Abstract
Traditionally, collagen mimetic peptides (CMPs) have been used for elucidating the structure of the collagen triple helix and the factors responsible for its stabilization. The wealth of fundamental knowledge on collagen structure and cell-extracellular matrix (ECM) interactions accumulated over the past decades has led to a recent burst of research exploring the potential of CMPs to recreate the higher order assembly and biological function of natural collagens for biomedical applications. Although a large portion of such research is still at an early stage, the collagen triple helix has become a promising structural motif for engineering self-assembled, hierarchical constructs similar to natural tissue scaffolds which are expected to exhibit unique or enhanced biological activities. This paper reviews recent progress in the field of collagen mimetic peptides that bears both direct and indirect implications to engineering collagen-like materials for potential biomedical use. Various CMPs and collagen-like proteins that mimic either structural or functional characteristics of natural collagens are discussed with particular emphasis on providing helpful information to bioengineers and biomaterials scientists interested in collagen engineering.
1. Introduction
With more than 28 different types known to date and representing approximately 25% of total protein content, collagen is one of the most diverse and prevalent proteins in mammals.1-3 Fibrous collagens (types I, II, and III) are the most abundant class of collagens and form the basic scaffolds of mammalian connective tissues while other collagens make network-like structures (type IV and VIII) as part of the basement membrane which delineates epithelial cell organization. Lately, with the completion of human genome analysis, numerous new types of collagens collectively known as fibril-associated collagen with interrupted triple helix (FACIT) have been identified.4,5 Unlike other classes of collagens, FACITs do not assemble into higher order structures by themselves and are often found attached to the surfaces of fibrous collagens in specific tissues. Although collagens’ supramolecular architectures and specialized roles within the extra-cellular matrix (ECM) vary widely, all collagen molecules share a similar basic structure; they are composed of three identical or similar protein strands with the repeating sequence Gly-X-Y and display extensive post-translational modification (primarily hydroxylation and glycosylation). Depending on the collagen type, part or all of the trimeric strands fold into a right-handed triple helix which is the hallmark structural feature of collagen (Fig. 1).
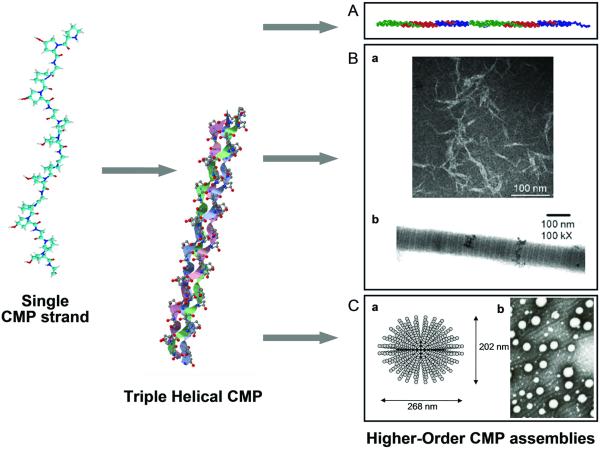
Fig. 1.
Triple helical structure and higher order assembly of collagen mimetic peptides. Collagen mimetic peptides have been designed to assemble into a long triple helical molecule by head-to-tail association (A, reproduced with permission from Ref. [21], © National Academy of Sciences), fibers by attractive interactions between CMP triple helices (B, reproduced with permission from Ref. [70] and [23], © Wiley and American Chemical Society, respectively), and micelle or vesicle-like structures by microphase separation of amphiphilic compounds (C, reproduced with permission from Ref. [79] and [80], © American Chemical Society).
The triple-helical structure of collagen has fascinated scientists for its unique multiplex architecture and its critical role in the formation of functional collagen molecules and their higher order assembly.2,6 The triple helical domains of collagens are rich in proline and hydroxyproline (Hyp), which confer rigidity to the backbone conformation similar to that of poly(proline). Periodic repeats of glycine at every third residue allow three collagen strands to pack closely and form a stable triple helix through interchain hydrogen bonds around a common helical axis. This multiplex structure bears striking similarity to the DNA double helix: both biopolymers have long semi-rigid backbones that are held together by periodic inter-chain hydrogen bonds formed across different polymer chains. The triple helical architecture is found in all 28 collagen types reported to date, as well as a handful of non-collagen proteins (e.g. complement factors7 and sugar binding proteins8). Type I and other fiber forming collagen helices (type II, III, V and XI) subsequently assemble into micro-meter long fibrils of great strength via staggered, side-by-side molecular interactions.9
Synthetic collagen model systems, also known as collagen mimetic peptides (CMPs) have been very useful in elucidating collagen structure and factors responsible for the stabilization of the triple helix (Fig. 1).10-13 Many research groups have prepared and studied polypeptides of Gly-X-Y sequences that fold into triple helical structure. Among them, CMPs based on GlyProPro and GlyProHyp trimers have been widely studied, and their collagen-like triple helical structure and melting behaviors are documented in the literatures. Unlike collagen molecules, CMPs exhibit reversible melting behavior due to their small size; when denatured (melted) collagen is cooled, the collagen regains only a fraction of its original triple helical content and turns into gelatin. In contrast, a CMP regains 100% of its original triple helical structure despite a relatively slow folding rate allowing full thermodynamic characterization of folding and melting processes. The small size of CMPs is also conducive to various structural analyses such as x-ray crystallography and NMR spectroscopy.
The thermal stability of CMPs is modulated by both the number of trimeric repeats and the amino acid compositions. Longer peptides exhibit higher thermal stability: Tm of (ProHypGly)x rises from 37°C for x=7 to 43°C and 69°C for x = 8 and 10, respectively. Using a host-guest system, Brodsky’s research group has determined the thermal stability of almost all possible X-Y amino acid combinations in the Gly-X-Y triplet which highlighted the stabilization effect of Hyp and other charged amino acids.14 Hyp’s ability to stabilize the triple helix has been recognized for several decades. Crystal structures of the CMP and other thermal studies by Brodsky seemed to suggest that the water bridges between Hyp’s hydroxyl groups molecules are responsible for increased thermal stability.15 Raines’s research lab at the University of Wisconsin reported that the thermal stability of a CMP can be enhanced by incorporation of trans 4-fluoroproline (Flp) in place of 4-hydroxyproline (Hyp).12,16 This was evidenced by the dramatic change in melting temperatures for the three model polypeptides; 41°C for (ProProGly)10, 69°C for (ProHypGly) 10, and 91°C for (ProFlpGly)10. In addition, 4-azidoproline was found to have conformational properties and stabilizing effect similar to Flp and Hyp.17,18 From these and the work of Brodsky and others, two distinct mechanisms by which amino acid side chains stabilize the collagen triple helix have been identified: i) the hydration layer around the triple helix promoted by Hyp and charged amino acids, and ii) the stereo-electronic effect of Hyp that stabilizes the trans peptide bond conformation of collagen strands. The use of CMPs for various biomedical applications may require that one be able to produce thermally stable CMPs in large quantity. Therefore understanding the origin of the structural stability within the collagen triple helix as well as the stabilization effect of various amino acids is critical for engineering synthetic collagens.
In addition to studying the structure and stability of CMPs as models for natural collagen, a number of research groups have developed new types CMPs and CMP derivatives that can potentially be used for biomedical applications. These include i) novel CMPs with new supramolecular architecture19,20 or CMPs tailored toward modulating the stability or composition of the triple helices,21,22 ii) CMPs that can self-assemble into higher order structures,23-25 iii) CMPs that can be triggered to fold or assemble into higher order structures,26-28 iv) CMPs that incorporate cell interactive sites as well as those that can specifically bind to natural collagen,29-34 and v) PEG-based hydrogels that display CMPs for facile assimilation with cellular ECMs.35-37 A significant portion of work in these applications is still at a fundamental level and the feasibility of CMP use in biomedical applications needs to be verified; however recent research activity in the design and synthesis of new CMPs suggests that the collagen triple helix has become a promising structural motif for engineering self-assembled, hierarchical constructs similar to natural tissue scaffolds which may exhibit unique or enhanced biological activity.
In this paper, we review recent advancements in the field of collagen mimetic peptides that bear both direct and indirect relations to engineering collagen-like materials for potential biomedical applications. The structure and stability of the collagen triple helix will be discussed within the context of developing new biomaterials, and readers seeking fundamental understanding of the collagen triple helix should refer to other recently published review papers.2,14,38 We will highlight various CMPs and collagen-like proteins that mimic either structural or functional characteristics of natural collagens, which should be helpful to bioengineers and biomaterials scientists interested in collagen engineering.
