SUMMARY
The Mycobacterium tuberculosis genome encodes five putative “alternative” ribosomal proteins whose expression is repressed at high Zn2+ concentration. Each alternative protein has a primary homolog that is predicted to bind Zn2+. We hypothesized that zinc triggers a switch between these paired homologous proteins and therefore chose one of these pairs, S18-1/S18-2, to study mechanisms of the predicted competition for their incorporation into ribosomes. As predicted, our data show that Zn2+-depletion causes accumulation of both S18-2 mRNA and protein. In contrast, S18-1 mRNA levels are unchanged to slightly elevated under Zn2+-limited conditions. However the amount of S18-1 protein is markedly decreased. We further demonstrate that both S18 proteins interact with ribosomal protein S6, a committed step in ribosome biogenesis. Zn2+ is absolutely required for the S18-1/S6 interaction, while it is dispensable for S18-2/S6 dimer formation. These data suggest a model in which the S18-1 is the dominant ribosome constituent in high zinc conditions, e.g. inside of phagosomes, but that it can be replaced by S18-2 when zinc is deficient, e.g. in the extracellular milieu. Consequently, Zn2+-depletion may serve as a signal for building alternative ribosomes when M. tuberculosis is released from macrophages, to allow survival in the extracellular environment.
Keywords: ribosomal proteins, zinc, Mycobacterium tuberculosis, transcription regulation, Zur, proteomics
Protein synthesis is an essential process in every living organism. It requires a large portion of a cell's energy, e.g., Escherichia coli uses 40% of its power for translation, and therefore it is expected to be tightly regulated (Wilson and Nierhaus, 2007). In bacteria, ribosomes are one of the major participants in the ppGpp-mediated stringent response, which allows adjustment of growth depending on nutrient availability (Magnusson et al., 2005). Ribosomes are dynamic, complex, and diverse machines; they are not, as they are often viewed, static “translators”, but possess tunable mRNA specificity (Gilbert, 2011). Being large complexes, ribosomes have countless potential points of regulation. In E. coli and other bacteria, rRNA can undergo many types of processing and the resulting modifications may affect ribosome biogenesis and/or confer drug resistance (Connolly and Culver, 2009). Ribosomal proteins (RPs) may also be modified, which can affect their function through interaction with other ribosomal components or ribosome-associated factors. For instance, E. coli has several known strategies to decrease translation during stationary phase or starvation: ribosome dimerization (Ueta et al., 2008), ribosomal protein L12 acetylation (Gordiyenko et al., 2008), and phosphorylation of glutamyl tRNA-synthetase (Germain et al., 2013; Kaspy et al., 2013) or translation elongation factor EF-Tu (Castro-Roa et al., 2013; Cruz et al., 2014).
Intriguingly, in addition to the full set of ribosomal proteins required to form a complete ribosome, around one half of all bacteria have at least one duplicated ribosomal protein (Koonin et al., 2012). These alternative ribosomal proteins (AltRPs) are likely to have a function in translation similar to that of homologous primary ribosomal proteins (PrimRPs), but may also have distinctive features. Therefore, substitution of AltRPs for PrimRPs may form alternative ribosomes with unique characteristics essential for adaptation to certain stresses, providing yet another layer of ribosome regulation. Importantly, in medically relevant bacteria, AltRPs may play a role in pathogenesis and thus may provide new drug targets for treating infectious diseases.
Mycobacterium tuberculosis, one of the deadliest bacterial pathogens in humans, is predicted, on the basis of its genomic sequence, to have five AltRPs (Table 1). M. tuberculosis encounters a complex and changing environment within the human host, to which it must adapt. Ribosome regulation through the incorporation of AltRPs may allow M. tuberculosis to survive and cause tuberculosis (TB) disease or prolonged asymptomatic latent infection. Thus, in order to develop new anti-TB strategies, it would be valuable to know whether alternative ribosomes in M. tuberculosis have a role in responses to stresses encountered during infection, such as those caused by the immune system or by antibiotics.
Table 1.
Paralogs of ribosomal protein genes in Mycobacterium tuberculosis
| Protein namesa | Gene namesb | Rv numbersc | Number of Cys (motif) |
|---|---|---|---|
| L28-1 / L28-2/ L28-3 | rpmB1 / rpmB2 /rpmB3 | Rv2975A / Rv2058c / Rv0105c | 4 (2 CxxC) / 1 / 1 |
| L33-1 / L33-2 | rpmG1 / rpmG2 | Rv0634B / Rv2057c | 4 (2 CxxC) / 0 |
| S14-1 / S14-2 | rpsN1 / rpsN2 | Rv0717 / Rv2056c | 4 (2 CxxC) / 0 |
| S18-1 / S18-2 | rpsR1 / rpsR2 | Rv0055 / Rv2055c | 3 (CxxC + CxxH ) / 1 |
There are some discrepancies in naming of the ribosomal proteins and their corresponding genes in the literature. For consistency, here we added suffix −1 and −2 (or −3 in case of L28) to both gene and protein names in order to distinguish between PrimRPs and AltRPs, respectively.
In the interest of simplicity, we use protein names to designate their corresponding genes in the text and figures.
Rv numbers correspond to gene numbering in M. tuberculosis H37Rv strain (http://www.tbdb.org/). These numbers roughly denote position on the chromosome. Lower case “c” stands for complemented strand. Note that L28-1 was only recently identified and therefore is not present in most databases and the literature (Koonin et al., 2012).
One of the common characteristics of the AltRP/PrimRP pairs is that the PrimRP contains a Cys-rich zinc binding motif (C+) whereas the alternative homolog lacks this motif (C−) (Makarova et al., 2001). Sequence analysis of the AltRPs and their PrimRP homologs in M. tuberculosis suggests that all of them are predicted to be matched C+/C− RPs; in the case of the L28 C+ protein, there are two distinct AltRP C− paralogs (Table 1). Four M. tuberculosis AltRPs genes are in a single operon (L28-3 is separate), while their PrimRP homologs are present at distinct loci in the genome. Expression of many bacterial AltRP genes, including the AltRP operon in M. tuberculosis, is repressed by a “zinc uptake regulator” (Zur), which binds to the promoter region when in complex with Zn2+ (Gaballa et al., 2002; Panina et al., 2003; Maciag et al., 2007; Owen et al., 2007; Shin et al., 2007; Li et al., 2009; Pawlik et al., 2012; Lim et al., 2013). It had been suggested that in some bacteria, such as Bacillus subtilis, E. coli, and Streptomyces coelicolor, C+ PrimRPs serve as zinc-storage proteins that are replaced by matching C− AltRPs when zinc is limited, so that this needed nutrient is released (Akanuma et al., 2006; Nanamiya et al., 2006; Natori et al., 2007; Owen et al., 2007; Gabriel and Helmann, 2009; Hensley et al., 2012). However, mechanisms that would allow this substitution are not known and it has not been investigated in mycobacteria. Of all predicted AltRPs in M. tuberculosis, S18-2 protein is particularly interesting as it has not been studied before and it is not a commonly found paralog (Koonin et al., 2012). Therefore, we initiated a study of alternative ribosomes in M. tuberculosis by focusing on the C− AltRP S18-2 and its primary C+ homolog S18-1.
Here, we show that M. tuberculosis grown in low-zinc medium accumulates S18-2 mRNA and protein, as predicted by the release of the Zur repression of the altRP operon. S18-1 mRNA levels are unchanged to slightly increased, while in contrast, the S18-1 protein amount decreases markedly in low-zinc medium at stationary phase. Our in vitro studies suggest that S18-1 requires Zn2+ binding for successful competition against S18-2 during ribosome biogenesis. Taken together, our in vivo and in vitro data suggest a model in which, under low zinc conditions, Zn2+-dependent post-translational regulation of the S18-1 protein in concert with de-repression of s18-2 gene expression, leads to a switch to alternative, i.e., S18-2-containing, ribosomes in M. tuberculosis.
RESULTS
Expression of S18 ribosomal protein genes during growth
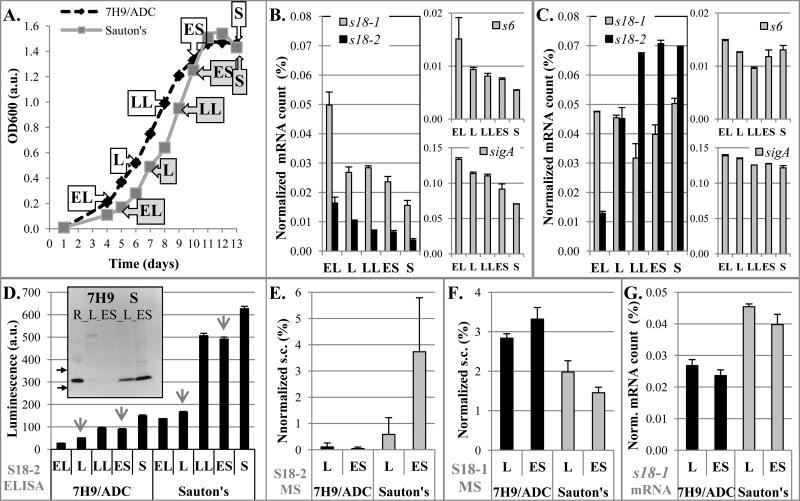
To first examine an unstressed, nutrient replete environment, we investigated expression of S18-1 and its alternative homolog S18-2 during growth in standard Middlebrook 7H9/ADC growth medium. Cultures were sampled at several growth stages and total RNA was isolated for quantification of specific gene expression (Figure 1A). As expected, when demand for protein synthesis and therefore ribosome biogenesis decreased as the culture was getting older, expression of the s18-1 gene decreased (Figure 1B). Expression of the alternative homolog s18-2 followed a similar pattern in this medium (Figure 1B). This pattern parallels the expression patterns of sigA, the primary sigma factor of M. tuberculosis responsible for transcription of many genes and of ribosomal protein S6, a putative interacting partner with the S18 proteins during ribosome biogenesis (Figure 1B).
FIGURE 1. Expression of S18 mRNAs and proteins in M. tuberculosis H37Rv during growth in 7H9/ADC vs. Sauton's medium.
A. Growth curves of M. tuberculosis in indicated media. Aliquots were sampled for analysis at different growth stages as labeled (dashed black line for 7H9/ADC, grey solid line for Sauton's). Abbreviations for growth stages: EL-early logarithmic, L- logarithmic, LL-late logarithmic, ES-early stationary, and S-stationary phase.
