Abstract
“Α picture is worth a thousand words.” This widely used adage sums up in a few words the notion that a successful visual representation of a concept should enable easy and rapid absorption of large amounts of information. Although, in general, the notion of capturing complex ideas using images is very appealing, would 1000 words be enough to describe the unknown in a research field such as the life sciences? Life sciences is one of the biggest generators of enormous datasets, mainly as a result of recent and rapid technological advances; their complexity can make these datasets incomprehensible without effective visualization methods. Here we discuss the past, present and future of genomic and systems biology visualization. We briefly comment on many visualization and analysis tools and the purposes that they serve. We focus on the latest libraries and programming languages that enable more effective, efficient and faster approaches for visualizing biological concepts, and also comment on the future human-computer interaction trends that would enable for enhancing visualization further.
Keywords: Biological data visualization, Network biology, Genomics, Systems biology, Multivariate analysis
Review
Introduction
In the current ‘big data’ era [1], the magnitude of data explosion in life science research is undeniable. The biomedical literature currently includes about 27 million abstracts in PubMed and about 3.5 million full text articles in PubMed Central. Additionally, there are more than 300 established biological databases that store information about various biological entities (bioentities) and their associations. Obvious examples include: diseases, proteins, genes, chemicals, pathways, small molecules, ontologies, sequences, structures and expression data. In the past 250 years, only 1.2 million eukaryotic species (out of the approximately 8.8 million that are estimated to be present on earth) [2] have been identified and taxonomically classified in the Catalog of Life and the World Register of Marine Species [3]. The sequencing of the first human genome (2002) took 13 years and cost over $3 million to complete. Although the cost for de novo assembly of a new genome to acceptable coverage is still high, probably at least $40,000, we can now resequence a human genome for $1000 and can generate more than 320 genomes per week [4]. Notably, few species have been fully sequenced, and a large fraction of their gene function is not fully understood or still remains completely unknown [5]. The human genome is 3.3 billion base pairs in length and consists of over 20,000 human coding genes organized into 23 pairs of chromosomes [6, 7]. Today over 60,000 solved protein structures are hosted in the Protein Data Bank [8]. Nevertheless, many of the protein functions remain unknown or are partially understood.
Shifting away from basic research to applied sciences, personalized medicine is on the cusp of a revolution allowing the customization of healthcare by tailoring decisions, practices and/or products to the individual patient. To this end, such information should be accompanied by medical history and digital images and should guarantee a high level of privacy. The efficiency and security of distributed cloud computing systems for medical health record organization, storage and handling will be one of the big challenges during the coming years.
Information overload, data interconnectivity, high dimensionality of data and pattern extraction also pose major hurdles. Visualization is one way of coping with such data complexity. Implementation of efficient visualization technologies is necessary not only to present the known but to also reveal the unknown, allowing inference of conclusions, ideas and concepts [9]. Here we focus on visualization advances in the fields of network and systems biology, present the state-of-the-art tools and provide an overview of the technological advances over time, gaining insights into what to expect in the future of visualization in the life sciences.
In the section on network biology below, we discuss widely used tools related to graph visualization and analysis, we comment on the various network types that often appear in the field of biology and we summarize the strengths of the tools, along with their citation trends over time. In this section we also distinguish between tools for network analysis and tools designed for pathway analysis and visualization. In a section on genomic visualization, we follow the same approach by distinguishing between tools designed for genome browsing and visualization, genome assembly, genome alignments and genome comparisons. Finally, in a section on visualization and analysis of expression data, we distinguish between tree viewers and tools implemented for multivariate analysis.
Network biology visualization
In the field of systems biology, we often meet network representations in which bioentities are interconnected with each other. In such graphs, each node represents a bioentity and edges (connections) represent the associations between them [10]. These graphs can be weighted, unweighted, directed or undirected. Among the various networks types within the field, some of the most widely used are protein-protein interaction networks, literature-based co-occurrence networks, metabolic/biochemical, signal transduction, gene regulatory and gene co-expression networks [11–13]. As new technological advances and high-throughput techniques come to the forefront every few years, such networks can increase dramatically in size and complexity, and therefore more efficient algorithms for analysis and visualization are necessary. Notably, a network consisting of a hundred nodes and connections is incomprehensible and impossible for a human to visually analyze. For example, techniques such as tandem affinity purification (TAP) [14], yeast two hybrid (Y2H) [15] and mass spectrometry [16] can nowadays generate a significant fraction of the physical interactions of a proteome. As network biology evolves over time, we indicate standard procedures that were developed over the past 20 years and highlight key tools and methodologies that had a crucial role in this maturation process (Fig. 1).
Fig. 1.
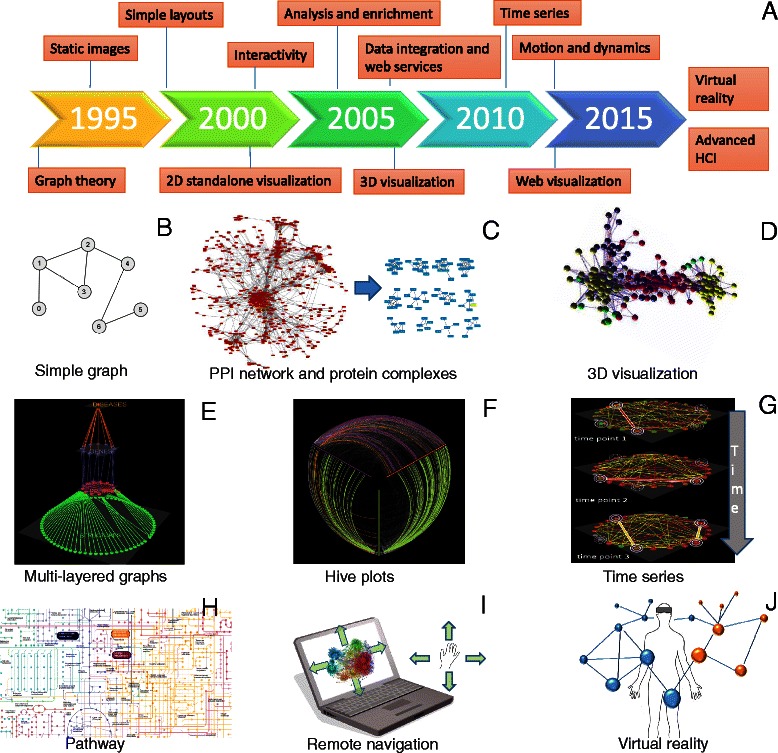
Visualization for network biology. a Timeline of the emergence of relevant technologies and concepts. b A simple drawing of an undirected unweighted graph. c A 2D representation of a yeast protein-protein interaction network visualized in Cytoscape (left) and potential protein complexes identified by the MCL algorithm from that network (right). d A 3D view of a protein-protein interaction network visualized by BiolayoutExpress3D. e A multilayered network integrating different types of data visualized by Arena3D. f A hive plot view of a network in which nodes are mapped to and positioned on radially distributed linear axes. g Visualization of network changes over time. h Part of lung cancer pathway visualized by iPath. i Remote navigation and control of networks by hand gestures. j Integration and control of 3D networks using VR devices
In the 1990s, two-dimensional (2D) static graph layouts were developed for visualizing networks. Topological analysis, layout and clustering were pre-calculated and results were captured in a single static image. Clustering analysis was performed to detect cliques or highly connected regions within a graph, layout techniques such as Fruchterman-Reingold [17] were implemented to place nodes in positions where the crossovers between the edges are minimized and topological analysis was used for detecting important nodes of the network such as hubs or nodes with high betweenness centrality. The typical visual encoding consisted of using arrows for directed graphs, adjusting the thickness of an edge to show the importance of a connection, using the same color for nodes that belong to the same cluster or modifying the node’s size to show its topological features, such as its neighbor connectivity. As integrative biology and high-throughput techniques advanced over the years, the necessity to move away from static images and add interactivity and navigation for easier data exploration became clearer.
Bridging between analysis and visualization became necessary, and tools that incorporated both increased the standards in the field. In clustering analysis, for example, new computational methods such as MCL [18] and variations [19], Cfinder [20], MCODE [21], Clique [22] and others were applied to biological networks to find highly connected regions of importance. DECAFF [23], SWEMODE [24] or STM [25], for example, were developed to predict protein complexes [26] incorporating graph annotations, whereas others such as DMSP [27], GFA [28] and MATISSE [29] were focused on gene-expression data. Most of these algorithms were command-line-based and only few tools such as jClust [30], GIBA [31], ClusterMaker [32] or NeAT [33] have been developed to integrate data in visual environments. These aforementioned techniques along with others are thoroughly discussed elsewhere [26, 34–36].
Although most network visualization tools are standalone applications, they guarantee efficient data exploration and the manipulation of visualization with mouse-hovering supporting actions. Such tools are for example the Pajek [37], Osprey [38], VisANT [39] and others. Next-generation visualization tools took advantage of standard file formats such as BioPAX [40, 41], SBML [42], PSI-MI [43] and CellML [44]; modern, more sophisticated layouts such as Hive-Plots [45]; and the available web services and data integration techniques to directly retrieve and handle information from public repositories on the fly. Functional enrichment of genes using the Gene Ontology (GO) repository [46] is a typical example. Among others, current state-of-the-art tools are the Ondex [47], Cytoscape [48] or Gephi [49], while tools such as iPath [50], PATIKA [51], PathVisio [52] and others [53] are pathway specific.
As biological networks became larger over time, consisting of thousands of nodes and connections, the so-called ‘hairball’ effect, where many nodes are densely connected with each other became very difficult to cope with. A partial solution to this was to shift from 2D representations to three-dimensional (3D) representations. Tools such as Arena3D [54, 55] or BioLayout Express 3D [56] take advantage of 3D space to show data in a virtual 3D universe. BioLayout Express uses whole 3D space to visualize networks, whereas Arena3D implements a multilayered concept to present 2D networks in a stack. Although a 2D network allows immediate visual feedback, a 3D rendering usually requires the user to interact more with the data in a more explorative mode, but can help reveal interesting features potentially hidden in a 2D representation. Although it is debatable whether 3D rendering is better than 2D visualization, hardware acceleration and performance still need to be taken into account when planning 3D visualizations (Fig. 1).
Tables 1 and 2 present currently freely available network and pathway visualization tools and their main characteristics. However, it is not the purpose of this review to perform a deeper comparative analysis of all available 2D and 3D visualization tools, as this is available elsewhere [53, 57–59]. Nevertheless, as network biology is gaining ground over the years, we sought to investigate the impact of the current tools in the field. To accomplish this, we tracked the tools that appeared after year 2000 and whose respective articles are indexed by Scopus (Fig. 2). We chose to keep track of the citations of only the first original publication for each tool. Although the number of citations is a reasonable indicator of popularity, it can sometimes be misleading as several tool versions appear in different articles that we have not yet tracked. Nevertheless, some immediate conclusions can be reached, such as that Cytoscape seems to be by far the biggest player for network visualization, as it comes with more than 200 plugins [60] implemented by an active module community (Fig. 1b). Similarly, MapMan [61] and Reactome SkyPainter [62] are the most used tools for pathway visualization (Fig. 2b).
Table 1.
Visualization tools for network biology
| Standalone applications for network analysis | ||
|---|---|---|
| Tool and references | Description | URL |
| Arena 3D [54, 55] | 3D visualization of multi-layer networks | http://www.arena3d.org |
| Biana [146] | Data integration and network management | http://sbi.imim.es/web/BIANA.php |
| BioLayout Express 3D [147] | 2D/3D network visualization | http://www.biolayout.org/ |
| BiologicalNetworks [148, 149] | Efficient integrated multi-level analysis of microarray, sequence, regulatory and other data | http://www.biologicalnetworks.org |
| BioMiner [150] | Modeling, analyzing and visualizing biochemical pathways and networks | http://www.zbi.uni-saarland.de/chair/projects/BioMiner |
| Cell Illustrator [151] | Petri nets for modeling and simulating biological networks | http://www.cellillustrator.com |
| COPASI [152] | Analysis of biochemical networks and their dynamics | http://www.copasi.org/ |
| Cytoscape [48, 153] | Network visualization and analysis. Over 200 plugins [60] | http://www.cytoscape.org/ |
| Dizzy [154] | Chemical kinetics stochastic simulation software | http://magnet.systemsbiology.net/software/Dizzy/ |
| DyCoNet [155] | Gephi plugin that can be used to identify dynamic communities in networks | https://github.com/juliemkauffman/DyCoNet |
| GENeVis [156, 157] | Network and pathway visualization | http://tinyurl.com/genevis/ |
| GEPHI [49] | Interactive visualization and exploration for any network and complex system, dynamic and hierarchical graph. | https://gephi.org |
| Igraph [158] | Collection of network analysis tools with the emphasis on efficiency, portability and ease of use | http://igraph.sourceforge.net |
| Medusa [159, 160] | Semantic and multi-edged simple networks | https://sites.google.com/site/medusa3visualization/ |
| NAViGaTOR [161, 162] | Visualizing and analyzing protein-protein interaction networks | http://tinyurl.com/navigator1/ |
| N-Browse [163] | Interactive graphical browser for biological networks | http://www.gnetbrowse.org/ |
| NeAT [33] | Topological and clustering analysis of networks | http://rsat.ulb.ac.be/neat/ |
| Ondex [47] | Data integration and visualization of large networks | http://www.ondex.org/ |
| Osprey [38] | Visualization and annotation of biological networks | http://biodata.mshri.on.ca/osprey/servlet/Index |
| Pajek [37] | Analysis and visualization of large networks and social network analysis | http://vlado.fmf.uni-lj.si/pub/networks/pajek/ |
| PathwayAssist [164] | Navigation and analysis of biological pathways, gene regulation networks and protein interaction maps. | http://www.ariadnegenomics.com/downloads/ |
| PIVOT [165] | Layout algorithms for visualizing protein interactions and families | http://acgt.cs.tau.ac.il/pivot/ |
| ProCope [166] | Prediction and evaluation of protein complexes from purification data experiments | http://www.bio.ifi.lmu.de/Complexes/ProCope/ |
| ProViz [167] | Visualization and exploration of interaction networks. Gene Ontology and PSI-MI formats supported | http://cbi.labri.fr/eng/proviz.htm |
| SpectralNET [168] | Network analysis and visualizations. Scatter plots and dimensionality reduction algorithms | https://www.broadinstitute.org/software/spectralnet |
| Tulip [169] | Enables the development of algorithms, visual encodings, interaction techniques, data models and domain-specific visualizations | http://tulip.labri.fr/TulipDrupal/ |
| VANESA [170] | Automatic reconstruction and analysis of biological networks and Petri nets based on life-science database information | http://agbi.techfak.uni-bielefeld.de/vanesa/ |
| VANTED [171] | Network reconstruction, data visualization, integration of various data types, network simulation | http://tinyurl.com/vanted/ |
| yEd | Creation of diagrams manually and import external data | http://tinyurl.com/yEdGraph/ |
| Web tools for network analysis | ||
| APID [172] | Unified protein-protein interactions from BIND, BioGRID, DIP, HPRD, IntAct and MINT | http://bioinfow.dep.usal.es/apid/ |
| Arcadia [173] | Translates text-based descriptions of biological networks (SBML files) into standardized diagrams (Systems Biology Graphical Notation Process Description maps) | http://arcadiapathways.sourceforge.net/ |
| AVIS [174] | Viewer for signaling networks | http://actin.pharm.mssm.edu/AVIS2 |
| bioPIXIE [175] | Discovery of biological networks from diverse functional genomic data | http://pixie.princeton.edu/pixie |
| CellPublisher [176] | Interactive representations of biochemical processes | http://cellpublisher.gobics.de/ |
| Graphle [177] | Distributed network exploration and visualization of interactive large, dense graphs | http://tinyurl.com/graphle/ |
| GraphWeb [178] | Web server for graph-based analysis of biological networks | http://biit.cs.ut.ee/graphweb/ |
| Hubba [179] | Web-based service to explore the essential nodes in a network | http://hub.iis.sinica.edu.tw/Hubba |
| NetworkBLAST [180] | Analysis of protein interaction networks across species to infer protein complexes that are conserved in evolution | http://www.cs.tau.ac.il/~bnet/networkblast.htm |
| Pathview [181] | Tool set for pathway-based data integration and visualization | http://Pathview.r-forge.r-project.org/ |
| PINA [182] | Integrated platform for protein interaction network construction, filtering, analysis, visualization and management | http://cbg.garvan.unsw.edu.au/pina/home.do |
| ReMatch [183] | Web-based tool for integration of user-given stoichiometric metabolic models into a database collected from public data sources | http://www.cs.helsinki.fi/group/sysfys/software/rematch/ |
| SNOW [184] | Gene mapping on a reference or human protein-protein interaction network that SNOW hosts | http://snow.bioinfo.cipf.es |
| STITCH [185] | Resource to explore known and predicted interactions of chemicals and proteins | http://stitch.embl.de/ |
| STRING [186] | Protein interaction networks and integration of data such as genomic context, high-throughput experiments, conserved coexpression and previous knowledge derived from the literature | http://string-db.org |
| TVNViewer [187] | An interactive visualization tool for exploring networks that change over time or space | http://www.sailing.cs.cmu.edu/main/?page_id=545 |
| tYNA [188] | System for managing, comparing and mining multiple networks | http://tyna.gersteinlab.org/tyna/ |
| VisANT [39, 189] | Visualization, mining, analysis and modeling of biological networks, metabolic networks and ecosystems | http://visant.bu.edu/ |
Table 2.
