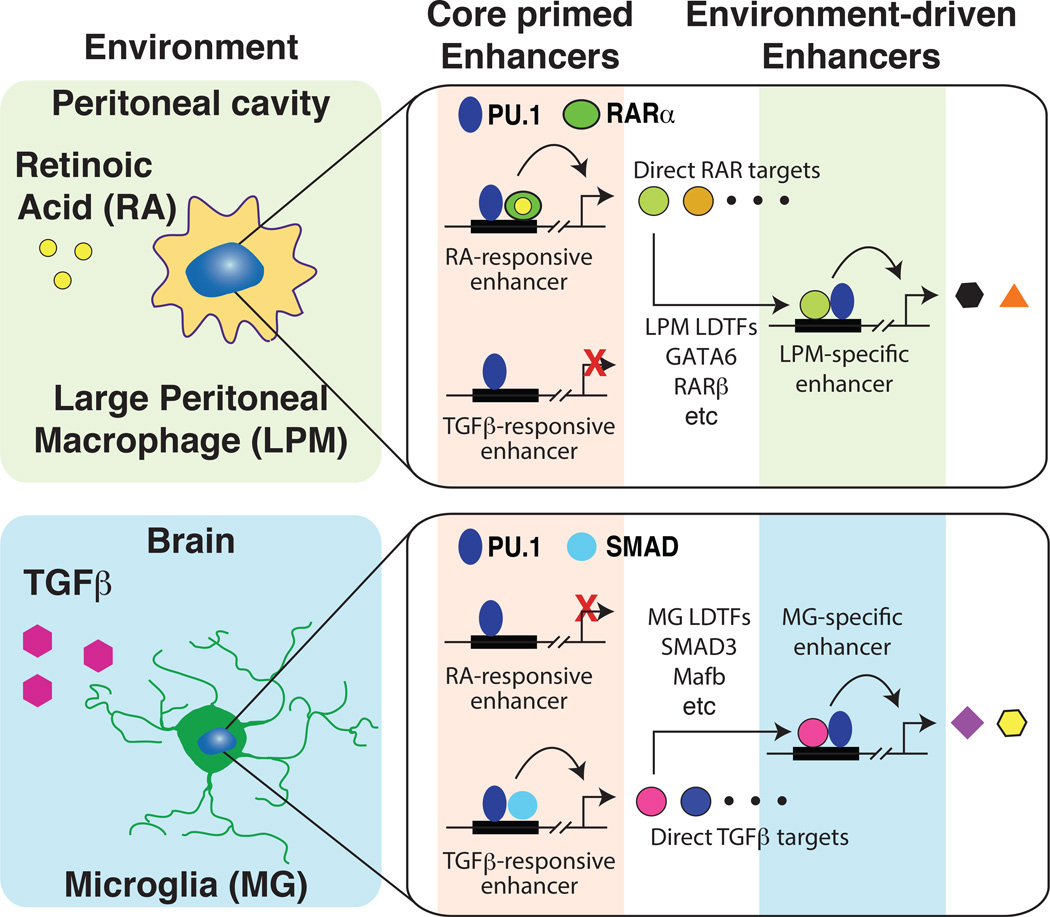
The use of genetic variation to identify motifs and corresponding transcription factors associated with both common and subset specific binding sites for PU.1 in LPMs and MG suggests a general model for the establishment of subset-specific enhancers and gene expression that builds upon the collaborative/hierarchical model initially described for macrophages and B cells. PU.1 and collaborative transcription factors that are common to many or all macrophage subsets prime a common set of enhancers that have the potential to respond to diverse internal and external signals. These enhancers become active in a context-dependent manner to drive downstream gene expression. For example, a common set of enhancers is primed to respond to retinoic acid signaling via retinoic acid receptors (Figure 5). However, these enhancers only become active in environments such as the peritoneal cavity, in which retinoic acid is present. Activation of these enhancers in LPMs leads to expression of retinoic acid target genes, which include transcription factors such as GATA6 that can collaborate with PU.1 to select a LPM-specific set of enhancers. An alternative set of common enhancers is primed to respond to TGFβ, but this only occurs in tissue environments that are characterized by high levels of TGFβ, such as the brain.