B, C. Quantification of RNA: Quantitative real time (qRT)-PCR of specific mRNA for genes encoding S18-1, S18-2, S6 proteins, and sigA in M. tuberculosis grown in 7H9/ADC (B) or Sauton's medium (C), for samples obtained at time points as indicated in panel A. Normalized mRNA count corresponds to the number of specific mRNAs as percentage of 16S rRNA.
D. Quantification of S18-2 protein using ELISA: S18-2 was quantified in total protein extracts from samples indicated in panel A using polyclonal anti-S18-2 antibodies. Relative luminescence is fold change over the background signal. Samples selected for western blotting and qMS are indicated with arrows. Insert: Western blot with same antibodies. R-recombinant S18-2 protein control. Arrows show position of 10 kDa and 15 kDa MW markers.
E, F. Quantification by qMS of S18-2 (E) and S18-1 (F) proteins in selected samples: Samples that were analyzed by mass spectrometry are indicated by arrows in panel D. Spectral count (s.c.) of peptides for each protein was normalized against s.c. of all identified ribosomal proteins in the same sample. Of note, S18-1 and S18-2 proteins do not share any tryptic peptides and detection efficiency of specific peptides may not be the same, so that s.c. cannot be compared between two different proteins. Error bars are standard deviation from two independent samples.
G. Amount of s18-1 mRNA from the same samples is shown for the comparison with the panel C.
Error bars in all graphs are ±1 standard deviation of technical replicates, unless noted differently.
At each time point, we noticed lower expression of the s18-2 gene compared to s18-1 and hypothesized that 6 μM ZnSO4 present in 7H9/ADC could be sufficient to repress expression of this gene via Zur activation. Therefore, we measured S18 protein expression during growth of M. tuberculosis in Sauton's medium prepared without adding zinc (Figure 1A). Interestingly, s18-1 gene expression remained high during growth and no decrease was observed in stationary phase, again matching sigA expression (Figure 1C). As observed in 7H9/ADC, the S6 protein gene followed the expression pattern of the S18-1 protein gene, indicating that they may be coordinately regulated during M. tuberculosis growth.
In contrast to s18-1, s18-2 gene expression steadily increased during logarithmic growth and remained elevated in stationary phase (Figure 1C). This increase in s18-2 expression may be the result of further depletion of zinc during growth and release of Zur repression. In addition, the increase in s18-2 gene expression during growth in Sauton's medium matches the expression of a fluorescent reporter under control of the altRP promoter (data not shown). This result indicates that the promoter activity, rather than changes in mRNA stability likely drove this increase in s18-2 mRNA amount under zinc-depleted conditions. When we added Zn2+ to 7H9/ADC medium to reach 500μM, a zinc concentration in the range experienced by intracellular M. tuberculosis (Wagner et al., 2005), s18-2 expression decreased up to fivefold compared to growth in medium containing 6 μM ZnSO4 (data not shown), showing that Zur repression may not be at its maximum in standard 7H9/ADC medium.
Quantification of the S18 proteins
The higher expression of both S18 genes in stationary phase in Sauton's medium lacking added zinc compared to zinc-replete 7H9/ADC led us to suspect that expression of one (or both) S18 proteins may not be regulated by transcription alone. Both S18 proteins are predicted from sequence homology to bind at the same place in the ribosome and it does not appear that ribosome production is increased overall in low zinc medium to accommodate both proteins. We therefore used anti-S18-2 antibodies to quantify expression of this protein in total protein preparations. ELISA experiments show that in Sauton's medium without added zinc, S18-2 protein roughly follows mRNA levels, increasing several-fold from early log phase to stationary phase (Figure 1D). We confirmed using western blotting that this signal comes mostly from a specific band matching the mobility of a recombinant S18-2 protein control (insert in Figure 1D). For cultures grown in 7H9/ADC, the ELISA results suggested that there was a small increase in S18-2 protein signal in late log through stationary phase, when mRNA levels were decreasing (Figure 1, panels B and D). However, these ELISA signals were very low and were not confirmed on western blotting (insert in Figure 1D).
We were unable to obtain high quality antibodies for quantification of the S18-1 protein. We therefore used quantitative mass spectrometry (qMS) to determine the amount of the S18 proteins in selected samples. Usually, an internal standard or protein labeling is needed to accurately quantify proteins using qMS. Conveniently, the S18 ribosomal proteins are part of a complex and therefore when incorporated into the ribosome, their ratio to other ribosomal proteins should be constant, making other ribosomal proteins a suitable internal control. The large number of ribosomal proteins and their abundance allow for a robust quantification. We optimized this method to reproducibly quantify the S18 proteins from total protein preparations. Proteins migrating in a range between 10-15 kDa on SDS-PAGE were digested, and fragmentation spectra from the two S18 proteins were counted (spectral count - s.c.) and normalized to the total s.c. of all identified ribosomal proteins (Supplemental table 1). This method recapitulated the ELISA results for the S18-2 protein, i.e., showed increased protein levels during later stages of growth in Sauton's medium without added zinc, and minimal change in 7H9/ADC medium (Figure 1E). Strikingly, quantification by mass spectrometry showed that the S18-1 protein concentration is lower in Sauton's medium without added zinc compared to 7H9/ADC, despite mRNA levels being higher (Figure 1F and 1G).
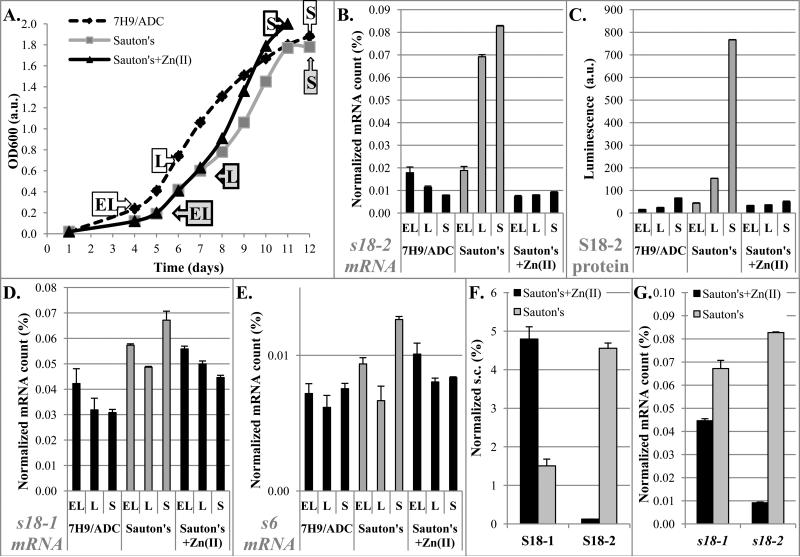
To determine whether the differences in S18 mRNA and protein levels between 7H9-ADC and Sauton's medium are caused by different zinc concentrations, we grew M. tuberculosis in Sauton's medium with or without added Zn2+ and sampled cultures at different growth stages (Figure 2A). ELISA of the S18-2 protein and qRT-PCR of s18-1, s18-2, and s6 mRNAs, showed similar trends in 7H9/ADC and Sauton's medium supplemented with 6 μM Zn2+ (Figure 2B-E). Using the qMS assay, we found that simply leaving out zinc salts from Sauton's medium caused a significant decrease of S18-1 protein, together with dramatic elevation of the S18-2 protein (Figure 2F, Supplemental table 2). These results indicate that zinc is likely to be the key factor that leads to the differences in S18-1 and S18-2 gene expression and protein amounts. The increase in the S18-2 protein levels in medium without added zinc can be explained at the level of transcription by the release of the Zur repression, which was evident by a drop in Zur concentration (Supplemental tables 1 and 2) and a large increase of the s18-2 mRNA (Figure 2G) when cells are grown in low zinc medium. However, the decrease of the S18-1 protein amount in the context of stable to increased levels of the corresponding mRNA in the low zinc environment (Figure 2D and 2G) suggested that regulation of the S18-1 protein in low zinc (Sauton's) medium is post-transcriptional, possibly tied to its putative zinc-binding properties.
FIGURE 2. Changes in mRNA and protein levels of S18-1 and S18-2 with varied zinc concentrations.
A. Growth curves of M. tuberculosis H37Rv. Aliquots were sampled for analysis at different growth stages as labeled: EL-early logarithmic, L-logarithmic, and S-stationary phase (dashed black line - 7H9/ADC, grey solid line - Sauton's medium, black solid line - Sauton's with 6 μM ZnSO4)
B, D, E. Quantification of mRNA: qRT-PCR of specific mRNA for genes encoding S18-2 (B), S18-1 (D), and S6 proteins (E) normalized to 16S rRNA in M. tuberculosis grown in 7H9/ADC or Sauton's medium prepared with or without added ZnSO4, as indicated in panel A.
C. ELISA quantification of S18-2 protein in total protein preparations. Relative luminescence is fold change over the background.
F. Mass spectrometry quantification of S18 proteins at stationary growth phase. Spectral count (s.c.) for identified peptides normalized against s.c. of all identified ribosomal proteins.
G. Normalized specific mRNA quantified by qRT-PCR at stationary growth phase. Data from panels B and D shown to highlight effects of Zn2+ on S18 gene expression.
Error bars in all graphs are ±1 standard deviation of technical replicates.
S18-1 binds zinc ion with high affinity
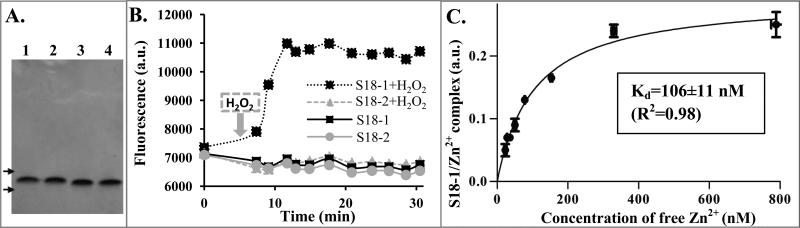
To investigate the disconnect between the S18-1 mRNA and protein levels in low-zinc Sauton's medium, we decided to study these two proteins in vitro. The first question we addressed was whether the S18 proteins, which are predicted to be a C+/C− pair, interact differently with zinc (Table 1). We quantified zinc bound to these proteins during dialysis in the presence of Zn2+, followed by more detailed analysis of protein-Zn2+ interactions. We expressed recombinant S18-1 and S18-2 proteins in E. coli and purified them from inclusion bodies under denaturing conditions, using engineered His tags. Refolding was done in ribosome buffer or, alternatively, in MST buffer with either added Zn2+ or EDTA to allow supplementation or removal of zinc ions, respectively. Preparations were of high purity and yield, as validated by SDS-PAGE (Figure 3A). In order to test if these protein preparations bind Zn2+, we exploited the zinc-binding ability of a fluorescent dye, Mag-fura 2 (MF-2). This fluorescent dye has two distinct excitation maxima, depending on zinc-binding (Simons, 1993). A shorter excitation wavelength maximum (325 nm) indicates MF-2/Zn2+ complex, while the free dye exhibits excitation at a longer wavelength (370 nm), with the isosbestic point of these two forms at 347 nm. Therefore this dye, if added in excess, can be used to quantify free zinc in solution by monitoring fluorescence of the Zn2+-bound dye. In addition, more detailed study of zinc binding can be achieved in a competition assay with a lower concentration of MF-2, as described below.
