Abstract
The mammalian intestine is colonized by beneficial commensal bacteria and is a site of infection by pathogens, including helminth parasites. Helminths induce potent immuno-modulatory effects, but whether these effects are mediated by direct regulation of host immunity or indirectly through eliciting changes in the microbiota is unknown. We tested this in the context of virus-helminth co-infection. Helminth co-infection resulted in impaired antiviral immunity and was associated with changes in the microbiota and STAT6-dependent helminth-induced alternative activation of macrophages. Notably, helminth-induced impairment of antiviral immunity was evident in germ-free mice but neutralization of Ym1, a chitinase-like molecule that is associated with alternatively-activated macrophages, could partially restore antiviral immunity. These data indicate that helminth-induced immuno-modulation occurs independently of changes in the microbiota but is dependent on Ym1.
The microbiota of the mammalian gastrointestinal (GI) tract is composed of trillions of beneficial commensal bacteria that promote nutrient metabolism and regulate multiple physiological processes (1, 2). The GI tract is also a common site of infection by pathogenic viruses, bacteria, protozoa and helminths. The dynamic cross-regulation that exists between the host, the microbiota and enteric pathogens regulates intestinal homeostasis (3-9), indicating that these are highly co-evolved relationships. Signals derived from commensal bacteria and helminth parasites can influence the mammalian immune response (1, 2, 10-12), and helminth infection can elicit alterations in the composition of commensal bacteria that have been associated with limiting inflammation in multiple tissues (13-16). However, despite speculation regarding whether helminth-induced immuno-modulation is mediated through direct effects on the mammalian immune system or indirectly via changes in the microbiota (14, 17, 18), this fundamental question has not been addressed. Given the impact of helminth-elicited immuno-modulation both as a risk factor for bacterial, viral and protozoan co-infection (19-22) and as a potential therapeutic strategy for multiple inflammatory diseases including asthma, multiple sclerosis and inflammatory bowel disease (23, 24), it is critical to define the regulatory mechanisms by which helminth parasites can influence innate and adaptive immunity.
To test whether helminth infection elicits immuno-modulation through direct effects on the mammalian immune system or via alterations in the microbiota, we developed a model of enteric co-infection employing the helminth parasite Trichinella spiralis (Ts), which inhabits the small intestine for approximately 2-3 weeks before progressing to a persistent extra-intestinal phase, and a murine norovirus that acutely infects the ileum (MNV CW3). As expected, Trichinella infection induced type 2 immune responses (fig. S1). In the presence of the microbiota, Trichinella-MNV co-infection (fig. S2A), was associated with decreased frequencies and numbers of MNV-specific CD8+ T cells within the small intestine, Peyer's patches and spleen compared to mono-infected controls (Fig. 1A and B, fig. S2B and C). Further, MNV-specific CD8+ T cells isolated from helminth co-infected mice exhibited diminished polyfunctional effector function as compared to mono-infected controls (Fig. 1C). Analysis of MNV-specific CD4+ T cells using MHCII IAb tetramers specific for an epitope within the MNV capsid (P1496) (table S1), identified that helminth co-infected mice also had fewer antigen-specific CD4+ T cells (fig. S2D) and impaired accumulation of virus-specific CD4+ T cells expressing both interferon (IFN)γ and tumor necrosis factor (TNF)α compared to mono-infected controls (Fig. 1D). Notably, the defective T cell responses were associated with elevated viral loads in intestinal tissue of Trichinella/MNV CW3-infected mice compared to mono-infected controls (Fig. 1E).
Fig. 1. Helminth co-infection impairs immunity to enteric viral infection.