Visualization tools for pathways
| Standalone applications | ||
|---|---|---|
| Tool and references | Description | URL |
| BiNA [190] | Drawings of metabolic networks supporting hiding of cofactors and drawing of chemical structures | http://bina.unipax.info/ |
| BioTapestry [191] | Interactive tool for building, visualizing and sharing gene regulatory network models over the web | http://www.biotapestry.org/ |
| Caleydo [192] | Visual analysis framework targeted at biomolecular data. Visualization of interdependencies between multiple datasets | http://www.caleydo.org/ |
| CellDesigner [193] | A modeling tool for biochemical networks | http://www.celldesigner.org/ |
| Edinburgh Pathway Editor [194] | Edit and draw pathway diagrams | http://epe.sourceforge.net/SourceForge/EPE.html |
| GenMAPP [195] | Visualization of gene expression and other genomic data on maps representing biological pathways and groupings of genes | http://www.genmapp.org/ |
| Ingenuity IPA | Data integration platform and manually annotated pathways | http://tinyurl.com/IngenuityPath |
| JDesigner [196] | Graphical modeling environment for biochemical reaction networks | http://jdesigner.sourceforge.net/Site/JDesigner.html |
| KaPPA View [197] | Plant pathways | http://kpv.kazusa.or.jp/ |
| KEGG Atlas [198] | Interactive Kyoto Encyclopedia of Genes and Genomes pathways | http://www.genome.jp/kegg/ |
| Omix [199] | Visualizing multi-omics data in metabolic networks | https://www.omix-visualization.com |
| PathVisio [52] | Biological pathway analysis software that allows drawing, editing and analysis of biological pathways | http://www.pathvisio.org/ |
| VitaPad [200] | Application to visualize biological pathways and map experimental data to them | http://tinyurl.com/vitapad/ |
| Web tools for pathways | ||
| ArrayXPath [201] | Mapping and visualizing microarray gene-expression data and integrated biological pathway resources using SVG | http://tinyurl.com/ArrayXPath/ |
| GEPAT [202] | Integrated analysis of transcriptome data in genomic, proteomic and metabolic contexts | http://gepat.sourceforge.net/ |
| iPath [50, 203] | Web-based tool for the visualization, analysis and customization of pathway maps | http://pathways.embl.de/ |
| Kegg-Based Viewer [204] | KEGG-based pathway visualization tool for complex high-throughput data | http://www.g-language.org/data/marray/ |
| MapMan [61] | User-driven tool that displays large datasets onto diagrams of metabolic pathways or other processes | http://mapman.gabipd.org/web/guest/mapman |
| MetPA [205] | Analysis and visualization of metabolomic data within the biological context of metabolic pathways | http://metpa.metabolomics.ca |
| Omics Viewer [206] | Data mapping on BioCyc pathways (collection of 5500 pathway/genome databases) | http://www.biocyc.org/ |
| Pathway Explorer [207] | Interactive Java drawing tool for the construction of biological pathway diagrams in a visual way and the annotation of the components and interactions between them | http://genome.tugraz.at/pathwayexplorer/pathwayexplorer_description.shtml |
| Pathway projector [208] | Zoomable pathway browser using KEGG atlas and Google Maps API | http://www.g-language.org/PathwayProjector/ |
| PATIKA [51] | Integrated environment composed of a central database and a visual editor, built around an extensive ontology and an integration framework | http://www.cs.bilkent.edu.tr/~patikaweb/ |
| Reactome SkyPainter [62] | Visualization of over-represented pathways and reactions from gene lists | http://www.reactome.org/skypainter-2 |
| WikiPathways [209] | Wiki-based, open, public platform dedicated to the curation of biological pathways by and for the scientific community | http://www.wikipathways.org/ |
Fig. 2.
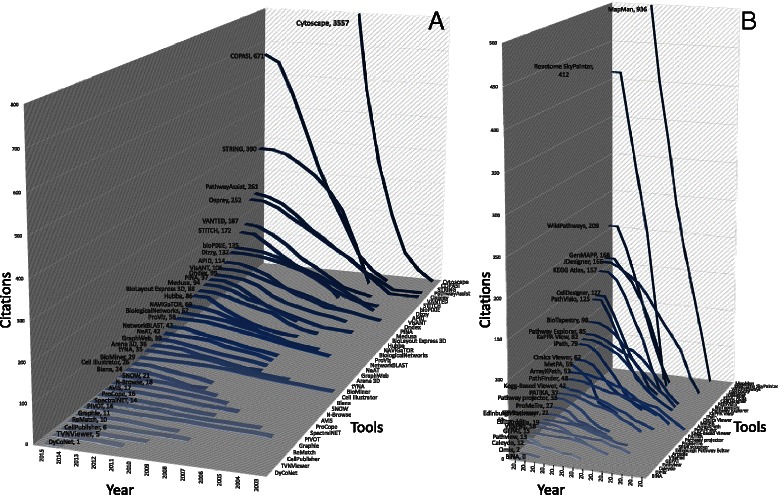
Citation trends and key player tools in network biology. a Citations of network visualization tools based on Scopus. b Citations of pathway visualization tools based on Scopus. The numbers of citations of each tool in 2015 are shown after its name
Over the past 5 years, the data visualization field has become more and more competitive. There is a trend away from standalone applications towards the integration of visualization implementations within web browsers. Therefore, libraries and new programming languages have been dedicated to this task (see the final section below). The greater visibility provided by web implementation means that advanced visualization can more easily become available to non-experts and to the broader community. Finally, one of the biggest visualization challenges today is to capture the dynamics of networks and the way in which topological properties change over time [63]. For this, motion or other sophisticated ideas, along with new human-computer interaction (HCI) techniques, should be taken into consideration. Although serious efforts on this are on the way [54, 64, 65], there are still much to expect in the future as HCI techniques and virtual reality (VR) devices (such as Oculus Rift) become cheaper and more advanced over time (Fig. 1).
Visualization in genomics
There remain many open challenges for advanced visualization for genome assemblies, alignments, polymorphisms, variations, synteny, single nucleotide polymorphisms (SNPs), rearrangements and annotations [66, 67]. To better follow progress in the visualization field, we first need to follow the way in which new technologies, questions and trends have been shaped over the years (Fig. 3).
Fig. 3.
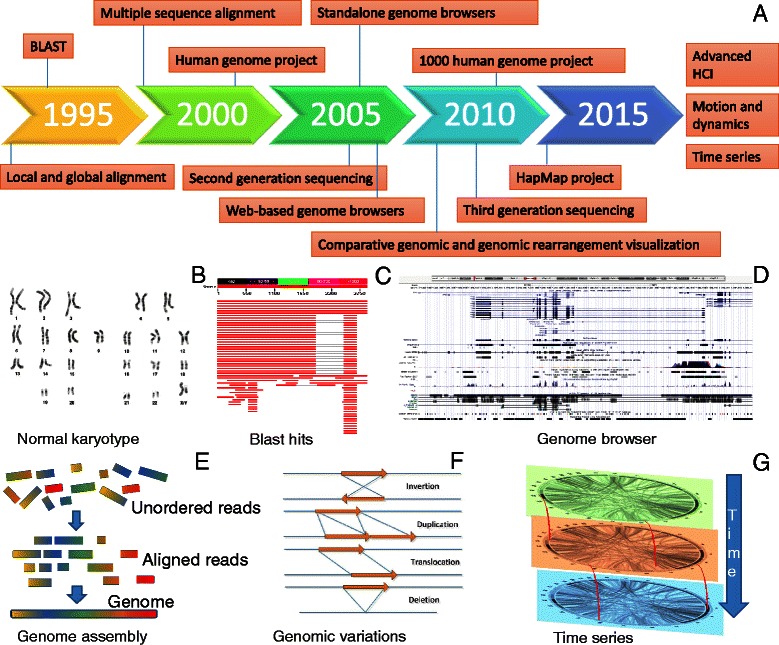
Visualization for genome biology. a Timeline of the emergence of relevant technologies and concepts. b A typical normal human karyotype. c Visualization of BLAST hits and alignment of orthologous genes for the human TP53 gene. d The human TP53 gene and its annotations visualized by the UCSC genome browser. e Visualization of a de novo genome assembly from its DNA fragments. f Examples of balanced and unbalanced genomic rearrangements. g Hypothetical visualization of genomic structural variations across time
Up to the 1990s, local and global pairwise and multiple sequence alignment algorithms such as Smith-Waterman [68], Needleman-Wunsch [69], FASTA [70] and BLAST [71] were the focus of bioinformatics methods development. Multiple sequence alignment tools such as the ClustalW/Clustal X [72], MUSCLE [73], T-Coffee [74] and others [75] used basic visualization schemes, in which sequences were represented as strings placed vertically in stacks. Colors were used to visually encode base conservation and to indicate matching, non-matching and similar nucleotides [76, 77].
Although these tools were successful for small numbers of nucleotide or protein sequences, a question was raised regarding their applicability to whole-genome sequencing and comparison. A few years later (2002), the Sanger (dideoxy) first generation sequencing, particularly capillary approaches, allowed the sequencing of the first whole human genome, consisting of about 3 billion base pairs and over 20,000 human genes [78, 79]. Shortly after that, second-generation (Illumina [80], Roche/454 [81], Biosystems/SOLiD [82]) and third-generation techniques (Helicos BioSciences [83], Pacific Biosciences [84], Oxford Nanopore [85] and Complete Genomics [86]) high-throughput sequencing techniques [87–91] allowed the sequencing of a transcriptome, an exome or a whole genome at a much lower cost and within reasonable timeframes.
Projects such as the 1000 Genomes Project, for comprehensive human genetic variation analysis [92–94], and the International HapMap Project [95–99], for the identification of common genetic variations among people from different countries, are just a few examples of the data explosion that has taken place in the era of comparative genomics, after 2005. Such large-scale genomic datasets necessitate powerful tools to link genomic data to its source genome and across genomes. Therefore, among others [66], widely used standalone and web-based genome browsers were dedicated to information handling, genome visualization, navigation, exploration and integration with annotations from various repositories. At present, many specialized tools for comparative genomic visualization are available and are widely used.
To follow trends in the field, we summarize the tools into four categories: genome alignment visualization tools (Table 3); genome assembly visualization tools (Table 4); genome browsers (Table 5); and tools to directly compare different genomes with each other for efficient detection of SNPs and genomic variations (Table 6). Following the same approach used for network biology (above), we examine the citation progress of the first article that was published for each tool using the Scopus repository (Fig. 4). Consed [76] and Gap [100, 101] seem to be the most widely used assembly viewers, while SAMtools tview [102] is the favorite tool for genomic assembly visualization. In addition, the University of California, Santa Cruz (UCSC) Genome Browser [103], Artemis [104] and Ensembl [105, 106] seem to be the go-to genome browsers, while Circos [107], VISTA [108] and cBio [109] are the most widely used tools for comparative genomics.
Table 3.
Visualization tools for genome alignments
| Tool and references | Description | URL |
|---|---|---|
| ABySS Explorer [210] | Interactive Java application that uses a novel graph-based representation to display a sequence assembly and associated metadata | http://www.bcgsc.ca/platform/bioinfo/software/abyss-explorer |
| BamView [211] | Genome browser and annotation tool that allows visualization of sequence features, next-generation sequencing (NGS) data and the results of analyses within the context of the sequence, and also its six-frame translation | http://www.sanger.ac.uk/resources/software/artemis/ |
| DNannotator [212] | Annotation web toolkit for regional genomic sequences | http://bioapp.psych.uic.edu/DNannotator.htm |
| JVM [213] | Java Visual Mapping tool for NGS reads | http://www.springer.com/cda/content/document/cda_downloaddocument/9789401792448-c2.pdf?SGWID=0-0-45-1487072-p176815501 |
| LookSeq [214] | Web-based visualization of sequences derived from multiple sequencing technologies. Low- or high-depth read pileups and easy visualization of putative single nucleotide and structural variation | http://lookseq.sourceforge.net |
| MagicViewer [215] | Visualization of short read alignment, identification of genetic variation and association with annotation information of a reference genome | http://bioinformatics.zj.cn/magicviewer/ |
| MapView [216] | Alignments of huge-scale single-end and pair-end short reads | http://omictools.com/mapview-s1367.html |
| MultiPipMaker [217] | Computes alignments of similar regions in two DNA sequences. The resulting alignments are summarized with a ‘percent identity plot’ (pip) | http://pipmaker.bx.psu.edu/pipmaker/ |
| PileLineGUI [218] | Handling genome position files in NGS studies | http://sing.ei.uvigo.es/pileline/pilelinegui.html |
| SAMtools tview [102] | Simple and fast text alignment viewer; NGS compatible | http://www.htslib.org/ |
| SEWAL [219] | Uses a locality-sensitive hashing algorithm to enumerate all unique sequences in an entire Illumina sequencing run | http://www.sourceforge.net/projects/sewal |
| STAR [220] | A web-based integrated solution to management and visualization of sequencing data | http://wanglab.ucsd.edu/star/browser |
| SVA [221] | Software for annotating and visualizing sequenced human genomes | http://www.svaproject.org |
| Viewer (IGV) [222] | Visualization of large heterogeneous datasets, providing a smooth and intuitive user experience at all levels of genome resolution | https://www.broadinstitute.org/igv/ |
| ZOOM Lite [223] | NGS data mapping and visualization software | http://bioinfor.com/zoom/lite/ |
Table 4.
Visualization tools for assemblies
| Tool and references | Description | URL |
|---|---|---|
| Archive Viewer [224] | Web graphical interface to make contigs and trace data changes in the National Center for Biotechnology Information (NCBI) | http://www.ncbi.nlm.nih.gov/Traces/assembly/assmbrowser.cgi? |
| CBrowse [225] | SAM/BAM-based contig web browser for transcriptome assembly visualization and analysis | http://bioinfolab.muohio.edu/CBrowse/ |
| Consed [76] | Assembly finishing package; NGS compatible | http://www.phrap.org/ |
| ContigScape [226] | A Cytoscape plugin facilitating microbial genome gap closing | http://sourceforge.net/projects/contigscape/ |
| DNASTAR Lasergene [227] | Analysis suite with an assembly package | http://www.dnastar.com/ |
| EagleView [228] | An information-rich viewer for genome assemblies with data integration capability | http://www.niehs.nih.gov/research/resources/software/biostatistics/eagleview/ |
| Gap [100, 101] | A fully developed set of DNA sequence assembly (Gap4 and Gap5), editing and analysis tools | http://staden.sourceforge.net/ |
| Hawkeye [229] | An interactive visual analytics tool for genome assemblies. Detection of anomalies in data and visual identification and correction of assembly errors | http://amos.sourceforge.net/wiki/index.php?title=Hawkeye |
| Tablet [230] | A lightweight, high-performance graphical viewer for NGS assemblies and alignments | http://bioinf.scri.ac.uk/tablet |
Table 5.