2. Hydroxyproline-Free Collagens
One of the most exciting developments in the past several years in the research of collagen as related to triple helical structure is the surprising discovery that electrostatic interactions between charged amino acids can stabilize the triple helix almost as well as hydroxyproline. Numerous past investigations have revealed the existence of collagen-like sequences featuring long stretches of Gly-X-Y trimer repeats in bacteria and viruses.39,40 Two distinct types of such sequences have been identified, one rich in GlyProThr repeats and the other rich in charged amino acids with less frequent Pro occurrence.41 Glycosylating the GlyProThr sequence has been reported to dramatically increase the triple helical stability in model collagen peptides and it is speculated that the GlyProThr(o-glycosylate) is the main component responsible for triple helix formation rather than the un-glycosylated precursor protein.42 Bacillus collagen-like protein (BclA), which forms hair-like filaments on the outermost surface layer of B. anthracis spores, is a good example of a bacterial protein with GlyProThr repeats. Sylvestre and coworkers discovered polymorphisms in the collagen-like region of BclA whereby it contained one to eight copies of the GlyProThr-rich 21 amino acid sequence (GPT)5GDTGTT;43 this variation in the length of the collagen-like region correlated to the change in the length of dense filaments on the spore surface as visualized by transmission electron microscopy (TEM, Fig. 2, A and B). The exact function of BclA remains unknown but the results indicate that the collagen-like sequence is integral to the surface structure of the B. anthracis spores.
Fig. 2.
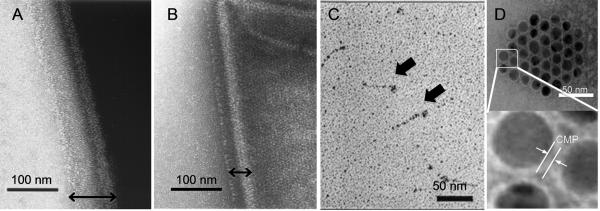
Transmission electron micrographs of B. anthracis spores after negative staining (A and B, reproduced with permission from Ref. [43], © American Society for Microbiology), Scl1 protein after rotary shadowing (C, reproduced with permission from Ref. [44], © American Society for Biochemistry and Molecular Biology), and gold nanoparticles with self-assembled CMP monolayers (D). B. anthracis spores show densely packed collagen-like layers on their surfaces (double headed arrows): Strain 7702 which has 8 copies of (GPT)5GDTGTT sequence displays a thick layer (A), while strain 5725R with only 1 copy of the same sequence displays thin collagen like layer (B). Sc1 protein (streptococcal collagen like protein with charged amino acids) exhibits lollipop morphology similar to complement factors (C, block arrows). Self-assembled CMP layer provides colloidal stability to gold nanoparticles (D).
In contrast to the GlyProThr rich sequence which has been the target of significant research, the effect of charged amino acids in stabilization of the collagen triple helix has not been well understood in part due to the lack of investigation; natural collagen-like sequences with charged amino acids typically lack the prolines considered to be critical for triple helix folding. Only recently have these collagen-like sequences with charged amino acids been shown to form the collagen triple helix.44,45 Some of the best studied charged collagen-like sequences are streptococcal Scl1 and Scl2 which are expressed on the surface of group A Streptococcus bacteria. These surface proteins are believed to be involved in adhesion to host cells and tissues. Both Scl1 and Scl2 contain three major domains: the central collagen-like domain, the C-terminal cell membrane domain, and the N-terminal variable domain. Under TEM, they exhibit a lollipop morphology similar to complement factors (Fig. 2C). Lukomski’s group produced various recombinant forms of streptococcal Scl1 and Scl2 in E. coli 44,46 and studied their triple helix folding behavior using SDS-PAGE, CD, and TEM. In collaboration with Brodsky’s group, additional pH dependent thermal stability experiments were performed using DSC and CD.45 All of the data indicated that these proteins are indeed in triple helical structure which led to a conclusion that the charge-charge interactions play a major role in stabilizing bacterial collagen-like proteins. In mammalian collagens, prolines in position Y of Gly-X-Y triplets are mostly hydroxylated. The relative lack of prolines in this position for bacterial and viral collagen-like sequences seems to reflect their inability to hydroxylate proline residues. Instead, these organisms seem to have found an alternative way to stabilize the triple helix through electrostatic interactions among the long Gly-X-Y repeat pattern.
As the triple helical nature of Scl1 and Scl2 were being verified and the discussion on the significance of electrostatic interactions among charged amino acids was taking place, Hartgerink’s research group reported a surprising discovery that was in agreement with the existence of electrostatic stabilization in collagen triple helices.13,47-49 They observed that a ABC type heterotrimer containing one positively charged CMP (ProArgGly)10, one negatively charged CMP (GluHypGly)10, and one neutral CMP (ProHypGly)10 forms a triple helix with Tm (54°C) comparable to a (ProHypGly)10 homotrimer (67.5°C) even though Arg and Glu have lower triple helical propensity when compared to Hyp and Pro. In contrast, AAB type heterotrimers containing two similarly charged and one opposite charged CMP exhibited significantly lower Tm, while homotrimers consisting entirely of either positively or negatively charged CMPs showed no signs of triple helical folding. Similar results were obtained when Lys and Asp were substituted for Arg and Glu, respectively, for the above CMPs. These results showed that the electrostatic interactions between the collagen strands do contribute to the stabilization of triple helix and although the sequences tested were largely artificial, it strengthens the possibility that nature has used this mechanism to produce stable collagen-like triple helices for various structural and biofunctional purposes.
The obvious excitement in the biomaterials community regarding Hyp-free collagen is based on the possibility of producing collagen-like protein from bacteria. The mammalian collagens are stabilized mainly by the Hyp, which is the product of a post-translational protein modification that is available only in eukaryotes. Although genetically engineered Hyp-containing collagen has been produced in yeast cells and other eukaryotic systems50-52 by co-expression of prolyl-4-hydroxylase, the ability to produce a stable collagen-like triple helix by bacterial synthesis will allow more facile manipulation of protein sequence, as well as production and purification of large amounts collagen analogues for various biomaterials applications. Recombinant Scl proteins expressed in E. coli as described above represent one of the first collagen-like proteins artificially produced in bacteria that form stable triple helices. Recently, Peng and co-workers reported that the recombinant Scl2 proteins are non-cytotoxic to fibroblasts and do not elicit immune response in mice.53 It remains to be seen whether such charged collagen-like proteins and peptides engineered in bacteria are able to assemble into higher order structures that could mimic natural collagen scaffolds.
3. Template-Tethered CMP
One of the well-known methods to stabilize a collagen triple helix is to link individual CMPs on a single template. An ideal template should have three functional groups that can covalently link three peptide chains, and be flexible enough to allow proper packing of the three chains with correct amino acid register. Templates that have been used in the past are peptide templates with reactive amino acids (e.g. Lys or Glu), Kemp’s tri-acid, and tris(2-aminoethyl) amine (TREN) succinic acid derivatives.54-56 When various CMPs were conjugated to these templates, their Tms were elevated mainly because of reduction in entropy differences between folded and unfolded states of CMPs. Lately, Goodman’s research group has developed a TRIS-based template with an additional amino group that can be further utilized for attaching fluorescent tags or therapeutic agents for biomedical applications.57 Khew and Tong used a peptide template containing two terminal glutamic acids to form templated CMP homotrimers via the three carboxylic acids, one at the C terminus and two at the Glu side chains.58 By reacting the peptide template directly to a fully grown CMP on solid resin, they were able to produce the templated CMPs with a remarkable 85% reaction yield. When cell binding motif GFOGER was inserted into the middle of the CMPs, the templated peptide formed a stable triple helix which showed structure-dependent cell adhesion activity.
More than half of human collagen types with known collagen composition are heterotrimers, however, despite few efforts in preparing non-tethered model heterotrimers by heating and cooling of CMP mixtures,59,60 almost all covalently templated CMPs developed to date are comprised of homotrimers. This is mainly due to the difficulties in selectively reacting CMPs to one or two reactive groups on the template. Previously, Moroder’s lab was the only group that produced covalently linked heterotrimers by the use of an orthogonal Cys protection/deprotection scheme. Recently, we introduced a simple strategy for CMP heterotrimer synthesis which involves serial solid phase and solution coupling of two different CMPs onto the TREN template.22 First, a single type A CMP was conjugated to the TREN-(suc-OH)3 template by reacting excess amount of template with the CMP directly on the resin. Due to the low density of reactive sites characteristic to the resin employed in this study, only one of the template’s three carboxylic acid moieties reacted with the resin-bound CMP. This process allowed easy synthesis and purification of single CMP-template conjugates, to which full length type B CMP was added in solution to produce a template-tethered ABB type trimer. Recently, another two-step tethering approach mediated by click chemistry has been reported for ABB type heterotrimer synthesis.61 We believe that these stepwise tethering approaches are versatile means to prepare heterotrimeric CMPs of defined peptide chain composition which will be useful not only for studying the thermal stability and folding behavior of heterotrimeric collagens and collagen-like proteins, but also for developing CMPs that mimic the high-order structure or biological function of natural collagens.
