Summary
The role of pyruvate kinase M2 (PKM2) in cell proliferation is controversial. A unique function of PKM2 proposed to be important for the proliferation of some cancer cells involves the direct activity of this enzyme as a protein kinase; however, a detailed biochemical characterization of this activity is lacking. Using [32P]-phosphoenolpyruvate (PEP) we examine the direct substrates of PKM2 using recombinant enzyme and in vitro systems where PKM2 is genetically deleted. Labeling of some protein species from [32P]-PEP can be observed, however most were dependent on the presence of ADP, and none were dependent on the presence of PKM2. In addition, we also failed to observe PKM2-dependent transfer of phosphate from ATP directly to protein. These findings argue against a role for PKM2 as a protein kinase.
Introduction
The M2 isoform of pyruvate kinase (PKM2) is expressed in cancer cells and many normal cells with proliferative capacity and/or anabolic functions. Several lines of evidence argue that PKM2 is important for cancer cell proliferation in some contexts. PKM2 is expressed in cancers and cell lines, and regulation of PKM2 enzymatic activity can influence cell proliferation (Anastasiou et al., 2012; Chaneton et al., 2012; Israelsen et al., 2013; Lunt et al., 2015). Two models, which are not mutually exclusive, seek to explain the importance of PKM2 expression in cancer (McKnight, 2014). One proposes that PKM2 exerts its effects through its canonical activity, which is regulated to control cell metabolism and macromolecular synthesis. The other proposes that PKM2 is a protein kinase involved in signaling and regulating gene expression.
Pyruvate kinase catalyzes the final step in glycolysis, transferring a phosphate group from phosphoenolpyruvate (PEP) to ADP to produce pyruvate and ATP. There are four mammalian isoforms, and alternative splicing of the PKM gene produces the isoforms PKM1 and PKM2. The exon unique to PKM2 is reported to confer protein kinase activity as well as regulatory properties that are lacking in PKM1. For example, PKM2 requires its allosteric activator fructose-1,6-bisphosphate (FBP) to be fully active, while PKM1 activity is constitutively high (Dombrauckas et al., 2005; Morgan et al., 2013). In support of a metabolic role for pyruvate kinase in proliferation the glycolytic enzyme activity of PKM2, but not PKM1, can be inhibited by growth signaling in rapidly proliferating cells, and the resulting low levels of enzyme activity facilitate tumor growth (Christofk et al., 2008; Hitosugi et al., 2009). PKM2 is also allosterically activated by the metabolites serine and SAICAR, enabling regulation of this enzyme by nutrient conditions and coordination with metabolic pathways outside of glycolysis (Chaneton et al., 2012; Keller et al., 2012; Kung et al., 2012). Synthetic activators of PKM2 impair tumor growth by raising PKM2 activity to PKM1 levels (Anastasiou et al., 2012), and high pyruvate kinase activity impairs mouse embryonic fibroblast (MEF) proliferation by limiting precursors for DNA replication (Lunt et al., 2015). Deletion of PKM2 does not prevent tumor growth, and results in compensatory PKM1 expression in non-proliferating cells in the tumor, arguing that the ability of PKM2 to be active or inactive allows this isoform to support different metabolic cell states in tissues (Israelsen et al., 2013).
The protein kinase activity reported for PKM2 is distinct from its role in glycolysis. Transfer of a phosphate from PEP to proteins, which also generates pyruvate, has been reported to occur as a result of a kinase activity that is unique to PKM2 among pyruvate kinase isoforms. PEP-dependent phosphorylation is uncommon in cells, but has been observed in both bacterial and mammalian systems (Deutscher et al., 2006; Vander Heiden et al., 2010). In both cases, phosphate is transferred from PEP is to a histidine residue on a protein, and the bacterial PEP-dependent histidine kinases have no homology to PKM2 or other enzymes in glycolysis. PKM2 has been reported to function as a dual-specificity kinase, transferring phosphate to serine/threonine and tyrosine residues on different proteins (Gao et al., 2012; Yang et al., 2012a). When studied, PKM2 protein kinase activity is catalyzed by dimeric PKM2, a state with much lower glycolytic enzyme activity when compared with fully active tetrameric PKM2. In particular, dimeric PKM2 localizes to the nucleus where it acts as a Stat3 kinase with an R399E mutant reported to be an obligate dimer with elevated protein kinase activity (Gao et al., 2012). Subsequent work suggested that PKM2 also phosphorylates histone H3. EGFR signaling to ERK1/2 can result in nuclear translocation of PKM2, where PKM2 protein kinase activity leads to c-Myc and cyclin D1 expression to drive cell cycle progression beyond the G1/S checkpoint (Yang et al., 2012a; Yang et al., 2012b). PKM2 is also reported to be a kinase of spindle assembly checkpoint protein Bub3, playing an important role in ensuring faithful chromosome segregation (Jiang et al., 2014). Finally, SAICAR, the allosteric activator of canonical PKM2 activity, was reported to synergize with ERK2 to stimulate protein kinase activity of PKM2, enabling it to phosphorylate a large number of proteins (Keller et al., 2012).