Genome browsers
| Tool and references | Description | URL | Web-based or standalone |
|---|---|---|---|
| ABrowse [231] | A customizable genome browser framework | http://www.abrowse.org/ | Web-based |
| AnnoJ [232] | A web 2.0 application designed for visualizing deep sequencing data and other genome annotation data | http://www.annoj.org/ | Web-based |
| Argo | Java tool for visualizing and manually annotating whole genomes | http://www.broadinstitute.org/annotation/argo/ | Standalone |
| Artemis [104] | Browser and annotation tool that allows visualization of sequence features, data and the results of analyses within the context of the sequence, and also its six-frame translation | https://www.sanger.ac.uk/resources/software/artemis/ | Standalone |
| CGView [233] | Static and interactive graphical maps of circular genomes using a circular layout | https://www.gview.ca/wiki/GView/ | Standalone |
| Combo [234] | Dynamic browser to visualize alignments of whole genomes and their associated annotations | http://www.broad.mit.edu/annotation/argo/ | Standalone |
| Ensembl [105, 106] | Annotation, analysis and display of vertebrates and other eukaryotic species | http://www.ensembl.org/ | Web-based |
| Family Genome Browser [235] | Visualizing genomes with pedigree information | http://mlg.hit.edu.cn/FGB/ | Web-based |
| Gaggle [236] | Genome browser within an analysis framework; good microarray support | http://gaggle.systemsbiology.net/ | Standalone |
| GBrowse [237, 238] | A combination of database and interactive web pages for manipulating and displaying annotations on genomes | http://gmod.org/wiki/Gbrowse | Web-based |
| GenoMap [239] | A circular genome data viewer | http://nsato4.c.u-tokyo.ac.jp/old/GenoMap/GenoMap.html | Standalone |
| Genome Projector [240] | Circular genome maps, traditional genome maps, plasmid maps, biochemical pathways maps and DNA walks. Google API | http://www.g-language.org/GenomeProjector/ | Web-based |
| GenomeView [241] | Designed to visualize and manipulate a multitude of genomics data | http://genomeview.org/content/integration | Standalone |
| GenPlay [242] | A multipurpose genome analyzer and browser | http://www.genplay.net | Standalone |
| IGB [243] | Optimized to achieve maximum flexibility and high quality genome visualization | http://genoviz.sourceforge.net/ | Standalone |
| IGV [222] | A high-performance visualization tool for interactive exploration of large, integrated genomic datasets | http://www.broadinstitute.org/igv/ | Standalone |
| JBrowse [244] | A fast, embeddable genome browser built completely with JavaScript and HTML5 | http://jbrowse.org/ | Web-based |
| JGI | Supports live annotation; primary portal for DOE Joint Genomics Institute genome projects | http://genome.jgi-psf.org/ | Web-based |
| NCBI Genome Workbench | An integrated application for viewing and analyzing sequence data | http://www.ncbi.nlm.nih.gov/tools/gbench/ | Standalone |
| NCBI Map Viewer [245] | Vertically oriented viewer; integrated with NCBI resources and tools | http://www.ncbi.nlm.nih.gov/mapview/ | Web-based |
| Phytozome [246] | A comparative platform for green plant genomics | http://www.phytozome.net | Web-based |
| Savant [247] | It was primarily developed for visualizing sequencing data, although it can be used to visualize almost any genome-based sequence, point, interval or continuous dataset | http://compbio.cs.toronto.edu/savant | Standalone |
| Scribl [248] | An HTML5 Canvas-based graphics library for visualizing genomic data over the web | http://chmille4.github.com/Scribl/ | Web-based |
| The HuRef Browser [249] | A web resource for individual human genomics | http://huref.jcvi.org | Web-based |
| The personal genome browser [250] | Visualizing functions of genetic variants | http://www.pgbrowser.org/ | Web-based |
| UCSC Cancer Genomics Browser [251, 252] | Integration of clinical data | http://genome-cancer.ucsc.edu/ | Web-based |
| UCSC Genome Browser [103] | Rapid linear visualization, examination and querying of the data at many levels | http://genome.ucsc.edu/cgi-bin/hgGateway | Web-based |
| UTGB [253] | Open-source software for developing personalized genome browsers that work in web browsers | http://utgenome.org/ | Web-based |
| X:map [254] | Mappings between genomic features and Affymetrix microarrays | http://xmap.picr.man.ac.uk/ | Web-based |
Table 6.
Visualization tools for comparative genomics
| Tool and references | Description | URL | Web-based or standalone |
|---|---|---|---|
| ACT [255] | A tool for displaying pairwise comparisons between two or more DNA sequences | http://www.sanger.ac.uk/Software/ACT/ | Standalone |
| cBio [109] | An open-access resource for interactive exploration of multidimensional cancer genomics datasets | http://cbioportal.org | Web-based |
| Cinteny [256] | Detection of syntenic regions across multiple genomes and measuring the extent of genome rearrangement using reversal distance as a measure | http://cinteny.cchmc.org/ | Web-based |
| Circos [107] | A software package for visualizing data and information. It visualizes data in a circular layout | http://mkweb.bcgsc.ca/circos | Standalone |
| CMap [257] | A browser-based tool for the visual comparison of various maps (sequence, genetic, etc.) from any number of species | http://gmod.org/wiki/CMap | Standalone |
| CoGe SynMap [258] | Generates a syntenic dot-plot between two organisms and identifies syntenic regions | https://genomevolution.org/coge/SynMap.pl | Web-based |
| Combo [234] | Dot-plot and linked-track views. Integration of annotation in both views | http://www.broadinstitute.org/annotation/argo/ | Standalone |
| DHPC [259] | Visualization of large-scale genome sequences by mapping sequences into a 2D using the space-filling function of Hilbert-Peano mapping | http://www.hpcurve.com | Standalone |
| DNAPlotter [260] | A Java application for generating circular and linear representations of genomes. Makes use of the Artemis libraries | http://www.sanger.ac.uk/resources/software/dnaplotter/ | Standalone |
| FilooT [261] | A visualization tool for exploring genomic data | No URL | Standalone |
| GBrowsesyn [262] | GBrowse-based synteny browser designed to display multiple genomes, with a central reference species compared with two or more additional species | http://gmod.org/wiki/GBrowse_syn | Standalone |
| GenomeComp [263] | A tool for summarizing, parsing and visualizing the genome-wide sequence comparison results derived from voluminous BLAST textual output | http://www.mgc.ac.cn/GenomeComp/ | Standalone |
| GenomeMatcher [264] | A dot-plot-based viewer for DNA sequence comparison | http://tinyurl.com/genomematcher/ | Web-based |
| GenPlay Multi-Genome [265] | A tool to compare and analyze multiple human genomes in a graphical interface | http://genplay.einstein.yu.edu | Standalone |
| ggbio [266] | R library to visualize particular genomic regions and genome-wide overviews | http://www.bioconductor.org/packages/2.11/bioc/html/ggbio.html | Standalone |
| Gramene [267, 268] | A comparative genome mapping database for grasses and a community resource for Oryza sativa | http://ensembl.gramene.org/genome_browser/index.html | Web-based |
| HilbertVis [269] | Functions to visualize long vectors of integer data by means of Hilbert curves | http://www.ebi.ac.uk/huber-srv/hilbert/ | Standalone |
| In-GAVsv [270] | Integrative genome analysis pipeline (inGAP), which uses a Bayesian principle to detect SNPs and small insertion/deletions (indels) | http://ingap.sourceforge.net/ | Standalone |
| Meander [271] | Hilbert plots to visually discover and explore structural variations in a genome based on read-depth and pair-end information | https://sites.google.com/site/meanderviz/ | Standalone |
| MEDEA [272] | Genomic feature densities and genome alignments of circular genomes. Comparative genomic visualization with Adobe Flash | http://www.broadinstitute.org/annotation/medea/ | Web-based |
| MizBee [273] | A multiscale synteny browser for exploring conservation relationships in comparative genomics data. Using side-by-side linked views, it enables efficient data browsing across a range of scales, from the genome to the gene | http://www.cs.utah.edu/~miriah/mizbee | Web-based |
| MuSiC [274] | Identifying mutational significance in cancer genomes | http://gmt.genome.wustl.edu | Standalone |
| ngs.plot [275] | Quick mining and visualization of NGS data by integrating genomic databases | https://github.com/shenlab-sinai/ngsplot | Standalone |
| PhIGs [276] | Ideogram-style interactive display of orthologs across >75 genomes | http://phigs.org | Web-based |
| PSAT [277] | A web tool to compare genomic neighborhoods of multiple prokaryotic genomes | http://www.nwrce.org/psat | Web-based |
| Seevolution [278] | Interactive 3D environment that enables visualization of diverse genome evolution processes | http://seevolution.org | Standalone |
| Sybil [279] | Comparative genome data, particularly protein and gene clustered data | http://sybil.sourceforge.net/ | Web-based |
| SynView [238] | A GBrowse-compatible approach to visualizing comparative genome data | http://gmod.org/wiki/SynView | Standalone |
| TREAT [280] | A bioinformatics tool for variant annotations and visualizations in targeted and exome sequencing data | http://ndc.mayo.edu/mayo/research/biostat/stand-alone-packages.cfm | Standalone |
| UCSC Genome Browser [281] | Conservation tracks within the popular UCSC genome browser | http://genome.ucsc.edu/cgi-bin/hgGateway/ | Web-based |
| Vanno [282] | A visualization-aided variant annotation tool | http://cgts.cgu.edu.tw/vanno | Web-based |
| Variant View [283] | Features an information-dense visual encoding that provides maximal information at the overview level, in contrast to the extensive navigation required by currently prevalent genome browsers | http://www.cs.ubc.ca/labs/imager/tr/2013/VariantView/ | Web-based |
| VISTA [108] | A comprehensive suite of programs and databases for comparative analysis of genomic sequences | http://genome.lbl.gov/vista/index.shtml | Web-based |
| VSV, VISTA-Dot [284, 285] | Three-scale viewer for synteny and dynamic, interactive dot plots for whole-genome DNA alignments | http://genome.jgi-psf.org/synteny/ | Web-based |
Fig. 4.
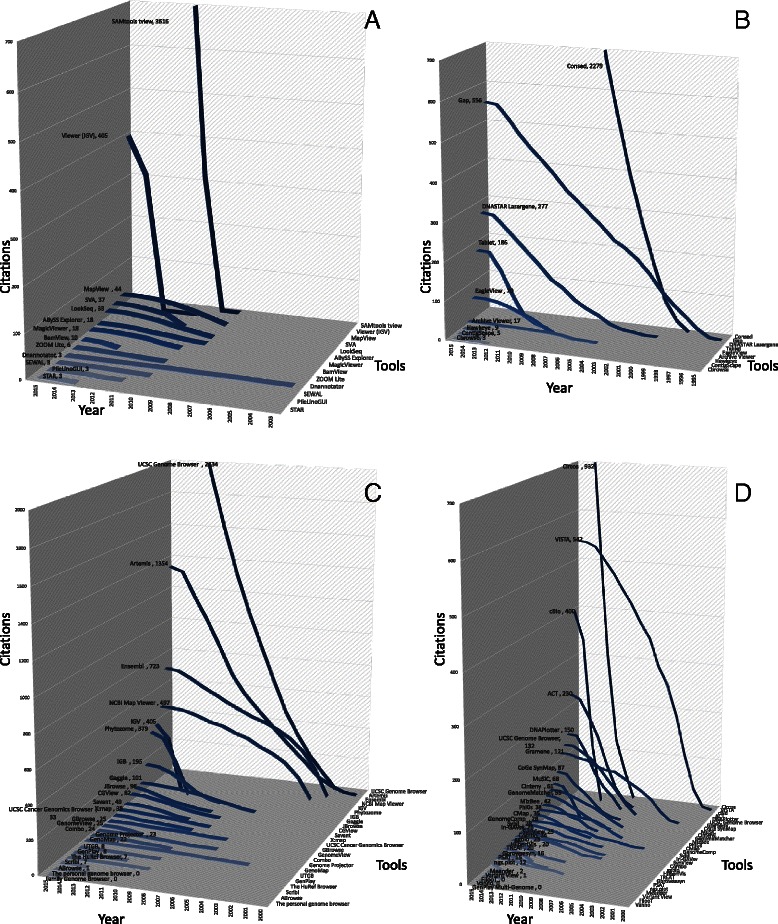
Citation trends and key players in genome biology. a Citations of genome alignment visualization tools based on Scopus. b Citations of genome assembly visualization tools based on Scopus. c Citations of genome browsers based on Scopus. d Citations of comparative genomics visualization tools based on Scopus. The numbers of citations of each tool in 2015 are shown after its name
Although tremendous progress has been made in genomic visualization and very large amounts of money have been invested in such projects, genome browsers [110] still need to address major problems. One of the biggest challenges is the integration of data in different formats (such as genomic and clinical data) as society enters the personalized medicine era. Furthermore, navigation at different resolution or granularity levels and smooth scaling are necessary as long as simultaneous comparisons across millions of elements [111] remains a bottleneck. Newer infrastructure and software that allow on-the-fly calculations both in the front end and the back end would definitely be a step forward. Finally, similarly to network biology, time-series data visualization is one of the great challenges. For example, in a hypothetical scenario in which it is required to follow genomic rearrangements over time during tumor development, time-series data visualization would be an invaluable tool. Motion integration and visualization using additional dimensions could be possible solutions. Overall, it would be unrealistic to expect an ideal universal genome browser that serves all the possible purposes in the field.
Visualization and analysis of expression data
Microarrays [112] and RNA sequencing [87] are the two main high-throughput techniques for measuring expression levels of large numbers of genes simultaneously. Both methods are revolutionary as one can simultaneously monitor the effects of certain treatments, diseases and developmental stages on gene expression across time (Fig. 5a) and for multiple transcript isoforms. Although microarrays and RNAseq technologies are comparable to each other [113], the latter tends to dominate, especially as sequencing technologies have improved, and there now are robust statistics to model the particular noise characteristics of RNAseq, particularly for low expression [114]. Microarrays are still cheaper and in some contexts may be more convenient as their analysis is still simpler and requires less computing infrastructure.
Fig. 5.
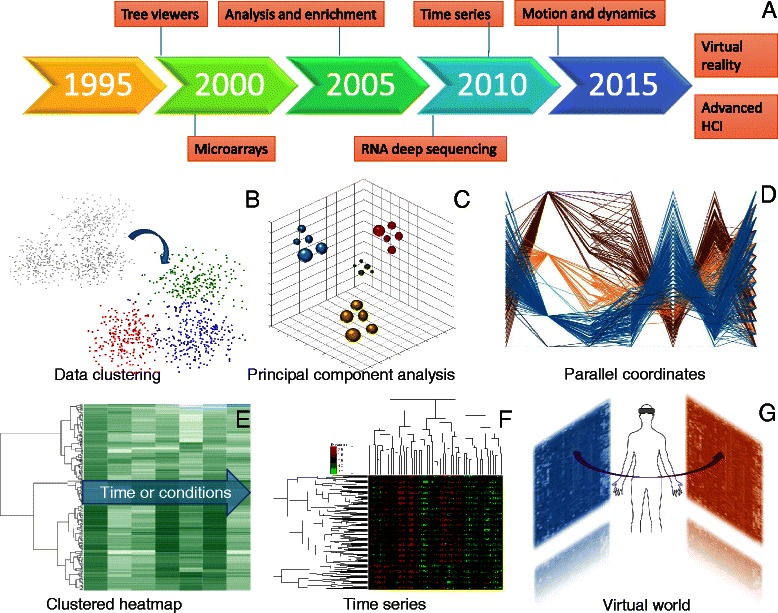
Multivariate analyses and visualization. a Timeline of the emergence of relevant technologies and concepts. b Visualization of k-means partitional clustering algorithm. c 3D visualization of a principal component analysis. d Visualization of gene-expression measures across time using parallel coordinates. e Visualization of gene-expression clustering across time. f 2D hierarchical clustering to visualize gene expressions against several time points or conditions. g Hypothetical integration of analyses and expression heatmaps and the control of objects by VR devices
In both cases, a typical analysis procedure is first to normalize experimental and batch differences between samples and then to identify up- and downregulated genes based on a fold-change level when comparing across samples, such as between a healthy and a non-healthy tissue. Statistical approaches are used to assess how reliable fold-change measurements are for each transcript of interest by modeling variation across transcripts and experiments. Subsequently, functional enrichment is performed to identify pathways and biological processes in which the up- and downregulated genes may be involved. Although there are numerous functional enrichment suites [115], David [116], Panther [117] and WebGestalt [118] are among the most widely used.