FIGURE 3. S18-1 protein binds Zn2+.
A. SDS-PAGE of purified proteins. Lanes: 1) S18-1 dialyzed in presence of Zn2+, 2) S18-1 dialyzed in presence of EDTA, 3) S18-2 dialyzed in presence of Zn2+, 4) S18-2 dialyzed in presence of EDTA; arrows depict 10 kDa and 15 kDa marker position
B. Zinc ion release: Protein preparations mixed with the zinc-binding dye MF-2 were exposed to hydrogen peroxide and fluorescence at Ex/Em (325 nm/490 nm) was measured over time.
C. S18-1-Zn2+ binding curve: MF-2 dye was used in a competition assay with S18-1 protein to measure the fraction of Zn2+ bound to S18-1 and calculate the dissociation constant (Kd). Error bars are ±1 standard deviation for technical replicates for both axes.
To analyze zinc-binding of S18-1 and S18-2, the proteins were dialyzed against buffer containing Zn2+ ions, which was then exchanged for zinc-free buffer prior to analysis. The proteins were equilibrated with MF-2 and then challenged with hydrogen peroxide in order to release Zn2+-bound to the S18 proteins (Lee and Helmann, 2006). When the S18-1 protein was exposed to peroxide, increased fluorescence was observed, indicating an increase of zinc ion bound by MF-2 (Figure 3B). Presumably, the predicted Cys-rich zinc-binding motif in the S18-1 protein underwent oxidation and lost its ability to chelate zinc. We calculated from a standard curve using serial dilutions of Zn2+ and a constant concentration of MF-2, that Zn2+ was bound to the S18-1 protein in a 1:1 ratio. There was a modest increase in fluorescence even for preparations that were dialyzed against ribosome buffer alone, which does not have any zinc added, but contains other salts, in particular MgSO4, which might contain zinc as an impurity. We calculated that approximately 5% of the S18-1 protein dialyzed against ribosome buffer without added zinc contains bound metal ion, indicating that binding affinity of the S18-1 protein for zinc is high enough to bind even traces of this metal ion.
In contrast to the S18-1 protein, the S18-2 protein, which has only one Cys that does not appear to be a part of a zinc-binding motif, does not release any free zinc upon exposure to peroxide (Figure 3B). Our finding that S18-1 retained zinc during dialysis against this metal ion, while S18-2 protein did not confirms that this pair of ribosomal protein orthologs fits the paradigm of Zn2+ binding by the C+ protein and not by the C− homolog.
We then investigated whether the observed interaction of the S18-1 with Zn2+ is specific and quantified this interaction. In these experiments proteins were dialyzed against ribosome buffer without added zinc, rather than against buffer containing EDTA, because we could not completely remove this chelator from samples to allow experiments with MF-2. As noted above, 5% of the S18-1 protein preparation has bound metal in the absence of added zinc. Therefore this percentage of bound protein was included in all calculations rather than assuming 100% Zn2+-free protein. We mixed MF-2 with either S18-1 or S18-2 protein to allow competition for binding to zinc ions. Zn2+ concentration was varied and the protein and MF-2 were kept constant at low concentrations. Fluorescence of the bound form of MF-2 was used to determine the fraction of Zn2+ bound to the dye, as described in Experimental Procedures. When MF-2 was mixed with S18-1 protein, the signal from MF-Zn2+ complex was lower compared to mixing with buffer alone or S18-2 protein (Figure S1A). This difference indicates that, as expected, S18-1 but not S18-2 competes with MF-2 for binding of Zn2+. This decrease in the fluorescence of the MF-2/Zn2+ complex in the presence of a non-fluorescent zinc-binding protein was shown previously to enable calculation of the fraction of Zn2+ bound to a competitor (Simons, 1993; Walkup and Imperiali, 1997). We first determined the dissociation constant of MF-2/Zn2+ complex (KMF) from the binding curve in buffer alone (224 nM) (Figure S1B). This constant was then used to determine the concentration of the S18-1/Zn2+ complex, and to calculate a dissociation constant Kd of 106 nM (Figure 3C), indicating high affinity of S18-1 for Zn2+ ion. This constant is likely the upper limit of Kd that may occur in vivo, considering the lack of chaperones and other factors that may facilitate binding of Zn2+ to S18-1 protein.
Both S18-1 and S18-2 interact with S6, but zinc is required only for S18-1 / S6 dimerization
We next determined whether both S18 proteins are components of the ribosome and if binding of zinc to the S18-1 protein has any effect on its function. From their sequence homology, both proteins appear to be typical S18 proteins, but have only 50% sequence identity (Figure 4), and thus may have distinct biochemical properties. Because they are a part of a large complex their individual contribution to overall ribosome function is difficult to probe. We therefore evaluated the interaction the M. tuberculosis S18 proteins with another ribosomal component.
FIGURE 4. M. tuberculosis S18-2 show sequence similarity to S18-1 and both are able to bind S6.
Upper panel: Alignment of M. tuberculosis S18-1 and S18-2 protein sequences. Peptide sequences used to raise polyclonal antibodies are underlined.
Lower panel: S18 proteins dialyzed against EDTA or 10 μM ZnSO4 were titrated into a fluorescently labeled S6 protein. Binding curves are shown as a fraction of S6 bound to S18-1 in presence of Zn2+ (A) or S18-2 in presence of EDTA (B)
Of note, we tested both His-tagged and the untagged versions of the S18 proteins and found that the tag did not affect binding to S6 over the range of protein concentrations relevant for the MST assays. Binding curves of the untagged S18 proteins are shown here.
M. tuberculosis ribosome structure and biogenesis have not been investigated. However, in other bacteria the S18 protein binds to another ribosomal protein, S6, before it enters the 30S subunit. This is a required step for ribosome assembly and the ability to form this heterodimer is a prerequisite for S18 incorporation into ribosomes (Mizushima and Nomura, 1970; Held et al., 1974; Recht and Williamson, 2001). Considering the conservation in ribosomal proteins and ribosome function between mycobacteria and E.coli (Bruell et al., 2008), we hypothesized that M. tuberculosis S18 proteins bind to M. tuberculosis S6. To test this hypothesis we asked if both S18-1 and S18-2 interact with S6 and if there are any differences in these interactions.
Microscale thermophoresis (MST) was used to investigate binding of S6 to S18 proteins. In MST, one binding partner is fluorescently labeled and its concentration is kept constant, while the concentration of an unlabeled interacting component is varied (Wienken et al., 2010). Fluorescent signal is measured over time after a temperature gradient is formed with an infrared (IR) laser. Soon after the IR laser is turned on, there is a sharp change in fluorescence, called MST temperature jump, followed by slower change caused by thermophoresis. The MST temperature jump depends mostly on the fluorophore's close environment, while thermophoresis depends on the hydration shell, shape, charge, and size of the labeled molecule (Seidel et al., 2013). Thus, changes in fluorescence in both regions of the response curve may be affected by binding of ligands to a fluorescently labeled molecule and can be used for quantification of that interaction (Seidel et al., 2013).
We labeled recombinant S6 protein and mixed it with increasing concentrations of S18-1 or S18-2 proteins in MST buffer. Time-dependent changes in fluorescence of the labeled S6 protein after the IR laser was turned on varied with different concentrations of S18-1, consistent with binding of those two proteins (Figure S2A). We found, however, that thermophoresis was an unreliable readout for S6 binding, likely because the S18-1 protein was undergoing conformational changes during its movement in the temperature gradient. Temperature jump, however, responded consistently to addition of the S18-1 or S18-2 protein to labeled S6 (Figure S2). Therefore, we were able to use MST temperature jump to analyze the interaction between labeled S6 protein and the S18 proteins under various conditions.
After optimization of the binding assay, we found that both S18 proteins from M. tuberculosis bind to the S6 protein, with S18-2 having higher affinity than S18-1 when dialyzed against ribosome buffer (data not shown). As a control, we established that S6 did not form homodimers up to 26 μM, the highest concentrations tested (data not shown). These data confirmed formation of S6 heterodimers with both S18 proteins from M. tuberculosis, which suggest that both are likely incorporated into ribosomes during ribosome assembly, as predicted by Nomura's assembly map (Mizushima and Nomura, 1970).
Since S18-1 is a zinc-binding protein, we next investigated the possible role of zinc in regulating the formation of S18-S6 heterodimers. To this end, S18-1 and S18-2 proteins were dialyzed against buffers containing EDTA or Zn2+ to achieve a minimal or maximal Zn2+ content in protein preparations. We found that S18-1 had a very high affinity for S6 in the presence of Zn2+ (Figure 4A), while binding was completely lost in presence of EDTA (data not shown). Together with our previous finding that the S18-1 was a zinc-binding protein, these data indicate that the S18-1 protein requires Zn2+ to make a heterodimer with the S6 protein.
In contrast to the S18-1 protein, the S18-2 protein bound S6 with high affinity in presence of EDTA (Figure 4B). The S18-2 protein consistently aggregated when refolded in the presence of Zn2+, so that we were unable to obtain a good binding curve with S18-2 that was re-folded against buffer containing this ion. In contrast, S18-2 showed much greater stability when it was refolded in presence of EDTA. However, if the S18-2 protein was refolded in ribosome buffer, which does not contain Zn2+, there was no difference in the binding of S18-2 to the S6 protein in the presence of Zn2+ versus EDTA (data not shown), suggesting that Zn2+ does not disrupt interaction of the properly folded S18-2 protein with S6.
The binding constants determined from MST predict that S18-1 protein is competent to compete with S18-2 for binding to S6 only in presence of zinc. To test this hypothesis, we coupled tagged S6 protein to Ni-NTA plate and incubated with the untagged S18-2 protein, which was dialyzed against EDTA. After the unbound protein was washed off, increasing concentration of the untagged S18-1 protein, which was either dialyzed against EDTA or zinc ions was added to the plate to allow for displacement of the S18-2 protein. Following incubation and washing, we performed ELISAs with anti-S18-2 antibodies to detect the amount of S18-2 bound to S6 protein. As expected from MST binding studies, the displacement assay showed that S18-2 binding to S6 decreased with increasing S18-1 concentration only in the presence of Zn2+ ions, while in the presence of EDTA binding was not affected, even with 7 fold higher concentration of S18-1 protein vs. S18-2 protein (Figure S3).