C57BL/6 mice were either left naïve or infected with 500 Trichinella spiralis (Ts) larvae per os (po). At day 12 post-Trichinella infection, mice were infected with 106 pfu MNV CW3 (po) and sacrificed at day 8 post-CW3 infection. (A to B) Flow cytometric detection of antigen-specific MNV CW3 P1519Y tetramer+ CD8+ T cells in naïve, Trichinella-infected (Ts), CW3-infected or Ts/CW3 co-infected mice in the (A) small intestine intraepithelial lymphocytes (IEL) and (B) spleen. Numbers in flow plots represent mean frequencies ± s.e.m. of CW3 Kb-P1519Y tetramer+ cells within the CD8+ lymphocyte gate of each group. (C) Splenocytes from CW3 and Ts/CW3 mice were stimulated with MNV CW3 P1519Y peptide and assayed for production of IFNγ, TNFα, CCL3, CD107a and Granzyme B. Pie charts show fractions of peptide-responsive CD8+ T cells producing any combination of these effector molecules. (D) Splenocytes from mono- and co-infected mice were stimulated with a pool of MHCII IAb-restricted MNV-specific peptides and assayed for production of IFNγ and TNFα. Numbers in flow plots represent mean frequencies ± s.e.m. of dual producing IFNγ+ TNFα+ cells within the CD4+ CD44hi lymphocyte gate of each group. (E) MNV genome copies in ileal tissue were quantified by RT-PCR. All data are representative of 2-4 independent experiments with a minimum of 3-5 mice per group. Statistics compare mono-infected versus co-infected groups using the Student's t-test. * P < 0.05. LoD: limit of detection.
We generated a recombinant gp33-expressing strain of MNV CW3 (CW3gp33) to track the activation and early proliferation of MNV-specific CD8+ T cells in vivo. Adoptively transferred congenic gp33-specific P14 CD8+ T cells accumulated and exhibited polyfunctional effector function in the intestine and spleen following MNV infection (fig. S3). Spatial and kinetic analysis of gp33-specific P14 CD8+ T cell activation and proliferation revealed that helminth co-infection resulted in delayed virus-specific CD8+ T cell proliferation and activation (Fig. 2A), as well as reduced or delayed accumulation of virus-specific T cells in the draining mesenteric lymph nodes (MLN) and spleen (Fig. 2B). Similar to oral infection, systemic viral infection stimulated robust antiviral P14 CD8+ T cell proliferation in mono-infected mice, which was dampened in helminth co-infected mice (fig. S4), suggesting that helminth infection can impair systemic antiviral immune responses to MNV independent of inflammatory or tissue repair processes operating in the intestine.
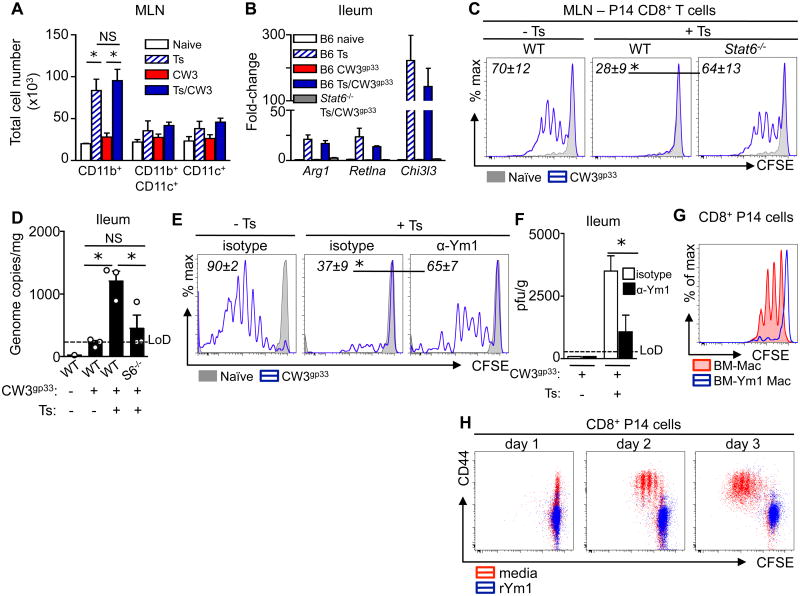
Fig. 2. Impaired virus-specific CD8+ T cell activation and proliferation in the presence of helminth co-infection.