When gene expression is measured across many time points or conditions so as to observe, for example, the expression patterns following treatment, various analyses can be taken into consideration. Principal component analysis or partitional clustering algorithms such as k-means [119] can be used to group together genes with similar behavior patterns. Scatter-plotting is the typical visualization to represent such groupings. Thus, each point on a plane represents a gene and the closer two genes appear, the more similar they are (Fig. 5b, c).
When one wants to categorize genes with similar behavior patterns across time (Fig. 5d), hierarchical clustering based on expression correlation can be performed. Average linkage, complete linkage, single linkage, neighbor joining [120] and UPGMA [121] are the most widely used methods. In such approaches, an all-against-all distance or correlation matrix that shows the similarities between each pair of genes is required and genes are placed as leaves in a tree hierarchy. The two most widely used correlation metrics for expression data are the Spearman and Pearson correlation metrics. A list of tree viewers for hierarchical clustering visualization is presented in Table 7. A more advanced visualization method is combining trees with heatmaps (Fig. 5e): genes are grouped together according to their expression patterns in a tree hierarchy and the heat map is a graphical representation of individual gene-expression values represented as colors. Darker colors indicate a higher expression value and vice versa. An even more complex visualization of a 2D hierarchical clustering is shown in Fig. 5f, in which genes are clustered based on their expression patterns across several conditions (vertical tree on the left) and conditions are clustered across genes (horizontal tree). The heatmap shows the correlations between gene groups and conditions by allowing the researcher to come to conclusions about whether a group of genes is affected by a set of conditions or not. Heatmaps do, however, have significant drawbacks with regards to color perception. Perception of the color of a cell in a heatmap is shaped by the color of the surrounding cells, so two cells with identical color can look very different depending on their position in the heatmap.
Table 7.
Tree viewers and phylogenies
| Tool and references | Description | URL |
|---|---|---|
| ARB [123] | A graphically oriented package comprising various tools for sequence database handling and data analysis | http://www.arb-home.de/ |
| Bio.Phylo [286] | A unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython | http://biopython.org |
| Dendroscope [125] | Software for visualizing phylogenetic trees and rooted networks | http://ab.inf.uni-tuebingen.de/software/dendroscope/ |
| ETE Toolkit [287] | Python programming toolkit that assists in the automated manipulation, analysis and visualization of phylogenetic and other types of trees | http://etetoolkit.org/ |
| EvolView [288] | Tool for displaying, managing and customizing phylogenetic trees | http://www.evolgenius.info/evolview.html |
| iTOL [126] | Online tool for the display and manipulation of phylogenetic trees | http://itol.embl.de/ |
| MEGA [122] | Integrated tool for phylogenetic analysis and visualization | http://www.megasoftware.net/ |
| NJplot [124] | A tree drawing program able to draw any phylogenetic tree expressed in the Newick phylogenetic tree format | http://doua.prabi.fr/software/njplot |
| OneZoom [289] | Committed to heightening awareness about the diversity of life on earth and its evolutionary history | http://www.onezoom.org/ |
| Paloverde [290] | 3D visualization of phylogenetic structure of moderately large trees on the scale of 100–2500 leaf nodes | http://loco.biosci.arizona.edu/paloverde/paloverde.html |
| PhyloDraw [291] | Drawing tool for creating phylogenetic trees | http://jade.cs.pusan.ac.kr/phylodraw/ |
| PhyloExplorer [292] | Tool to facilitate assessment and management of phylogenetic tree collections | http://www.ncbi.orthomam.univ-montp2.fr/phyloexplorer/ |
| PhyloWidget [293] | Program for viewing, editing and publishing phylogenetic trees online | http://www.phylowidget.org/ |
| TreeDyn [294] | TreeDyn links unique leaf labels to lists of variables/values pairs of annotations, independently of the tree topologies | http://www.treedyn.org/ |
| TreeGraph [295] | A graphical editor for phylogenetic trees that allows many graphical formats for the elements of the tree | http://treegraph.bioinfweb.info/ |
| TreeQ-Vista [296] | Designed for presenting functional annotations in a phylogenetic context | http://genome.lbl.gov/vista/TreeQVista/ |
| TreeVector [297] | Web utility to create and integrate phylogenetic trees as Scalable Vector Graphics (SVG) files | http://supfam.cs.bris.ac.uk/TreeVector/ |
| TreeVolution [298] | Java tool to support visual analysis of phylogenetic trees | http://vis.usal.es/treevolution |
| T-REX [299] | Web server dedicated to the reconstruction of phylogenetic trees and reticulation networks and to the inference of horizontal gene transfer events | http://www.trex.uqam.ca/ |
| ViPhy [300] | Comparison of multiple phylogenetic trees | http://www.gris.tu-darmstadt.de/research/vissearch/projects/ViPhy/ |
Although RNAseq analysis is still an active field, microarray analysis has matured a lot over the past 15 years and many suites for analyzing such data are currently available (Table 8). To identify the key players in the field of microarray/RNAseq visualization we followed the citation patterns of the available tools from Scopus (Fig. 6). MEGA [122], ARB [123], NJplot [124], Dendroscope [125] and iTOL [126] are the most widely used tree viewers to visualize phylogenies and hierarchical clustering results. MultiExperiment Viewer [127], Genesis [128], GenePattern [129] and EXPANDER [130] are advanced suites that can perform various multivariate analyses such as the ones discussed in this section. Nevertheless, the commercial GeneSpring platform and the entire R/BioConductor framework [131, 132] are mostly used in such analyses.
Table 8.
Microarray and RNAseq analysis viewers
| Tool and references | Description | URL |
|---|---|---|
| ArrayXPath [201] | Mapping and visualizing microarray gene-expression data with integrated biological pathway resources using scalable vector graphics | http://www.snubi.org/software/ArrayXPath/ |
| BicOverlapper [301, 302] | Supports visual analysis of gene expression by means of biclustering | http://vis.usal.es/bicoverlapper/ |
| BiGGEsTS [303] | Tool providing an integrated environment for the biclustering analysis of time-series gene-expression data | http://tinyurl.com/BiGGEsTS/ |
| eRNA [304] | RNA data analysis tool for high-throughput RNA sequencing experiments | https://sourceforge.net/projects/erna/?source=directory |
| EXPANDER [130] | A Java-based tool for analysis of gene-expression and NGS data | http://acgt.cs.tau.ac.il/expander/ |
| ExpressionProfiler [305] | Web-based platform for microarray gene-expression and other functional-genomics-related data analysis | http://www.ebi.ac.uk/expressionprofiler |
| GenePattern [129] | Modular analysis web platform; several visualization modules available | http://genepattern.broadinstitute.org/gp/pages/login.jsf |
| Genesis [128] | Java package of tools to simultaneously visualize and analyze a whole set of gene-expression experiments | http://genome.tugraz.at/genesisclient/genesisclient_description.shtml |
| GeneVAnD [306] | Linked heatmaps, dendrograms and 2D/3D scatter plots | http://tinyurl.com/GeneVAnD/ |
| geWorkbench [307] | A Java-based open-source platform for integrated genomics. It allows individually developed plugins to be configured into complex bioinformatic applications. Currently more than 70 available plugins supporting the visualization and analysis | http://wiki.c2b2.columbia.edu/workbench/index.php/Home |
| Gitools [308] | Analysis and visualization of genomic data using interactive heatmaps | http://www.gitools.org |
| HCE [309] | Linked heat map, profile and scatter plots; systematic exploration | http://www.cs.umd.edu/hcil/hce/ |
| HeatmapGenerator [310] | Create customized gene-expression heatmaps from RNAseq and microarray data | http://sourceforge.net/projects/heatmapgenerator/ |
| HeatMapViewer [311] | Interactive display of microarray experiments or the outcome of mutational studies and the study of SNP-like sequence variants | http://dx.doi.org/10.5281/zenodo.7706 |
| Mayday [312] | A graphical user interface that supports the development and integration of existing and new analysis methods. Many linked visualizations | http://it.informatik.uni-tuebingen.de/?page_id=248/wp/ |
| MultiExperiment Viewer [127] | Analysis suite. Heatmaps, dendrograms, profile and scatter plots | http://www.tm4.org/ |
| PointCloudXplore [313] | Visualization of transcription data in Drosophila embryos. Multiple views to ease analysis of complex gene-expression data. This type of interaction blends high-dimensional information exploration with interactive, 3D visualization | http://tinyurl.com/PointCloudXplore/ |
| RNASeqBrowser [314] | A genome browser for simultaneous visualization of raw strand specific RNAseq reads and UCSC genome browser custom tracks | http://www.australianprostatecentre.org/research/software/rnaseqbrowser |
| RNAseqViewer [315] | Visualization of the various data from the RNAseq analyzing process, for single or multiple samples | http://bioinfo.au.tsinghua.edu.cn/software/RNAseqViewer/ |
| TimeSearcher [316] | Interactive querying and exploration of time-series data | http://www.cs.umd.edu/hcil/timesearcher/ |
| TraV [317] | Visualization and analysis of multiple transcriptome sequencing experiments | http://appmibio.uni-goettingen.de/index.php?sec=serv |
Fig. 6.
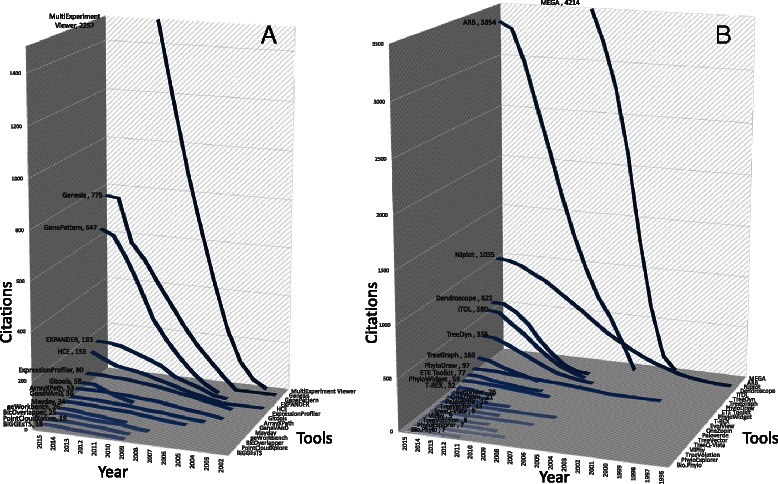
Citation trends and tools for gene-expression analysis. a Citations of microarray/RNAseq visualization tools based on Scopus. b Citations of tree viewers based on Scopus. The numbers of citations of each tool in 2015 are shown after its name
Concerning the future of multivariate data visualization, new HCI techniques and VR devices could allow parallel visualizations, analyses and data integration simultaneously (Fig. 5g).
Programming languages and complementary libraries for building visual prototypes
Although the field of biological data visualization has been active for 25 years, it is still evolving rapidly today, as the complexity and the size of results produced by high-throughput approaches increase. Although most of the current software is offered in the form of standalone distributions, a shift towards web visualization is growing. Important features of modern visualization tools include: interactivity; interoperability; efficient data exploration; quick visual data querying; smart visual adjustment for different devices with different dimensions and resolutions; fast panning; fast zooming in or out; multilayered visualization; visual comparison of data; and smart visual data filtering. As functions and libraries implementing these features for standalone applications become available, similar libraries for web visualizations immediately follow. Therefore, in this section we discuss the latest programming languages, libraries and application program interfaces (APIs) that automate and simplify many of the aforementioned features, enabling higher-quality visualization implementations. It is not in the scope of this review to extensively describe all programming language possibilities for data visualization; therefore, we focus on the five languages that are mostly used for high-throughput biological data. Nevertheless, Table 9 summarizes other languages, along with generic and language-specific libraries (for R, Perl and Python), that target specific problems and make the implementation of biological data visualization more practical.
Table 9.
Programming languages and libraries to build visual prototype
| Language/Library | Description | URL |
|---|---|---|
| Adobe Edge | Animated, interactive web content for projects that previously required Flash | https://creative.adobe.com/products/animate |
| Arbor.js | Efficient, force-directed layout algorithm plus abstractions for graph organization and screen refresh handling | http://arborjs.org/ |
| Biojs | BioJS enables a full-featured biological workbench directly in your browser | http://biojs.net/ |
| Bonsai.js | Lightweight graphics library with an intuitive graphics API and an SVG renderer | https://bonsaijs.org/ |
| Chart.js | Object oriented client side graphs. Data visualization in six animated, fully customizable chart types | http://www.chartjs.org/ |
| Cube | Time-series data, built on MongoDB, Node and D3. Real-time visualizations of aggregate metrics | https://square.github.io/cube/ |
| Cubism | D3 plugin for visualizing time series | https://square.github.io/cubism/ |
| Cytoscape Web | Easily embed interactive networks in your website | http://cytoscapeweb.cytoscape.org/ |
| D4 | Friendly charting domain-specific language for D3 | https://github.com/heavysixer/d4 |
| Easeljs | API to work with rich graphics and interactivity with HTML5 Canvas | http://www.createjs.com/EaselJS |
| Ember Charts | Ember.js and d3.js based time series, bar, pie and scatter charts that are easy to extend and modify | http://addepar.github.io/ |
| Envision | Fast, dynamic and interactive HTML5 visualizations | http://www.humblesoftware.com/envision |
| Flare | Interactive data visualizations in Flash (ActionScript) | http://flare.prefuse.org/ |
| Foamtree | Tree map visualization with innovative layout algorithms and animations such as Voronoi Treemaps | http://carrotsearch.com/foamtree-overview |
| Highcharts.js | HTML5/JavaScript-based line, spline, area, area-spline, column, bar, pie, scatter, angular gauges, area-range, area-spline-range, column-range, bubble, box plot, error bars, funnel, waterfall and polar charts | http://www.highcharts.com/ |
| Infovis Toolkit | A comprehensive range of tools for creating Interactive Data Visualizations for the Web | http://philogb.github.io/jit/ |
| Jgrapht | A free Java graph library that provides mathematical graph-theory objects and algorithms | http://jgrapht.org/ |
| Kartograph | Kartograph is a simple and lightweight framework for creating beautiful, interactive vector maps | http://kartograph.org/ |
| Matplotlib | A Python 2D plotting library that produces publication quality figures | http://matplotlib.org/ |
| Miso | Interactive storytelling and data visualization content | http://misoproject.com/ |
| Netadvantage | Charts with a range of frameworks including asp.net and Silverlight. Visualization options include bar, bubble, Gantt, line, radial, scatter, spline and doughnut charts | http://www.infragistics.com/products |
| Orange | Data mining through visual programming or Python scripting. Components for machine learning. Add-ons for bioinformatics and text mining. Packed with features for data analytics | http://orange.biolab.si/ |
| Paper.js | A vector graphics scripting framework that runs on top of the HTML5 Canvas | http://paperjs.org/ |
| Pivotviewer | A Silverlight control that makes it easier to interact with massive amounts of data on the web | http://www.microsoft.com/silverlight/pivotviewer/ |
| Polychart.js | A JavaScript graphing library capable of producing a wide array of graphics fairly easily | http://www.polychartjs.com/ |
| Prefuse | Java-based interactive data. Data structures for tables, graphs and trees, a host of layout and visual encoding techniques, animation, dynamic queries, integrated search and database connectivity | http://prefuse.org/ |
| Prefuse Flare | Visualization and animation for ActionScript. From basic charts and graphs to complex interactive graphics. Data management, visual encoding, animation and interaction techniques | http://flare.prefuse.org/ |
| Ractive.js | It transforms templates into blueprints for apps that are interactive by default. Two-way binding, animations, SVG support and more | http://www.ractivejs.org/ |
| Raphael.js | JavaScript library for vector graphics on the web. To create a specific chart or image. Crop and rotate widget | http://raphaeljs.com/ |
| Rcharts | R package to create, customize and publish interactive JavaScript visualizations from R using a familiar lattice style plotting interface | http://rcharts.io/ |
| Seaborn | A Python visualization library based on matplotlib. It provides a high-level interface for drawing attractive statistical graphics | http://stanford.edu/~mwaskom/software/seaborn/ |
| Shiny | A web application framework for R to turn an analysis into interactive web applications. No HTML, CSS or JavaScript knowledge required | http://shiny.rstudio.com/ |
| Sigma.js | A JavaScript library dedicated to graph drawing. It makes easy to publish networks on Web pages and allows developers to integrate network exploration in rich web applications | http://sigmajs.org/ |
| Three.js | A lightweight cross-browser JavaScript library/API used to create and display animated 3D computer graphics on a web browser that supports WebGL | http://threejs.org/ |
| Timeline.js | Visually rich interactive timelines, available in 40 languages | http://timeline.knightlab.com/ |
| Variance | Build powerful data visualizations for the web without writing JavaScript. Wide range of visualizations | https://variancecharts.com/ |
| Vega | A visualization grammar, a declarative format for creating, saving and sharing visualization designs. Data visualizations in JSON format and interactive views using either HTML5 Canvas or SVG | http://trifacta.github.io/vega/ |
| Vida.io | A way to build reusable cloud visualizations: clone visualization templates, customize without coding skills and embed or share in the cloud | https://vida.io/ |
| Vis | A data visualization platform designed to assist investigative journalists, activists and others in mapping complex business or crime networks | http://vis.occrp.org/ |
| Visual Sedimentation | A JavaScript library for visualizing streaming data, inspired by the process of physical sedimentation. jQuery (to facilitate HTML and JavaScript development) and Box2DWeb (for physical world simulation) | http://www.visualsedimentation.org/ |
| WebGL | A JavaScript API for rendering interactive 3D computer graphics and 2D graphics within any compatible web browser without the use of plugins | https://www.khronos.org/webgl/ |
Processing
‘Processing’ is a programming language and a development platform for writing generative, interactive and animated standalone applications. Basic shapes such as lines, triangles, rectangles and ellipses, inner/outer coloring and basic operations such as transformations, translations, scaling and rotations can be implemented in a single line of code and each shape can be drawn within a canvas of a given dimension and a given refresh rate. It is designed for easier implementations of 2D dynamic visualizations but it also supports 3D rendering, although not optimized. Its core library is now extended by more than 100 other libraries and it is one of the best documented languages in the field. The integrated development environment allows exporting of executable files for all Windows, MacOS and Linux operating systems as well as Java applet .jar files. Finally, it can be used as an excellent educational tool for computer programming fundamentals in a visual context. It is free for download, can easily be plugged in a Java standalone application, and is fully cooperative with the NetBeans and Eclipse environments. Code examples and tutorials can be found at [133].