Figure 5. Environment differentially activates common primed enhancers to drive selection of tissue specific enhancers.
Top panel, LPMs residing in the peritoneal cavity respond to retinoic acid, which activates primed retinoic acid responsive enhancers. These in turn induce the expression of direct retinoic acid target genes that include transcription factors (e.g., GATA6, RARβ) that collaborate with PU.1 to select and activate LPM-specific enhancers. Lower panel, MG residing in the brain respond to TGFβ, which activates primed SMAD-responsive enhancers. These in turn induced the expression of direct SMAD target genes that include transcription factos (e.g., Mef2b) that collaborate with PU.1 to select and activate MG-specific enhancers.
Implications for human disease
Recent advances in the field of gene regulation on the genome-wide scale, such as emergent properties of enhancer selection and activation by different classes of transcription factors, have valuable applications in the field of human genetics. The observation that the majority (~88%) of risk loci for common diseases in genome-wide association studies (GWAS) are outside of the protein-coding genome [110] certainly necessitates insightful strategies for elucidating the functional sequence variants, perturbed regulatory mechanisms, affected genes, and affected cell types. Toward this goal, studies integrating chromatin modifications and/or DNA accessibility, transcription factor binding and computational predictions are demonstrating substantial progress [29, 42, 107, 111–115].
For immune cells in particular, the Immune Variation (ImmVar) project has mapped the effect of common human genetic variation on gene expression programs in healthy innate and adaptive immune cells at baseline and upon exposure to activating agents like influenza, LPS or interferon [116–118]. When the abundance of a given transcript measured across the population statistically associates with genotype, an expression quantitative trait locus, or eQTL, is observed (Cookson et al., 2009). ImmVar has robustly demonstrated that, in accordance with a similar study in human monocytes [119] and others, gene expression is genetically determined for thousands of expression traits by thousands of genetic variants. One common theme of these and other studies is that some eQTLs are apparent only for particular environmental exposures [120, 121]. For example, mRNA expression of the cytokine INF-β from the IFNB1 locus does not have an eQTL with any proximal SNPs (i.e., cis-eQTL) in unstimulated monocytes or those stimulated with LPS for 24 hours. Upon LPS stimulation for 2 hours, however, a significant cis-eQTL was observed at rs2275888 [119]. The expression of many additional transcripts also mapped to rs2275888 in trans only after 24 hours of LPS treatment. These eQTLs are termed trans-eQTLs, which means that the loci from which the genes are transcribed are far from the SNP to which they associate. The temporal relationship between IFN-β regulation in cis at 2 hours, and trans genes at 24 hours is consistent with transient signaling whereby the altered cis-regulated gene (IFNB1) transduces to its downstream targets in a defined timeframe. In general, overlapping cis- and trans-eQTL has proven a helpful way to pair direct targets of genetic variation with downstream targets. For example, in dendritic cells, this approach identified a cis-eQTL for IRF7 upon influenza exposure. Several genes co-mapped to the same SNP, rs12805435, in trans. Overexpression of IRF7 subsequenctly validated the predicted targets, overall summarizng a genetically-determined response during influenza infection in humans [116]
The observation that genetic variation affects gene expression with cell type and cell state context-specificity justifies a need to annotate enhancer elements in many cellular states. Several examples now exist where suggested functional variants for GWAS loci reside in signal-dependent enhancers [112, 116–118, 122]. For example, GWAS loci for Crohn’s disease, multiple sclerosis and rheumatoid arthritis are highly enriched for enhancers that gain epigenetic activation marks upon ex vivo stimulation of CD4+ T cells with phorbol myristate acetate (PMA)/ionomycin or Th0, Th1, and Th2 stimulation with anti-CD3/CD28 [112, 123]. These observations suggest that a significant fraction of disease-causing mechanisms originate in context-dependent regulatory states and highlight the importance of acquiring enhancer data from pure in vivo cell types and disease-relevant contexts. Looking ahead, tremendous opportunities will exist for geneticists and human immunologists to uncover the mechanistic underpinnings of human disease.