Although tumors express PKM2 this enzyme is not essential for growth of all tumors. PKM2 deletion does not prevent tumorigenesis in a mouse model of breast cancer and loss of PKM2 can promote tumor progression (Israelsen et al., 2013). PKM2 is also not required for hematopoietic stem cell or leukemia cell proliferation, but PKM2 expression enhances transplantation of hematopoietic stem cells (Wang et al., 2014). Finally, knockdown of PKM2 using RNA interference does not alter formation or maintenance of xenograft tumors, although it partially obstructs proliferation in vitro (Cortes-Cros et al., 2013). These data argue against any model where PKM2 is essential for regulating cell cycle mediators or other events that are important for eukaryotic cell proliferation.
To date, studies examining PKM2 functions in cancer have not resolved differential roles for PKM2 in signaling and regulation of metabolism. We investigated the protein kinase activity of PKM2; however, we were unable to demonstrate PKM2-dependent phosphorylation of any proteins in vitro using either PEP or ATP as a phosphate donor. Instead, all PKM2-dependent phosphotransfer events observed involved regeneration of ATP from ADP and PEP, the known function of PKM2 in glycolysis. This study also establishes methods to track PEP-dependent protein phosphorylation in cells.
Results
PEP-dependent phosphorylation events in cell lysates are independent of PKM2
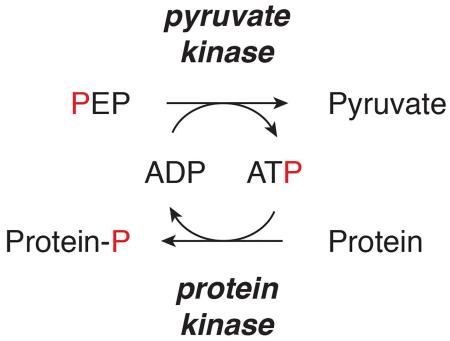
PEP-dependent phosphorylation events are well described as part of two-component signaling in bacteria (Deutscher et al., 2006). Transfer of phosphate from [32P]-PEP to phosphoglycerate mutase (PGAM) has been shown in eukaryotic cell lysates, but this activity is not dependent on PKM2 (Vander Heiden et al., 2010). To identify other PEP-dependent phosphorylation events in mammalian cells, we tracked [32P]-phosphate transfer to proteins as a sensitive measure of protein phosphorylation. The phosphorylation reaction was performed in the presence of vanadate to inhibit phosphatase activity, and also to minimize PGAM1 labeling as vanadate promotes PGAM1-His11 dephosphorylation (Carreras et al., 1982). PGAM1 labeled from PEP is the dominant species generated in this reaction (Vander Heiden et al., 2010), and we sought to minimize this to avoid masking phosphorylation events involving less abundant proteins. Vanadate was included as a phosphatase inhibitor in past studies of PKM2 protein kinase activity and thus is not expected to inhibit this enzyme activity of PKM2 (Gao et al., 2012; Yang et al., 2012a). [32P]-PEP was added to hypotonic cell lysates prepared to minimize disruption of endogenous protein complexes and protein phosphorylation was visualized by SDS-PAGE and autoradiography (Figure 1). PGAM1 labeling was observed when phosphatase inhibitors were not included (Figure S1A). To determine whether the phosphotransfer events were dependent on ATP or PEP as a phosphate donor, 1 mM unlabeled PEP or ATP was added as a competitor for the reaction, a control lacking from published demonstrations of PKM2 protein kinase activity that rely heavily on antibodies to detect phosphorylation events. Most protein labeling events observed are competed by non-radioactive ATP and not non-radioactive PEP regardless of whether vanadate is included in the reaction (Figures 1, S1B). This finding is consistent with ATP serving as the direct phosphate donor for most phosphorylation events and is consistent with previous reports (Vander Heiden et al., 2010). Interestingly, diffuse bands of variable intensity that migrated on SDS-PAGE with the 15-20 kDa and 100-120 kDa molecular weight markers were labeled from [32P]-PEP that were sensitive to competition by non-radioactive PEP but not non-radioactive ATP. A previous study had observed non-enzymatic labeling of proteins upon boiling (Schieven and Martin, 1988), so to determine if these bands were dependent on transfer to protein in cell lysates, [32P]-PEP or [γ-32P]-ATP were added to loading buffer in the absence of added protein (Figure S1C). The same bands were observed when [32P]-PEP, but not [γ-32P]-ATP, was subjected to SDS-PAGE, and were sensitive to the addition of non-radioactive PEP to the loading dye. The same bands are observed when [32P]-PEP is loaded with glycerol alone, arguing that these events reflect an artifact and are not protein phosphorylation events.
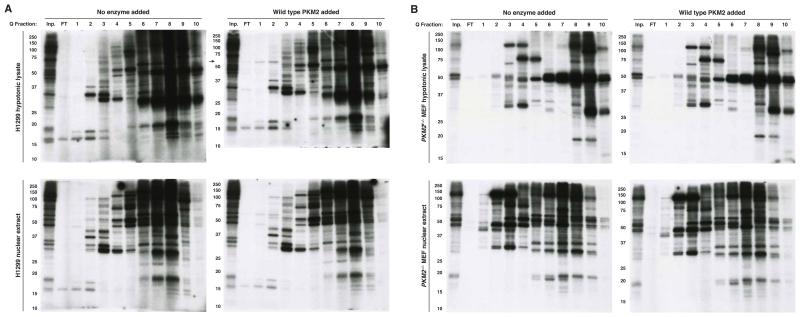
Figure 1.