Processing.js
Java applets were an easy way to run standalone applications within web browsers. This technology has now mainly been abandoned because of security considerations. To avoid JavaScript’s complexity and compensate for applet limitations, Processing.js was implemented, as the sister project of the popular Processing programming language, to allow interactive web visualization. It is a mediator between HTML5 and Processing and is designed to allow visual prototypes, digital arts, interactive animations, educational graphs and so on to run immediately within any HTML5-compatible browser, such as Firefox, Safari, Chrome, Opera or Internet Explorer. No plugins are required and one can code any visualization directly in the Processing language, include it in a web page, and let Processing.js bridge the two technologies. Processing.js brings the best of visual programming to the web, both for Processing and web developers. Code examples and tutorials can be found at [134].
D3
D3 is the main competitor of Processing/Processing.js and has gained ground over recent years. It was initially used to generate scalable vector graphics (SVG). Like Processing.js, it is designed for powerful interactive web visualizations and it comes with its own JavaScript-like syntax. It is a JavaScript library for manipulating document object model objects and a programming interface for HTML, XML and SVG. The idea behind this approach is to load data into a browser and then generate document object model elements based on that data. Subsequently, one can apply data-driven transformations on the document. This avoids proprietary representation and affords extraordinary flexibility. With minimal overhead, D3 is extremely fast and supports large datasets and dynamic behaviors for interaction and animation. D3’s functional style allows code reuse through a diverse collection of components and plugins. It is extensively documented and code examples can be found at [135].
Flash
Adobe Flash was once the industry standard for authoring innovative, interactive content. In conjunction with the platform’s programming language, ActionScript, Flash allows designers to implement dynamic visualization, opening up many possibilities for creativity. Some of the most pioneering, best practice visualizations built in Flash can be found with online news and media sites, introducing interactivity to supplement and enhance the presentation of information. Because of the lack of support for Flash across Apple’s suite of devices and the emergence of competing developments, demanding less computational power, including D3 and HTML5, this technology is now fading.
Java3D
Java 3D is an API, acting as a mediator between OpenGL and Java and enables the creation of standalone 3D graphics applications and internet-based 3D applets. It is easy to use and provides high-level functions for creating and manipulating 3D objects in space and their geometry. Programmers initially create a virtual world and then place any 3D object anywhere in this world. Rotation in three axes, zooming in or out and translation of the whole canvas are functions are offered by default, and the hierarchy of the transformation groups define the 3D transformations that can be applied individually to an object or a set of objects. Java3D code can be compiled under any of the Windows, MacOS and Unix Systems.
Conclusion
The future of biological data visualization
Biological data visualization is a rapidly evolving field. Nevertheless, it is still in its infancy. Hardware acceleration, standardized exchangeable file formats, dimensionality reduction, visual feature selection, multivariate data analyses, interoperability, 3D rendering and visualization of complex data at different resolutions are areas in which great progress has been achieved. Additionally, image processing combined with artificial-intelligence-based pattern recognition, new libraries and programming languages for web visualization, interactivity, visual analytics and visual data retrieval, storing and filtering are still ongoing efforts with remarkable advances over the past years [58, 136, 137]. Today, many of the current visualization tools serve as front ends for very advanced infrastructures dedicated to data manipulation and have driven significant advances in user interfaces. Although the implementation of sophisticated graphical user interfaces is necessary, the effort to minimize back-end calculations is of great importance. Unfortunately, only a limited number of visualization tools today take advantage of libraries designed for parallelization. Multi-threading, for example, allows the distribution of computational tasks in terminals over the network, and CUDA (available on Nvidia graphic cards) allows parallel calculations at multiple graphical processing units.
Despite the fact that multiple screens, light and laser projectors and other technologies partially solve the space limitation problem, HCI techniques are changing the rules of the game and biological data visualization is expected to adjust to these trends in the longer term. 3D control can be achieved without intermediate devices such as mouse, keyboards or touch screens [138] in modern perceptual input systems. Sony’s EyeToy, Playstation Eye and Artag, for example, use non-spatial computer vision to determine hand gestures. Similarly, the Nintendo Wii and Sony Move devices support object manipulation in 3D space. These actions are mediated through the detection of the position in space of physical devices held by the user or, even more impressively, through immediate tracking of the human body or parts of the human body. Equally impressive is the prospect of ocular tracking, one implementation of which has recently been introduced by the VR startup Fove. The Fove headset tracks eye movement and translates into spatial movement or even other types of action within the simulated 3D space. The recently implemented Molecular Control Toolkit [139] is a characteristic example of a new API based on the Kinect and Leap Motion devices (which track the human body and human fingers, respectively) to control molecular graphics such as 3D protein structures. Moreover, large screens, tiled arrays or VR environments should be taken into consideration by programmers and designers as they become more and more affordable over time. A great benefit of such technologies is that they allow the representation of complete datasets without the need for algorithms dedicated to dimensionality reduction, which might lead to information loss.
VR environments are expected to bring a revolution in biological data visualization as one could integrate metabolomics networks and gene expression in virtual worlds, as in MetNet3D [140], or create virtual universes of living systems such as a whole cell [59, 141–144]. A visual representation of the whole cell with its components in an immense environment in which users can visually explore the location of molecules and their interaction in space and time could lead to a better understanding of the biological systems. Oculus Rift (which promoted the reemergence of VR devices), Project Morpheus, Google Cardboard, Sony Smart Eyeglass, HTC Vive, Samsung Gear VR, Avegant Glyph, Razer OSVR, Archos VR Headset and Carl Zeiss VR One are state-of-the-art commercial devices that offer VR experiences. All of them overlay the user’s eyesight with some kind of screen and aim to replace the field of view with a digital 3D alternative. Between them, those devices use many technologies and new ideas such as the monitoring of the position of the head (allowing for more axes of movement), the substitution of the VR screen with smartphones (thus harnessing efficient modern smartphone processors), eye tracking and projection of images straight onto the retina.
Approaching the problem from a different angle, Google Glass, HoloLens and Magic Leap offer an augmented reality experience (the latter is rumored to achieve that by projecting a digital light field into the user’s eye). Augmented reality can facilitate the learning process of the biological systems because it builds on exploratory learning. This allows scientists to visualize existing knowledge, whereas the unstructured nature of augmented reality could allow them to construct knowledge themselves by making connections between information and their own experiences or intuition and thus offer novel insights to the studied biological system [145]. Efforts such as the Visible Cell [141] and CELLmicrocosmos have already begun. The Visible Cell project aims to inform advanced in silico studies of cell and molecular organization in 3D using the mammalian cell as a unitary example of an ordered complex system; the CELLmicrocosmos integrative cell modeling and stereoscopic 3D visualization project is a typical example of the use of 3D vision.
Finally, starting from a living entity, the process of digitizing it, visualizing it, placing it in virtual worlds or even recreating it as a physical object using 3D printing is no longer the realm of science fiction. Data visualization and biological data visualization are rapidly developing in parallel with advances in the gaming industry and HCI. These efforts are complementary and there are already strong interactions developing between these fields, something that is expected to become more obvious in the future.
Funding
This work was supported by the European Commission FP7 programs INFLA-CARE (EC grant agreement number 223151), ‘Translational Potential’ (EC grant agreement number 285948).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GAP was the main writer of the article and the one who was inspired about the topic. DM collected all the necessary information about the tools and their citation trends. NP and TT provided fruitful feedback about the recent HCI technologies. AE provided information about the latest technological trends in the areas of genomics. II was the main supervisor of the project. All authors read and approved the final manuscript.
Contributor Information
Georgios A. Pavlopoulos, Email: g.pavlopoulos@med.uoc.gr
Dimitris Malliarakis, Email: dim-mal@hotmail.com.
Nikolas Papanikolaou, Email: papnikol@med.uoc.gr.
Theodosis Theodosiou, Email: theodosios.theodosiou@gmail.com.
Anton J. Enright, Email: aje@ebi.ac.uk
Ioannis Iliopoulos, Email: iliopj@med.uoc.gr.
References
- 1.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, et al. Big data: the future of biocuration. Nature. 2008;455:47–50. doi: 10.1038/455047a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liolios K, Mavromatis K, Tavernarakis N, Kyrpides NC. The Genomes On Line Database (GOLD) in 2007: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–9. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Census of Marine Life. How many species on Earth? About 8.7 million, new estimate says. ScienceDaily. 24 August 2011. http://www.sciencedaily.com/releases/2011/08/110823180459.htm. Accessed 27 July 2015.
- 4.May M. Life Science Technologies: Big biological impacts from big data. Science. 2014; doi:10.1126/science.opms.p1400086.
- 5.Reddy TB, Thomas AD, Stamatis D, Bertsch J, Isbandi M, Jansson J, et al. The Genomes OnLine Database (GOLD) v. 5: a metadata management system based on a four level (meta)genome project classification. Nucleic Acids Res. 2015;43:D1099–1106. doi: 10.1093/nar/gku950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 7.Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum Mol Genet. 2014;23:5866–78. doi: 10.1093/hmg/ddu309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 9.Pavlopoulos GA, Iacucci E, Iliopoulos I, Bagos PG. Interpreting the Omics 'era' data. Multimedia Services in Intelligent Environments vol. 25. Heidelber: Springer; 2013. pp. 79–100. [Google Scholar]
- 10.Pavlopoulos GA, Secrier M, Moschopoulos CN, Soldatos TG, Kossida S, Aerts J, et al. Using graph theory to analyze biological networks. BioData Min. 2011;4:10. doi: 10.1186/1756-0381-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moschopoulos CN, Pavlopoulos GA, Likothanassis SD, Kossida S. Analyzing protein-protein interaction networks with web tools. Curr Bioinform. 2011;6:389–97. doi: 10.2174/157489311798072972. [DOI] [Google Scholar]
- 12.Papanikolaou N, Pavlopoulos GA, Theodosiou T, Iliopoulos I. Protein-protein interaction predictions using text mining methods. Methods. 2015;74:47–53. doi: 10.1016/j.ymeth.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Pavlopoulos GA, Promponas VJ, Ouzounis CA, Iliopoulos I. Biological information extraction and co-occurrence analysis. Methods Mol Biol. 2014;1159:77–92. doi: 10.1007/978-1-4939-0709-0_5. [DOI] [PubMed] [Google Scholar]
- 14.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–29. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 15.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 17.Fruchterman T, Reingold E. Graph drawing by force-directed placement. Softw Pract Exp. 1991;21:1129–64. doi: 10.1002/spe.4380211102. [DOI] [Google Scholar]
- 18.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–84. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moschopoulos CN, Pavlopoulos GA, Likothanassis SD, Kossida S. An enhanced Markov clustering method for detecting protein complexes. 8th IEEE International Conference on Bioinformatics and Bioengineering. 2008. doi:10.1109/BIBE.2008.4696656.
- 20.Adamcsek B, Palla G, Farkas IJ, Derenyi I, Vicsek T. CFinder: locating cliques and overlapping modules in biological networks. Bioinformatics. 2006;22:1021–3. doi: 10.1093/bioinformatics/btl039. [DOI] [PubMed] [Google Scholar]
- 21.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci USA. 2003;100:12123–12128. doi: 10.1073/pnas.2032324100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XL, Foo CS, Ng SK. Discovering protein complexes in dense reliable neighborhoods of protein interaction networks. Comput Syst Bioinformatics Conf. 2007;6:157–68. doi: 10.1142/9781860948732_0019. [DOI] [PubMed] [Google Scholar]
- 24.Lubovac Z, Gamalielsson J, Olsson B. Combining functional and topological properties to identify core modules in protein interaction networks. Proteins. 2006;64:948–59. doi: 10.1002/prot.21071. [DOI] [PubMed] [Google Scholar]
- 25.Cho YR, Hwang W, Ramanathan M, Zhang A. Semantic integration to identify overlapping functional modules in protein interaction networks. BMC Bioinformatics. 2007;8:265. doi: 10.1186/1471-2105-8-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moschopoulos CN, Pavlopoulos GA, Iacucci E, Aerts J, Likothanassis S, Schneider R, et al. Which clustering algorithm is better for predicting protein complexes? BMC Res Notes. 2011;4:549. doi: 10.1186/1756-0500-4-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maraziotis IA, Dimitrakopoulou K, Bezerianos A. Growing functional modules from a seed protein via integration of protein interaction and gene expression data. BMC Bioinformatics. 2007;8:408. doi: 10.1186/1471-2105-8-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng J, Jiang R, Jiang T. A max-flow-based approach to the identification of protein complexes using protein interaction and microarray data. IEEE/ACM Trans Comput Biol Bioinform. 2011;8:621–34. doi: 10.1109/TCBB.2010.78. [DOI] [PubMed] [Google Scholar]
- 29.Ulitsky I, Shamir R. Identification of functional modules using network topology and high-throughput data. BMC Syst Biol. 2007;1:8. doi: 10.1186/1752-0509-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlopoulos GA, Moschopoulos CN, Hooper SD, Schneider R, Kossida S. jClust: a clustering and visualization toolbox. Bioinformatics. 2009;25:1994–6. doi: 10.1093/bioinformatics/btp330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moschopoulos CN, Pavlopoulos GA, Schneider R, Likothanassis SD, Kossida S. GIBA: a clustering tool for detecting protein complexes. BMC Bioinformatics. 2009;10(Suppl 6):S11. doi: 10.1186/1471-2105-10-S6-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G, et al. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics. 2011;12:436. doi: 10.1186/1471-2105-12-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brohee S, Faust K, Lima-Mendez G, Sand O, Janky R, Vanderstocken G, et al. NeAT: a toolbox for the analysis of biological networks, clusters, classes and pathways. Nucleic Acids Res. 2008;36:W444–51. doi: 10.1093/nar/gkn336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Wu M, Kwoh CK, Ng SK. Computational approaches for detecting protein complexes from protein interaction networks: a survey. BMC Genomics. 2010;11(Suppl 1):S3. doi: 10.1186/1471-2164-11-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brohee S, Faust K, Lima-Mendez G, Vanderstocken G, van Helden J. Network analysis tools: from biological networks to clusters and pathways. Nat Protoc. 2008;3:1616–29. doi: 10.1038/nprot.2008.100. [DOI] [PubMed] [Google Scholar]
- 36.Brohee S, van Helden J. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinformatics. 2006;7:488. doi: 10.1186/1471-2105-7-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batagelj V, Mrvar A. Pajek - Program for large network analysis. Connections. 1998;21:47–57. [Google Scholar]
- 38.Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Z, Hung JH, Wang Y, Chang YC, Huang CL, Huyck M, et al. VisANT 3.5: multi-scale network visualization, analysis and inference based on the gene ontology. Nucleic Acids Res. 2009;37:W115–21. doi: 10.1093/nar/gkp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luciano JS, Stevens RD. e-Science and biological pathway semantics. BMC Bioinformatics. 2007;8(Suppl 3):S3. doi: 10.1186/1471-2105-8-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luciano JS. PAX of mind for pathway researchers. Drug Discov Today. 2005;10:937–42. doi: 10.1016/S1359-6446(05)03501-4. [DOI] [PubMed] [Google Scholar]
- 42.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–31. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 43.Hermjakob H, Montecchi-Palazzi L, Bader G, Wojcik J, Salwinski L, Ceol A, et al. The HUPO PSI's molecular interaction format--a community standard for the representation of protein interaction data. Nat Biotechnol. 2004;22:177–83. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 44.Lloyd CM, Halstead MD, Nielsen PF. CellML: its future, present and past. Prog Biophys Mol Biol. 2004;85:433–50. doi: 10.1016/j.pbiomolbio.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 45.Krzywinski M, Birol I, Jones SJ, Marra MA. Hive plots--rational approach to visualizing networks. Brief Bioinform. 2012;13:627–44. doi: 10.1093/bib/bbr069. [DOI] [PubMed] [Google Scholar]
- 46.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohler J, Baumbach J, Taubert J, Specht M, Skusa A, Ruegg A, et al. Graph-based analysis and visualization of experimental results with ONDEX. Bioinformatics. 2006;22:1383–90. doi: 10.1093/bioinformatics/btl081. [DOI] [PubMed] [Google Scholar]
- 48.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bastian M, Heymann S, Jacomy M. Gephi: an open source software for exploring and manipulating networks. In: International AAAI Conference on Weblogs and Social Media. 2009. https://www.aaai.org/ocs/index.php/ICWSM/09/paper/view/154. Accessed 27 July 2015.