Concluding remarks
Rapid progress is being made with respect to how enhancers function; nonetheless, many challenges remain (see Outstanding Questions). For example, the ability to predict transcription factor binding and enhancer selection based on DNA sequence and knowledge of expressed transcription factors is a distant goal. Predicting the consequences of transcription factor binding is also problematic. One challenging observation, for instance, is that the binding of NF-κB to an enhancer can result in an increase, decrease, or no change in enhancer activity, as measured by histone acetylation, eRNA production, or other surrogate measures of activity. How spatial organization of transcription factor binding motifs, specific combinations of sequence-specific transcription factors, and associated co-regulators are integrated to specify different enhancer activity states remains poorly understood. An additional challenge is to link specific enhancers to target genes. Chromatin conformation capture assays indicate that these interactions may occur over megabases [124, 125] and multiple enhancers may regulate genes with unexpected effects. However, chromatin interactions detected by conformation capture assays do not necessary predict functional interactions [126]. Mutational analysis therefore remains the most reliable method for determining whether a putative enhancer element is of functional importance. Limitations to systematic mutagenesis have been greatly reduced by the development of CRISPR/Cas9- and TALEN-based methods, which will be a mainstay in interrogating enhancer function going forward [28]. However, these methods at present are most amenable to enhancer deletions, with specific point mutations being more difficult to generate. The tens of millions of SNPs provided by natural genetic variation in the mouse, and the greater than 10 million SNPs present in human populations, thus provide a valuable substrate for investigation of mechanisms that control enhancer selection, activity and target gene expression.
Box 2. Outstanding Questions.
Can we predict TF binding from genomic sequence alone? Many binding motifs are not bound by transcription factors, and conversely, many binding events do not involve canonical motifs.
How can we identify the repertoire of TFs with predominant activity in a given cell type? Many transcription factor family members bind the same motifs, different homo/heterodimer combinations complicate matters, and compensation among family members can occur.
What are the cell fate and state-determining signals that orchestrate enhancer selection and activity in different cell types? Signal integration involves numerous receptors, transduction molecules and transcription factors.
How can we predict gene targets of enhancers, and conversely, all enhancers for a gene? The nomination of nearest genes as enhancer targets is often incorrect. The resolution required to pair enhancers with promoters in genome-wide chromosome conformation experiments are cost-prohibitive for most laboratories, and physical interaction is not sufficient to determine enhancer activity.
What are the consequences of genetic mutations in respective enhancers? Enhancers may be redundant and genetic variation affecting enhancers is often buffered at the level of gene expression.
Highlights.
Enhancers are major determinants of cell specific gene expression
Small sets of lineage-determining factors prime the majority of macrophage enhancers
Tissue environment drives selective activation of macrophage enhancers
Genetic variation can be exploited to discover mechanisms of enhancer activation
Glossary
- C/EBP
a family of basic-leucine zipper (bZIP) transcription factors that bind DNA and form homo-and hetero-dimer interactions. C/EBPα and C/EBPβ are LDTFs in macrophages.
- ChIP-Seq
chromatin immunoprecipitation followed by high-throughput sequencing. This assay identifies the genomic location and frequency with which a particular protein or histone modification associates with DNA.
- Chromatin
DNA that is wrapped around nucleosomes. Chromatin compaction is dynamic with spatiotemporal patterns dependent on the cell cycle, developmental state, and chromosomal location. Chromatin provides a regulatory barrier between DNA and DNA-interacting proteins.
- cis-eQTL
An eQTL where the SNP and the gene locus for the associated transcript are close in linear genomic space (usually <1 megabase). cis-eQTLs typically quantify effects of genetic variation in the coding, promoter or enhancer for the given gene.
- De novo enhancer/latent enhancer
an enhancer that transitions from a closed chromatin state to an open and active state by interactions involving SDTFs and LDTFs.
- DNase-Seq
high-throughput DNA sequencing of accessible, often regulatory, regions of chromatin that result from chromatin digestion with DNase I.
- E2A
encoded by the TCF3 locus, binds to E-box sequences in DNA and forms homo- and hetero-dimers with other transcription factors. E2A is a critical LDTF for B cell development.
- EBF
a transcription factor expressed exclusively in the B cell lineage and directs B cell fate.