In summary, these in vitro binding studies indicate that S18-1 requires bound zinc to interact with S6, whereas S18-2 is able to bind S6 with high affinity in the absence of zinc. Moreover, these data suggest that S18-2 can successfully compete with the S18-1 protein for binding to the S6 protein during ribosome biogenesis when Zn2+ is depleted.
Expression of S18-1 in the absence of S18-2
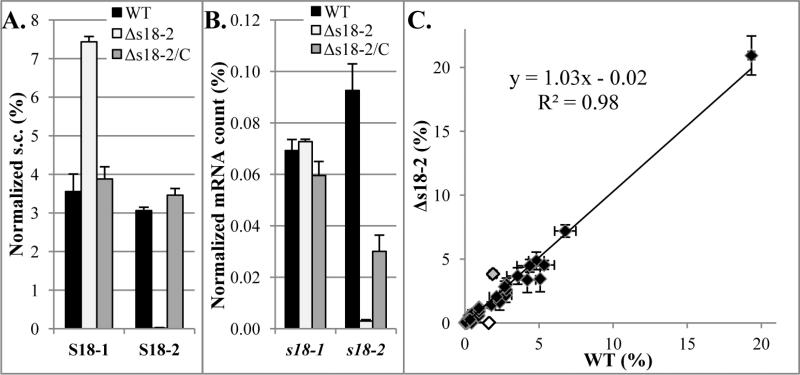
Our in vitro data suggest that both S18-1 and S18-2 proteins are likely to compete for the S6 protein during ribosome biogenesis. Because un-incorporated S18 protein is likely degraded, we hypothesized that the absence of one S18 protein (e.g. S18-2) would allow increased incorporation into ribosomes and protection from proteolysis of the other (e.g. S18-1). To test this hypothesis, we deleted the s18-2 gene and used qMS to measure the amount of the S18-1 protein in this strain compared to the wild type. Consistent with this hypothesis, our data showed a two-fold increase of the S18-1 protein in the Δs18-2 deletion mutant (Figure 5A and Supplemental table 3). Considering that negligible amounts of ribosomal proteins are expected to be outside of the ribosomal complex (Ulbrich and Niearhaus, 1975; Chen et al., 2012), this increase in protein amount likely reflects an increased incorporation of S18-1 in the ribosome in the absence of competing S18-2 protein in this mutant strain.
FIGURE 5. Changes in protein and mRNA levels of S18-1 in the presence or absence of S18-2.
Wild type (WT), deletion mutant (Δs18-2), and complemented (Δs18-2/C) strains of M. tuberculosis H37Rv were grown to stationary phase in Sauton's medium and harvested for protein and RNA quantification.
A. Protein: Mass spectrometry analysis Spectral count (s.c.) for identified peptides normalized against s.c. of all identified ribosomal proteins
B. mRNA: qRT-PCR of S18 genes mRNA normalized against 16S rRNA. Note that the complemented strain does not have wild type levels of s18-2 mRNA, but complements well at the protein level.
C. Comparison of proteins in wild type (WT) and deletion mutant (Δs18-2) strains
Spectral counts of identified proteins were normalized to total spectral counts in each sample. Gray data point is the S18-1 protein and white is the S18-2 protein. The other three AltRPs from the same operon were also detected and their amount was not changed in the absence of the S18-2 protein. Note that only the 10-15kDa range was analyzed. Error bars in all graphs are ±1 standard deviation of technical duplicates.
To test if deletion of s18-2 has off-target effects, e.g., changes in expression of adjacent genes, we reintroduced the s18-2 gene expressed under its native promoter into the deletion mutant. This complemented strain (Δs18-2/C) has S18-2 protein levels comparable to the wild type, which restored the amount of S18-1 protein to the wild type level (Figure 5A), indicating that the absence of the S18-2 protein caused the increase in the S18-1 protein amount.
Although the S18-1 protein amount was increased, deletion of the s18-2 gene did not have any effect on s18-1 mRNA (Figure 5B), indicating translational or post-translational regulation. While translational regulation cannot be excluded, our binding studies described above point to the latter, i.e., direct competition of the two proteins for incorporation into ribosomes, with increased incorporation of S18-1 in the absence of S18-2. The excess S18-1 is likely degraded via proteosomes (Festa et al., 2010).
The effect on S18-1 protein abundance was specific, i.e. we did not observe significant perturbation of other proteins, determined by qMS of all proteins in the 10-15 kDa range, when s18-2 was deleted (Figure 5C). Because the small MW range used for qMS excluded many other proteins, resulting in detection of less than 200 proteins in each sample, we also digested a total protein extract and compared identified proteins in the deletion and parental strains (Supplemental table 4). Using this method, we identified over 1400 proteins in each sample, including all ribosomal proteins. We found a 2.4 fold increase in S18-1 protein in the Δs18-2 mutant compared to the wild type. No other protein showed a two-fold increase, confirming minimal perturbation of other proteins the Δs18-2 strain.
S18-2 protein is incorporated into ribosomes
The S18 protein is essential for growth in both Gram positive and Gram negative model organisms, e.g. E. coli and B. subtilis (Shoji et al., 2011; Akanuma et al., 2012). However, both S18 genes in M. tuberculosis are predicted to be non-essential by high-density transposon mutagenesis (Zhang et al., 2012). Consistent with this result, we were able to construct a Δs18-2 deletion mutant in M. tuberculosis. However, despite repeated attempts, we failed to delete the S18-1 protein encoding gene, suggesting that the S18-2 protein cannot substitute for S18-1 under the in vitro growth conditions used to construct the deletion mutant. Even traces of Zn2+ in the growth media may be sufficient to repress expression of the s18-2 gene and thus prevent effective substitution of S18-2 for the S18-1 protein. Our observation that increased expression of the S18-2 protein does not occur until stationary phase even in medium without added Zn2+ suggests that cell division is required to deplete Zn2+ from the medium to allow Zur de-repression. That S18-1 appears to be essential for growth in M. tuberculosis while S18-2 is not raised the possibility that the S18-2 protein may not be incorporated into ribosomes. Although this is unlikely, considering its tight binding to the S6 protein and homology to other S18 proteins, we decided to isolate ribosomes and determine if they contain the S18-2 protein.
Because cell lysis methods required for ribosome isolation are not compatible with our BSL3 protocols, we used an attenuated auxotroph of M. tuberculosis that can be grown in BSL2 laboratory for these experiments (Sambandamurthy et al., 2002; Sampson et al., 2004). This strain could not be grown in Sauton's medium from low OD to allow for Zn2+ depletion as for the wild type strain. Therefore we grew cultures to mid-log phase, spun down, and then re-suspended in Sauton's medium to OD 0.2. To verify Zn2+ depletion, we transformed the strain with a fluorescent reporter, mCherry, expressed under control of the altRP promoter, so we could follow Zur de-repression. After two rounds of re-growth in Sauton's medium, we observed increased fluorescence, i.e., increase in the altRP promoter activity (Figure S4) in cultures grown without added Zn2+, thus identifying a Zn2+ depletion condition were S18-2 protein should be abundant and available for incorporation into ribosomes.
Ribosomes were isolated from the M. tuberculosis auxotroph using Monolith chromatography, as previously described for M. smegmatis (Trauner et al., 2011). It was shown using M. smegmatis, that ribosomes bind to a strong anion-exchange matrix and elute mostly in elution peak 2 (E2) (Trauner et al., 2011). We got very similar elution profile with M. tuberculosis (Figure 6A). Basic proteins, such as ribosomal proteins, are expected in the unbound fraction (FT), unless they are in a negatively charged complex, i.e., the ribosome. FPLC traces were very similar between two lysates prepared from cultures grown with or without added Zn2+. The sharp increase in absorbance of FT for the Zn2+-depleted sample is most likely from the mCherry reporter (Figure 6A and Figure S5A).
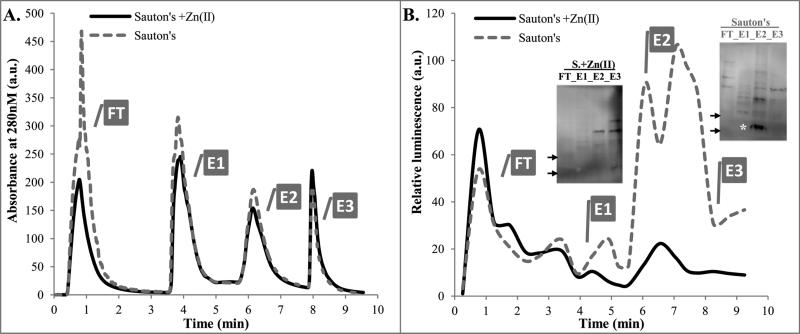
FIGURE 6. Ribosome isolation from M. tuberculosis mc26206 grown in Sauton's medium with or without added Zn2+.
A. FPLC UV traces of two lysates from cultures grown with or without Zn2+, as indicated. FT fraction from growth in Sauton's medium without Zn2+ was pink, indicating presence of mCherry. BugBuster extraction buffer alone strongly absorbs at 280nm in FT (data not shown).
B. ELISA of FPLC fractions using anti-S18-2 antibodies: Each fraction was diluted 4× in 4M urea/TBS, proteins were coupled to wells, and then processed as described in Experimental Procedures.
Insert: Proteins from each peak fraction were precipitated with TCA (20%, overnight at 4°C), washed with cold ethanol, and solubilized in urea/CHAPS buffer. 12μg of each sample was analyzed by Western blot. * band corresponding to MW of the S18-2 protein. Arrows depict 10kDa and 15kDa markers.
The western blot of TRIzol-purified proteins resulted in a smaller number of non-specific bands (see insert in Figure 1D) than sonication in BugBuster buffer (panel B).