- 50.Letunic I, Yamada T, Kanehisa M, Bork P. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem Sci. 2008;33:101–3. doi: 10.1016/j.tibs.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Dogrusoz U, Erson EZ, Giral E, Demir E, Babur O, Cetintas A, et al. PATIKAweb: a web interface for analyzing biological pathways through advanced querying and visualization. Bioinformatics. 2006;22:374–5. doi: 10.1093/bioinformatics/bti776. [DOI] [PubMed] [Google Scholar]
- 52.van Iersel MP, Kelder T, Pico AR, Hanspers K, Coort S, Conklin BR, et al. Presenting and exploring biological pathways with PathVisio. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–6. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Secrier M, Pavlopoulos GA, Aerts J, Schneider R. Arena3D: visualizing time-driven phenotypic differences in biological systems. BMC Bioinformatics. 2012;13:45. doi: 10.1186/1471-2105-13-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pavlopoulos GA, O'Donoghue SI, Satagopam VP, Soldatos TG, Pafilis E, Schneider R. Arena3D: visualization of biological networks in 3D. BMC Syst Biol. 2008;2:104. doi: 10.1186/1752-0509-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Freeman TC, Goldovsky L, Brosch M, van Dongen S, Maziere P, Grocock RJ, et al. Construction, visualisation, and clustering of transcription networks from microarray expression data. PLoS Comput Biol. 2007;3:2032–42. doi: 10.1371/journal.pcbi.0030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gehlenborg N, O'Donoghue SI, Baliga NS, Goesmann A, Hibbs MA, Kitano H, et al. Visualization of omics data for systems biology. Nat Methods. 2010;7(Suppl 3):S56–68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 58.Pavlopoulos GA, Wegener AL, Schneider R. A survey of visualization tools for biological network analysis. BioData Min. 2008;1:12. doi: 10.1186/1756-0381-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suderman M, Hallett M. Tools for visually exploring biological networks. Bioinformatics. 2007;23:2651–9. doi: 10.1093/bioinformatics/btm401. [DOI] [PubMed] [Google Scholar]
- 60.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, et al. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–76. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–39. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 62.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–22. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klein C, Marino A, Sagot MF, Vieira Milreu P, Brilli M. Structural and dynamical analysis of biological networks. Brief Funct Genomics. 2012;11:420–33. doi: 10.1093/bfgp/els030. [DOI] [PubMed] [Google Scholar]
- 64.Secrier M, Schneider R. PhenoTimer: software for the visual mapping of time-resolved phenotypic landscapes. PloS One. 2013;8:e72361. doi: 10.1371/journal.pone.0072361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Secrier M, Schneider R. Visualizing time-related data in biology, a review. Brief Bioinform. 2014;15:771–82. doi: 10.1093/bib/bbt021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nielsen CB, Cantor M, Dubchak I, Gordon D, Wang T. Visualizing genomes: techniques and challenges. Nat Methods. 2010;7(Suppl 3):S5–15. doi: 10.1038/nmeth.1422. [DOI] [PubMed] [Google Scholar]
- 67.Procter JB, Thompson J, Letunic I, Creevey C, Jossinet F, Barton GJ. Visualization of multiple alignments, phylogenies and gene family evolution. Nat Methods. 2010;7(Suppl 3):S16–25. doi: 10.1038/nmeth.1434. [DOI] [PubMed] [Google Scholar]
- 68.Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–7. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 69.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–53. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 70.Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–41. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 71.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 72.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 73.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 75.Daugelaite J, O' Driscoll A, Sleator R. An overview of multiple sequence alignments and cloud computing in bioinformatics. ISRN Biomathematics. 2013. doi:10.1155/2013/615630.
- 76.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 77.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–85. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 78.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 79.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 80.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–8. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 81.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Keravala A, Lee S, Thyagarajan B, Olivares EC, Gabrovsky VE, Woodard LE, et al. Mutational derivatives of PhiC31 integrase with increased efficiency and specificity. Mol Ther. 2009;17:112–20. doi: 10.1038/mt.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chiang DY, Getz G, Jaffe DB, O'Kelly MJ, Zhao X, Carter SL, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim TM, Luquette LJ, Xi R, Park PJ. rSW-seq: algorithm for detection of copy number alterations in deep sequencing data. BMC Bioinformatics. 2010;11:432. doi: 10.1186/1471-2105-11-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morin R, Bainbridge M, Fejes A, Hirst M, Krzywinski M, Pugh T, et al. Profiling the HeLa S3 transcriptome using randomly primed cDNA and massively parallel short-read sequencing. Biotechniques. 2008;45:81–94. doi: 10.2144/000112900. [DOI] [PubMed] [Google Scholar]
- 89.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 90.Hall N. Advanced sequencing technologies and their wider impact in microbiology. J Exp Biol. 2007;210:1518–25. doi: 10.1242/jeb.001370. [DOI] [PubMed] [Google Scholar]
- 91.Li H, Homer N. A survey of sequence alignment algorithms for next-generation sequencing. Brief Bioinform. 2010;11:473–83. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Karchin R. Next generation tools for the annotation of human SNPs. Brief Bioinform. 2009;10:35–52. doi: 10.1093/bib/bbn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods. 2009;6(Suppl 11):S13–20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 95.Buchanan CC, Torstenson ES, Bush WS, Ritchie MD. A comparison of cataloged variation between International HapMap Consortium and 1000 Genomes Project data. J Am Med Inform Assoc. 2012;19:289–94. doi: 10.1136/amiajnl-2011-000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tanaka T. [International HapMap project] Nihon Rinsho. 2005;63(Suppl 12):29–34. [PubMed] [Google Scholar]
- 97.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project web site. Genome Res. 2005;15:1592–3. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.International HapMap Consortium Integrating ethics and science in the International HapMap Project. Nat Rev Genet. 2004;5:467–75. doi: 10.1038/nrg1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 100.Bonfield JK, Smith K, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–9. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dear S, Staden R. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 1991;19:3907–11. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102.ArticlepublishedonlinebeforeprintinMay2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–5. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 105.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–7. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 109.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang J, Kong L, Gao G, Luo J. A brief introduction to web-based genome browsers. Brief Bioinform. 2013;14:131–43. doi: 10.1093/bib/bbs029. [DOI] [PubMed] [Google Scholar]
- 111.Pavlopoulos GA, Oulas A, Iacucci E, Sifrim A, Moreau Y, Schneider R, et al. Unraveling genomic variation from next generation sequencing data. BioData Min. 2013;6:13. doi: 10.1186/1756-0381-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Quackenbush J. Computational analysis of microarray data. Nat Rev Genet. 2001;2:418–27. doi: 10.1038/35076576. [DOI] [PubMed] [Google Scholar]
- 113.Mantione KJ, Kream RM, Kuzelova H, Ptacek R, Raboch J, Samuel JM, et al. Comparing bioinformatic gene expression profiling methods: microarray and RNA-Seq. Medical science monitor basic research. 2014;20:138–42. doi: 10.12659/MSMBR.892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.MAQC Consortium. Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–61. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 117.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–86. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.MacQueen J. Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Volume 1. Berkeley: University of California Press; 1967. Some methods for classification and analysis of multivariate observations; pp. 281–97. [Google Scholar]
- 120.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 121.Li Y, Xu L. Unweighted multiple group method with arithmetic mean. 2010 IEEE Fifth International Conference on Bio-Inspired Computing: Theories and Applications (BIC-TA). 2010;830–4.
- 122.Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–5. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 123.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–71. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Perriere G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–9. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- 125.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 127.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 128.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–8. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 129.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 130.Shamir R, Maron-Katz A, Tanay A, Linhart C, Steinfeld I, Sharan R, et al. EXPANDER--an integrative program suite for microarray data analysis. BMC Bioinformatics. 2005;6:232. doi: 10.1186/1471-2105-6-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 132.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fry B, Reas C. Processing. 2015. http://processing.org. Accessed 30 July 2015.
- 134.Fry B, Reas C, Resig, J. Processing.js. 2015. http://processingjs.org. Accessed 30 July 2015.
- 135.Bostock M. Data-Driven Documents. 2015. http://d3js.org/. Accessed 30 July 2015.
- 136.O'Donoghue SI, Gavin AC, Gehlenborg N, Goodsell DS, Heriche JK, Nielsen CB, et al. Visualizing biological data-now and in the future. Nat Methods. 2010;7(Suppl 3):S2–4. doi: 10.1038/nmeth.f.301. [DOI] [PubMed] [Google Scholar]
- 137.Thomas J, Cook KA. Illuminating the path: the research and development agenda for visual analytics. National Visualization and Analytics Center. 2005. http://vis.pnnl.gov/pdf/RD_Agenda_VisualAnalytics.pdf. Accessed 27 July 2015.
- 138.Karam M, Schraefel . Electronics and Computer Science. Southampton: University of Southampton; 2005. A taxonomy of gestures in human computer interactions; pp. 1–45. [Google Scholar]
- 139.Sabir K, Stolte C, Tabor B, O'Donoghue SI. The molecular control toolkit: controlling 3D molecular graphics via gesture and voice. IEEE Symposium on Biological Data Visualization (BioVis) 2013;2013:49–56. doi: 10.1109/BioVis.2013.6664346. [DOI] [Google Scholar]
- 140.Yang Y, Engin L, Wurtele ES, Cruz-Neira C, Dickerson JA. Integration of metabolic networks and gene expression in virtual reality. Bioinformatics. 2005;21:3645–50. doi: 10.1093/bioinformatics/bti581. [DOI] [PubMed] [Google Scholar]
- 141.Burrage K, Hood L, Ragan MA. Advanced computing for systems biology. Brief Bioinform. 2006;7:390–8. doi: 10.1093/bib/bbl033. [DOI] [PubMed] [Google Scholar]
- 142.McComb T, Cairncross O, Noske AB, Wood DL, Marsh BJ, Ragan MA. Illoura: a software tool for analysis, visualization and semantic querying of cellular and other spatial biological data. Bioinformatics. 2009;25:1208–10. doi: 10.1093/bioinformatics/btp125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Loew LM, Schaff JC. The Virtual Cell: a software environment for computational cell biology. Trends Biotechnol. 2001;19:401–6. doi: 10.1016/S0167-7799(01)01740-1. [DOI] [PubMed] [Google Scholar]
- 144.McClean P, Johnson C, Rogers R, Daniels L, Reber J, Slator BM, et al. Molecular and cellular biology animations: development and impact on student learning. Cell Biol Educ. 2005;4:169–79. doi: 10.1187/cbe.04-07-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kaufmann H. Collaborative Augmented Reality in Education. Imagina Conference 2003;TUW-137414.
- 146.Garcia-Garcia J, Guney E, Aragues R, Planas-Iglesias J, Oliva B. Biana: a software framework for compiling biological interactions and analyzing networks. BMC Bioinformatics. 2010;11:56. doi: 10.1186/1471-2105-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Theocharidis A, van Dongen S, Enright AJ, Freeman TC. Network visualization and analysis of gene expression data using BioLayout Express(3D) Nat Protoc. 2009;4:1535–50. doi: 10.1038/nprot.2009.177. [DOI] [PubMed] [Google Scholar]
- 148.Baitaluk M, Sedova M, Ray A, Gupta A. BiologicalNetworks: visualization and analysis tool for systems biology. Nucleic Acids Res. 2006;34:W466–71. doi: 10.1093/nar/gkl308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kozhenkov S, Sedova M, Dubinina Y, Gupta A, Ray A, Ponomarenko J, et al. BiologicalNetworks--tools enabling the integration of multi-scale data for the host-pathogen studies. BMC Syst Biol. 2011;5:7. doi: 10.1186/1752-0509-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sirava M, Schafer T, Eiglsperger M, Kaufmann M, Kohlbacher O, Bornberg-Bauer E, et al. BioMiner--modeling, analyzing, and visualizing biochemical pathways and networks. Bioinformatics. 2002;18(Suppl 2):S219–30. doi: 10.1093/bioinformatics/18.suppl_2.S219. [DOI] [PubMed] [Google Scholar]
- 151.Nagasaki M, Saito A, Jeong E, Li C, Kojima K, Ikeda E, et al. Cell Illustrator 4.0: a computational platform for systems biology. In Silico Biol. 2010;10:5–26. doi: 10.3233/ISB-2010-0415. [DOI] [PubMed] [Google Scholar]
- 152.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, et al. COPASI--a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–74. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 153.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ramsey S, Orrell D, Bolouri H. Dizzy: stochastic simulation of large-scale genetic regulatory networks. J Bioinform Comput Biol. 2005;3:415–36. doi: 10.1142/S0219720005001132. [DOI] [PubMed] [Google Scholar]
- 155.Kauffman J, Kittas A, Bennett L, Tsoka S. DyCoNet: a Gephi plugin for community detection in dynamic complex networks. PloS One. 2014;9:e101357. doi: 10.1371/journal.pone.0101357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Westenberg MA, van Hijum SAFT, Kuipers OP, Roerdink JBTM. Visualizing genome expression and regulatory network dynamics in genomic and metabolic context. Comput Graph Forum. 2008;27:887–94. doi: 10.1111/j.1467-8659.2008.01221.x. [DOI] [Google Scholar]
- 157.Baker C, Carpendale S, Prusinkiewicz P, Surette M. GeneVis: simulation and visualization of genetic networks. Information Visualization. 2003;2:201–17. doi: 10.1057/palgrave.ivs.9500055. [DOI] [Google Scholar]
- 158.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Syst. 2006;1695.