(A) CFSE dilution and CD69 expression of adoptively transferred gp33-specific P14 CD8+ T cells isolated from the MLN and spleen of MNV CW3gp33 mono- and Ts/MNV CW3gp33 co-infected mice. Red, mono-infected; blue, co-infected. Numbers in flow plots represent frequencies of cells in divisions 0, 1-5 or 6+ (right to left). (B) Quantification of total P14 cells in the MLN (top row) and spleen (bottom row) in mono- and co-infected mice. (C to E) C57BL/6 mice were infected with 500 Ts larvae po. At day 12 post-Ts infection, mice were infected with 105 TCID50 influenza X31gp33 intranasally (in) and sacrificed at day 7 post-X31 infection. (C) Lung lymphocytes from influenza X31gp33 mono-infected (X31) and co-infected (Ts/X31) mice were stimulated with gp33 peptide and assayed for production of IFNγ. Total gp33-specific IFNγ+ CD8+ T cells in the lung. (D) Frequency of gp33-specific CD8+ T cells in the lungs. Numbers in flow plots represent mean frequencies ± s.e.m. of Kb-gp33 tetramer+ cells within the CD8+ lymphocyte gate of each group. (E) Quantification of Influenza PA gene in lung tissue, normalized to Hprt1. (F to H) C57BL/6 mice were infected with 200 Heligmosomoides polygyrus bakeri (Hp) larvae po. At day 12 post-Hp infection, mice were infected with 106 pfu MNV CW3 po and sacrificed at day 7 post-CW3 infection. (F) Total splenic Kb-P1519Y tetramer+ CD8+ T cells from CW3 mono-infected and Hp/CW3 co-infected mice. (G) Splenocytes were stimulated with P1519Y peptide and assayed for production of IFNγ and TNFα. Numbers in flow plots represent mean frequencies ± s.e.m. of dual producing IFNγ+ TNFα+ cells within the CD8+ CD44hi lymphocyte gate of each group. (H) MNV genome copies in ileal tissue were quantified by RT-PCR. All data are representative of 2 independent experiments with a minimum of 3-5 mice per group. Graphs represent means ± s.e.m. Statistics compare mono-infected versus co-infected groups using a two-way ANOVA with Bonferroni's post-testing (B, C and E) or the Student's t-test (D, F to H). * P < 0.05. MLN, mesenteric lymph node.
The immuno-modulatory effects of Trichinella infection on antiviral immunity were long-lived (fig. S5), were able to influence established infection with MNV CR6, a related strain that persists in the colon of immune-competent mice (fig. S6) and were evident in the lung following respiratory influenza infection (Fig. 2C to E), indicating that helminth-elicited immuno-modulation is operational at extra-intestinal tissues and can influence immunity to multiple viral pathogens. Further, the immuno-modulatory effects of helminth infection on antiviral immunity were not restricted to Trichinella and were also evident following infection with Heligmosomoides polygyrus bakeri (Hp) (Fig. 2F to H).
Despite ongoing clinical trials testing the potential efficacy of helminth immunotherapy to treat inflammatory diseases and the detrimental effects of helminth co-infection on protective immune responses to other human pathogens (19, 21, 23, 24), the mechanisms underlying helminth-elicited immuno-modulation remain poorly understood. Helminth infections can induce shifts in the composition of commensal bacterial communities and these alterations have been proposed to play a role in modulating the host immune response (13-18). Further, commensal bacteria-derived signals can modulate lymphocyte responses to infection (3, 4, 9), including antiviral immunity (25-27). To test whether Trichinella infection altered the composition of the intestinal microbiota, we performed sequencing and phylogenetic analysis of bacterial 16S rRNA genes of small intestine and colon luminal contents. These analyses revealed alterations in commensal bacterial communities in both the small and large intestine following Trichinella infection (Fig. 3A to D). Notable changes included a reduction in the relative abundance of the Turicibacteraceae family and an increase in the relative abundance of Lactobacillacaeae in the small intestine (Fig. 3C) and increased colonic representation of Clostridiales (Fig. 3D).
Fig. 3. Helminth-induced immuno-modulation of antiviral immunity is independent of changes in the microbiota.