In addition to covalent tethering, non-covalent affinity interactions (e.g. ligand-metal coordination and biological interaction) can also be used to template CMPs. One of the benefits of non-covalent templates is that the templating effect is reversible and that the templating process can be triggered. Since CMP templating typically leads to formation of stabile helices, one can also trigger triple helix folding by use of non-covalent templating processes. Koide’s group conjugated 2,2'-bipyridyl (bpy) to the C-termini of CMPs that can coordinate to Fe (II) allowing formation of templated CMP in the form of FeII (bpy-CMP)3.62 Cai and coworkers developed a similar system using catechol-Fe3+ metal-ligand interactions by attaching dihydroxy-phenylalanine to the C-termini.63 Both systems can be considered an environmentally sensitive folding system where presence of the metal ion triggers trimerization which in turn leads to triple helix folding. Biological interactions have been also explored for nucleating CMP folding processes. In order to produce stable triple helical CMPs using recombinant bacterial synthesis, Frank and coworkers designed a fusion protein of (ProProGly)10 and bacteriophage T4 fibritin foldon domain64 that folds into trimeric α-helical coiled coils. As a result of the templating effect of the foldon domain, the Tm of the (ProProGly)10 jumped from 35°C to 65°C. This opens up a possibility of designing other triple helix folding systems controlled by biological interactions. We note that the host-guest systems developed by Brodsky’s group and Hartgerink’s group can also be considered non-covalent templates for stable homo- and hetero-trimeric triple helical CMPs.6,47.
4. Triggered Triple Helix Folding
Folding of the collagen triple helix can be triggered not only by association of the strands with the templates but also by a change in the helical propensity of the peptide strands themselves. Recently, photo-triggered folding has been achieved by cis-trans photo-isomerization of an azobenzene group that is clamped intra-molecularly to the collagen mimetic peptides.27,65 Using a rigid acetylene linker, Kusebauch and coworkers conjugated a single azobenzene chromophore to two 4-thioprolines in the middle of a CMP that were seven amino acids apart, and studied photo-induced folding/unfolding via CD, IR and NMR. Similar studies were conducted using related CMPs that were clamped with azobenzene at two cysteines instead of 4-thioproline. It was unclear if photo-isomerization of the azobenzene group can lead to complete disruption of the triple helical structure; however these results demonstrated at least local photo-induced unfolding of the triple helix which could be exploited further for ultrafast folding and unfolding experiments. Chmielewski’s research group developed new CMPs that show pH sensitive folding characteristics.26 In order to produce a pH sensitive CMP that is also conducive to formation of stable triple helix, the carboxylic acid derivative of Hyp, 4-propiate Hyp (PE), was synthesized and used in place of Hyp. CD melting and folding experiments indicated that substitution of two or three Hyps in the middle of (ProHypGly)7 with PE does not result in pH-triggered triple helix folding but it dramatically increases the folding rate at acidic pH (pH = 2.7) compared to neutral pH (pH = 7.2). A pH-triggered folding was only observed for a peptide with all seven Hyps substituted with PE.
5. Higher-Order Assembly of CMPs
Most collagens exist in nature in a form of supramolecular assembly with precisely defined molecular compositions and structures. In recent years, a variety of CMP designs have been developed within the collagen community to produce CMPs that not only fold into stable triple helices but also self-assemble into higher order structures. Such assemblies typically range from nano- to micro- meter scales and are especially important from the perspective of developing collagen-like materials for biomedical applications.
Head-to-tail triple helical folding and assembly
The simplest higher order assembly of CMPs is the long collagen triple helix first designed by Raines’ research group.21 Two cysteine knots were used to link three CMPs with one CMP out of register so that it produced sticky ends during triple helix assembly. For such preorganized CMP constructs, the only way to satisfy full triple helical structure was to configure themselves in a head-to-tail fashion resulting in a long single collagen triple helix (Fig. 1A). Using this approach, the Raines group produced collagen-like fibrils of 1-nm diameter and 400 nm length which is significantly longer than the natural type I collagen molecule (300 nm). Considering the small diameter of the fibril which is commensurate with the diameter of triple helix (1.5 nm), it is very likely that these fibrils consisted of a long, single triple helix with an overall structure similar to the type I collagen molecule.
Koide and coworkers developed a similar CMP system that can self assemble in a head-to-tail manner.66 In this case, the three CMPs were preorganized in a staggered configuration locked in place by two cysteine knots. Head-to-tail organization would also lead to full realization of the triple helix. Although the overall structure of the self-assembly is similar, it is unclear whether this trimeric unit peptide faithfully assembles into a single triple helix analgous to that of Kotch and Raines21 or if it also produces aggregates by lateral interactions, since the dynamic light scattering data indicated bimodal size distribution that peaks at 600 nm and 14 μm. Attempts to produce hydrogel out of this collagen fibril were not successful because the fibrils precipitates at high CMP concentration. However, by placement of hydrophilic Arg at the end of the knotted CMPs, Koide and coworkers were able to produce self-supporting hydrogel where the melting of triple helix correlated with the length of overlap between the CMP assemblies as determined by the CD and the sol-gel transition temperatures estimated by the ball drop method.67 Recently, Koide’s group incorporated the integrin-binding sequence GFOGER into one CMP strand of the same knotted trimeric base unit.68 In this publication, the authors did not present any direct evidence for controlled supramolecular assembly; however the culture plates coated with the new peptide exhibited cell adhesion activity almost as high as natural collagens while those coated with simple CMPs that still incorporate the GFOGER sequence exhibited almost no cell binding activity.
Association of CMP triple helices
Higher order assembly of CMPs can also lead to formation of macroscopic fibers that are similar to natural collagen fibers in terms of assembly mechanisms and overall fiber morphology. Brodsky’s lab reported that a CMP as simple as (ProHypGly)10 can form macroscopic precipitates when concentrated solution (1 mM) is subjected to a temperature just below CMP’s melting temperature.25 Although the morphology of the precipitates lacked long-fiber like appearance, the turbidity experiments indicated that the triple helices self-associate by a nucleation and growth mechanism under conditions similar to those of collagen fibril formation such as neutral pH and 37°C. A morphology that is more fiber-like was observed when the central domain of (ProHypGly)10 sequence was replaced with a Hyp-rich hydrophobic sequence derived from type IV collagen.69 Kishimoto and coworkers synthesized poly(ProHypGly)x by polycondensation of monomeric CMP oligomers (ProHypGly)10.70 This process yielded high but polydisperse molecular weight samples which formed long nanofibrils when investigated under TEM (Fig. 1B-a). In contrast, only short and aggregated elliposides were seen for the monomeric peptide (ProHypGly)10 which was the same morphology observed by the Brodsky’s group.25 These results indicate that fiber-like aggregates can be formed by the anisotropic shape and the hydrophobicity of the CMP based on simple ProHypGly sequence.
A number of research groups have looked into controlled association of CMP triple helices in the hopes of producing synthetic collagen fibers that are more akin to natural fibrous collagens in terms of fibrillogenesis mechanisms, fiber morphology, and ultimately their biological role as native ECM. The structures of CMP triple helices can be tailored to further assemble into fibrous structure either by end to end interactions or by side to side interactions. Cejas and coworkers synthesized collagen mimetic peptides with a core (GlyProHyp)10 sequence flanked by a C-terminal phenylalanine, which has an electron-rich phenyl ring, and N-terminal pentafluorophenyl alanine, which has an electron-poor phenyl ring.24,71 The two aromatic groups at the opposite ends of the peptide are designed to interact by π−π stacking resulting in head-to-tail association of the full length CMPs. Using this approach, the authors were able to produce macroscopic fibers that are several micrometers in length and hundreds of nanometers in thickness which, according to authors, display periodic banding morphology similar to that seen in natural collagen fibers. The size of the fibers is much thicker than would be expected for a single triple helix, and it is likely that the fibers are formed not only by end-to-end but also by side-to-side interactions of assembled triple helices. According to platelet aggregation experiments, these artificial collagen fibers are almost as good as type I collagen in activating human blood platelets and therefore may be used as hemostatic biomaterials.24
In addition to biological applications, the collagen-like fibrils assembled by end to end π−π stacking interactions were explored as a nano-template for the formation of metallic nanowires. Popularized by Stupp’s research group,72 this is a bottom-up nanofabrication method that uses a well-defined supramolecular assembly composed of peptides, polymers, or amphiphilic molecules. The assemblies are impregnated with metals, metal precursor molecules, or inorganics that are subsequently sintered into continuous nanostructures under mild conditions. Using a similar approach, Gottlieb and coworkers conjugated two gold nanoparticles (1.4 nm) to single CMP strands of sequence pentafluoro-F(GPO)4GPK(GPO)5F; one to the amino group at the N terminus and the other to the lysine group in the middle of the peptide.73 Even after the gold nanoparticle conjugation, the peptides were able to assemble into micrometer-long fibers, and electroless silver plating of the fiber resulted in continuous nanowires that conduct electricity. These CMP-derived synthetic nanowires exemplify how CMP molecular and supramolecular assembly can be exploited for producing complex metal or inorganic nanostructures.74,75
The hallmark feature of the native type I collagen fiber is the 67 nm periodic banding morphology that is observed by TEM and atomic force microscopy (AFM). The periodic bands, also known as D-periodicity, originate from the precisely defined axial stagger of the collagen molecules during the fiber assembly. Until recently, artificial collagen fibers with such banding features have not been produced in part due to difficulty in designing peptide sequences with an optimal balance between electrostatic interactions and hydrophobic interactions. Conticello and Chaikof’s research labs reported production of the first synthetic collagen fibers clearly featuring the collagen-like banding morphology (Fig. 1B-b).23 Here, instead of π−π stacking interactions,71 they took advantage of ion pair interactions to assemble CMPs into collagen-like fibers. This was achieved by placing multiple positive charges (Arg) at the N-terminus and negative charges (Glu) at the C-terminus within the ubiquitous Gly-X-Y triad sequence in a form of (PRG)4(POG)4(EOG)4. Even under dilute conditions (<1.0 mg/mL), this peptide showed a strong tendency to form fibers following a nucleation-growth mechanism. Surprisingly, TEM of this fiber revealed clear periodic banding very similar to that of natural collagen fibers. This indicated that the charge-charge interactions resulted in a well-defined arrangement of the CMPs during the fiber formation process which was unattainable with simple POG-based CMPs.70 One point to note is the fact that the banding periodicity of the peptide fibers (18 nm) is significantly longer than the length of the peptide itself (10 nm). This suggests that the basic operative units that undergo self-assembly are not the individual triple helical peptides but oligomers of multiple triple helices that are preorganized in such a way that their subsequent assembly into mature fibers lead to periodic packing density and charge distribution as visualized by TEM. This is in drastic contrast to native type I collagen fiber where staggered assembly of individual collagen molecules is responsible for periodic banding pattern and alludes to the fact that true mimicry of natural collagen assembly process is yet to be achieved.