FT- flow-through, E1, E2, E3- elution peaks
SDS-PAGE analysis of the FPLC fractions showed that the E2 peak contained low concentration of protein (Figure S5A). Spectra of elution E2 had an absorbance peak at 260nm, rather than 280nm, indicating presence of nucleic acids, most likely rRNA from ribosomes. DNA is unlikely to be present, because of DNase treatment during ribosome preparation. In agreement with this observation, two peaks with size of 16S and 23S rRNA were identified using Bioanalyzer (Agilent Technologies) in E2 fraction (Figure S5B). Despite the relatively low amounts of protein in E2, mass spectrometry analysis of proteins in either high or low Zn2+ samples confirmed presence of all ribosomal proteins, except L28-3 (Supplemental table 5). In agreement with qMS of the total protein (Figures 1 and 2), S18-2 protein amount increased in ribosomal fraction extracted from low Zn2+ cultures, while S18-1 amount slightly decreased (Supplemental table 5). ELISA using anti-S18-2 antibodies showed that the strongest signal came in the E2 fraction only for the Zn2+ depleted sample, also in agreement with our hypothesis that S18-2 is ribosome-associated at low Zn2+ concentration (Figure 6B). Binding of antibodies to proteins from the unbound fractions was also observed. However, this binding was equally strong for Zn2+-containing and Zn2+-depleted cultures and therefore is likely due to a non-specific signal from these polyclonal antibodies. We concentrated and normalized samples from the FPLC fractions (Figure S5C). Western blot from the concentrated samples verifies that a strong band corresponding to the molecular weight of the S18-2 protein was present only in the E2 fraction of the Zn2+-depleted sample (insert in Figure 6B), thus confirming the presence of the S18-2 protein in the ribosome fraction.
Calprotectin activates the altRP promoter in vitro
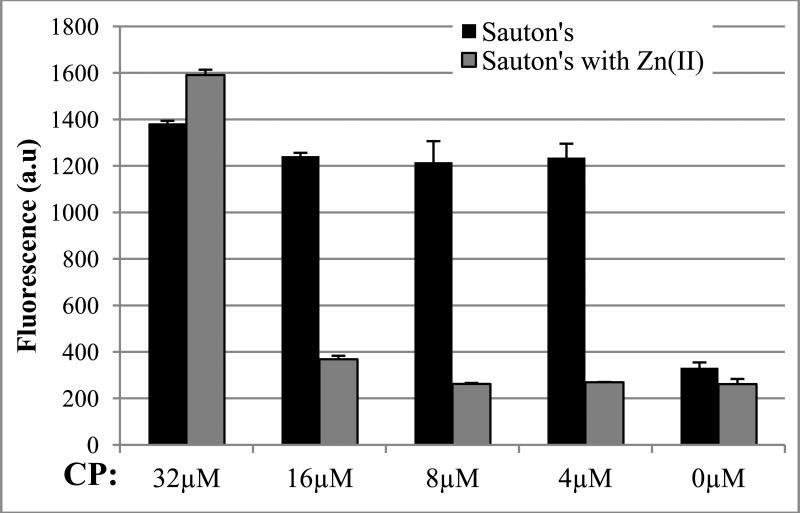
Although zinc overload experienced by phagosomal M. tuberculosis is addressed in several studies (Wagner et al., 2005; Botella et al., 2011), a key question is how/if depletion of Zn2+ is achieved in vivo during TB infection to allow expression of AltRPs. A physiologically plausible mechanism is that the presence of large amounts of calprotectin (CP), which binds Zn2+ and Mn2+ and is released from lysed neutrophils and macrophages in necrotic granulomas, triggers de-repression of the altRP operon in extracellular bacteria. In order to test whether extracellular CP is capable of inducing expression of the AltRPs, we grew the M. tuberculosis auxotroph containing an mCherry reporter expressed under control of the altRP promoter in the presence of recombinant human CP. In Sauton's medium, exogenously added CP over a range of concentrations (4-32μM) markedly increased the activity of the altRP promoter, as shown by increased fluorescence of the reporter (Figure 7), consistent with CP binding residual zinc in this medium. Adding Zn2+ to the growth medium decreased mCherry fluorescence at all but 32 μM CP, which is a concentration sufficient to bind effectively all of the excess Zn2+. In contrast to other organisms tested so far (Damo et al., 2013), we did not observe significant growth inhibition of M. tuberculosis in the presence of CP (data not shown). It is possible that additional stresses are required to show a direct inhibitory effect of CP on a slow growing organism such as M. tuberculosis. Nevertheless, these data suggest that CP may increase expression of AltRPs in vivo by a mechanism involving depletion of zinc at the site of M. tuberculosis infection.
FIGURE 7. altRP promoter activity in M. tuberculosis mc26206 grown in presence of calprotectin.
Serial dilutions of calprotectin (CP) were added to cultures in absence or presence of 50 μM Zn2+. Expression of an mCherry reporter under control of the altRP promoter was measured by fluorescence at 590 nm/635 nm after 4 days of growth. Error bars are ±1 standard deviation for technical replicates.
DISCUSSION
We present the first study of a C+/C− pair of homologous ribosomal proteins from M. tuberculosis and demonstrate a link between their abundance in the cell and extracellular zinc availability. Zinc was shown previously to inhibit expression of genes encoding alternative ribosomal proteins including the object of this study, s18-2, through activation of the transcriptional repressor Zur (Maciag et al., 2007). Here, we confirm up-regulation of s18-2 expression in M. tuberculosis grown in low zinc medium and demonstrate that this leads to increased amounts of S18-2 protein in the cell. We also examined the effect of zinc on its primary homolog, S18-1. Compared to the S18-2 protein, we show that zinc has an opposite effect on the S18-1 protein amount, i.e., S18-1 protein concentration is decreased with lower zinc availability in growth medium. However, in contrast to S18-2 regulation, this change in the S18-1 protein amount under zinc limitation is not explained by transcriptional control. Rather, our studies suggest that zinc is required for binding of the S18-1 to the S6 ribosomal protein. Consequently, the inability of S18-1 to bind S6 in the absence of zinc decreases S18-1 incorporation into ribosomes. Zinc-dependent control of the S18-1 protein abundance is therefore post-translational. Further, under zinc-limiting conditions, even Zn2+-replete S18-1 must compete with increased amounts of S18-2 protein for binding to S6 and incorporation into ribosomes.
S18 paralogs are relatively rare compared to more common duplications of L31, L33, L36, and S14 (Makarova et al., 2001; Koonin et al., 2012). S18-2 homologs are present in all mycobacteria, except M. avium 104 and M. leprae; the latter species is missing all five AltRPs (Koonin et al., 2012). Ribosomal proteins in bacteria and archea are classified either as zinc-binding (C+) or homologs that are missing the zinc-binding motif (C−) (Makarova et al., 2001). Among 58 predicted ribosomal proteins, M. tuberculosis has six C+ ribosomal proteins, four of which have C− AltRP paralogs (Table 1); the two that do not have paralogs are L31 and L36. The existence of Cys-rich Zn2+-binding motifs in C+ proteins may have advantages under some conditions. Because thermophilic bacteria and hyperthermophilic archea have a higher frequency of C+ proteins, it was suggested that zinc-binding provides higher protein stability at elevated temperatures and may be more common in early evolution of life (Makarova et al., 2001). However, the function of C+ ribosomal proteins present in mesophilic bacteria and their C− paralogs is not well understood.
Zinc storage is one of the proposed roles for C+ ribosomal proteins (Gabriel and Helmann, 2009; Nanamiya and Kawamura, 2010). It had been suggested that one of the alternative ribosomal proteins, L31-2 in Bacillus subtilis is able to displace its zinc binding homolog and allow release of zinc from L31-1 in zinc depleted environments (Akanuma et al., 2006; Nanamiya et al., 2006; Natori et al., 2007). A similar function of the L31-1/L31-2 pair has been proposed for E.coli and S. coelicolor (Owen et al., 2007; Graham et al., 2009). However, this function may not be common for all alternative ribosomal proteins. L31 is loosely associated with ribosomes and may be more readily replaced than AltRPs, which are core proteins. It has been therefore speculated that alternative ribosomal proteins, such as the core ribosomal protein S14 in B. subtilis, substitute for primary homologs in de novo ribosome assembly when zinc is limited (Natori et al., 2007). It is not known if such ribosomes exhibit any additional or distinct function. Furthermore, there are no published data suggesting a role of any M. tuberculosis AltRP. In terms of zinc storage, it has been shown that M. tuberculosis has a copper and zinc binding protein MymT that has a role in resistance to toxicity form high copper and zinc concentrations encountered in macrophages (Gold et al., 2008; Rowland and Niederweis, 2012). This protein may also serve for metal ion storage after the bacterium is released from phagosomes, so that M. tuberculosis may not rely on ribosomal proteins for this purpose.
Our in vitro data show that S18-1 protein binds zinc with high affinity and thus it is indeed functional C+ protein. This experimental demonstration of the predicted binding of Zn2+ by the C+ S18 protein has not been reported before in any species, but it is consistent with the finding that a 26-mer peptide from Thermus thermophilus L36 containing the CxxC-CxxxxH motif is capable of binding Zn2+ (Boysen and Hearn, 2001), as well as studies on zinc-binding L31 from B. subtilis and E. coli (Nanamiya et al., 2004; Hensley et al., 2012) and S14 from T. thermophilus (Tsiboli et al., 1998). However, binding affinities for those C+ proteins have not been published and we do not know if the Kd of 106 nM that we obtained for S18-1/Zn2+ complex formation in vitro is physiologically relevant. Though the affinity for zinc is relatively high, this dissociation constant is substantially higher than the 20 pM of free Zn2+ measured in E.coli grown without zinc supplementation (Wang et al., 2011). However, the intracellular concentration of free zinc ion in E. coli can increase to 40 nM in the presence of 2.5 μM zinc salt in growth medium (Wang et al., 2011). This concentration is 200 times lower than the 500 μM concentration experienced by intracellular M. tuberculosis (Wagner et al., 2005). Thus, the free zinc concentration in M. tuberculosis residing in macrophages is likely to be high enough for binding to the S18-1 protein. M. tuberculosis may also have other mechanisms for decreasing the Kd in vivo, such as incorporation of zinc during translation.
The S18-1/Zn2+ complex and the S18-2 protein have comparable affinity for the S6 protein. Based on our observation of lower expression of the S18-2 protein when zinc concentration is high, S18-1/Zn2+ is likely to be the major S18 protein in ribosomes in M. tuberculosis residing in phagosomes (Figure 8, upper panel). When zinc is depleted, however, for example in the extracellular milieu, S18-2 expression will be up-regulated and the increased amount of S18-2 protein likely competes with S18-1 for binding to the S6 protein (Figure 8, lower panel). Further, our in vitro data show that S18-1 cannot bind the S6 protein in the absence of zinc. Thus, during ribosome biogenesis under zinc-limiting conditions, S18-1 would be out-competed by S18-2, which does bind S6 in the absence of zinc (Figure 8, lower panel).