Assessement of phosphoenolpyruvate (PEP) as a phosphate donor for protein phosphorylation. Hypotonic lysates and nuclear extracts from (A) H1299, (B) PKM2-null MEF, (C) U87, and (D) U87 EGFR-VIII cells were incubated for 1 h with impure [32P]-PEP, and phosphorylated proteins analyzed by SDS-PAGE and autoradiography. Reactions were carried out with the addition of no enzyme, recombinant wild type (WT) or mutant (R399E) his-tagged PKM2 (each at 10 μg/mL) as indicated. In addition, no competitor or excess (1 mM) non-radioactive competitor PEP or ATP was also included where indicated. See also Figure S1.
Consistent with PEP-dependent phosphorylation being uncommon, most other phosphorylation events from [32P]-PEP were competed by 1 mM ATP and not 1 mM PEP; however phosphorylation of a species migrating at ~45 kDa that was not blocked by excess ATP was observed in both H1299 (human non-small cell lung carcinoma) (Figure 1A) and mouse embryonic fibroblast (MEF) (Figure 1B) cell lysates. No reported substrates of PKM2-dependent protein phosphorylation are consistent with this molecular weight, suggesting that this event is a previously uncharacterized target of PEP-dependent phosphorylation.
To determine whether addition of PKM2 would result in additional phosphorylation events, we included recombinant PKM2 (rPKM2) in the reaction. rPKM2 has been used previously to show PKM2-dependent phosphorylation (Gao et al., 2012; Keller et al., 2014; Yang et al., 2012a); however, we did not observe any PEP-dependent phosphorylation events upon addition of either wild type (WT) rPKM2 or a mutant PKM2 (R399E) that is reported to have higher protein kinase activity (Gao et al., 2012) (Figure 1A). Both WT- and R399E-rPKM2 enzymes exhibit pyruvate kinase activity as a glycolytic enzyme as both can support synthesis of [γ-32P]-ATP from [32P]-PEP and cold ADP; although consistent with reports that the R399E mutant favors a less active form of the enzyme, this protein is less efficient than WT PKM2 in transferring [32P]-phosphate from PEP to ADP (Figure S1D). Importantly, the WT PKM2 generated for these studies had a similar specific activity for conversion of PEP and ADP to pyruvate and ATP as reported previously (Figure S1E) (Dombrauckas et al., 2005). Because most substrates of PKM2-dependent phosphorylation are nuclear, we also tested the ability of PKM2 to phosphorylate proteins in a nuclear extract and again failed to observe any PEP-dependent phosphorylation events with or without rPKM2 included in the reaction (Figure 1).
Because PKM2 is an abundant protein (Beck et al., 2011), we considered the possibility that endogenous PKM2 present in the H1299 hypotonic and nuclear lysates (Figure S1F) could be sufficient to catalyze PKM2-dependent phosphorylation, and that this might explain why addition of rPKM2 did not produce any additional phosphorylated species. To definitively assess a requirement for PKM2 in any PEP-dependent phosphotransfer events, we utilized PKM2-deleted MEFs (Lunt et al., 2015) and confirmed that PKM2-specific exon 10 was deleted from the genome and that PKM2 protein is absent from these cells (Figure S1G,H). Upon deletion of this exon, PKM1 is expressed and the cells continue to possess pyruvate kinase activity (Lunt et al., 2015). Phosphotransfer from PEP was similar to that in H1299 cells, and we did not observe phosphorylation events dependent on the addition of rPKM2 to PKM2−/− lysates (Figure 1B). PKM2 protein kinase activity is reported to be important specifically for glioblastoma development downstream of EGFR activation (Gao et al., 2012; Keller et al., 2014; Yang et al., 2012a; Yang et al., 2012b), however, we also failed to observe PEP-dependent phosphorylation events in hypotonic and nuclear lysates of U87 glioblastoma multiforme cells and U87 cells expressing constitutively active EGFR-VIII except for the species at 45 kDa, and this was not dependent on PKM2 addition (Figure 1C, D).