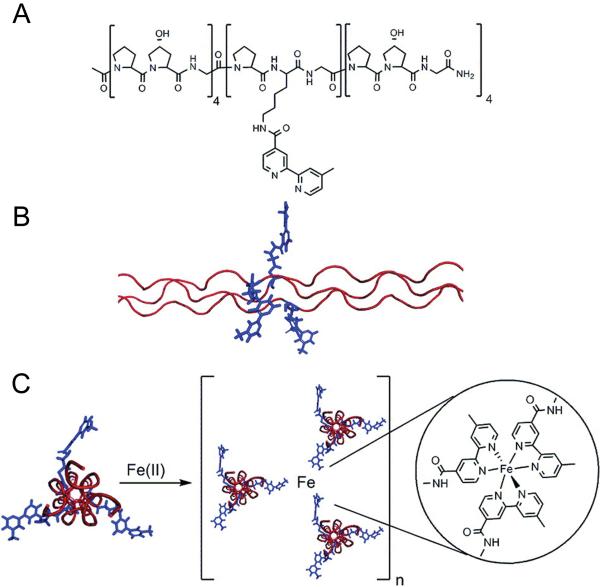
One of the most innovative approaches in controlled CMP fiber assembly is the metal-triggered side-to-side assembly reported recently by Przybyla and Chmielewski (Fig. 3).28 Instead of using electrostatic interactions, they applied coordination chemistry of bipyridyl ligands (bpy) and metal ions to induce CMP association. By coupling a single bpy group to the amino group of the unique Lys in (POG)4PKG(POG)4, they synthesized a CMP that can be triggered to form micrometer-long fibers by addition of metal ions (Fe2+). Although the lack of periodic banding morphology suggests that such assembly is not as structurally defined as that of Conticello and Chaikof’s,23 this work mimics the intermolecular crosslinks among lysine and hydroxylysine side chains in natural fibril collagens,76 and demonstrates that modification of collagen backbone and the use of artificial amino acids within the CMP sequence can lead to new types of engineered collagen fibers.
Fig. 3.
Fiber-like association of CMPs mediated by metal-ligand coordination. Fe2+ brings together three CMP triple helices by coordinating with Bpys incorporated into the CMP’s central amino acids: Structures of single strand CMP with central Bpy (A) and corresponding homotrimeric triple helix (B), and lateral CMP association by Fe2+ coordinating to three CMP triple helices (C). Reprinted with permission from Ref. [28], © American Chemical Society.
Non-fibrous CMP assembly
Scientists and engineers have also explored CMP derivatives as the basic building blocks of various supramolecular assemblies that do not exhibit fibrous morphology (e.g. micelles, liposomes, and dendrimers). This research effort began as early as 1996, when the groups of Fields and Tirrell developed amphiphilic fatty acid-CMP conjugates that can assemble into micelles under appropriate conditions (Fig. 1C-a).77-79 They synthesized single and double tail amphiphiles with variations in the hydrophobic tail lengths (C12-20) and composition of the hydrophilic CMP head groups. In aqueous solution, single-tail amphiphiles and double-tail amphiphiles with short tails (C12 and C14) self-assembled into spherical micelles whereas double-tail amphiphiles with long C16 tails formed disk-like micelles that showed a tendency to further aggregate into stacked disks. Due to the hydrophobic tail, the CMP amphiphiles adhered strongly to conventional cell culture plates. When coated with CMP amphiphiles, the culture plates became cell adhesive as evidenced by strong attachment of melanoma cells.78
Recently, Sarkar and coworkers reported formation of liposomes from fatty acid-CMP conjugates.80 Specifically, stearic acid was conjugated to the N- termini of a series of CMPs that incorporated a MMP (matrix metalloprotease) recognition site within or next to the (ProHypGly)x triple helical domain. When mixed with phosphocholine derivatives that readily form liposomes (30 mol% CMP derivatives and 70% phospholipids), the amphiphiles resided within the lipid bilayer of the liposome (Fig. 1C-b). Although the details of liposome structure and the conditions that favor liposome assembly are not presented in the report, the authors demonstrated that the content of the liposome could be released by the enzymatic activity of MMP-9, which may lead to future drug delivery vehicles for the detection and treatment of human diseases that are related to the over-expression of matrix metalloproteases (e.g. cancer).
Poly(amidoamine)-based dendrimers (PAMAM) that display CMP on the outer periphery have been prepared by a number of research groups. Goodman’s research lab20 conjugated (GlyProNleu)6 to the 8 carboxylic termini of a 0.5 generation PAMAM dendrimer and investigated its triple helical stability. Tong’s research group conjugated a 1.5 generation PAMAM dendrimer with a CMP that contained three types of domains: a triple helical domain [(GPO)x], cell binding domain (GFOGER), and crosslinking domain (APQQEA or EDGFFKI) that is activated by transglutaminase (tTGase).81 The CMP-conjugated dendrimer showed enhanced triple helical stability due to the templating effect of the dendritic core and when treated with tTGase, formed high molecular weight product that preserved the stable triple helical structure. Both the dendrimer and cross-linked dendrimers induced adhesion of human hepatocarcinoma cells (Hep3B) via the interactions between the CMP cell binding domain and integrins on cell surfaces as evidenced by cell binding and competitive binding inhibition studies as well as by immunofluorescent microscopic observation of focal adhesions.
One recent intriguing report related to this topic involves the use of non-triple helical CMP dendrimers for controlled release. Kojima and coworkers synthesized a 4 generation PAMAM dendrimer with (ProProGly)5 decorating the surface.82,83 Although this dendrimer exhibited CD trace characteristics of a collagen triple helix, absence of melting transition in the CD melting experiment suggested that the CMPs are more likely in a form of poly(proline)-II helix rather than triple helix. This is fully understandable considering the short length of the peptide and the lack of triple helix stabilizing Hyp. Even without triple helix assembly, the CMP dendrimer showed sustained release behavior at low temperature. This could be explained by the dynamic interactions of the CMPs on the dendrimer surface which could act as a barrier to releasing encapsulated contents. Overall, CMP-based dendrimers and other non-fibrous supramolecular assemblies show great promise in drug delivery and formation of synthetic hydrogels for bioactive systems.
6. Study and Application of CMP-Collagen Hybridization
CMP-type I collagen hybridization
In contrast to majority of CMP research that studies the triple helical structure and higher order assembly of the CMPs themselves, our research lab has been investigating the binding interactions between CMP and natural collagen and the application of such interactions for encoding cellular cues onto collagen scaffolds. In the biomedical community, there has been widespread interest in immobilizing bioactive components (such as growth factors, antimicrobial agents, or adhesion molecules) and structural stabilizers (small molecule and polymeric crosslinking agents) to natural collagen and other ECM components.84,85 However, due to loose network structure, natural collagen is ineffective at retaining passively adsorbed materials. Chemical coupling reactions of the amino acid side chains have been used to immobilize exogenous components to collagen,86 but chemical reactions on collagen are difficult to control and can compromise the biochemical features of natural collagen. More importantly, chemical reactions are not ideal for modifying integrated collagen matrices that contain bioactive compounds, and/or live cells and tissues.