- Enhancer
a region of DNA that can amplify RNA PolII transcription at associated promoters. Enhancers are largely cell type-specific and are bound by sequence-specific transcription factors.
- Epigenetic marks
includes methylation of DNA as well as modifications such as acetylation, methylation, phosphorylation, and ubiquitinylation of amino acids on histone tails. Certain patterns of these epigenetic marks provide information about the function of the associated DNA.
- eQTL
expression quantitative trait locus, which results when the abundance of a transcript associates to the genotypes at a given genetic variant (usually at SNPs). eQTL studies require measuring transcripts across many individuals and can be classified as cis- or trans- (also in glossary).
- GATA6
a member of the GATA family of zinc finger transcription factors that is induced by retinoic acid signaling in LPMs.
- GWAS
genome wide association study. In most common form, uses specific SNPs (alleles) to link genomic loci to disease risk using cohorts of individuals with disease and healthy controls.
- H3K27ac
acetylation of the lysine at position 27 of the histone tail of histone H3. H3K27ac marks active enhancers and promoters.
- H3K4me1
mono-methylation of the lysine at position 4 of the histone tail of histone H3. H3K4me1 marks primed and active enhancers.
- H3K4me2
di-methylation of the lysine at position 4 of the histone tail of histone H3. H3K4me2 marks primed and active enhancers and promoters.
- H3K4me3
tri-methylation of the lysine at position 4 of the histone tail of histone H3. H3K4me3 marks promoters.
- Histone tails
peptides that extend from histones that can be modified with epigenetic marks.
- LDTF
lineage determining transcription factor: A transcription factor required for the development of a specific cell type. LDTFs typically have the ability to select enhancers in concert with other LDTFs or collaborative factors. Examples PU.1, C/EBPs in macrophages; E2A in B cells.
- LPM
large peritoneal macrophage - a macrophage population resident in the peritoneal cavity that is dependent on retinoic acid.
- LXRs
SDTF nuclear receptors that translocate to the nucleus and bind DNA in response to endogenous oxysterols to regulate cholesterol efflux and biosynthetic genes.
- Macrophage
innate immune phagocytic cells of myeloid origin that reside in every organ of the body and perform diverse functions in health and disease.
- MG
microglia - major resident macrophage population of the brain that is dependent on TGFβ.
- NF-κB
a SDTF transcription factor complex that binds DNA upon toll-like receptor signaling to activate inflammatory genes in macrophages. During B and T cell differentiation NF-κB has important function more in line with a LDTF role.
- Nucleosome
the unit of chromatin, that is composed of two copies each of histone proteins H2A, H2B, H3, and H4. 147 base pairs of DNA wrap around one nucleosome.
- Promoter
The region of DNA that contains binding sequences necessary to assemble the minimal transcriptional machinery and ultimately load RNA PolII at gene start sites.
- PU.1
encoded by the SPI1 human locus (Sfpi1 in mus musculus), is a member of the ETS family of sequence-specific transcription factors that is a LDTF critical for macrophage and B cell differentiation.
- RNA-Seq
high-throughput sequencing of RNA that is used to measure gene expression genome-wide.
- SDTF
signal dependent transcription factor: A transcription factor that becomes active in response to an internal or external signal. Examples: NF-κB, LXRs.
- SNP
single nucleotide polymorphism.
- Super-enhancer
clusters of enhancers that are densely occupied by master regulators and the Mediator co-regulator complex, which frequently occur at genes that define the identity of a given cell. Super-enhancers can alternatively be defined by tracking epigenetic marks indicative of enhancer activity, such as H3K27ac.