FIGURE 8. “Zinc-switch” model.
Competition and differential incorporation of S18-1 or S18-2 proteins into ribosomes when Zn2+ is present at high (upper panel) or at low concentration (lower panel). Line thickness represents level of expression, shift in equilibrium, or degree of incorporation into ribosomes. Unincorporated proteins are not shown in the model because they are likely degraded. This model is consistent with findings first reported for the L31 ribosomal protein in B. subtilis (Nanamiya et al., 2004).
We have shown that the level of S18-1 protein is decreased in M. tuberculosis grown to stationary phase in low zinc medium, despite slightly higher levels of S18-1 mRNA. This result suggests that un-incorporated S18-1 is likely degraded, as suggested previously for B. subtilis L31, L33, and S14 (Moore and Helmann, 2005). This degradation of S18-1 may result from highly conserved mechanisms of protein quality control, such as chaperone-mediated degradation of misfolded proteins. Notably, S18-1 was shown to be a part of the “pupylome” (Festa et al., 2010), suggesting that free S18-1 protein is actively targeted for degradation by M. tuberculosis proteasome. In agreement with this model of S18 competition and degradation of unincorporated S18-1, when the competing S18-2 protein is absent, we see an increase in the S18-1 protein levels, which is likely protected from degradation in ribosomes.
A low zinc environment that would release the repression of AltRPs expression during M. tuberculosis infection, as presented in the model in Figure 8, has not yet been identified. However, it has been shown in several infectious diseases that the host metal-binding protein calprotectin (CP) inhibits growth of pathogens via “nutritional immunity”, i.e., sequestration of essential metals such as zinc and manganese (Corbin et al., 2008; Damo et al., 2012). Interestingly, mice infected with M. tuberculosis have increased expression of CP (Shepelkova et al., 2013). CP is also found at elevated levels in plasma from TB patients and was positively correlated with disease severity, implying that it has a role in TB pathogenesis (Pechkovsky et al., 2000). Neutrophils, a major source of extracellular CP, which is released upon their lysis, are found in M. tuberculosis-infected necrotic granulomas and greater numbers of neutrophils correlate with more severe disease and larger number of extracellular bacteria (Berry et al., 2012). Our in vitro data indicate that presence of CP stimulates expression of AltRPs. Therefore, release of intracellular M. tuberculosis from macrophages into the extracellular environment may provide new challenges for the bacterium, including low availability of zinc, which may be overcome with a switch to AltRP-containing ribosomes (Figure 8, lower panel).
Beyond allowing translation to continue in zinc-depleted conditions in vivo, what other functions might Alt ribosomes have in M. tuberculosis? One possibility is that AltRPs may provide for flexible adaptation to stresses. The S18 proteins have a predicted role in translation initiation based on their homology to the S18 protein from E. coli and T. thermophilus (Wimberly et al., 2000; Schuwirth et al., 2005; Yusupova et al., 2006; Marzi et al., 2007). The M. tuberculosis S18-1 and S18-2 proteins share less than 50% sequence similarity and therefore incorporation of one versus the other into ribosomes may be a determinant of ribosomal specificity and/or activity.
In conclusion, our data indicate that zinc regulates the switch between S18-1 and S18-2 expression and incorporation into ribosomes in M. tuberculosis via transcriptional and post-translational mechanisms. These mechanisms are likely to apply to other C+/C− pairs in mycobacteria and in other bacteria. Based on our data and prior results on zinc concentrations in different in vivo environments, PrimRP-containing ribosomes are likely to be dominant in the intracellular M. tuberculosis population. AltRP-containing ribosomes may constitute a greater proportion of ribosomes in the extracellular, zinc-depleted milieu and contribute to survival of the extracellular M. tuberculosis population. In this low zinc environment, translation regulation by AltRPs may have effects on important aspects of TB pathogenesis such as transmission and drug tolerance.
EXPERIMENTAL PROCEDURES
Strains, media, and cloning
M. tuberculosis H37Rv was grown either in Middlebrook 7H9 liquid culture medium (Difco) supplemented with ADC (0.5% albumin, 0.2% glucose, 0.085% NaCl and 0.05% Tween 80) or Sauton's medium prepared without zinc salts (0.05% KH2PO4, 0.05% MgSO47H2O, 0.2% citric acid, 0.005% ferric ammonium citrate, 6% glycerol, 0.4% Asparagine, 0.05% Tween 80, pH 7.4). Zinc sulfate was added to medium at 6 μM or 500 μM final concentrations, as indicated for the different experiments.
A deletion mutant for the s18-2 gene was constructed in M. tuberculosis H37Rv background using a suicide plasmid as described before (Raman et al., 2004), with some modifications. Flanking regions of the gene were cloned into pDONR221 vector (Life Technologies), followed by inserting a hygromycin resistance marker cassette. The construct was then transferred into pDONR-1351 plasmid, a vector derived from pRH1351 into a donor vector using Gateway conversion system (Life Technologies). Double cross-over containing clones were selected as described previously (Raman et al., 2004) and proper insertion was confirmed with PCR and sequencing. Complementation of the Δs18-2 strain (Δs18-2/C) was achieved by transforming the mutant with an integrating plasmid containing another copy of the s18-2 gene and a native promoter. The plasmid used to transform the Δs18-2 strain (pMV306altRP-s18-2) was constructed by cloning the s18-2 gene with the altRP promoter into pMV306 vector (Stover et al., 1991). Transformants were confirmed by PCR and sequencing.
In order to be able to monitor atlRP promoter activity during growth, M. tuberculosis H37Rv and an auxotroph for Leu and pantothenate biosynthesis M. tuberculosis mc26206 (ΔleuCD panCD) (Sampson et al., 2004) was transformed with a fluorescent reporter, mCherry, expressed under the altRP promoter in integrating vector (pMV306altRP-mCherry). The protein is optimized for expression in mycobacteria and was transferred from pVV16-mCherry plasmid (Abramovitch et al., 2011).
Gateway system (Life Technologies) was used for cloning and expression of ribosomal proteins. In short, genes encoding proteins S18-1, S18-2 (see Table 1), and S6 (rpsF / Rv0053) were amplified using genomic DNA from M. tuberculosis H37Rv with primers containing attB sites and TEV cleavage site at the N-terminus. After cloning into pDONR221 and verifying the absence of mutations by sequencing, the inserts were transferred into pDEST17, an expression vector that provides a poly-His tag at the N-terminus, as described in the manufacturer's manual (Life Technologies). The constructed plasmids were used to transform OverExpress C41 E. coli host cells (Lucigen) for recombinant protein expression.
Quantitative RT-PCR
Total RNA was extracted from M. tuberculosis cultures at different growth stages by breaking cell pellets in a bead-beater in the presence of glass beads and TRIzol reagent and then following the manufacturer's protocol for RNA purification (Life Technologies). Remaining genomic DNA was removed with Turbo DNase (Ambion) and final purification of RNA was done using RNeasy columns (Qiagen). qRT-PCR was performed using One-Step QuantiFast (Qiagen) in an ABI 7000 instrument (Life Technologies), following the recommended protocol. Total RNA was 100ng per reaction, except for 16S rRNA where it was decreased to 0.1 ng. Relative specific mRNA abundance, i.e., copy number per 100 ng of total RNA, for each gene was calculated from standard curves, which were obtained from amplification of genes from M. tuberculosis genomic DNA (gDNA) dilutions. Ct (cycle threshold) as given by ABI PRISM software (default settings) vs. ln(gDNA in pg) was plotted for each set of primers. Copy numbers were calculated by using following formula:
| (1) |
with a and b being slopes and intercepts from specific standard curves, respectively, NA is Avogadro constant, and MW is molecular weight of gDNA calculated to be 2726326000 g/mol. Specific mRNA was then normalized to 16S rRNA amount. All analyses were done in Excel.
ELISA and western blotting
Polyclonal rabbit antibodies were obtained from NeoBioSci (Cambridge, MA). Antibodies were raised against conjugated peptides. For S18-1, a peptide close to the N-terminus was chosen and for S18-2, the peptide sequence that was the most unique was from the very C-terminus (Figure 4). After obtaining antibodies from immunized rabbits, the same peptides were used for affinity purification. Proteins were extracted from M. tuberculosis cultures by beating the cell pellets with glass beads in TRIzol (Life Technologies), followed by the instructions from the manufacturer's manual for protein preparation. Final ethanol pellets were solubilized in 9.5 M urea 2% CHAPS pH 9.1 buffer, as it was shown to be favorable for solubilization of TRIzol protein preparations previously (Man et al., 2006). Proteins were quantified with the DC assay (Biorad) and 96-well Optiplates HB (Perkin Elmer) were coated with 100 μL of 0.01 μg mL−1 protein in Tris Buffered Saline (TBS) overnight at 4°C. After blocking in 3% BSA in TBS with 0.05% Tween 20 (TBS-T), 100 μL of 1:1000 dilution of antibodies were incubated overnight at 4°C. Unbound antibodies were removed by washing in TBS-T and plates were subsequently incubated with 100 μL of anti-rabbit HRP-conjugated secondary antibodies at a 1:5000 dilution (Cell Signaling) for 1 hour at room temperature. SuperSignal Pico (Pierce) was used for chemiluminescent detection.
Western blotting was performed in parallel for selected samples to evaluate specificity of the antibodies. Urea/CHAPS protein preparations from TRIzol extraction were mixed with reducing Laemmli sample buffer (Bio-Rad), separated on SDS-PAGE (TGX Any kD precast gel, Biorad), and transferred onto SequiBlot PVDF (Biorad). The same antibody dilutions and incubation times were used as for ELISA. After incubation with LumiGLO (Cell Signaling), membrane was scanned with Kodak Image Station. Untagged recombinant proteins were used as positive controls both for ELISA and western blotting.
Quantitative mass spectrometry (qMS)
Selected protein preparations from TRIzol extraction were analyzed by qMS. Proteins in urea/CHAPS (total of 15 μg) were separated by SDS-PAGE as described above for western blotting. After staining with GelCode Blue (Pierce) the 10-15 kDa gel region was excised and cut into four strips. The proteins in each strip were separately digested as described previously, except using 10 times lower reagent volumes (Prisic et al., 2010). The digested peptides were resuspended in 5% acetonitrile/5% formic acid and analyzed by LC-MS/MS. For more details about runs, see Supplemental tables. Spectral count analysis and ribosomal protein-based normalization was carried out using Excel.