PKM2-dependent regeneration of ATP can account for PEP-dependent phosphorylation
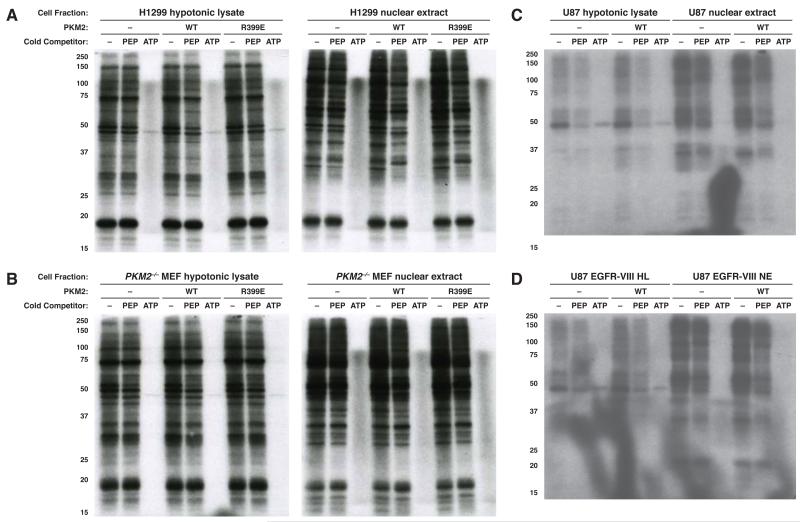
Most labeling from [32P]-PEP was eliminated by addition of excess cold ATP, suggesting that [γ-32P]-ATP was either generated from [32P]-PEP, or present as a contaminant in the reaction to serve as the phosphate group donor for most of the observed phosphorylation events. Indeed, [32P]-PEP is synthesized from commercial [32P]-ATP using pyruvate kinase (Mattoo and Waygood, 1983; Vander Heiden et al., 2010), and despite a step to enrich for [32P]-PEP, these preparations are contaminated by small quantities of [γ-32P]-ATP (Figure 2A). Preparative HPLC of [32P]-PEP allowed for quantitative separation from the contaminating [γ-32P]-ATP. To test the contribution of each phosphate donor to protein phosphorylation in a cell lystate, we incubated cells with crude [32P]-PEP, HPLC purified [32P]-PEP, and the HPLC fraction containing [32P]-ATP present as a contaminant in crude [32P]-PEP. Most proteins phosphorylated by the impure [32P]-PEP were phosphorylated by the contaminating [γ-32P]-ATP fraction but not the purified [32P]-PEP fraction (Figure 2B). Consistent with competition by cold ATP and PEP allowing determination of direct phosphate donors, the ~45 kDa protein identified previously was labeled from purified [32P]-PEP. Addition of ADP to purified [32P]-PEP restored phosphorylation of many proteins, and the resulting phosphorylation events could be competed by both cold ATP and cold PEP. These data argue that pyruvate kinase activity present in these reactions is sufficient to allow [γ-32P]-ATP synthesis from ADP and [32P]-PEP (Figure 2B, lanes 10-12). Since [γ-32P]-ATP is the phosphate donor for most protein kinase activity in cells, these data suggest that the known role of PKM2 as a glycolytic enzyme might explain PKM2-dependent phosphorylation whereby PKM2 transfers phosphate from [32P]-PEP to ADP present in cell lysate to synthesize [γ-32P]-ATP and result in phosphorylation seemingly dependent on PEP and the addition of PKM2. Consistent with this possibility, a hypotonic cell lysate contains sufficient ADP to enable synthesis of [γ-32P]-ATP from [32P]-PEP when added to the lystate, and this ability is diminished if size exclusion chromatography is used to deplete metabolites in the lysate (Figure S2).
Figure 2.
Contaminating [γ-32P]-ATP is the predominant phosphoryl donor when using crude [32P]-PEP. (A) [32P]-PEP synthesized from [γ-32P]-ATP is contaminated by small amounts of [γ-32P]-ATP, which can be separated by preparative HPLC. The purity of each fraction was analyzed by thin-layer chromatography and autoradiography. (B) The contaminating [γ-32P]-ATP component of impure [32P]-PEP is the phosphoryl donor for the majority of proteins phosphorylated by impure [32P]-PEP. An H1299 hypotonic lysate was incubated for 1 h with recombinant PKM2 and different 32P-containing metabolites as well as no competitor or excess (1 mM) non-radioactive competitor PEP or ATP as indicated. Phosphorylated proteins were analyzed by SDS-PAGE and autoradiography. See also Figure S2.
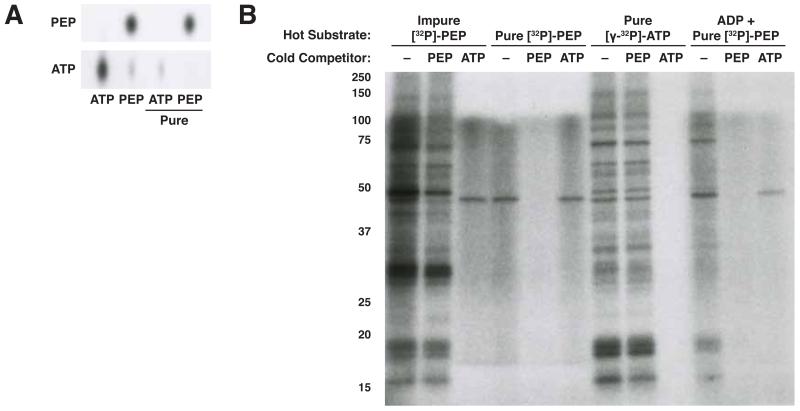
We next considered the possibility that our failure to observe protein phosphorylation dependent on both [32P]-PEP and PKM2 in hypotonic or nuclear lysates may have been due to the absence of relevant protein kinase substrates. For instance, histones, a putative target of PEP-dependent PKM2 protein kinase activity (Yang et al., 2012a), are not efficiently extracted by many protocols used to prepare nuclear extracts (Figure S1F) (Shechter et al., 2007). To determine whether PKM2 can phosphorylate histones in our assay, we incubated WT- or R399E-rPKM2 and commercially prepared histones from calf thymus with impure [32P]-PEP. Although we failed to observe any phosphorylation upon addition of either rPKM2 or PKM2 (R399E), a nuclear extract of H1299 cells incubated with impure [32P]-PEP was able to phosphorylate proteins that run at the expected size of histones when histones are added (Figure 3A), suggesting that the presence of PKM2 in a nuclear extract could enable indirect transfer of phosphate from PEP to histones. The inability of PKM2 to do this directly confirms that any dependence on PKM2 is to transfer the phosphate to generate ATP rather than PKM2 acting directly as a protein kinase. We also conducted the assay in a purely recombinant system using two putative protein substrates of PKM2 using antibodies to detect protein phosphorylation and did not observe direct PKM2 protein kinase activity (Figure S3A,B).