(A to D) Luminal contents of the small and large intestine isolated from naïve or day 12 Trichinella (Ts)-infected C57/BL6 mice were subjected to 16S sequencing using Illumina MiSeq. (A, B) Principal coordinate analysis of bacterial populations in the small intestine (A) and large intestine (B) of naïve and Ts-infected mice. Axes represent weighted UniFrac distances between naïve and Trichinella-infected groups, and are annotated with the percentage of total variation explained. Statistical analyses were performed on groups of 4 mice, using PERMANOVA to test for differences in community composition according to weighted UniFrac centroid position. A, P = 0.034; B, P = 0.037. (C, D) Family level phylogenetic analysis of bacterial sequences in the small (C) and large (D) intestine of naïve and Ts-infected mice. Legend on right: bold, italicized taxa exhibited differences in relative abundance between naïve and Ts-infected mice. (E to G) Naïve and Trichinella (Ts)-infected conventional (CNV) or germ-free (GF) C57BL/6 mice were infected with MNV CW3 and sacrificed at day 7 post-CW3. (E) MNV CW3 P1519Y-specific CD8+ T cells in the Peyer's patches of mono- and co-infected CNV and GF mice. (F) Frequency of MNV CW3 P1519Y-specific CD8+ T cells in the spleens of mono- and co-infected CNV and GF mice. Numbers in flow plots represent mean frequencies ± s.e.m. of CW3 Kb-P1519Y tetramer+ cells within the CD8+ lymphocyte gate of each group. (G) MNV genome copies in ileal tissue were quantified by RT-PCR. Data in (E to G) are representative of 3 independent experiments with a minimum of 3-5 mice per group. Statistical analyses in (E to G) were performed using two-way ANOVA with Bonferroni's post-testing. * P < 0.05. LoD, limit of detection. NS, not significant.
To test whether these alterations contributed to helminth-induced immuno-modulation, we compared the influence of helminth co-infection on antiviral immunity in the presence or absence of the microbiota using germ-free (GF) mice. Establishment of Trichinella in the intestine and peripheral tissues and induction of helminth-induced TH2 cell responses were similar in conventional (CNV) and GF mice (fig. S7). The diminished antiviral responses seen in helminth co-infected mice were evident in both CNV and GF mice (Fig. 3E to G). These data indicate that helminth-elicited immuno-modulation of antiviral immunity can occur independently of changes in the microbiota and suggest that co-evolution may have resulted in mechanisms that allow helminths to directly regulate host immune responses.
Helminth infection elicits the induction of type 2 cytokines and is associated with STAT6-dependent alternative activation of macrophages (AAMacs), but these responses are not typically associated with protective antiviral immune responses. Therefore, we tested whether STAT6-dependent differentiation of AAMacs may underlie the dysregulated antiviral immune response in the context of helminth co-infection. Ex vivo analysis revealed a significant population expansion of macrophages (CD11b+ CD11c- cells) in the MLN of Trichinella-infected mice that was unaltered by MNV infection (Fig. 4A). Up-regulation of known AAMac-associated genes, including Arginase-1 (Arg1), RELMα (Retnla) and Ym1 (Chi3l3) was detected in the ileum of WT but not STAT6- or IL-4Rα-deficient mice (Fig. 4B, fig. S8A), indicating that AAMac responses were evident in co-infected mice at the site of MNV CW3 infection. Critically, deletion of STAT6 was sufficient to restore MNV CW3gp33-induced CD8+ T cell proliferation (Fig. 4C) and mice lacking STAT6 or IL-4Rα exhibited reduced viral loads compared to co-infected WT controls following MNV infection (Fig. 4D, fig. S8B). Further, delivery of IL-4/anti-IL-4 complexes was sufficient to induce expression of AAMac-associated genes and impair antiviral immunity in vivo compared to PBS treated mice (fig. S9). Collectively, these data suggest that AAMacs may limit the induction of host-protective antiviral immune responses in a type 2 cytokine- and STAT6-dependent manner.