Filter-aided sample preparation (FASP) was used to obtain total proteome as described before (Wisniewski et al., 2009). In short, the wild type, Δs18-2 mutant, and the complemented strain were grown in Sauton's medium until stationary phase. Proteins were isolated using TRIzol and solubilized in urea/CHAPS, as described for ELISA. One hundred μg of total protein was digested following published protocol (Wisniewski et al., 2009) and the resulting peptides were analyzed as described above.
Protein expression and purification
Expression of recombinant proteins S6, S18-1, and S18-2 was induced with 1 mM IPTG in E. coli grown to a log phase in LB medium. After 2.5 hour shaking at 37 °C, cell pellets were lysed and inclusion bodies were purified with BugBuster (Novagen). Proteins were purified on HisPur Cobalt columns (Pierce) in presence of 8M urea using manufacturer's protocol for purification under denaturing conditions. Proteins were then re-folded by step-wise removal of urea during dialysis in either ribosome buffer (20 mM HEPES pH 7.6, 10% glycerol, 330 mM KCl, 20 mM MgCl2, 1 mM DTT) or microscale thermophoresis (MST) buffer (20 mM HEPES pH 7.6, 330 mM KCl, 0.05% Tween 20, 14 mM β mercaptoethanol), in presence of EDTA (100 μM) or ZnSO4 (10 μM). Untagged S18-1 and S18-2 proteins were obtained by digestion with ProTEV protease (Promega) for 2 days at 4 °C in presence of 1 M urea, denatured by adding 4 M urea, and refolded as described above.
Re-natured and dialyzed proteins were checked for quality on SDS-PAGE gel and their concentration was determined by measuring absorbance at 280 nm in NanoDrop using extension coefficients as calculated by VectorNTI (Life Technologies). Proteins were used immediately or stored at −80 °C in small aliquots. Proteins dialyzed in ribosome buffer were slightly more stable, but buffer had to be exchanged into MST buffer before MST experiments using spin buffer exchange columns (Pierce) and used immediately.
Mag-fura 2 binding
Purified recombinant proteins were transferred into 20 mM HEPES pH 7.6 330 mM KCl, 1 mM TCEP with a buffer exchange spin column (Pierce). For competition assay, each of the S18-1 or S18-2 protein was mixed with a zinc-binding fluorescent dye Mag-fura 2 tetrapotassium salt (MF-2) (Life Technologies) to a final concentration of 1 μM of protein and 0.4 μM of the dye. No protein, i.e., MF-2 in buffer was used to determine zinc binding affinity of MF-2. Samples were mixed 1:1 with the increased concentration of zinc sulfate in black narrow-well 96 well plates to a total volume of 50 μL. After incubation at room temperature for 15 minutes fluorescence was measured at two excitation / emission combinations: 325 nm/505 nm (MF-2/Zn2+) and 347 nm/505 nm (isosbestic point).
The following steps were applied to calculate dissociation constants, using an approach similar to that previously described by Walkup and Imperiali (Walkup and Imperiali, 1997). The main difference was measurement of fluorescence, not absorbance, as suggested by Simons (Simons, 1993).
First, the dissociation constant for MF-2/Zn2+ complex was calculated from mixing MF-2 at 0.2 μM concentration with a range of ZnSO4 two-fold serial dilutions. Fraction of MF-2 bound to Zn2+, i.e., [MF]b was determined from:
| (2) |
with [MF]t being a total concentration of MF-2 (200 nM) and R was a ratio of fluorescence for bound MF-2 normalized to isosbestic point calculated from this formula:
| (3) |
Rmin and Rmax were R values in presence of 100mM EGTA and 10μM Zn2+, respectively. Background fluorescence of the buffer (with no added Zn2+) was used to calculate traces of Zn2+ present in buffer and this value was added to known concentrations of titrated ZnSO4. This total Zn2+ concentration [Zn]t was plotted against MFb obtained from formula (2) in GraphPad Prism to calculate zinc dissociation constant for MF-2 (KMF) from one site binding model:
| (4) |
Second, when zinc-binding protein was added to MF-2, they both were competing for zinc binding and therefore total Zn2+ concentration was distributed between free, MF-bound and protein-bound form:
| (5) |
Each component of the equation (5) can be transformed to calculate Zn2+ bound to a protein, [P · Zn], by introducing another variable, a ratio between bound and free MF-2:
| (6) |
From steady-state equilibrium, this can be converted to
| (7) |
| (8) |
Therefore, concentration of protein that binds zinc is readily calculated from:
| (9) |
for each varied total zinc concentration, [Zn]t. The fraction of Zn2+ that is already bound to the protein from buffer at “zero” Zn2+ concentration was determined from Zn2+ release experiments (see below), was added to the [P · Zn] values. Finally, the protein fraction bound to zinc was fit into a competitive binding model vs. [Zn]:
| (10) |
with [P]t being 500 nM, [Zn] and [P · Zn] calculated from (7) and (9), respectively, and Kd was the dissociation constant for the tested protein, in this case, S18-1.
For zinc release experiments, recombinant proteins at 50 μM concentration in ribosome buffer lacking Mg2+ and DTT was mixed 1:1 with 100 μM MF-2 in total volume of 40 μL in black narrow 96 well plates and fluorescence was read at 325/490 nm to maximize sensitivity. Proteins were oxidized by adding 20 mM H2O2. This experiment was done with proteins dialyzed against zinc or to determine binding of zinc to proteins from impurities in ribosome buffer. Zinc standard curves were used to calculate amount of released zinc from the proteins.
Microscale Thermophoresis (MST)
Proteins were fluorescently labeled using NT protein labeling kit RED-NHS (NanoTemper Technologies) and un-reacted dye was removed according to the manufacturer's manual. Labeled S6 protein (10 nM) was mixed with a two-fold serial dilution of a binding partner (S18-1 or S18-2), incubated at room temperature for 20 minutes and filled into capillaries for measurements. Labeling of the S18 proteins resulted in inactivation and therefore all experiments presented here were done with the labeled S6 protein. MST runs were performed on Monolith NT.115 (NanoTemper Technologies) with 30s laser on and 5s laser off and with 70% LED and 80% laser power. All data were initially analyzed using NT Analysis software and then exported into Excel and GraphPad software for further analysis. Raw data were transformed into percentage of bound S6 proteins by setting the unbound signal to 0% and value at the saturation to 100% in Excel. GraphPad was used to calculate dissociation constants using one site specific binding model.
Displacement assay
All steps were done at room temperature and 200 μL per well volume. MST buffer was used for washing, protein and antibody dilutions. His-tagged S6 protein (30 pmol/well) was bound to Ni-NTA coated plates (5Prime) in MST buffer for 40 minutes and washed 4 times with MST buffer. Fifty pmol of the untagged S18-2 protein (dialyzed against 100 μM EDTA as described above) was added to the plate and incubated for 30 minutes. The unbound protein was washed 4 times again and increasing concentration of the untagged S18-1 protein (dialyzed against 100 μM EDTA or 10 μM Zn2+) was applied to the pre-bound S6/S18-2 complex on the plate. After incubation for 20 minutes, wells were washed 4 times with MST buffer (containing EDTA or Zn2+) and S18-2 was detected with anti-S18-2 antibodies as described for ELISA above, but with shorter incubation times (30 minutes for primary and 30 minutes for secondary antibodies) and diluting antibodies in MST buffer with EDTA or Zn2+.
Ribosome purification
Ribosomes were purified from M. tuberculosis auxotroph, as described previously for Mycobacterium smegmatis, with few modifications (Trauner et al., 2011). The auxotroph with altRP-mCherry reporter was grown in 7H9/OADC (Hardy Diagnostics) supplemented with Leu (50μg/mL), pantothenate (24μg/mL), glycerol (0.5%), and Tyloxapol (0.05%) to mid-log phase and then transferred into Sauton's medium supplemented with Leu, pantothenate, and Tween 80 (0.05%) at OD 0.2. When the culture reached late stationary phase, it was re-inoculated to OD 0.2 with or without 500μM ZnSO4. Fluorescence at 590nm/635nm and absorbance at 600nm was measured in a clear-bottom 96-well plate until maximal fluorescence was reached and cultures were at stationary phase. Cell pellets of the attenuated M. tuberculosis strain were resuspended in lysis buffer containing 10× BugBuster (Novagen) diluted to 1× with FPLC buffer A (70mM KCl, 10mM MgCl2, 10mM TrisHCl pH7.5) and containing 0.5mM CaCl2. One mL of lysis buffer per each mg of wet cells was used. Baseline-ZERO DNase (Epicentre) was added at 10μL per 1mL of lysate and incubated with shaking at room temperature for 15 minutes. Lysates were then sonicated with cooling in Bioruptor (Diagenode) 8 times for 7.5 minutes (30s off and 30s at highest setting). Unbroken cells and debris were removed by centrifugation at 21000g for 1 hour at 4°C. Supernatants (0.8mL) were then purified on FPLC and 1mL fractions were collected, as described previously (Trauner et al., 2011).
altRP promoter activity in presence of calprotectin
M. tuberculosis mc26206 with altRP-mCherry reporter was grown in 7H9/OADC (Hardy Diagnostics) supplemented with Leu (50 μg mL−1), pantothenate (24 μg mL−1), glycerol (0.5%), and Tyloxapol (0.05%) to mid-log phase and then transferred into Sauton's medium supplemented with Leu, pantothenate, and Tween 80 (0.05%) at OD 0.2. The culture was grown until late stationary phase (OD~2) and then diluted 10 times into same medium supplemented with 2 mM CaCl2, with or without added 50 μM ZnSO4. Recombinantly expressed and purified human calprotectin (CP) (Hunter and Chazin, 1998) was dialyzed against Tris buffer (25 mM, 75 mM NaCl, pH 7.4) in presence of 2 mM CaCl2 and mixed with the diluted culture (1:4). Cultures were grown and fluorescence of mCherry reporter (590 nm/635 nm) or OD (590 nm) was measured in Tecan plate reader at 37 °C for 4 days.
Supplementary Material
Acknowledgements
We thank Dr. Yang Shi from Boston Children's Hospital for allowing us to use MST machine and Dr. Roman Alpatov for the initial training. We are also grateful to Dr. William Jacobs Jr. from Albert Einstein College of Medicine for providing the M. tuberculosis mc26206 strain and Dr. David Russell from Cornell University and Dr. Robert Abramovitch from Michigan State for pVV16-mCherry plasmid used in this study. Four supplemental tables are available. This work is supported by Harvard University Center for AIDS Research (CFAR) Scholar Award to S.P., an NIH funded program (P30 AI060354) and NIH grant R01AI099204 and R01AI099204 to R.N.H. and R01 AI101171 to W.J.C.