Figure 3.
(A) Calf thymus histones were incubated for 1 h with impure [32P]-PEP and no enzyme, recombinant wild type (WT) or mutant (R399E) PKM2 (each at 10 μg/mL) as indicated. An H1299 nuclear extract was also incubated with impure [32P]-PEP with and without calf thymus histones added. Phosphorylated proteins were analyzed by SDS-PAGE and autoradiography. Arrows indicate phosphorylated proteins with apparent molecular weights similar to histones (core histones, 11-17 kDa, many histone H1 isoforms 25-35 kDa) (Shechter et al., 2007). (B) H1299 cell hypotonic lysates and nuclear extracts and calf thymus histones were incubated for 1 h with [32P]-PEP and 10 μg/mL rPKM2 with or without 0.5 mM SAICAR as well as no competitor or excess (1 mM) cold competitor PEP or ATP. (C) Hypotonic lysates and nuclear extracts from H1299 cells and PKM2-expressing (flox/flox) or PKM2-null (−/−) MEFs were desalted and incubated for 1 h with HPLC-purified [32P]-PEP with no additional enzymes or with added recombinant wild type or mutant (R399E) rPKM2 (each at 10 μg/mL). See also Figure S3.
The purine precursor SAICAR may be an activator of PKM2 protein kinase activity (Keller et al., 2014). In particular, it has been suggested that absence of SAICAR from in vitro phosphorylation reactions could account for an apparent disparity in protein kinase activity between rPKM2 produced in E. coli and PKM2 obtained from mammalian cell nuclear extracts. To determine if the absence of SAICAR explained why we failed to observe PKM2-mediated protein kinase activity in cell and nuclear extracts, we tested if addition of SAICAR to a reaction with rPKM2 and cell lysate, nuclear extract, or purified histones and other substrates could reconstitute PKM2-depedent protein kinase activity (Figure 3B, S3A-C). In no case was SAICAR addition sufficient to cause protein phosphorylation that was competed by PEP addition but not ATP addition, or that was dependent on the presence of PKM2. These data suggest that any increase in PEP-dependent protein phosphorylation following SAICAR addition that involves PKM2 is likely explained by the ability of SAICAR to activate the glycolytic enzyme function of PKM2 (Keller et al., 2012), and promote generation of ATP as a phosphate donor for other protein kinases.
Because ATP is present at millimolar concentrations in cells (Marcussen and Larsen, 1996), it is unlikely that addition of 1 mM cold competitor ATP is sufficient to inhibit physiologically relevant PKM2-dependent phosphorylation. However, we considered the possibility that addition of cold ATP might inhibit apparent PKM2 protein kinase activity in vitro. To test this possibility, we incubated cell lysates with 5′-(4-fluorosulfonobenzoyl)-adenosine (FSBA) to covalently inhibit endogenous ATP-dependent kinases aid identification of PEP-dependent phosphorylation event without adding cold ATP. FSBA was confirmed to be a partial inhibitor of ATP-dependent kinases in cell lysates and nuclear extracts (Figure S3D), however when the same lysates and nuclear extracts were assayed with only PEP as a phosphate donor, we failed to observe any FSBA insensitive protein phosphorylation (Figure S3E).
To further search for PEP-dependent phosphorylation events that require PKM2 functions other than its known action in generating ATP, we tested HPLC purified [32P]-PEP in a reaction with protein extracts that were first desalted to remove contaminating ADP, and with or without the addition of rPKM2 or rPKM2-R399E. These reactions were performed with whole cell hypotonic lysates and nuclear extracts prepared from PKM2-expressing H1299 cells and MEFs as well as PKM2-null MEFs (Figure 3C). Using this approach, we observed several phosphorylated species, including the ~45 kDa species noted previously (Figure 1A); however no events were dependent on the addition of WT- or R399E-rPKM2, including events in extracts prepared from PKM2-null MEFs. These data argue that a PKM2-dependent protein phosphorylation activity is not present in these cells.