Fig. 4. Helminth-induced STAT6-dependent alternatively-activated macrophages and Ym1 are associated with diminished virus-specific CD8+ T cell responses.
(A) C57BL/6 mice were either left naive, mono-infected (Ts or CW3) or co-infected (Ts/CW3). MLN antigen presenting cell subsets were quantified ex vivo at day 15 post-Ts or day 3 post-CW3. All cells gated on live lineage-negative (CD3-CD19-NK1.1-Ly6G-CD49b-Siglec-F-) cells. Macrophages (CD11b+CD11c-); myeloid DCs (CD11b+CD11c+MHCIIhi); conventional DCs (CD11b-CD11c+MHCII+). (B to D) Naïve and Ts infected C57BL/6 (WT) and Stat6-/- mice received an adoptive transfer of P14 cells iv (day 11 post-Ts), were infected with MNV CW3gp33 (day 12 post-Ts), and sacrificed at day 3 post-CW3gp33 infection. (B) AAMac signature genes Arg1, Reltna, Chi3l3 in the ileum. (C) CFSE dilution of P14 cells in the MLN. (D) MNV genome copies in ileal tissue. (E and F) Naïve and Ts-infected mice received daily treatments of 300 μg isotype or anti-Ym1 mAb starting at day 11 post-Ts infection and were MNV CW3gp33 infected at day 12 post-Ts infection. (E) CFSE dilution of adoptively transferred P14 CD8+ T cells in the MLN at day 3 post-CW3gp33 infection. (F) Viral load in the ileum as determined by plaque assay at day 7 post-CW3gp33 infection. (G) BM-Mac were infected with CW3gp33 and cultured with CFSE-labeled P14 CD8+ T cells in the presence (blue trace) or absence (red fill) of rYm1 for 3 days. (H) Purified CFSE-labeled P14 CD8+ T cells were stimulated with plate-bound αCD3 and soluble αCD28 in the presence (red) or absence (blue) of rYm1. Data in A to F are representative of 1-3 independent experiments with a minimum of 3-5 mice per group. Graphs represent means ± s.e.m. C and E: Grey solid histogram, naïve mice; blue trace, CW3gp33-infected. Numbers in flow plots represent mean frequencies ± s.e.m. of CFSE-dividing P14 cells of each group. Data in G and H are representative of at least 2 independent experiments with technical replicates. Statistical analysis was performed using two way ANOVA with Bonferroni post-testing (A, F), Student's t-test (C, E) or one-way ANOVA with Tukey's post-test (D). *, P <0.05. LoD, limit of detection. NS, not significant.
Since MNV has tropism for macrophages (28), we tested the ability of bone marrow-derived macrophages (BM-Mac) to control viral replication. Following recombinant IL-4 (rIL-4) treatment, BM-IL4 Mac undergo STAT6-dependent alternative activation and BM-IL4 Mac infected with MNV CW3gp33 exhibited STAT6-dependent increases in viral burdens compared to control BM-Mac (fig. S10). Furthermore, while MNV CW3gp33-infected control BM-Mac could elicit robust CD8+ T cell proliferation in vitro, BM-IL4 Mac demonstrated STAT6-dependent inhibition of MNV-specific CD8+ T cell proliferation (fig. S11A). To test whether AAMac could modulate early antiviral immune responses in vivo, control BM-Mac or BM-IL4 Mac were adoptively transferred into mice along with MNV CW3gp33. Mice receiving BM-IL4 Macs exhibited fewer antiviral CD8+ T cells and increased viral burdens (fig. S11B, C). Together, these data indicate that helminth-elicited STAT6-dependent induction of AAMacs limits innate and adaptive immune responses to enteric viral infection.