As an alternative to the conventional “chemical” modification method, we developed a novel “physical” collagen modification technique that is based on collagen’s native ability to associate into triple-helical molecular architecture.34 We discovered that biochemically inert synthetic CMPs with the sequence (ProHypGly)x exhibit structure-dependent binding affinity to natural collagen. The binding took place when melted CMPs were allowed to fold in the presence of regenerated type I collagen fibers. The binding experiments indicated that type I collagen films and gels attract only single stranded CMPs, and not triple helical CMP nor control peptides comprising scrambled peptide sequences. The exact mechanism of binding remains to be elucidated but we speculate that the binding occurs by CMP(s) hybridizing into the collagen triple helix by a strand exchange mechanism as shown in Fig. 4. In 1973, Heidemann and coworkers reported hybrid formation between (ProGlyPro)25 and chymotrypsin treated α1 collagen chain (atelopeptide). In addition, a number of researchers believe that type I collagen has domains that are unfolded, also known as micro-unfolded domains, which could be receptive to CMP invasion and hybrid triple helix formation.87,88 It is also possible that the CMPs are hybridizing with partially denatured collagens since denaturation can occur during collagen fiber regeneration and subsequent film fabrication steps.
Fig. 4.
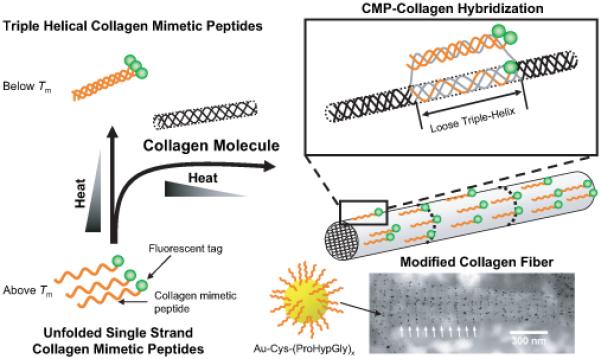
Schematic depiction of the hypothetical mechanism of CMP-collagen hybridization. Allowing melted CMP to fold in the presence of natural collagen triggers CMP hybridization with unstructured domains, either in the middle (shown in this figure) or end of the collagen molecules. Bottom right photo: transmission electron micrograph of reconstituted type I collagen fiber (mouse tail tendon) showing periodic localization of CMP conjugated gold nanoparticles (reproduced with permission from Ref. [33], © American Chemical Society).
Recently, we investigated the thermal stability of model peptides that simulate the potential hybrid heterotrimers that might form when CMP binds to type I collagen.22 This was made possible by the use of a tri-functional template (TREN derivative, discussed in the Template-Tethered CMP section) that can covalently tether three predefined CMPs and force the formation of ABB type CMP triple helices. For a potential target CMP-binding domain in collagen, we selected an unstable domain in the α2 chain of type I collagen (Gly268 to Gly286: GPKGELGPVGNPGPAGY). Due to the lack of Hyp in peptide sequence, this peptide exhibited no melting behavior even after it was covalently linked to the TREN template. In contrast, a templated heterotrimer comprising this peptide and two (ProHypGly)6 showed a well-defined melting transition at 43°C. We acknowledge that the templated heterotrimers are only our first attempt at modeling the CMP-collagen hybrid products. Although the model heterotrimers do not take into consideration the kinetic competition by homotrimer formation or neighboring effects during collagen hybridization, the results clearly demonstrated that the synthetic/natural CMP hybrid complexes can fold into stable triple helices, and that unstable sequences derived from natural collagen can fold into stable triple helices by hybridization with the synthetic CMPs.
When a small peptide (e.g. CMP) forms a complex with a large biopolymer (e.g. collagen), the molecular structure and binding location of the peptide is difficult to determine even by modern spectroscopic and diffraction techniques. We decided to employ transmission electron microscopy (TEM) to visualize the CMP-collagen complex using electron dense gold (Au) nanoparticles (NP) that are functionalized with Cys-CMPs.89 To our surprise, the CMP-conjugated NPs showed excellent colloidal stability in aqueous conditions. The NPs remained soluble in a wide range of pH and salt concentration and could be resuspended after lyophilization (Fig. 2D). This is in contrast to the CMP-conjugated dendrimers that tend to aggregate into a gel at high concentration.82,83 We believe that the densely aligned CMPs on the NP surface form a hydrophilic barrier preventing particle aggregation since CMPs from different NPs can only come in contact in anti-parallel configurations that cannot associate into triple helices. In contrast, the floppy nature of the dendrimers seems to permit parallel assembly of a CMP triple helix between adjacent dendrimers which results in attractive inter-particle interaction and hydrogel formation. Room temperature incubation of type I collagen fibers with gold NPs displaying Cys-(ProHypGly)7 produced intact fibers decorated with a large number of NPs.33,89 For some collagen fibers, NPs were present on the collagen fiber with a marked periodicity along the long fiber axes (Fig. 4, bottom right transmission electron micrograph, white arrows). The TEM results indicated that intact collagen fibers attract CMPs by presenting well-defined domains that are susceptible to CMP binding.
Targeting pathologic collagens by CMP hybridization
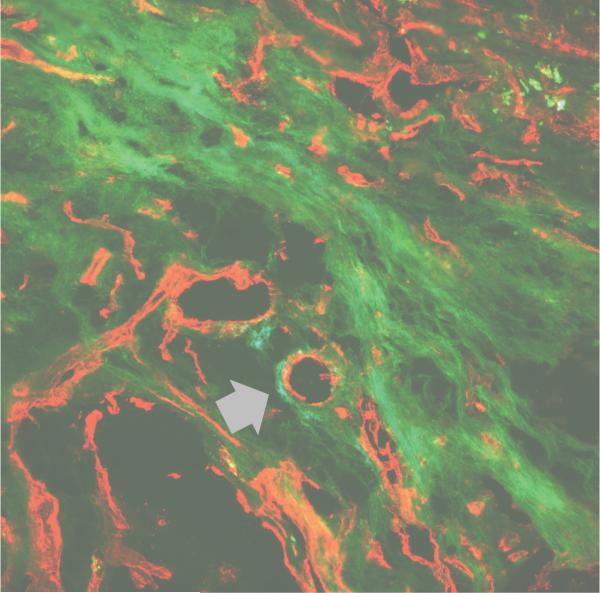
Many debilitating diseases are caused by structural or metabolic abnormalities in collagen. In addition, overproduction of collagen has been linked to tumor growth, atherosclerosis, and formation of pathological scar tissues.1 Fig. 5 shows fluorescence histology of human liver tissue treated with fluorescence tag labeled CMP (stains collagen in green) and CD-31 (stains blood vessels in red). In the confocal microscopic image, the green colors are seen throughout the tissue sample, sometimes in a mesh-like network or in fibrous textures. It can be also seen around the cross sections of blood vessels (arrow). Previously, we had also shown that images of human tissue stained by CMP are nearly identical to those stained by collagen antibodies.33 This simple work demonstrates that CMP derivatives can be used to image collagens in human tissue samples. In contrast to collagen antibodies used commonly for collagen imaging, CMP is a small synthetic peptide that can be readily delivered to vascularized ECM. We believe that the CMPs can not only be used to stain the collagens of tissue samples in vitro but also be used to image collagens in vivo, especially by conjugation of contrast agents for modern imaging systems such as MRI or PET. We also envision linking therapeutic compounds to CMP so that the drugs can be targeted specifically to pathogenic collagens. Therefore, CMP derivatives have immense potential for therapeutic and diagnostic applications in treating a wide range of diseases.
Fig. 5.
Fluorescence confocal microscopy of a frozen, unfixed human liver carcinoma slice stained with carboxyfluorescene-CMP (in green) and CD31 antibody (in red).
Encoding insoluble factors in collagen scaffolds
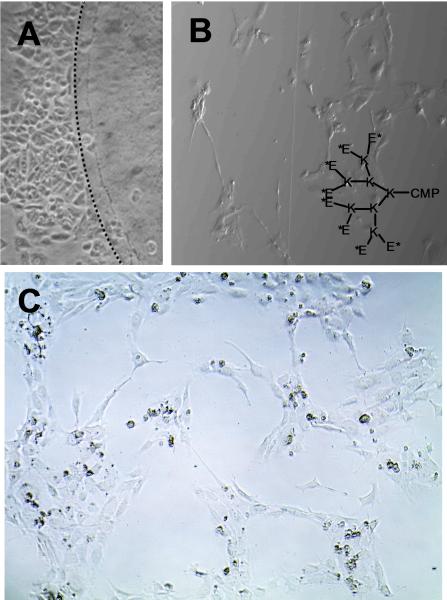
Collagen is inherently a highly adhesive substrate for cell attachment. Many pathological conditions are caused by uncontrolled migration and proliferation of cells through collagen scaffolds.1 We prepared poly-(ethyleneglycol)2000-CMP (PEG-CMP), that was designed to reduce cell adhesiveness when applied to prefabricated collagen film.34 As presented in Fig. 6A, the areas (upper right side of dotted lines) of collagen film treated with PEG-CMP are devoid of cells confirming the effectiveness of our approach in modifying collagen with spatial control.