Lack of evidence for an ATP-dependent PKM2 protein kinase activity
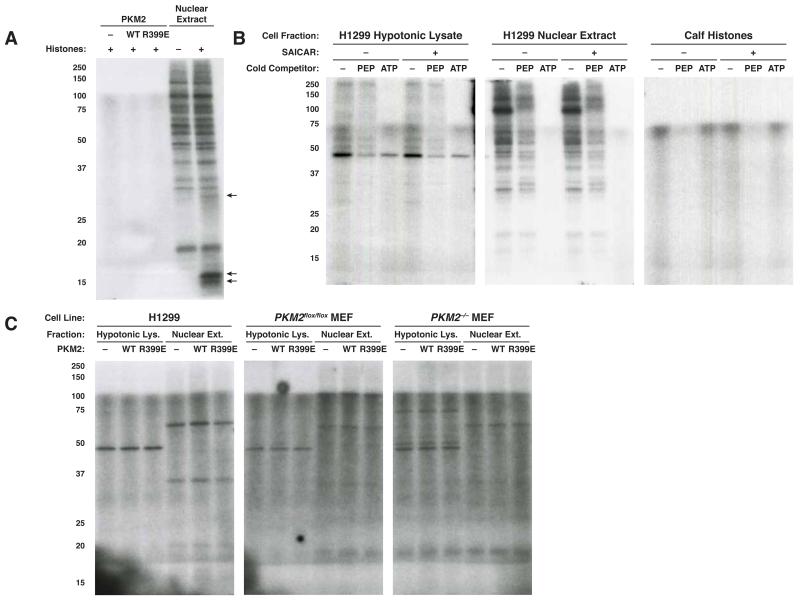
Failure to observe PEP-dependent protein phosphorylation that is dependent on PKM2 does not rule out the possibility that PKM2 possesses an ATP-dependent protein kinase activity. Both PEP and ATP can act as phosphate donors for normal PKM2 enzymatic activity, so an alternative explanation for reports of PKM2 protein kinase activity might involve PKM2 as an ATP-dependent protein kinase. The ability to detect ATP-dependent phosphorylation events dependent on the addition of rPKM2 to a cell lysate is complicated by the fact that many proteins undergo ATP-dependent phosphorylation. To mitigate this complication, and to control for the presence or absence of PKM2 in the lysates, we fractionated hypotonic and nuclear extracts from H1299 cells and from PKM2-expressing and PKM2-null MEFs by anion exchange chromatography. We assayed each fraction for phosphorylation dependent on the addition of rPKM2 (Figure 4, S4A). One phosphorylated species (Figure 4A, H1299 hypotonic lysate fractions 1-2, MW ~60 kDa) showed dependence on the addition of rPKM2. rPKM2 also has a MW of ~60 kDa, raising the possibility that phosphorylation of rPKM2 by kinases present in the lysate rather than production of a PKM2-phosphorylated species explains this result. Prior work has suggested that PKM2 can be a substrate for phosphorylation (Hitosugi et al., 2009). In support of rPKM2 being phosphorylated by a kinase in the lysate, we were able to purify this species by Ni-NTA affinity chromatography (Figure S4B), indicating that this represents phosphorylated rPKM2 rather than a protein phosphorylated by PKM2. Taken together, these data argue against PKM2 having direct protein kinase activity.
Figure 4.
Lack of evidence for an ATP-dependent protein kinase activity for PKM2. (A) H1299 and (B) PKM2-null MEF hypotonic lysates and nuclear extracts were used to identify substrates for PKM2 kinase activity. Lysates were fractionated on a 1mL Q Sepharose HP column (Inp., 1:10 dilution of column input; FT, flow through; 1-10, fractions 1-10), and incubated for 1 h with [γ-32P]-ATP with or without the addition of 10 μg/mL rPKM2. Phosphorylated proteins were analyzed by SDS-PAGE and autoradiography. See also Figure S4.
Discussion
We were unable to find evidence for PKM2 activity as a protein kinase utilizing either [32P]-PEP or [γ-32P]-ATP as a substrate. While negative results cannot be definitive, sensitive 32P-labeling failed to identify any proteins phosphorylated in a PKM2-dependent manner. Our findings demonstrate the importance of depleting contaminating adenine nucleotides from cell lystates and purification of enzymatically synthesized [32P]-PEP to reduce [γ-32P]-ATP generation in situ, which might otherwise lead to ATP-dependent phosphorylation events being misinterpreted as PEP-dependent. Omitting these steps can result in pyruvate kinase mediated regeneration of ATP and produce data that appear to be PEP-dependent phosphorylation events.
To have activity as a protein kinase, PKM2 would have to adopt a conformation in which both PEP and its protein substrate are simultaneously bound. To allow phosphotransfer reactions, water must be excluded from the enzyme active site or hydrolysis of high-energy phosphate anhydrides would be favored over the phosphate transfer reaction. To accommodate a large protein substrate in place of the much smaller ADP, the active site would need to be dramatically altered to exclude water and allow transfer of phosphate to a residue on the target protein. An alternative mechanism for phosphotransfer could involve a covalent phospho-enzyme intermediate; however, the known mechanism of the pyruvate kinase reaction involves direct phosphate transfer (Dombrauckas et al., 2005), and we did not observe labeling of PKM2 from [32P]-PEP alone.