AAMac effector molecules Arginase-1, RELMα and Ym1 have been implicated in regulating immunity and inflammation (29-31). However, antiviral CD8+ T cell activation remained impaired in helminth co-infected mice following inhibition of Arginase-1 (fig. S12A) or deletion of RELMα (fig. S12B), indicating these factors are dispensable for helminth-induced impairment of antiviral immunity. The chitinase-like molecule Ym1 is one of the most highly expressed AAMac effector molecules in multiple settings of acute and chronic type 2 cytokine-associated inflammation (32-36). Although Ym1 has been implicated in promoting chemoattraction and inflammation (37, 38), the biological functions of this molecule are not well understood (39). To test whether Ym1 contributes to helminth-induced immuno-modulation, virus-helminth co-infected mice were treated with isotype control or anti-Ym1 monoclonal antibodies (mAbs). Neutralization of Ym1 in co-infected animals significantly enhanced virus-specific CD8+ T cell proliferation (Fig. 4E), the polyfunctional capacity of antiviral CD8+ T cells and the number of virus-specific CD4+ T cells expressing effector cytokines (fig. S13). Anti-Ym1 mAb treatment of co-infected mice was associated with enhanced control of viral replication (Fig. 4F). Further, incubation of CW3gp33-infected BM-Macs with recombinant Ym1 (rYm1) resulted in impaired P14 CD8+ T cell proliferation as compared to control BM-Macs (Fig. 4G). Following TCR stimulation, rYm1 also inhibited activation and proliferation of purified CD8+ T cells (Fig. 4H), indicating that Ym1 can directly inhibit CD8+ T cell responses. Taken together, these findings reveal a role for helminth-induced AAMacs and Ym1 in immuno-modulation of protective immune responses to concomitant viral infection.
In summary, these data identify that helminth co-infection can inhibit antiviral immunity via a pathway of innate immuno-modulation. This was independent of changes in the microbiota, but was associated with induction of potent AAMac responses and Ym1-dependent inhibition of antiviral T cell responses (fig. S14). Therefore, although helminth parasites and commensal bacteria colonize the same niche within the intestine, co-evolution of helminths with their mammalian hosts may have resulted in mechanisms that allow direct regulation of the host innate immune system and antiviral immunity independently of changes the microbiota. These findings, coupled with a report by Reese et al. identifying a critical role for helminth-induced IL-4/IL-13 and STAT6 activity in reactivation of latent γ-herpes virus infection in macrophages (40), indicate a conserved mechanism of innate immuno-modulation in the context of virus-helminth co-infection. The identification of an immuno-modulatory role for the AAMac-Ym1 pathway could have implications for the development of helminth-based therapies for multiple chronic inflammatory diseases as well as improved vaccination strategies in helminth-infected individuals.
Supplementary Material
Acknowledgments
We thank members of the Artis laboratory for discussions and critical reading of the manuscript. The authors would like to thank F. Finkelman for providing anti-IL-4 clone 1D11.2 and the NIH Tetramer Core Facility (contract HHSN272201300006C) for providing MHCII IAb tetramers. The authors would like to thank the University of Pennsylvania Matthew J. Ryan Veterinary Hospital Pathology Lab, the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306) and its core facilities (Molecular Pathology and Imaging; Molecular Biology; Cell Culture; Transgenic and Chimeric Mouse) and acknowledge assistance and support from the Penn Center for AIDS Research (P30-AI045008). We also thank the Abramson Cancer Center Flow Cytometry and Cell Sorting Resource Laboratory for technical advice and support. The ACC Flow Cytometry and Cell Sorting Shared Resource is partially supported by NCI Comprehensive Cancer Center Support Grant (2-P30 CA016520). The data tabulated in this paper are reported in the main paper and in the supplementary materials. Illumina MiSeq data can be found in the NCBI Sequence Read Archive with accession number SRP043964. Research in the Artis lab is supported by the National Institutes of Health (AI061570, AI074878, AI087990, AI095466, AI095608, AI097333, AI106697 and AI102942 to D.A.; AI083022 and AI082630 to E.J.W.), T32-AI007532 to L.A.M., F32-AI085828 to M.C.S., K08-DK097301 to V.T.T., the Crohn's and Colitis Foundation of America (D.A. and M.R.H.), the Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease Award (D.A.) and the Edmond J. Safra Foundation/Cancer Research Institute Irvington Fellowship (L.C.O.). T.E.S and J.E.A were supported by the Medical Research Council UK (MR/J001929/1). TJN was supported by NIH training grant 5T32A100716334 and postdoctoral fellowships from the Cancer Research Institute and American Cancer Society. H.W.V. was supported by grant R01 AI 084887, the Crohn's and Colitis Genetics Initiative Grant #274415, and the Broad Foundation grant #IBD-0357. Washington University and H.W.V. receive income based on licenses for MNV technology.