- trans-eQTL
An eQTL where the SNP and the gene locus for the associated transcript are far in linear genomic space (usually >1 megabase). trans-eQTLs typically quantify effects of the genetic variant on the associated gene through an intermediate product, such as by altered expression of a transcription factor or signaling molecule that perpetuate expression differences on the target gene.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional Materials
Text Box
Glossary
Outstanding questions
References
- 1.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327(5966):656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496(7446):445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucas T, et al. Differential roles of macrophages in diverse phases of skin repair. J Immunol. 2010;184(7):3964–3977. doi: 10.4049/jimmunol.0903356. [DOI] [PubMed] [Google Scholar]
- 4.Gordon S, Pluddemann A, Martinez Estrada F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol Rev. 2014;262(1):36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paolicelli RC, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333(6048):1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 6.Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289(5484):1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 7.Odegaard JI, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447(7148):1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu Y, et al. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell. 2014;157(6):1292–1308. doi: 10.1016/j.cell.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright JR. Clearance and recycling of pulmonary surfactant. Am J Physiol. 1990;259(2 Pt 1):L1–L12. doi: 10.1152/ajplung.1990.259.2.L1. [DOI] [PubMed] [Google Scholar]
- 10.Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011;145(3):341–355. doi: 10.1016/j.cell.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4(1):71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 12.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41(1):49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 14.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3(1):23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 15.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8(12):958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9(10):692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 17.Smale ST. Transcriptional regulation in the innate immune system. Curr Opin Immunol. 2012;24(1):51–57. doi: 10.1016/j.coi.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takeda K, Akira S. STAT family of transcription factors in cytokine-mediated biological responses. Cytokine Growth Factor Rev. 2000;11(3):199–207. doi: 10.1016/s1359-6101(00)00005-8. [DOI] [PubMed] [Google Scholar]
- 19.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13(11):1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159(6):1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159(6):1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butovsky O, et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17(1):131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157(4):832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyce BF, Xing L. Biology of RANK, RANKL, and osteoprotegerin. Arthritis Res Ther. 2007;9(Suppl 1):S1. doi: 10.1186/ar2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 26.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507(7493):455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roadmap Epigenomics C, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Core LJ, et al. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet. 2014;46(12):1311–1320. doi: 10.1038/ng.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 32.Heinz S, et al. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015 doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15(4):272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 34.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489(7414):83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buenrostro JD, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 37.May D, et al. Large-scale discovery of enhancers from human heart tissue. Nat Genet. 2012;44(1):89–93. doi: 10.1038/ng.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dickel DE, et al. Function-based identification of mammalian enhancers using site-specific integration. Nat Methods. 2014;11(5):566–571. doi: 10.1038/nmeth.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith RP, et al. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet. 2013;45(9):1021–1028. doi: 10.1038/ng.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arnold CD, et al. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339(6123):1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 41.Vanhille L, et al. High-throughput and quantitative assessment of enhancer activity in mammals by CapStarr-seq. Nat Commun. 2015;6:6905. doi: 10.1038/ncomms7905. [DOI] [PubMed] [Google Scholar]
- 42.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42(4):343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8(5):e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam MT, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498(7455):511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaukowitch K, et al. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56(1):29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsieh CL, et al. Enhancer RNAs participate in androgen receptor-driven looping that selectively enhances gene activation. Proc Natl Acad Sci U S A. 2014;111(20):7319–7324. doi: 10.1073/pnas.1324151111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaikkonen MU, et al. Remodeling of the Enhancer Landscape during Macrophage Activation Is Coupled to Enhancer Transcription. Mol Cell. 2013;51(3):310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bonn S, et al. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44(2):148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- 54.Kieffer-Kwon KR, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155(7):1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25(21):2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114(Pt 13):2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 58.Heinz S, et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Molecular Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scott EW, et al. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265(5178):1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 60.McKercher SR, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15(20):5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 61.Lara-Astiaso D, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345(6199):943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kazemian M, et al. Widespread evidence of cooperative DNA binding by transcription factors in Drosophila development. Nucleic Acids Res. 2013;41(17):8237–8252. doi: 10.1093/nar/gkt598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng Y, et al. Principles of regulatory information conservation between mouse and human. Nature. 2014;515(7527):371–375. doi: 10.1038/nature13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stergachis AB, et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515(7527):365–370. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McConnell SC, et al. Human globin knock-in mice complete fetal-to-adult hemoglobin switching in postnatal development. Mol Cell Biol. 2011;31(4):876–883. doi: 10.1128/MCB.00725-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peterson KR, et al. Transgenic mice containing a 248-kb yeast artificial chromosome carrying the human beta-globin locus display proper developmental control of human globin genes. Proc Natl Acad Sci U S A. 1993;90(16):7593–7597. doi: 10.1073/pnas.90.16.