It has been suggested that protein substrates of PKM2 bind to the ADP binding site of this enzyme, supported by the apparent competitive inhibition of PKM2 protein kinase activity by ADP (Gao et al., 2012; Keller et al., 2014). This observation could also be explained by end-product inhibition of a contaminating ATP-dependent kinase by ADP. The ability of protein substrates to occupy the ADP-binding site of PKM2 is particularly surprising given that PKM2 has been reported to phosphorylate 149 protein substrates lacking a clear phosphorylation motif, suggesting degenerate specificity of the active site (Keller et al., 2014). By contrast, nucleotide binding is highly specific as PKM2 is unable to efficiently phosphorylate diverse nucleotide substrates, instead showing strong selectivity for ADP (Mazurek et al., 1998). In addition, PKM1 has the same binding site for ADP as PKM2, and shows similar selectivity for ADP (Morgan et al., 2013; Plowman and Krall, 1965). In fact, PKM2 and PKM1 have identical active sites. These isoforms differ by the alternative inclusion of one exon, which encodes only regulatory components of the protein further arguing against a specific kinase function for PKM2. A more plausible explanation is that PKM2 serves to regenerate ATP from PEP for other kinases as an explanation for reports of PKM2 protein kinase activity.
Although we did not detect protein kinase activity by PKM2, several protein species were labeled by [32P]-PEP independent of the presence of PKM2: an unknown 45 kDa protein as well as PGAM, which has been previously identified as a target of PEP-dependent phosphorylation. While PGAM is labeled by the phosphate from PEP, it is important to note that PEP may be transferring more than just a phosphate to the unknown 45 kDa protein. Interestingly, another glycolytic intermediate, 1,3-bisphosphoglycerate, can post-translationally modify proteins on lysine to produce 3-phosphoglyceryl-lysine via a non-enzymatic mechanism (Moellering and Cravatt, 2013). Depletion of enolase from the reaction would distinguish between PEP-dependent and 2-PG-dependent phosphorylation, and will help clarify whether PEP-dependent labeling of the ~45 kDa protein is mediated by PEP or an enzymatic product of PEP other than ATP. Nevertheless, it appears that PEP-dependent phosphorylation is rare in mammalian cells and that PKM2 is not required for any of the events observed in this study. The methods to separate PEP-dependent phosphorylation in mammalian cells from ATP-dependent events with high sensitivity will allow identification of both the targets and enzymes involved.
Experimental Procedures
Cell Culture
H1299 cells, MEFs, U87 cells, and U87 EGFR-VIII cells were cultured in RPMI 1640 or DMEM, supplemented with 10% heat-inactivated fetal bovine serum, penicillin (100 U/mL), and streptomycin (100 μg/mL). MEFs expressing Cre-ER and homozygous for a PKM2 conditional allele were generated from mice as described previously (Lunt et al., 2015) and immortalized by knockdown of p53. To delete PKM2, the MEFs were treated with 1 μM 4-hydroxytamoxifen for at least 3 days.
Protein Extraction and Purification
Hypotonic lysates and nuclear extracts were prepared using standard techniques described previously (Wang et al., 1994). Where noted, cell lysates were fractionated by binding to a 1 mL HiTrap Q HP column (GE Healthcare) in 20 mM bis-tris-propane-HCl pH 8.7, 5 mM KCl, 1 mM MgCl2 and eluting with increasing concentrations of KCl.
Expression and Purification of Recombinant PKM2
N-terminally 6×His-tagged recombinant PKM2 and PKM2-R399E were expressed from the pET28a(+) vector in E. coli and batch purified using Ni-NTA agarose beads (Qiagen) as described previously (Anastasiou et al., 2012).
[32P]-PEP Synthesis and Purification
[32P]-PEP was synthesized as described previously (Mattoo and Waygood, 1983; Vander Heiden et al., 2010). Further purification was accomplished by reverse-phase ion-pairing high performance liquid chromatography. [32P]-PEP (containing contaminating [γ-32P]-ATP) was mixed with 100 μM of each of cold PEP and ATP. This mixture was loaded on to a ZORBAX SB-C18 Analytical column (Agilent), and eluted in a linear gradient from 5% (v/v) methanol, 15 mM acetic acid, and 10 mM tributylamine to 100% methanol. 500 μL fractions were collected. Three fractions containing the highest amount of PEP or ATP (as determined by TLC and autoradiography) were pooled and dried under nitrogen gas. Samples were resuspended in water for use in in vitro reactions.
In Vitro Phosphorylation Reactions
Reactions were carried out at 30 °C for 1 hr in a previously published buffer (Gao et al., 2012). Cell extracts and calf histones (Worthington Biochemical) were assayed at 100 μg/mL and PKM2 was used at 10 μg/mL. In some reactions, 1 mM Na3VO4, 0.5 mM SAICAR, or 1 mM cold competitor ATP or PEP were included in the reaction. Radioactive substrates, prepared as above were included at ~10 μCi/mL. Reactions were quenched by addition of Lamelli sample buffer and analyzed by SDS-PAGE and autoradiography.