Footnotes
The authors declare no other potential conflicts of interest.
References and Notes
- 1.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol. 2004;4:478. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe. 2009;6:187. doi: 10.1016/j.chom.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall JA, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hayes KS, et al. Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science. 2010;328:1391. doi: 10.1126/science.1187703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raetz M, et al. Parasite-induced TH1 cells and intestinal dysbiosis cooperate in IFN-gamma-dependent elimination of Paneth cells. Nat Immunol. 2013;14:136. doi: 10.1038/ni.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen JE, Maizels RM. Diversity and dialogue in immunity to helminths. Nat Rev Immunol. 2011;11:375. doi: 10.1038/nri2992. [DOI] [PubMed] [Google Scholar]
- 12.Elliott DE, Weinstock JV. Helminth-host immunological interactions: prevention and control of immune-mediated diseases. Ann N Y Acad Sci. 2012;1247:83. doi: 10.1111/j.1749-6632.2011.06292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rausch S, et al. Small Intestinal Nematode Infection of Mice Is Associated with Increased Enterobacterial Loads alongside the Intestinal Tract. PLoS One. 2013;8:e74026. doi: 10.1371/journal.pone.0074026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walk ST, Blum AM, Ewing SA, Weinstock JV, Young VB. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm Bowel Dis. 2010;16:1841. doi: 10.1002/ibd.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broadhurst MJ, et al. Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS Pathog. 2012;8:e1003000. doi: 10.1371/journal.ppat.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li RW, et al. Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis. Infect Immun. 2012;80:2150. doi: 10.1128/IAI.00141-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bancroft AJ, Hayes KS, Grencis RK. Life on the edge: the balance between macrofauna, microflora and host immunity. Trends Parasitol. 2012;28:93. doi: 10.1016/j.pt.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Leung JM, Loke P. A role for IL-22 in the relationship between intestinal helminths, gut microbiota and mucosal immunity. Int J Parasitol. 2013;43:253. doi: 10.1016/j.ijpara.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stelekati E, Wherry EJ. Chronic bystander infections and immunity to unrelated antigens. Cell Host Microbe. 2012;12:458. doi: 10.1016/j.chom.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salgame P, Yap GS, Gause WC. Effect of helminth-induced immunity on infections with microbial pathogens. Nat Immunol. 2013;14:1118. doi: 10.1038/ni.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metenou S, Babu S, Nutman TB. Impact of filarial infections on coincident intracellular pathogens: Mycobacterium tuberculosis and Plasmodium falciparum. Curr Opin HIV AIDS. 2012;7:231. doi: 10.1097/COH.0b013e3283522c3d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potian JA, et al. Preexisting helminth infection induces inhibition of innate pulmonary anti-tuberculosis defense by engaging the IL-4 receptor pathway. J Exp Med. 2011;208:1863. doi: 10.1084/jem.20091473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolff MJ, Broadhurst MJ, Loke P. Helminthic therapy: improving mucosal barrier function. Trends Parasitol. 2012;28:187. doi: 10.1016/j.pt.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McSorley HJ, Maizels RM. Helminth infections and host immune regulation. Clin Microbiol Rev. 2012;25:585. doi: 10.1128/CMR.05040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011;108:5354. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abt MC, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganal SC, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 28.Wobus CE, et al. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2:e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pesce JT, et al. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 2009;5:e1000371. doi: 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pesce JT, et al. Retnla (relmalpha/fizz1) suppresses helminth-induced Th2-type immunity. PLoS Pathog. 2009;5:e1000393. doi: 10.1371/journal.ppat.