- 159.Hooper SD, Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21:4432–3. doi: 10.1093/bioinformatics/bti696. [DOI] [PubMed] [Google Scholar]
- 160.Pavlopoulos GA, Hooper SD, Sifrim A, Schneider R, Aerts J. Medusa: a tool for exploring and clustering biological networks. BMC Res Notes. 2011;4:384. doi: 10.1186/1756-0500-4-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Brown KR, Otasek D, Ali M, McGuffin MJ, Xie W, Devani B, et al. NAViGaTOR: network analysis. Visualization and Graphing Toronto. Bioinformatics. 2009;25:3327–9. doi: 10.1093/bioinformatics/btp595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Djebbari A, Ali M, Otasek D, Kotlyar M, Fortney K, Wong S, et al. NAViGaTOR: large scalable and interactive navigation and analysis of large graphs. Internet Math. 2011;7:314–47. doi: 10.1080/15427951.2011.604289. [DOI] [Google Scholar]
- 163.Kao HL, Gunsalus KC. Browsing multidimensional molecular networks with the generic network browser (N-Browse). Curr Protoc Bioinf. 2008: Chapter 9:Unit 9.11. [DOI] [PMC free article] [PubMed]
- 164.Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio--the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–7. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- 165.Orlev N, Shamir R, Shiloh Y. PIVOT: protein interacions visualizatiOn tool. Bioinformatics. 2004;20:424–5. doi: 10.1093/bioinformatics/btg426. [DOI] [PubMed] [Google Scholar]
- 166.Krumsiek J, Friedel CC, Zimmer R. ProCope--protein complex prediction and evaluation. Bioinformatics. 2008;24:2115–16. doi: 10.1093/bioinformatics/btn376. [DOI] [PubMed] [Google Scholar]
- 167.Iragne F, Nikolski M, Mathieu B, Auber D, Sherman D. ProViz: protein interaction visualization and exploration. Bioinformatics. 2005;21:272–4. doi: 10.1093/bioinformatics/bth494. [DOI] [PubMed] [Google Scholar]
- 168.Forman JJ, Clemons PA, Schreiber SL, Haggarty SJ. SpectralNET--an application for spectral graph analysis and visualization. BMC Bioinformatics. 2005;6:260. doi: 10.1186/1471-2105-6-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Auber D. A huge graph visualization framework. In: Mutzel P, Jünger M, editors. Graph Drawing Software (Mathematics and Visualization) Heidelberg: Springer; 2004. pp. 105–26. [Google Scholar]
- 170.Brinkrolf C, Janowski SJ, Kormeier B, Lewinski M, Hippe K, Borck D, et al. VANESA - a software application for the visualization and analysis of networks in system biology applications. J Integr Bioinform. 2014;11:239. doi: 10.2390/biecoll-jib-2014-239. [DOI] [PubMed] [Google Scholar]
- 171.Junker BH, Klukas C, Schreiber F. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics. 2006;7:109. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Prieto C, De Las Rivas J. APID: agile protein interaction DataAnalyzer. Nucleic Acids Res. 2006;34:W298–302. doi: 10.1093/nar/gkl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Villeger AC, Pettifer SR, Kell DB. Arcadia: a visualization tool for metabolic pathways. Bioinformatics. 2010;26:1470–1. doi: 10.1093/bioinformatics/btq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Berger SI, Iyengar R, Ma'ayan A. AVIS: AJAX viewer of interactive signaling networks. Bioinformatics. 2007;23:2803–5. doi: 10.1093/bioinformatics/btm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Myers CL, Robson D, Wible A, Hibbs MA, Chiriac C, Theesfeld CL, et al. Discovery of biological networks from diverse functional genomic data. Genome Biol. 2005;6:R114. doi: 10.1186/gb-2005-6-13-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Florez LA, Lammers CR, Michna R, Stulke J. Cell Publisher: a web platform for the intuitive visualization and sharing of metabolic, signalling and regulatory pathways. Bioinformatics. 2010;26:2997–9. doi: 10.1093/bioinformatics/btq585. [DOI] [PubMed] [Google Scholar]
- 177.Huttenhower C, Mehmood SO, Troyanskaya OG. Graphle: interactive exploration of large, dense graphs. BMC Bioinformatics. 2009;10:417. doi: 10.1186/1471-2105-10-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Reimand J, Tooming L, Peterson H, Adler P, Vilo J. GraphWeb: mining heterogeneous biological networks for gene modules with functional significance. Nucleic Acids Res. 2008;36:W452–9. doi: 10.1093/nar/gkn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lin CY, Chin CH, Wu HH, Chen SH, Ho CW, Ko MT. Hubba: hub objects analyzer--a framework of interactome hubs identification for network biology. Nucleic Acids Res. 2008;36:W438–43. doi: 10.1093/nar/gkn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kalaev M, Smoot M, Ideker T, Sharan R. NetworkBLAST: comparative analysis of protein networks. Bioinformatics. 2008;24:594–6. doi: 10.1093/bioinformatics/btm630. [DOI] [PubMed] [Google Scholar]
- 181.Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wu J, Vallenius T, Ovaska K, Westermarck J, Makela TP, Hautaniemi S. Integrated network analysis platform for protein-protein interactions. Nat Methods. 2009;6:75–7. doi: 10.1038/nmeth.1282. [DOI] [PubMed] [Google Scholar]
- 183.Pitkanen E, Akerlund A, Rantanen A, Jouhten P, Ukkonen E. ReMatch: a web-based tool to construct, store and share stoichiometric metabolic models with carbon maps for metabolic flux analysis. J Integr Bioinform. 2008;5. doi:10.2390/biecoll-jib-2008-102. [DOI] [PubMed]
- 184.Minguez P, Gotz S, Montaner D, Al-Shahrour F, Dopazo J. SNOW, a web-based tool for the statistical analysis of protein-protein interaction networks. Nucleic Acids Res. 2009;37:W109–14. doi: 10.1093/nar/gkp402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kuhn M, Szklarczyk D, Franceschini A, Campillos M, von Mering C, Jensen LJ, et al. STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Res. 2010;38:D552–6. doi: 10.1093/nar/gkp937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, et al. STRING 7--recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35:D358–62. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Curtis RE, Yuen A, Song L, Goyal A, Xing EP. TVNViewer: an interactive visualization tool for exploring networks that change over time or space. Bioinformatics. 2011;27:1880–1. doi: 10.1093/bioinformatics/btr273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yip KY, Yu H, Kim PM, Schultz M, Gerstein M. The tYNA platform for comparative interactomics: a web tool for managing, comparing and mining multiple networks. Bioinformatics. 2006;22:2968–70. doi: 10.1093/bioinformatics/btl488. [DOI] [PubMed] [Google Scholar]
- 189.Hu Z, Mellor J, Wu J, DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC Bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gerasch A, Faber D, Küntzer J, Niermann P, Kohlbacher O, Lenhof H-P, et al. BiNA: a visual analytics tool for biological network data. PloS One. 2014;9:e87397. doi: 10.1371/journal.pone.0087397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Longabaugh WJ, Davidson EH, Bolouri H. Computational representation of developmental genetic regulatory networks. Dev Biol. 2005;283:1–16. doi: 10.1016/j.ydbio.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 192.Streit M, Lex A, Kalkusch M, Zatloukal K, Schmalstieg D. Caleydo: connecting pathways and gene expression. Bioinformatics. 2009;25:2760–1. doi: 10.1093/bioinformatics/btp432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Funahashi A, Matsuoka Y, Akiya J, Morohashi M, Kikuchi N, Kitano H. Cell Designer 3.5: a versatile modeling tool for biochemical networks. Proc IEEE Inst Electr Electron Eng. 2008;96:1254–65. doi: 10.1109/JPROC.2008.925458. [DOI] [Google Scholar]
- 194.Sorokin A, Paliy K, Selkov A, Demin OV, Dronov S, Ghazal P, et al. The Pathway Editor: a tool for managing complex biological networks. IBM Journal of Research and Development. 2006;50:561–73. doi: 10.1147/rd.506.0561. [DOI] [Google Scholar]
- 195.Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, et al. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Sauro HM, Hucka M, Finney A, Wellock C, Bolouri H, Doyle J, et al. Next generation simulation tools: the Systems Biology Workbench and BioSPICE integration. OMICS. 2003;7:355–72. doi: 10.1089/153623103322637670. [DOI] [PubMed] [Google Scholar]
- 197.Tokimatsu T, Sakurai N, Suzuki H, Ohta H, Nishitani K, Koyama T, et al. KaPPA-view: a web-based analysis tool for integration of transcript and metabolite data on plant metabolic pathway maps. Plant Physiol. 2005;138:1289–300. doi: 10.1104/pp.105.060525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, et al. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36:W423–6. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Droste P, Nöh K, Wiechert W. Omix - a visualization tool for metabolic networks with highest usability and customizability in focus. Chemie Ingenieur Technik. 2013;85:849–62. doi: 10.1002/cite.201200234. [DOI] [Google Scholar]
- 200.Holford M, Li N, Nadkarni P, Zhao H. VitaPad: visualization tools for the analysis of pathway data. Bioinformatics. 2005;21:1596–602. doi: 10.1093/bioinformatics/bti153. [DOI] [PubMed] [Google Scholar]
- 201.Chung H-J, Kim M, Park CH, Kim J, Kim JH. ArrayXPath: mapping and visualizing microarray gene-expression data with integrated biological pathway resources using Scalable Vector Graphics. Nucleic Acids Res. 2004;32:W460–4. doi: 10.1093/nar/gkh476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Weniger M, Engelmann JC, Schultz J. Genome expression pathway analysis tool--analysis and visualization of microarray gene expression data under genomic, proteomic and metabolic context. BMC Bioinformatics. 2007;8:179. doi: 10.1186/1471-2105-8-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yamada T, Letunic I, Okuda S, Kanehisa M, Bork P. iPath2.0: interactive pathway explorer. Nucleic Acids Res. 2011;39:W412–15. doi: 10.1093/nar/gkr313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Arakawa K, Kono N, Yamada Y, Mori H, Tomita M. KEGG-based pathway visualization tool for complex omics data. In Silico Biol. 2005;5:419–23. [PubMed] [Google Scholar]
- 205.Xia J, Wishart DS. MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics. 2010;26:2342–4. doi: 10.1093/bioinformatics/btq418. [DOI] [PubMed] [Google Scholar]
- 206.Paley SM, Karp PD. The pathway tools cellular overview diagram and omics viewer. Nucleic Acids Res. 2006;34:3771–8. doi: 10.1093/nar/gkl334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mlecnik B, Scheideler M, Hackl H, Hartler J, Sanchez-Cabo F, Trajanoski Z. PathwayExplorer: web service for visualizing high-throughput expression data on biological pathways. Nucleic Acids Res. 2005;33:W633–7. doi: 10.1093/nar/gki391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kono N, Arakawa K, Ogawa R, Kido N, Oshita K, Ikegami K, et al. Pathway projector: web-based zoomable pathway browser using KEGG atlas and Google Maps API. PloS One. 2009;4:e7710. doi: 10.1371/journal.pone.0007710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Pico AR, Kelder T, van Iersel MP, Hanspers K, Conklin BR, Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008;6:e184. doi: 10.1371/journal.pbio.0060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Nielsen CB, Jackman SD, Birol I, Jones SJ. ABySS-Explorer: visualizing genome sequence assemblies. IEEE Trans Vis Comput Graph. 2009;15:881–8. doi: 10.1109/TVCG.2009.116. [DOI] [PubMed] [Google Scholar]
- 211.Carver T, Harris SR, Otto TD, Berriman M, Parkhill J, McQuillan JA. BamView: visualizing and interpretation of next-generation sequencing read alignments. Brief Bioinform. 2013;14:203–12. doi: 10.1093/bib/bbr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Liu C, Bonner TI, Nguyen T, Lyons JL, Christian SL, Gershon ES. DNannotator: annotation software tool kit for regional genomic sequences. Nucleic Acids Res. 2003;31:3729–35. doi: 10.1093/nar/gkg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yang Y, Liu J. JVM: Java Visual Mapping tool for next generation sequencing read. Adv Exp Med Biol. 2015;827:11–18. doi: 10.1007/978-94-017-9245-5_2. [DOI] [PubMed] [Google Scholar]
- 214.Manske HM, Kwiatkowski DP. LookSeq: a browser-based viewer for deep sequencing data. Genome Res. 2009;19:2125–32. doi: 10.1101/gr.093443.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hou H, Zhao F, Zhou L, Zhu E, Teng H, Li X, et al. MagicViewer: integrated solution for next-generation sequencing data visualization and genetic variation detection and annotation. Nucleic Acids Res. 2010;38:W732–6. doi: 10.1093/nar/gkq302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bao H, Guo H, Wang J, Zhou R, Lu X, Shi S. MapView: visualization of short reads alignment on a desktop computer. Bioinformatics. 2009;25:1554–5. doi: 10.1093/bioinformatics/btp255. [DOI] [PubMed] [Google Scholar]
- 217.Elnitski L, Riemer C, Burhans R, Hardison R, Miller W. MultiPipMaker: comparative alignment server for multiple DNA sequences. Curr Protoc Bioinf. 2005, Chapter 10: Unit 10.14. [DOI] [PubMed]
- 218.López-Fernández H, Glez-Peña D, Reboiro-Jato M, Gómez-López G, Pisano DG, Fdez-Riverola F. PileLineGUI: a desktop environment for handling genome position files in next-generation sequencing studies. Nucleic Acids Res. 2011;39:W562–6. doi: 10.1093/nar/gkr439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Pitt JN, Rajapakse I, Ferre-D'Amare AR. SEWAL: an open-source platform for next-generation sequence analysis and visualization. Nucleic Acids Res. 2010;38:7908–15. doi: 10.1093/nar/gkq661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wang T, Liu J, Shen L, Tonti-Filippini J, Zhu Y, Jia H, et al. STAR: an integrated solution to management and visualization of sequencing data. Bioinformatics. 2013;29:3204–10. doi: 10.1093/bioinformatics/btt558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ge D, Ruzzo EK, Shianna KV, He M, Pelak K, Heinzen EL, et al. SVA: software for annotating and visualizing sequenced human genomes. Bioinformatics. 2011;27:1998–2000. doi: 10.1093/bioinformatics/btr317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhang Z, Lin H, Ma B. ZOOM Lite: next-generation sequencing data mapping and visualization software. Nucleic Acids Res. 2010;38(Suppl 2):W743–8. doi: 10.1093/nar/gkq538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Salzberg SL, Church D, DiCuccio M, Yaschenko E, Ostell J. The genome Assembly Archive: a new public resource. PLoS Biol. 2004;2:E285. doi: 10.1371/journal.pbio.0020285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Li P, Ji G, Dong M, Schmidt E, Lenox D, Chen L, et al. CBrowse: a SAM/BAM-based contig browser for transcriptome assembly visualization and analysis. Bioinformatics. 2012;28:2382–4. doi: 10.1093/bioinformatics/bts443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tang B, Wang Q, Yang M, Xie F, Zhu Y, Zhuo Y, et al. ContigScape: a Cytoscape plugin facilitating microbial genome gap closing. BMC Genomics. 2013;14:289. doi: 10.1186/1471-2164-14-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Burland TG. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol. 2000;132:71–91. doi: 10.1385/1-59259-192-2:71. [DOI] [PubMed] [Google Scholar]
- 228.Huang W, Marth G. EagleView: a genome assembly viewer for next-generation sequencing technologies. Genome Res. 2008;18:1538–43. doi: 10.1101/gr.076067.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Schatz MC, Phillippy AM, Sommer DD, Delcher AL, Puiu D, Narzisi G, et al. Hawkeye and AMOS: visualizing and assessing the quality of genome assemblies. Brief Bioinform. 2013;14:213–24. doi: 10.1093/bib/bbr074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, et al. Tablet—next generation sequence assembly visualization. Bioinformatics. 2010;26:401–2. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kong L, Wang J, Zhao S, Gu X, Luo J, Gao G. ABrowse--a customizable next-generation genome browser framework. BMC Bioinformatics. 2012;13:2. doi: 10.1186/1471-2105-13-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Tonti-Filippini J. AnnoJ. http://www.annoj.org. Accessed: 27 July 2015.