Thin-Layer Chromatography
0.5 μL of each reaction or compound were spotted on to PEI-cellulose F (EMD Millipore), and metabolites were resolved with 0.25 M NH4HCO3. Plates were dried and then visualized by autoradiography.
Supplementary Material
Acknowledgements
U87 and U87 EGFR-VIII cells were generously provided by the White Lab at MIT. We acknowledge support from the HHMI International Student Research Fellowship, the Vertex Scholars Program, T32GM007287, and Koch Institute Graduate Student Fellowships, as well as funding from the NCI (R01CA168653, P30CA14051) and the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anastasiou D, Yu Y, Israelsen WJ, Jiang JK, Boxer MB, Hong BS, Tempel W, Dimov S, Shen M, Jha A, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nature chemical biology. 2012;8:839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Schmidt A, Malmstroem J, Claassen M, Ori A, Szymborska A, Herzog F, Rinner O, Ellenberg J, Aebersold R. The quantitative proteome of a human cell line. Molecular systems biology. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreras J, Climent F, Bartrons R, Pons G. Effect of vanadate on the formation and stability of the phosphoenzyme forms of 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase and of phosphoglucomutase. Biochimica et biophysica acta. 1982;705:238–242. doi: 10.1016/0167-4838(82)90183-2. [DOI] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, et al. Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature. 2012;491:458–462. doi: 10.1038/nature11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- Cortes-Cros M, Hemmerlin C, Ferretti S, Zhang J, Gounarides JS, Yin H, Muller A, Haberkorn A, Chene P, Sellers WR, et al. M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:489–494. doi: 10.1073/pnas.1212780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiology and molecular biology reviews: MMBR. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Science signaling. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Li X, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, Wei C, Guo F, Chen Y, et al. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol Cell. 2014;53:75–87. doi: 10.1016/j.molcel.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller KE, Doctor ZM, Dwyer ZW, Lee YS. SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells. Mol Cell. 2014;53:700–709. doi: 10.1016/j.molcel.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller KE, Tan IS, Lee YS. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. 2012;338:1069–1072. doi: 10.1126/science.1224409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung C, Hixon J, Choe S, Marks K, Gross S, Murphy E, DeLaBarre B, Cianchetta G, Sethumadhavan S, Wang X, et al. Small molecule activation of PKM2 in cancer cells induces serine auxotrophy. Chemistry & biology. 2012;19:1187–1198. doi: 10.1016/j.chembiol.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt Sophia Y., Muralidhar V, Hosios Aaron M., Israelsen William J., Gui Dan Y., Newhouse L, Ogrodzinski M, Hecht V, Xu K, Acevedo Paula N.M., et al. Pyruvate Kinase Isoform Expression Alters Nucleotide Synthesis to Impact Cell Proliferation. Molecular Cell. 2015 doi: 10.1016/j.molcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen M, Larsen PJ. Cell cycle-dependent regulation of cellular ATP concentration, and depolymerization of the interphase microtubular network induced by elevated cellular ATP concentration in whole fibroblasts. Cell motility and the cytoskeleton. 1996;35:94–99. doi: 10.1002/(SICI)1097-0169(1996)35:2<94::AID-CM2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Mattoo RL, Waygood EB. An enzymatic method for [32P]phosphoenolpyruvate synthesis. Analytical biochemistry. 1983;128:245–249. doi: 10.1016/0003-2697(83)90372-x. [DOI] [PubMed] [Google Scholar]
- Mazurek S, Grimm H, Wilker S, Leib S, Eigenbrodt E. Metabolic characteristics of different malignant cancer cell lines. Anticancer research. 1998;18:3275–3282. [PubMed] [Google Scholar]
- McKnight SL. Please keep me 2uned to PKM2. Mol Cell. 2014;53:683–684. doi: 10.1016/j.molcel.2014.02.022. [DOI] [PubMed] [Google Scholar]
- Moellering RE, Cravatt BF. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science. 2013;341:549–553. doi: 10.1126/science.1238327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HP, O’Reilly FJ, Wear MA, O’Neill JR, Fothergill-Gilmore LA, Hupp T, Walkinshaw MD. M2 pyruvate kinase provides a mechanism for nutrient sensing and regulation of cell proliferation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5881–5886. doi: 10.1073/pnas.1217157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman KM, Krall AR. A kinetic study of nucleotide interactions with pyruvate kinase. Biochemistry. 1965;4:2809–2814. doi: 10.1021/bi00888a035. [DOI] [PubMed] [Google Scholar]
- Schieven G, Martin GS. Nonenzymatic phosphorylation of tyrosine and serine by ATP is catalyzed by manganese but not magnesium. The Journal of biological chemistry. 1988;263:15590–15593. [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nature protocols. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- Wang YH, Israelsen WJ, Lee D, Yu VW, Jeanson NT, Clish CB, Cantley LC, Vander Heiden MG, Scadden DT. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell. 2014;158:1309–1323. doi: 10.1016/j.cell.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012a;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nature cell biology. 2012b;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.