Fig. 6.
The optical micrographs of human breast epithelial cells (3rd day culture) on collagen films treated with PEG-CMP (A, reproduced with permission from Ref. [34], © American Chemical Society), tube-like morphology of HUVEC cells in collagen gel treated with Glu8-CMP-8 and VEGF (B), and network morphology of group of HUVECs on collagen coating treated with QK-CMP (C).
We also synthesized an anionically charged CMP, Glu8-CMP-8 (N-acetylated form, structure shown in Fig. 6B) that can induce tubulogenesis of endothelial cells (ECs) by attracting vascular endothelial cell growth factor (VEGF) within a 3D collagen gel.90 Fig. 6B shows the micrograph of Glu8-CMP-8 treated collagen gels, 6 hr after human umbilical vascular endothelial cell (HUVEC) seeding, in which numerous tube-like structures are readily observed. The results clearly demonstrate the collagen binding affinity of Glu8-CMP and its ability to induce EC tubulogenesis by binding to VEGF.
We aim to study the effects of VEGF as an insoluble factor which requires strict control over VEGF’s associative state with CMP. Therefore we are currently employing two alternate types of CMP derivatives, CMP-VEGF and CMP conjugated to a 15 amino acid peptide sequence (QK-CMP) recently reported to mimic the angiogenic activity of VEGF.91 While the study of these two conjugates is currently in progress, the QK-CMP peptide has already been synthesized and its collagen affinity and tubulogenic activity has been confirmed (Fig. 6C, manuscript submitted).
PEG-based synthetic hydrogels incorporating CMP
We have also been working on fabricating a new PEG-based tissue engineering matrix. We prepared a novel poly(ethylene oxide) diacrylate (PEODA) hydrogel that carries CMP side chains, and investigated its ability to i) retain cell secreted collagen and promote maintenance of chondrocytes in a three-dimensional culture system,92 and ii) to direct the differentiation of stem cells into the chondrogenic pathway.35 The motivating hypothesis of this work was that the CMPs in the PEG hydrogel will sequester the cell secreted collagens and allow quick formation of an ideal microcellular environment. In a model retention experiment, diffusional loss of type I collagen that was added to the hydrogel was greatly limited when CMP was conjugated to the PEODA hydrogel. Chondrocytes encapsulated in the CMP-conjugated PEODA (CMP/PEODA) gel showed an 87% increase in glycosaminoglycan content and a 103% increase in collagen content compared to those of control PEODA hydrogels. The histology and immunohistochemistry analyses also showed increased staining of extracellular matrix. In the case of mesenchymal stem cell (MSC) encapsulation, the histological and biochemical analysis of the CMP/PEODA gel revealed twice as much glycosaminoglycan and collagen content as compared to control PEODA hydrogels. Moreover, MSCs cultured in CMP/PEODA hydrogel exhibited a lower level of hypertrophic markers, core binding factor alpha 1, and type X collagen than MSCs in PEODA hydrogel as revealed by gene expression and immunohistochemisty. These results indicated that the CMPs enhance the tissue production of cells encapsulated in the PEG-based hydrogel by providing cell-manipulated crosslinks and collagen binding sites that simulate natural extracellular matrix. Compared to PEODA hydrogel, the CMP/PEODA hydrogel provides a much more favorable microenvironment for encapsulated MSCs and even seem to regulate their downstream chondrogenic differentiation.
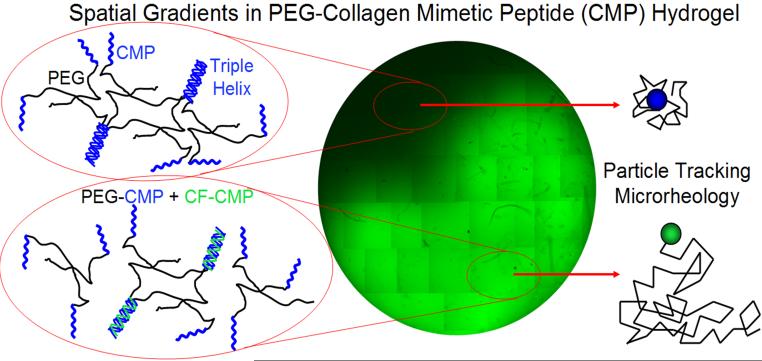
Encouraged by the results of the chondrocyte and MSC encapsulation experiments, we recently explored the possibility of controlling the mechanical properties of PEG hydrogels via triple helical CMP assembly.37 The mechanical properties of tissue scaffolds have major effects on the morphology and differentiation of cells. In contrast to two-dimensional substrates, local biochemical and mechanical properties of three-dimensional hydrogels are difficult to control due to the geometrical confinement. We prepared synthetic 3D hydrogels featuring complexes of four-arm PEG and CMPs that form hydrogels via thermally reversible triple helix-mediated crosslinks. The triple helical crosslinks could be disrupted by raising the temperature of the gel or by addition of hot CMPs that compete for triple helix formation. By injecting free fluorescently labeled CMPs directly into the preformed hydrogel, we were able to create a gradient of CMPs immobilized to the PEG-CMP hydrogel as evidenced by the reconstructed fluorescence microscopy.37 Particle tracking microrheology data showed modulation of local stiffness as well as the creation of stiffness gradients within the PEG-CMP hydrogel. This demonstrated CMP-based synthetic hydrogel’s potential for encoding spatially controlled physico-chemical signals in 3D tissue scaffold (Fig. 7).
Fig. 7.
Spatial gradients of CMP immobilized in 3D PEG-CMP hydrogel. CF-CMP that competes for triple helix was injected to the bottom right side of the gel. Particle tracking microrheology indicated gradual change in stiffness of the gel due to gradient of immobilized CMP that break-up the triple helical cross-links. Reproduced with permission from Ref. [37], © American Chemical Society.
7. Concluding Remarks
From an engineer’s perspective, it is encouraging to witness a burst of research activities in the use of collagen mimetic peptides for recreating the higher order assembly and biological function of natural collagens. This was made possible by a wealth of fundamental knowledge on collagen structure and cell-ECM interactions that has been accumulating over the past 15 years. This research activity comes somewhat late compared to other synthetic protein-based biomaterials, such as α-helix and β-sheet proteins and peptides whose supramolecular assembly is already being widely used for practical applications and clinical trials.93,94 Considering collagen’s critical role in tissue regeneration and its association with numerous pathologic conditions, we believe that continued research with CMPs95 will eventually lead to the design of artificial tissue scaffolds and collagen targeting system that will find practical biomedical use. Fibrous collagens are abundant in nature and are readily available in various forms (e.g. sheets, sponges, or tubes) from commercial sources. Therefore, the practical impact is relatively low for simply reproducing the structural or functional characteristics of natural collagens from CMPs although such efforts will help elucidate the physico-chemical factors responsible for functional ECM assembly. In this regard, we need and welcome new research efforts that go beyond normal production and assembly of collagens, like exploiting unnatural higher-order structures and assembly mechanisms as well as incorporating biological cues that do not belong to natural collagens.