- 233.Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–4. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Engels R, Yu T, Burge C, Mesirov JP, DeCaprio D, Galagan JE. Combo: a whole genome comparative browser. Bioinformatics. 2006;22:1782–3. doi: 10.1093/bioinformatics/btl193. [DOI] [PubMed] [Google Scholar]
- 235.Juan L, Liu Y, Wang Y, Teng M, Zang T, Wang Y. Family genome browser: visualizing genomes with pedigree information. Bioinformatics. 2015;31:2262–8. doi: 10.1093/bioinformatics/btv151. [DOI] [PubMed] [Google Scholar]
- 236.Shannon PT, Reiss DJ, Bonneau R, Baliga NS. The Gaggle: an open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Papanicolaou A, Heckel DG. The GMOD Drupal bioinformatic server framework. Bioinformatics. 2010;26:3119–24. doi: 10.1093/bioinformatics/btq599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wang H, Su Y, Mackey AJ, Kraemer ET, Kissinger JC. SynView: a GBrowse-compatible approach to visualizing comparative genome data. Bioinformatics. 2006;22:2308–9. doi: 10.1093/bioinformatics/btl389. [DOI] [PubMed] [Google Scholar]
- 239.Sato N, Ehira S. GenoMap, a circular genome data viewer. Bioinformatics. 2003;19:1583–4. doi: 10.1093/bioinformatics/btg195. [DOI] [PubMed] [Google Scholar]
- 240.Arakawa K, Tamaki S, Kono N, Kido N, Ikegami K, Ogawa R, et al. Genome Projector: zoomable genome map with multiple views. BMC Bioinformatics. 2009;10:31. doi: 10.1186/1471-2105-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Abeel T, Van Parys T, Saeys Y, Galagan J, Van de Peer Y. GenomeView: a next-generation genome browser. Nucleic Acids Res. 2012;40:e12. doi: 10.1093/nar/gkr995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lajugie J, Bouhassira EE. GenPlay, a multipurpose genome analyzer and browser. Bioinformatics. 2011;27:1889–93. doi: 10.1093/bioinformatics/btr309. [DOI] [PubMed] [Google Scholar]
- 243.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The integrated genome browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–1. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Skinner ME, Uzilov AV, Stein LD, Mungall CJ, Holmes IH. JBrowse: a next-generation genome browser. Genome Res. 2009;19:1630–8. doi: 10.1101/gr.094607.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–86. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Fiume M, Williams V, Brudno M. Savant: genome browser for high throughput sequencing data. Bioinformatics. 2010;26:1938–44. doi: 10.1093/bioinformatics/btq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Miller CA, Anthony J, Meyer MM, Marth G. Scribl: an HTML5 Canvas-based graphics library for visualizing genomic data over the web. Bioinformatics. 2013;29:381–3. doi: 10.1093/bioinformatics/bts677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Axelrod N, Lin Y, Ng PC, Stockwell TB, Crabtree J, Huang J, et al. The HuRef Browser: a web resource for individual human genomics. Nucleic Acids Res. 2009;37(Suppl 1):D1018–24. doi: 10.1093/nar/gkn939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Juan L, Teng M, Zang T, Hao Y, Wang Z, Yan C, et al. The personal genome browser: visualizing functions of genetic variants. Nucleic Acids Res. 2014;42:W192–7. doi: 10.1093/nar/gku361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Zhu J, Sanborn JZ, Benz S, Szeto C, Hsu F, Kuhn RM, et al. The UCSC cancer genomics browser. Nat Methods. 2009;6:239–40. doi: 10.1038/nmeth0409-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Goldman M, Craft B, Swatloski T, Cline M, Morozova O, Diekhans M, et al. The UCSC cancer genomics browser: update 2015. Nucleic Acids Res. 2015;43:D812–17. doi: 10.1093/nar/gku1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Saito TL, Yoshimura J, Sasaki S, Ahsan B, Sasaki A, Kuroshu R, et al. UTGB toolkit for personalized genome browsers. Bioinformatics. 2009;25:1856–61. doi: 10.1093/bioinformatics/btp350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Yates T, Okoniewski MJ, Miller CJ. X:Map: annotation and visualization of genome structure for Affymetrix exon array analysis. Nucleic Acids Res. 2008;36:D780–6. doi: 10.1093/nar/gkm779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–6. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sinha AU, Meller J. Cinteny: flexible analysis and visualization of synteny and genome rearrangements in multiple organisms. BMC Bioinformatics. 2007;8:82. doi: 10.1186/1471-2105-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Youens-Clark K, Faga B, Yap IV, Stein L, Ware D. CMap 1.01: a comparative mapping application for the internet. Bioinformatics. 2009;25:3040–2. doi: 10.1093/bioinformatics/btp458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lyons E, Pedersen B, Kane J, Alam M, Ming R, Tang H, et al. Finding and comparing syntenic regions among Arabidopsis and the outgroups papaya, poplar, and grape: CoGe with rosids. Plant Physiol. 2008;148:1772–81. doi: 10.1104/pp.108.124867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Deng X, Rayner S, Liu X, Zhang Q, Yang Y, Li N. DHPC: a new tool to express genome structural features. Genomics. 2008;91:476–83. doi: 10.1016/j.ygeno.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 260.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–20. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Zeinaly M, Soltangheis M, Shaw CD. FilooT: a visualization tool for exploring genomic data. SPIE 9017, Visualization and Data Analysis 2014. doi:10.1117/12.2042589.
- 262.McKay SJ, Vergara IA, Stajich JE. Using the Generic Synteny Browser (GBrowse_syn). Curr Protoc Bioinf. 2010, Chapter 9:Unit 9.12. [DOI] [PMC free article] [PubMed]
- 263.Yang J, Wang J, Yao ZJ, Jin Q, Shen Y, Chen R. GenomeComp: a visualization tool for microbial genome comparison. J Microbiol Methods. 2003;54:423–6. doi: 10.1016/S0167-7012(03)00094-0. [DOI] [PubMed] [Google Scholar]
- 264.Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008;9:376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lajugie J, Fourel N, Bouhassira EE. GenPlay Multi-Genome, a tool to compare and analyze multiple human genomes in a graphical interface. Bioinformatics. 2015;31:109–11. doi: 10.1093/bioinformatics/btu588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Yin T, Cook D, Lawrence M. ggbio: an R package for extending the grammar of graphics for genomic data. Genome Biol. 2012;13:R77. doi: 10.1186/gb-2012-13-8-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T, et al. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 2008;36:D947–53. doi: 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ware DH, Jaiswal P, Ni J, Yap IV, Pan X, Clark KY, et al. Gramene, a tool for grass genomics. Plant Physiol. 2002;130:1606–13. doi: 10.1104/pp.015248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Anders S. Visualization of genomic data with the Hilbert curve. Bioinformatics. 2009;25:1231–5. doi: 10.1093/bioinformatics/btp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Qi J, Zhao F. inGAP-sv: a novel scheme to identify and visualize structural variation from paired end mapping data. Nucleic Acids Res. 2011;39:W567–75. doi: 10.1093/nar/gkr506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Pavlopoulos GA, Kumar P, Sifrim A, Sakai R, Lin ML, Voet T, et al. Meander: visually exploring the structural variome using space-filling curves. Nucleic Acids Res. 2013;41:e118. doi: 10.1093/nar/gkt254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Broad Institute: MEDEA: Comparative Genomic Visualization with Adobe Flash. http://www.broadinstitute.org/annotation/medea/ (2015). Accessed 27 July 2015.
- 273.Meyer M, Munzner T, Pfister H. MizBee: a multiscale synteny browser. IEEE Trans Vis Comput Graph. 2009;15:897–904. doi: 10.1109/TVCG.2009.167. [DOI] [PubMed] [Google Scholar]
- 274.Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–98. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Shen L, Shao N, Liu X, Nestler E. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Dehal PS, Boore JL. A phylogenomic gene cluster resource: the Phylogenetically Inferred Groups (PhIGs) database. BMC Bioinformatics. 2006;7:201. doi: 10.1186/1471-2105-7-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Fong C, Rohmer L, Radey M, Wasnick M, Brittnacher MJ. PSAT: a web tool to compare genomic neighborhoods of multiple prokaryotic genomes. BMC Bioinformatics. 2008;9:170. doi: 10.1186/1471-2105-9-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Esteban-Marcos A, Darling AE, Ragan MA. Seevolution: visualizing chromosome evolution. Bioinformatics. 2009;25:960–1. doi: 10.1093/bioinformatics/btp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Crabtree J, Angiuoli SV, Wortman JR, White OR. Sybil: methods and software for multiple genome comparison and visualization. Methods Mol Biol. 2007;408:93–108. doi: 10.1007/978-1-59745-547-3_6. [DOI] [PubMed] [Google Scholar]
- 280.Asmann YW, Middha S, Hossain A, Baheti S, Li Y, Chai HS, et al. TREAT: a bioinformatics tool for variant annotations and visualizations in targeted and exome sequencing data. Bioinformatics. 2012;28:277–8. doi: 10.1093/bioinformatics/btr612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Miller W, Rosenbloom K, Hardison RC, Hou M, Taylor J, Raney B, et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Huang PJ, Lee CC, Tan BC, Yeh YM, Huang KY, Gan RC, et al. Vanno: a visualization-aided variant annotation tool. Hum Mutat. 2015;36:167–74. doi: 10.1002/humu.22684. [DOI] [PubMed] [Google Scholar]
- 283.Ferstay JA, Nielsen CB, Munzner T. Variant view: visualizing sequence variants in their gene context. IEEE Trans Vis Comput Graph. 2013;19:2546–55. doi: 10.1109/TVCG.2013.214. [DOI] [PubMed] [Google Scholar]
- 284.Nordberg H, Cantor M, Dusheyko S, Hua S, Poliakov A, Shabalov I, et al. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014;42:D26–31. doi: 10.1093/nar/gkt1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40:D26–32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Talevich E, Invergo BM, Cock PJ, Chapman BA. Bio. Phylo: a unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython. BMC Bioinformatics. 2012;13:209. doi: 10.1186/1471-2105-13-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Huerta-Cepas J, Dopazo J, Gabaldon T. ETE: a python environment for tree exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Zhang H, Gao S, Lercher MJ, Hu S, Chen WH. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012;40:W569–72. doi: 10.1093/nar/gks576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Smits SA, Ouverney CC. jsPhyloSVG: a javascript library for visualizing interactive and vector-based phylogenetic trees on the web. PloS One. 2010;5:e12267. doi: 10.1371/journal.pone.0012267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Sanderson MJ. Paloverde: an OpenGL 3D phylogeny browser. Bioinformatics. 2006;22:1004–6. doi: 10.1093/bioinformatics/btl044. [DOI] [PubMed] [Google Scholar]
- 291.Choi JH, Jung HY, Kim HS, Cho HG. PhyloDraw: a phylogenetic tree drawing system. Bioinformatics. 2000;16:1056–8. doi: 10.1093/bioinformatics/16.11.1056. [DOI] [PubMed] [Google Scholar]
- 292.Ranwez V, Clairon N, Delsuc F, Pourali S, Auberval N, Diser S, et al. PhyloExplorer: a web server to validate, explore and query phylogenetic trees. BMC Evol Biol. 2009;9:108. doi: 10.1186/1471-2148-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Jordan GE, Piel WH. PhyloWidget: web-based visualizations for the tree of life. Bioinformatics. 2008;24:1641–2. doi: 10.1093/bioinformatics/btn235. [DOI] [PubMed] [Google Scholar]
- 294.Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Stover BC, Muller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Gu S, Anderson I, Kunin V, Cipriano M, Minovitsky S, Weber G, et al. TreeQ-VISTA: an interactive tree visualization tool with functional annotation query capabilities. Bioinformatics. 2007;23:764–6. doi: 10.1093/bioinformatics/btl643. [DOI] [PubMed] [Google Scholar]
- 297.Pethica R, Barker G, Kovacs T, Gough J. TreeVector: scalable, interactive, phylogenetic trees for the web. PloS One. 2010;5:e8934. doi: 10.1371/journal.pone.0008934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Santamaria R, Theron R. Treevolution: visual analysis of phylogenetic trees. Bioinformatics. 2009;25:1970–1. doi: 10.1093/bioinformatics/btp333. [DOI] [PubMed] [Google Scholar]
- 299.Boc A, Diallo AB, Makarenkov V. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012;40:W573–9. doi: 10.1093/nar/gks485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Bremm S, von Landesberger T, Hess M, Schreck T, Weil P, Hamacherk K. Interactive visual comparison of multiple trees. 2011 IEEE Conference on Visual Analytics Science and Technology (VAST). 2011. doi:10.1109/VAST.2011.6102439.
- 301.Santamaria R, Theron R, Quintales L. BicOverlapper: a tool for bicluster visualization. Bioinformatics. 2008;24:1212–13. doi: 10.1093/bioinformatics/btn076. [DOI] [PubMed] [Google Scholar]
- 302.Santamaria R, Theron R, Quintales L. BicOverlapper 2.0: visual analysis for gene expression. Bioinformatics. 2014;30:1785–6. doi: 10.1093/bioinformatics/btu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Goncalves JP, Madeira SC, Oliveira AL. BiGGEsTS: integrated environment for biclustering analysis of time series gene expression data. BMC research notes. 2009;2:124. doi: 10.1186/1756-0500-2-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Yuan T, Huang X, Dittmar RL, Du M, Kohli M, Boardman L, et al. eRNA: a graphic user interface-based tool optimized for large data analysis from high-throughput RNA sequencing. BMC Genomics. 2014;15:176. doi: 10.1186/1471-2164-15-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kapushesky M, Kemmeren P, Culhane AC, Durinck S, Ihmels J, Korner C, et al. Expression Profiler: next generation--an online platform for analysis of microarray data. Nucleic Acids Res. 2004;32:W465–70. doi: 10.1093/nar/gkh470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Hibbs MA, Dirksen NC, Li K, Troyanskaya OG. Visualization methods for statistical analysis of microarray clusters. BMC Bioinformatics. 2005;6:115. doi: 10.1186/1471-2105-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Floratos A, Smith K, Ji Z, Watkinson J, Califano A. geWorkbench: an open source platform for integrative genomics. Bioinformatics. 2010;26:1779–80. doi: 10.1093/bioinformatics/btq282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Perez-Llamas C, Lopez-Bigas N. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PloS One. 2011;6:e19541. doi: 10.1371/journal.pone.0019541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Jinwook S, Shneiderman B. Interactively exploring hierarchical clustering results [gene identification] Computer. 2002;35:80–6. doi: 10.1109/MC.2002.1016905. [DOI] [Google Scholar]
- 310.Khomtchouk BB, Van Booven DJ, Wahlestedt C. HeatmapGenerator: high performance RNAseq and microarray visualization software suite to examine differential gene expression levels using an R and C++ hybrid computational pipeline. Source Code Biol Med. 2014;9:30. doi: 10.1186/s13029-014-0030-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Yachdav G, Hecht M, Pasmanik-Chor M, Yeheskel A, Rost B. HeatMapViewer: interactive display of 2D data in biology. F1000Research. 2014;3:48. doi: 10.12688/f1000research.3-48.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Dietzsch J, Gehlenborg N, Nieselt K. Mayday--a microarray data analysis workbench. Bioinformatics. 2006;22:1010–12. doi: 10.1093/bioinformatics/btl070. [DOI] [PubMed] [Google Scholar]
- 313.Weber GH, Rubel O, Huang MY, DePace AH, Fowlkes CC, Keranen SV, et al. Visual exploration of three-dimensional gene expression using physical views and linked abstract views. IEEE/ACM Trans Comput Biol Bioinform. 2009;6:296–309. doi: 10.1109/TCBB.2007.70249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.An J, Lai J, Wood DL, Sajjanhar A, Wang C, Tevz G, et al. RNASeqBrowser: a genome browser for simultaneous visualization of raw strand specific RNAseq reads and UCSC genome browser custom tracks. BMC Genomics. 2015;16:145. doi: 10.1186/s12864-015-1346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Roge X, Zhang X. RNAseqViewer: visualization tool for RNA-Seq data. Bioinformatics. 2014;30:891–2. doi: 10.1093/bioinformatics/btt649. [DOI] [PubMed] [Google Scholar]
- 316.Hochheiser H, Baehrecke EH, Mount SM, Shneiderman B. Dynamic querying for pattern identification in microarray and genomic data. International Conference on Multimedia and Expo 2003, ICME '03. 2003. doi:10.1109/ICME.2003.1221346.
- 317.Dietrich S, Wiegand S, Liesegang H. TraV: a genome context sensitive transcriptome browser. PloS One. 2014;9:e93677. doi: 10.1371/journal.pone.0093677. [DOI] [PMC free article] [PubMed] [Google Scholar]