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dowen JM, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159(2):374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155(4):934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153(2):307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parker SC, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110(44):17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47(1):8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 72.Forrester WC, et al. Evidence for a locus activation region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 1987;15(24):10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grosveld F, et al. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51(6):975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 74.Tuan D, et al. The "beta-like-globin" gene domain in human erythroid cells. Proc Natl Acad Sci U S A. 1985;82(19):6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.May G, et al. Dynamic analysis of gene expression and genome-wide transcription factor binding during lineage specification of multipotent progenitors. Cell Stem Cell. 2013;13(6):754–768. doi: 10.1016/j.stem.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doulatov S, et al. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10(2):120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 77.Gifford CA, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153(5):1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koche RP, et al. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8(1):96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsankov AM, et al. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518(7539):344–349. doi: 10.1038/nature14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu J, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23(24):2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang JA, et al. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149(2):467–482. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ziller MJ, et al. Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature. 2015;518(7539):355–359. doi: 10.1038/nature13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anguita E, et al. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 2004;23(14):2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grass JA, et al. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100(15):8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gao X, et al. Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J Exp Med. 2013;210(13):2833–2842. doi: 10.1084/jem.20130733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maurano MT, et al. Widespread site-dependent buffering of human regulatory polymorphism. PLoS Genet. 2012;8(3):e1002599. doi: 10.1371/journal.pgen.1002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tall AR, Yvan-Charvet L. Cholesterol, inflammation and innate immunity. Nat Rev Immunol. 2015;15(2):104–116. doi: 10.1038/nri3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1(2):135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 89.Heinz S, et al. Effect of natural genetic variation on enhancer selection and function. Nature. 2013 doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152(1–2):157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 91.Ghisletti S, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32(3):317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 92.Garber M, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47(5):810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9(2):279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 94.Mullen AC, et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147(3):565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151(1):153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43(3):264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barish GD, et al. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24(24):2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vahedi G, et al. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151(5):981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trompouki E, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147(3):577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brunkow ME, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27(1):68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 101.Khattri R, et al. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol. 2003;4(4):337–342. [PubMed] [Google Scholar]
- 102.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126(6):1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 103.Szabo SJ, et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100(6):655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 104.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151(2):289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li P, et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490(7421):543–546. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kasowski M, et al. Variation in transcription factor binding among humans. Science. 2010;328(5975):232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McVicker G, et al. Identification of genetic variants that affect histone modifications in human cells. Science. 2013;342(6159):747–749. doi: 10.1126/science.1242429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gautier EL, et al. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. J Exp Med. 2014;211(8):1525–1531. doi: 10.1084/jem.20140570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Makwana M, et al. Endogenous transforming growth factor beta 1 suppresses inflammation and promotes survival in adult CNS. J Neurosci. 2007;27(42):11201–11213. doi: 10.1523/JNEUROSCI.2255-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518(7539):337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Trynka G, et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet. 2012 doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kasowski M, et al. Extensive variation in chromatin states across humans. Science. 2013;342(6159):750–752. doi: 10.1126/science.1242510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kilpinen H, et al. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science. 2013;342(6159):744–747. doi: 10.1126/science.1242463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee MN, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343(6175):1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Raj T, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344(6183):519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ye CJ, et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345(6202):1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343(6175):1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Romanoski CE, et al. Systems genetics analysis of gene-by-environment interactions in human cells. Am J Hum Genet. 2010;86(3):399–410. doi: 10.1016/j.ajhg.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Orozco LD, et al. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell. 2012;151(3):658–670. doi: 10.1016/j.cell.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Harismendy O, et al. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470(7333):264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hawkins RD, et al. Global chromatin state analysis reveals lineage-specific enhancers during the initiation of human T helper 1 and T helper 2 cell polarization. Immunity. 2013;38(6):1271–1284. doi: 10.1016/j.immuni.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lieberman-Aiden E, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sanyal A, et al. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhou HY, et al. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 2014;28(24):2699–2711. doi: 10.1101/gad.248526.114. [DOI] [PMC free article] [PubMed] [Google Scholar]