References
- 1.Nimni ME. Collagen. CRC Press; Boca Raton: 1988. [Google Scholar]
- 2.Shoulders MD, Raines RT. Annu. Rev. Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birk DE, Bruckner P. In: Collagen. Brinckmann J, Notbohm H, Müller PK, editors. Vol. 247. Springer Berlin; Heidelberg, Berlin: 2005. pp. 185–205. [Google Scholar]
- 4.Kassner A, Tiedemann K, Notbohm M, Ludwig T, Morgelin M, Reinhardt DP, Chu ML, Bruckner P, Grassel S. J. Mol. Biol. 2004;339:835–853. doi: 10.1016/j.jmb.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 5.Koch M, Foley JE, Hahn R, Zhou PH, Burgeson RE, Gerecke DR, Gordon MK. J. Biol. Chem. 2001;276:23120–23126. doi: 10.1074/jbc.M009912200. [DOI] [PubMed] [Google Scholar]
- 6.Brodsky B, Persikov AV. Adv. Protein Chem. 2005;70:301–339. doi: 10.1016/S0065-3233(05)70009-7. [DOI] [PubMed] [Google Scholar]
- 7.Gaboriaud C, Thielens NM, Gregory LA, Rossi V, Fontecilla-Camps JC, Arlaud GJ. Trends Immunol. 2004;25:368–373. doi: 10.1016/j.it.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Lu J, Teh C, Kishore U, Reid KBM. Biochim. Biophys. Acta. 2002;1572:387–400. doi: 10.1016/s0304-4165(02)00320-3. [DOI] [PubMed] [Google Scholar]
- 9.Jones EY, Miller A. J. Mol. Biol. 1991;218:209–219. doi: 10.1016/0022-2836(91)90885-a. [DOI] [PubMed] [Google Scholar]
- 10.Goodman M, Bhumralkar M, Jefferson EA, Kwak J, Locardi E. Biopolymers. 1998;47:127–142. doi: 10.1002/(SICI)1097-0282(1998)47:2<127::AID-BIP2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 11.Stetefeld J, Frank S, Jenny M, Schulthess T, Kammerer RA, Boudko S, Landwehr R, Okuyama K, Engel J. Structure. 2003;11:339–346. doi: 10.1016/s0969-2126(03)00025-x. [DOI] [PubMed] [Google Scholar]
- 12.Holmgren SK, Bretscher LE, Taylor KM, Raines RT. Chem. Biol. 1999;6:63–70. doi: 10.1016/S1074-5521(99)80003-9. [DOI] [PubMed] [Google Scholar]
- 13.Gauba V, Hartgerink JD. J. Am. Chem. Soc. 2008;130:7509–7515. doi: 10.1021/ja801670v. [DOI] [PubMed] [Google Scholar]
- 14.Brodsky B, Thiagarajan G, Madhan B, Kar K. Biopolymers. 2008;89:345–353. doi: 10.1002/bip.20958. [DOI] [PubMed] [Google Scholar]
- 15.Bella J, Eaton M, Brodsky B, Berman HM. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 16.Holmgren SK, Taylor KM, Bretscher LE, Raines RT. Nature. 1998;392:666–667. doi: 10.1038/33573. [DOI] [PubMed] [Google Scholar]
- 17.Sonntag L-S, Schweizer S, Ochsenfeld C, Wennemers H. J. Am. Chem. Soc. 2006;128:14697–14703. doi: 10.1021/ja0654938. [DOI] [PubMed] [Google Scholar]
- 18.Erdmann RS, Wennemers H. J. Am. Chem. Soc. 132:13957–13959. doi: 10.1021/ja103392t. [DOI] [PubMed] [Google Scholar]
- 19.Berndt P, Fields GB, Tirrell M. J. Am. Chem. Soc. 1995;117:9515–9522. [Google Scholar]
- 20.Kinberger GA, Taulane JP, Goodman M. Tetrahedron. 2006;62:5280–5286. [Google Scholar]
- 21.Kotch FW, Raines RT. Proc. Natl. Acad. Sci. U. S. A. 2006;103:3028–3033. doi: 10.1073/pnas.0508783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Mo X, Kim D, Yu SM. Biopolymers. 2011;95:94–104. doi: 10.1002/bip.21536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rele S, Song YH, Apkarian RP, Qu Z, Conticello VP, Chaikof EL. J. Am. Chem. Soc. 2007;129:14780–14787. doi: 10.1021/ja0758990. [DOI] [PubMed] [Google Scholar]
- 24.Cejas MA, Kinnney WA, Chen C, Vinter JG, Almond HR, Balss KM, Maryanoff CA, Schmidt U, Breslav M, Mahan A, Lacy E, Maryanoff BE. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8513–8518. doi: 10.1073/pnas.0800291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kar K, Amin P, Bryan MA, Persikov AV, Mohs A, Wang YH, Brodsky B. J. Biol. Chem. 2006;281:33283–33290. doi: 10.1074/jbc.M605747200. [DOI] [PubMed] [Google Scholar]
- 26.Lee SG, Lee JY, Chmielewski J. Angew. Chem., Int. Ed. Engl. 2008;47:8429–8432. doi: 10.1002/anie.200802224. [DOI] [PubMed] [Google Scholar]
- 27.Kusebauch U, Cadamuro SA, Musiol HJ, Moroder L, Renner C. Chem. Eur. J. 2007;13:2966–2973. doi: 10.1002/chem.200601162. [DOI] [PubMed] [Google Scholar]
- 28.Przybyla DE, Chmielewski J. J. Am. Chem. Soc. 2008;130:12610–12611. doi: 10.1021/ja804942w. [DOI] [PubMed] [Google Scholar]
- 29.Reyes CD, Petrie TA, Burns KL, Schwartz Z, Garcia AJ. Biomaterials. 2007;28:3228–3235. doi: 10.1016/j.biomaterials.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyes CD, Garcia AJ. J. Biomed. Mater. Res. 2003;65:511–523. doi: 10.1002/jbm.a.10550. [DOI] [PubMed] [Google Scholar]
- 31.Rao GHR, Fields CG, White JG, Fields GB. J. Biol. Chem. 1994;269:13899–13903. [PubMed] [Google Scholar]
- 32.Khew ST, Zhu XH, Tong YW. Tissue Eng. 2007;13:2451–2463. doi: 10.1089/ten.2007.0063. [DOI] [PubMed] [Google Scholar]
- 33.Wang AY, Foss CA, Leong S, Mo X, Pomper MG, Yu SM. Biomacromolecules. 2008;9:1755–1763. doi: 10.1021/bm701378k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang AY, Mo X, Chen CS, Yu SM. J. Am. Chem. Soc. 2005;127:4130–4131. doi: 10.1021/ja0431915. [DOI] [PubMed] [Google Scholar]
- 35.Lee JH, Lee J-S, Chansakul T, Yu C, Elisseeff JH, Yu SM. Biomaterials. 2006;27:5268–5276. doi: 10.1016/j.biomaterials.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Lee HJ, Yu C, Chansakul T, Hwang NS, Varghese S, Yu SM, Elisseeff JH. Tissue Eng Part A. 2008;14:1843–1851. doi: 10.1089/ten.tea.2007.0204. [DOI] [PubMed] [Google Scholar]
- 37.Stahl PJ, Romano NH, Wirtz D, Yu SM. Biomacromolecules. 2010;11:2336–2344. doi: 10.1021/bm100465q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fallas JA, O'Leary LER, Hartgerink JD. Chem. Soc. Rev. 2010;39:3510–3527. doi: 10.1039/b919455j. [DOI] [PubMed] [Google Scholar]
- 39.Smith MCM, Burns N, Sayers JR, Sorrell JA, Casjens SR, Hendrix RW. Science. 1998;279:1834–1834. doi: 10.1126/science.279.5358.1831g. [DOI] [PubMed] [Google Scholar]
- 40.Bamford DH, Bamford JKH. Nature. 1990;344:497–497. doi: 10.1038/344497b0. [DOI] [PubMed] [Google Scholar]
- 41.Rasmussen M, Jacobsson M, Bjorck L. J. Biol. Chem. 2003;278:32313–32316. doi: 10.1074/jbc.M304709200. [DOI] [PubMed] [Google Scholar]
- 42.Bann JG, Peyton DH, Bachinger HP. FEBS Lett. 2000;473:237–240. doi: 10.1016/s0014-5793(00)01493-9. [DOI] [PubMed] [Google Scholar]
- 43.Sylvestre P, Couture-Tosi E, Mock M. J. Bacteriol. 2003;185:1555–1563. doi: 10.1128/JB.185.5.1555-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Y, Keene DR, Bujnicki JM, Hook M, Lukomski S. J. Biol. Chem. 2002;277:27312–27318. doi: 10.1074/jbc.M201163200. [DOI] [PubMed] [Google Scholar]
- 45.Mohs A, Silva T, Yoshida T, Amin R, Lukomski S, Inouye M, Brodsky B. J. Biol. Chem. 2007;282:29757–29765. doi: 10.1074/jbc.M703991200. [DOI] [PubMed] [Google Scholar]
- 46.Han RL, Zwiefka A, Caswell CC, Xu Y, Keene DR, Lukomska E, Zhao ZH, Hook M, Lukomski S. Appl. Microbiol. Biotechnol. 2006;72:109–115. doi: 10.1007/s00253-006-0387-5. [DOI] [PubMed] [Google Scholar]
- 47.Gauba V, Hartgerink JD. J. Am. Chem. Soc. 2007;129:2683–2690. doi: 10.1021/ja0683640. [DOI] [PubMed] [Google Scholar]
- 48.Gauba V, Hartgerink JD. J. Am. Chem. Soc. 2007;129:15034–15041. doi: 10.1021/ja075854z. [DOI] [PubMed] [Google Scholar]