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nair MG, et al. Alternatively activated macrophage-derived RELM-{alpha} is a negative regulator of type 2 inflammation in the lung. J Exp Med. 2009;206:937. doi: 10.1084/jem.20082048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loke P, et al. IL-4 dependent alternatively-activated macrophages have a distinctive in vivo gene expression phenotype. BMC Immunol. 2002;3:7. doi: 10.1186/1471-2172-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao J, et al. Ym1, an eosinophilic chemotactic factor, participates in the brain inflammation induced by Angiostrongylus cantonensis in mice. Parasitol Res. 2013;112:2689. doi: 10.1007/s00436-013-3436-x. [DOI] [PubMed] [Google Scholar]
- 34.Nair MG, et al. Chitinase and Fizz family members are a generalized feature of nematode infection with selective upregulation of Ym1 and Fizz1 by antigen-presenting cells. Infect Immun. 2005;73:385. doi: 10.1128/IAI.73.1.385-394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ford AQ, et al. Adoptive transfer of IL-4Ralpha+ macrophages is sufficient to enhance eosinophilic inflammation in a mouse model of allergic lung inflammation. BMC Immunol. 2012;13:6. doi: 10.1186/1471-2172-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang NC, et al. A macrophage protein, Ym1, transiently expressed during inflammation is a novel mammalian lectin. J Biol Chem. 2001;276:17497. doi: 10.1074/jbc.M010417200. [DOI] [PubMed] [Google Scholar]
- 37.Owhashi M, Arita H, Hayai N. Identification of a novel eosinophil chemotactic cytokine (ECF-L) as a chitinase family protein. J Biol Chem. 2000;275:1279. doi: 10.1074/jbc.275.2.1279. [DOI] [PubMed] [Google Scholar]
- 38.Arora M, et al. Simvastatin promotes Th2-type responses through the induction of the chitinase family member Ym1 in dendritic cells. Proc Natl Acad Sci U S A. 2006;103:7777. doi: 10.1073/pnas.0508492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutherland TE, Maizels RM, Allen JE. Chitinases and chitinase-like proteins: potential therapeutic targets for the treatment of T-helper type 2 allergies. Clin Exp Allergy. 2009;39:943. doi: 10.1111/j.1365-2222.2009.03243.x. [DOI] [PubMed] [Google Scholar]
- 40.Reese TA, et al. Helminth infection reactivates latent gamma-herpesvirus via cytokine competition at a viral promoter. Science. 2014 doi: 10.1126/science.1254517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osborne LC, et al. Persistent enteric murine norovirus infection is associated with functionally suboptimal virus-specific CD8 T cell responses. J Virol. 2013;87:7015. doi: 10.1128/JVI.03389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giacomin PR, et al. Thymic stromal lymphopoietin-dependent basophils promote Th2 cytokine responses following intestinal helminth infection. J Immunol. 2012;189:4371. doi: 10.4049/jimmunol.1200691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laidlaw BJ, et al. Cooperativity between CD8+ T cells, non-neutralizing antibodies, and alveolar macrophages is important for heterosubtypic influenza virus immunity. PLoS Pathog. 2013;9:e1003207. doi: 10.1371/journal.ppat.1003207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strong DW, Thackray LB, Smith TJ, Virgin HW. Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. J Virol. 2013;86:2950. doi: 10.1128/JVI.07038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon JJ, et al. Tracking epitope-specific T cells. Nat Protoc. 2009;4:565. doi: 10.1038/nprot.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The R-Core Team. R: A language and environment for statistical computing. 2014 [Google Scholar]
- 48.Aronesty E. Comparison of Sequencing Utility Programs. The Open Bioinformatics Journal. 2013;7:1. [Google Scholar]
- 49.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 50.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 55.Oksanen J, et al. vegan: Community Ecology Package R package version 2.0-10. 2013 [Google Scholar]
- 56.Sosnovtsev SV, et al. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J Virol. 2006;80:7816. doi: 10.1128/JVI.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vita R, et al. The immune epitope database 2.0. Nucleic Acids Res. 2012;38:D854. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.