- 49.Russell LE, Fallas JA, Hartgerink JD. J. Am. Chem. Soc. 132:3242–3243. doi: 10.1021/ja909720g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Myllyharju J. Methods Mol. Biol. 2009;522:51–62. doi: 10.1007/978-1-59745-413-1_3. [DOI] [PubMed] [Google Scholar]
- 51.Merle C, Perret S, Lacour T, Jonval V, Hudaverdian S, Garrone R, Ruggiero F, Theisen M. FEBS Lett. 2002;515:114–118. doi: 10.1016/s0014-5793(02)02452-3. [DOI] [PubMed] [Google Scholar]
- 52.Stein H, Wilensky M, Tsafrir Y, Rosenthal M, Amir R, Avraham T, Ofir K, Dgany O, Yayon A, Shoseyov O. Biomacromolecules. 2009;10:2640–2645. doi: 10.1021/bm900571b. [DOI] [PubMed] [Google Scholar]
- 53.Peng YY, Yoshizumi A, Danon SJ, Glattauer V, Prokopenko O, Mirochnitchenko O, Yu ZX, Inouye M, Werkmeister JA, Brodsky B, Ramshawa JAM. Biomaterials. 2010;31:2755–2761. doi: 10.1016/j.biomaterials.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fields CG, Mickelson DJ, Drake SL, McCarthy JB, Fields GB. J. Biol. Chem. 1993;268:14153–14160. [PubMed] [Google Scholar]
- 55.Feng Y, Melacini G, Taulane JP, Goodman M. J. Am. Chem. Soc. 1996;118:10351–10358. [Google Scholar]
- 56.Kwak J, De Capua A, Locardi E, Goodman M. J. Am. Chem. Soc. 2002;124:14085–14091. doi: 10.1021/ja0209621. [DOI] [PubMed] [Google Scholar]
- 57.Cai W, Wong D, Kinberger GA, Kwok SW, Taulane JP, Goodman M. Bioorg. Chem. 2007;35:327–337. doi: 10.1016/j.bioorg.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 58.Khew ST, Tong YW. Biochemistry. 2008;47:585–596. doi: 10.1021/bi702018v. [DOI] [PubMed] [Google Scholar]
- 59.Slatter DA, Miles CA, Bailey AJ. J. Mol. Biol. 2003;329:175–183. doi: 10.1016/s0022-2836(03)00380-2. [DOI] [PubMed] [Google Scholar]
- 60.Berisio R, Granata V, Vitagliano L, Zagari A. Biopolymers. 2004;73:682–688. doi: 10.1002/bip.20017. [DOI] [PubMed] [Google Scholar]
- 61.Byrne C, McEwan PA, Emsley J, Fischer PM, Chan WC. Chem. Commun. 2011;47:2589–2591. doi: 10.1039/c0cc04795c. [DOI] [PubMed] [Google Scholar]
- 62.Koide T, Yuguchi M, Kawakita M, Konno H. J. Am. Chem. Soc. 2002;124:9388–9389. doi: 10.1021/ja026182+. [DOI] [PubMed] [Google Scholar]
- 63.Cai WB, Kwok SW, Taulane JP, Goodman M. J. Am. Chem. Soc. 2004;126:15030–15031. doi: 10.1021/ja0442062. [DOI] [PubMed] [Google Scholar]
- 64.Frank S, Kammerer RA, Mechling D, Schulthess T, Landwehr R, Bann J, Guo Y, Lustig A, Bachinger HP, Engel J. J. Mol. Biol. 2001;308:1081–1089. doi: 10.1006/jmbi.2001.4644. [DOI] [PubMed] [Google Scholar]
- 65.Kusebauch U, Cadamuro SA, Musiol HJ, Lenz MO, Wachtveitl J, Moroder L, Renner C. Angew. Chem., Int. Ed. Engl. 2006;45:7015–7018. doi: 10.1002/anie.200601432. [DOI] [PubMed] [Google Scholar]
- 66.Koide T, Homma DL, Asada S, Kitagawa K. Bioorg. Med. Chem. Lett. 2005;15:5230–5233. doi: 10.1016/j.bmcl.2005.08.041. [DOI] [PubMed] [Google Scholar]
- 67.Yamazaki CM, Asada S, Kitagawa K, Koide T. Biopolymers. 2008;90:816–823. doi: 10.1002/bip.21100. [DOI] [PubMed] [Google Scholar]
- 68.Yamazaki CM, Kadoya Y, Hozumi K, Okano-Kosugi H, Asada S, Kitagawa K, Nomizu M, Koide T. Biomaterials. 2010;31:1925–1934. doi: 10.1016/j.biomaterials.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 69.Kar K, Wang YH, Brodsky B. Protein Sci. 2008;17:1086–1095. doi: 10.1110/ps.083441308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kishimoto T, Morihara Y, Osanai M, Ogata S, Kamitakahara M, Ohtsuki C, Tanihara M. Biopolymers. 2005;79:163–172. doi: 10.1002/bip.20348. [DOI] [PubMed] [Google Scholar]
- 71.Cejas MA, Kinney WA, Chen C, Leo GC, Tounge BA, Vinter JG, Joshi PP, Maryanoff BE. J. Am. Chem. Soc. 2007;129:2202–2203. doi: 10.1021/ja066986f. [DOI] [PubMed] [Google Scholar]
- 72.Hartgerink JD, Beniash E, Stupp SI. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 73.Gottlieb D, Morin SA, Jin S, Raines RT. J. Mater. Chem. 2008;18:3865–3870. doi: 10.1039/b807150k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dujardin E, Peet C, Stubbs G, Culver JN, Mann S. Nano Lett. 2003;3:413–417. [Google Scholar]
- 75.Mo X, Yu SM. Abstracts of Papers of the American Chemical Society. 2003;226:U421–U421. [Google Scholar]
- 76.Eyre DR, Wu J-J. In: Collagen. Brinckmann J, Notbohm H, Müller PK, editors. Vol. 247. Springer; Berlin / Heidelberg: 2005. pp. 207–229. [Google Scholar]
- 77.Yu YC, Berndt P, Tirrell M, Fields GB. J. Am. Chem. Soc. 1996;118:12515–12520. [Google Scholar]
- 78.Fields GB, Lauer JL, Dori Y, Forns P, Yu YC, Tirrell M. Biopolymers. 1998;47:143–151. doi: 10.1002/(SICI)1097-0282(1998)47:2<143::AID-BIP3>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 79.Gore T, Dori Y, Talmon Y, Tirrell M, Bianco-Peled H. Langmuir. 2001;17:5352–5360. [Google Scholar]
- 80.Sarkar N, Banerjee J, Hanson AJ, Elegbede AI, Rosendahl T, Krueger AB, Banerjee AL, Tobwala S, Wang R, Lu X, Mallik S, Srivastava DK. Bioconjug. Chem. 2008;19:57–64. doi: 10.1021/bc070081p. [DOI] [PubMed] [Google Scholar]
- 81.Khew ST, Yang QJ, Tong YW. Biomaterials. 2008;29:3034–3045. doi: 10.1016/j.biomaterials.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 82.Kojima C, Tsumura S, Harada A, Kono K. J. Am. Chem. Soc. 2009;131:6052–6053. doi: 10.1021/ja809639c. [DOI] [PubMed] [Google Scholar]
- 83.Suehiro T, Kojima C, Tsumura S, Harada A, Kono K. Biopolymers. 2010;93:640–648. doi: 10.1002/bip.21413. [DOI] [PubMed] [Google Scholar]
- 84.Zisch AH, Lutolf MP, Ehrbar M, Raeber GP, Rizzi SC, Davies N, Schmokel H, Bezuidenhout D, Djonov V, Zilla P, Hubbell JA. FASEB J. 2003;17:2260–2262. doi: 10.1096/fj.02-1041fje. [DOI] [PubMed] [Google Scholar]
- 85.Lee JY, Choo JE, Park HJ, Park JB, Lee SC, Jo I, Lee SJ, Chung CP, Park YJ. Biochem. Biophys. Res. Commun. 2007;357:68–74. doi: 10.1016/j.bbrc.2007.03.106. [DOI] [PubMed] [Google Scholar]
- 86.Tiller JC, Bonner G, Pan L-C, Kilbanov AM. Biotechnol. Bioeng. 2001;73:246–252. doi: 10.1002/bit.1057. [DOI] [PubMed] [Google Scholar]
- 87.Miles CA, Bailey AJ. Micron. 2001;32:325–332. doi: 10.1016/s0968-4328(00)00034-2. [DOI] [PubMed] [Google Scholar]
- 88.Leikina E, Mertts MV, Kuznetsova N, Leikin S. Proc. Natl. Acad. Sci. U. S. A. 2002;99:1314–1318. doi: 10.1073/pnas.032307099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mo X, An YJ, Yun CS, Yu SM. Angew. Chem., Int. Ed. Engl. 2006;45:2267–2270. doi: 10.1002/anie.200504529. [DOI] [PubMed] [Google Scholar]
- 90.Wang AY, Leong S, Liang YC, Huang RCC, Chen CS, Yu SM. Biomacromolecules. 2008;9:2929–2936. doi: 10.1021/bm800727z. [DOI] [PubMed] [Google Scholar]
- 91.D'Andrea LD, Iaccarino G, Fattorusso R, Sorriento D, Carannante C, Capasso D, Trimarco B, Pedone C. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14215–14220. doi: 10.1073/pnas.0505047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee HJ, Lee JS, Chansakul T, Yu C, Elisseeff JH, Yu SM. Biomaterials. 2006;27:5268–5276. doi: 10.1016/j.biomaterials.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 93.Horii A, Wang X, Zhang S. Nanotechnology and Tissue Engineering: the scaffold. CRC Press; 2008. pp. 283–292. [Google Scholar]
- 94.Omenetto FG, Kaplan DL. Science. 2010;329:528–531. doi: 10.1126/science.1188936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krishna OD, Kiick KL. Biomacromolecules. 2009;10:2626–2631. doi: 10.1021/bm900551c. [DOI] [PMC free article] [PubMed] [Google Scholar]