The abbreviations used are
- RP
ribosomal protein
- PrimRP
primary ribosomal protein
- AltRP
alternative ribosomal protein
- qRT-PCR
quantitative real-time PCR
- qMS
quantitative mass spectrometry
- MF-2
Mag-fura 2
- MST
microscale thermophoresis
- FASP
filter-aided sample preparation
- TB
tuberculosis
- CP
calprotectin
Footnotes
The authors declare that there are no conflicts of interest.
REFERENCES
- Abramovitch RB, Rohde KH, Hsu F-F, Russell DG. aprABC: A Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol Microbiol. 2011;80:678–694. doi: 10.1111/j.1365-2958.2011.07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akanuma G, Nanamiya H, Natori Y, Nomura N, Kawamura F. Liberation of Zinc-containing L31 ( RpmE ) from ribosomes by its paralogous gene product , YtiA , in Bacillus subtilis. J Bacteriol. 2006;188:2715–2720. doi: 10.1128/JB.188.7.2715-2720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akanuma G, Nanamiya H, Natori Y, Yano K, Suzuki S, Omata S, et al. Inactivation of ribosomal protein genes in Bacillus subtilis reveals importance of each ribosomal protein for cell proliferation and cell differentiation. J Bacteriol. 2012;194:6282–91. doi: 10.1128/JB.01544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MPR, Graham CM, Mcnab FW, Xu Z, Bloch AA, Oni T, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2012;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella H, Peyron P, Levillain F, Poincloux R, Poquet Y, Brandli I, et al. Mycobacterial p(1)-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe. 2011;10:248–59. doi: 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boysen RI, Hearn MT. The metal binding properties of the CCCH motif of the 50S ribosomal protein L36 from Thermus thermophilus. J Pept Res. 2001;57:19–28. doi: 10.1034/j.1399-3011.2001.00752.x. [DOI] [PubMed] [Google Scholar]
- Bruell CM, Eichholz C, Kubarenko A, Post V, Katunin VI, Hobbie SN, et al. Conservation of bacterial protein synthesis machinery: initiation and elongation in Mycobacterium smegmatis. Biochemistry. 2008;47:8828–39. doi: 10.1021/bi800527k. [DOI] [PubMed] [Google Scholar]
- Castro-Roa D, Garcia-Pino A, Gieter S, De, Nuland N. a J. van, Loris R, Zenkin N. The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat Chem Biol. 2013;9:811–7. doi: 10.1038/nchembio.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SS, Sperling E, Silverman JM, Davis JH, Williamson JR. Measuring the dynamics of E. coli ribosome biogenesis using pulse-labeling and quantitative mass spectrometry. Mol Biosyst. 2012;8:3325–3334. doi: 10.1039/c2mb25310k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly K, Culver G. Deconstructing ribosome construction. Trends Biochem Sci. 2009;34:256–63. doi: 10.1016/j.tibs.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science (80− ) 2008;319:962–5. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- Cruz JW, Rothenbacher FP, Maehigashi T, Lane WS, Dunham CM, Woychik N. a. Doc toxin is a kinase that inactivates elongation factor Tu. J Biol Chem. 2014;289:7788–7798. doi: 10.1074/jbc.M113.544429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damo S, Chazin WJ, Skaar EP, Kehl-fie TE. Inhibition of bacterial superoxide defense. Virulence. 2012;3:325–328. doi: 10.4161/viru.19635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damo SM, Kehl-Fie TE, Sugitani N, Holt ME, Rathi S, Murphy WJ, et al. Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc Natl Acad Sci. 2013 doi: 10.1073/pnas.1220341110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festa R. a, McAllister F, Pearce MJ, Mintseris J, Burns KE, Gygi SP, Darwin KH. Prokaryotic ubiquitin- like protein (Pup) proteome of Mycobacterium tuberculosis. PLoS One. 2010;5:e8589. doi: 10.1371/journal.pone.0008589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Wang T, Ye RW, John D, Helmann JD. Functional analysis of the Bacillus subtilis Zur regulon. J Bacteriol. 2002;184:6508–14. doi: 10.1128/JB.184.23.6508-6514.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SE, Helmann JD. Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J Bacteriol. 2009;191:6116–22. doi: 10.1128/JB.00802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain E, Castro-Roa D, Zenkin N, Gerdes K. Molecular Mechanism of Bacterial Persistence by HipA. Mol Cell. 2013;52:248–254. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- Gilbert WV. Functional specialization of ribosomes? Trends Biochem Sci. 2011;36:127–32. doi: 10.1016/j.tibs.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Deng H, Bryk R, Vargas D, Eliezer D, Roberts J, et al. Identification of a copper-binding metallothionein in pathogenic mycobacteria. Nat Chem Biol. 2008;4:609–16. doi: 10.1038/nchembio.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordiyenko Y, Deroo S, Zhou M, Videler H, Robinson CV. Acetylation of L12 increases interactions in the Escherichia coli ribosomal stalk complex. J Mol Biol. 2008;380:404–14. doi: 10.1016/j.jmb.2008.04.067. [DOI] [PubMed] [Google Scholar]
- Graham AI, Hunt S, Stokes SL, Bramall N, Bunch J, Cox AG, et al. Severe zinc depletion of Escherichia coli: roles for high affinity zinc binding by ZinT, zinc transport and zinc-independent proteins. J Biol Chem. 2009;284:18377–89. doi: 10.1074/jbc.M109.001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held W. a, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- Hensley MP, Gunasekera TS, Easton JA, Sigdel TK, Stacy A, Klingbeil L, et al. Characterization of Zn(II)- responsive ribosomal proteins YkgM and L31 in E. coli. J Inorg Biochem. 2012:164–172. doi: 10.1016/j.jinorgbio.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter MJ, Chazin WJ. High level expression and dimer characterization of the S100 EF-hand proteins, migration inhibitory factor-related proteins 8 and 14. J Biol Chem. 1998;273:12427–35. doi: 10.1074/jbc.273.20.12427. [DOI] [PubMed] [Google Scholar]
- Kaspy I, Rotem E, Weiss N, Ronin I, Balaban NQ, Glaser G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun. 2013;4:3001. doi: 10.1038/ncomms4001. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Yutin N, Puigbo P. Phylogenomics of prokaryotic ribosomal proteins. PLoS One. 2012;7:e36972. doi: 10.1371/journal.pone.0036972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-W, Helmann JD. Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J Biol Chem. 2006;281:23567–78. doi: 10.1074/jbc.M603968200. [DOI] [PubMed] [Google Scholar]
- Li Y, Qiu Y, Gao H, Guo Z, Han Y, Song Y, et al. Characterization of Zur-dependent genes and direct Zur targets in Yersinia pestis. BMC Microbiol. 2009;9:128. doi: 10.1186/1471-2180-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CK, Hassan K. a, Penesyan A, Loper JE, Paulsen IT. The effect of zinc limitation on the transcriptome of Pseudomonas protegens Pf-5. Environ Microbiol. 2013;15:702–15. doi: 10.1111/j.1462-2920.2012.02849.x. [DOI] [PubMed] [Google Scholar]
- Maciag A, Dainese E, Rodriguez GM, Milano A, Provvedi R, Pasca MR, et al. Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J Bacteriol. 2007;189:730–40. doi: 10.1128/JB.01190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nyström T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–42. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Ponomarev V. a, Koonin EV. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2001;2:1–14. doi: 10.1186/gb-2001-2-9-research0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man T-K, Li Y, Dang TA, Shen J, Perlaky L, Lau CC. Optimising the use of TRIzol-extracted proteins in surface enhanced laser desorption/ ionization (SELDI) analysis. Proteome Sci. 2006;4:3. doi: 10.1186/1477-5956-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi S, Myasnikov AG, Serganov A, Ehresmann C, Romby P, Yusupov M, Klaholz BP. Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Cell. 2007;130:1019–31. doi: 10.1016/j.cell.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Mizushima S, Nomura M. Assembly Mapping of 30S Ribosomal Proteins from E. coli. Nature. 1970;226:1214–1218. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- Moore CM, Helmann JD. Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol. 2005;8:188–95. doi: 10.1016/j.mib.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Nanamiya H, Akanuma G, Natori Y, Murayama R, Kosono S, Kudo T, et al. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol Microbiol. 2004;52:273–83. doi: 10.1111/j.1365-2958.2003.03972.x. [DOI] [PubMed] [Google Scholar]
- Nanamiya H, Kawamura F. Towards an elucidation of the roles of the ribosome during different growth phases in Bacillus subtilis. Biosci Biotechnol Biochem. 2010;74:451–461. doi: 10.1271/bbb.90859. [DOI] [PubMed] [Google Scholar]
- Nanamiya H, Kawamura F, Kosono S. Proteomic study of the Bacillus subtilis ribosome : Finding of zinc-dependent replacement for ribosomal protein L31 paralogues. J Gen Appl Microbiol. 2006;52:249–258. doi: 10.2323/jgam.52.249. [DOI] [PubMed] [Google Scholar]
- Natori Y, Nanamiya H, Akanuma G, Kosono S, Kudo T, Ochi K, Kawamura F. A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol Microbiol. 2007;63:294–307. doi: 10.1111/j.1365-2958.2006.05513.x. [DOI] [PubMed] [Google Scholar]
- Owen G. a, Pascoe B, Kallifidas D, Paget MSB. Zinc-responsive regulation of alternative ribosomal protein genes in Streptomyces coelicolor involves zur and sigmaR. J Bacteriol. 2007;189:4078–86. doi: 10.1128/JB.01901-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panina EM, Mironov A. a, Gelfand MS. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc Natl Acad Sci U S A. 2003;100:9912–7. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlik M-C, Hubert K, Joseph B, Claus H, Schoen C, Vogel U. The zinc-responsive regulon of Neisseria meningitidis comprises 17 genes under control of a Zur element. J Bacteriol. 2012;194:6594–603. doi: 10.1128/JB.01091-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechkovsky DV, Zalutskaya OM, Ivanov GI, Misuno NI. Calprotectin (MRP8/14 protein complex) release during mycobacterial infection in vitro and in vivo. FEMS Immunol Med Microbiol. 2000;29:27–33. doi: 10.1111/j.1574-695X.2000.tb01501.x. [DOI] [PubMed] [Google Scholar]
- Prisic S, Dankwa S, Schwartz D, Chou MF, Locasale JW, Kang C-M, et al. Extensive phosphorylation with overlapping specificity by Mycobacterium tuberculosis serine/threonine protein kinases. Proc Natl Acad Sci U S A. 2010;107:7521–6. doi: 10.1073/pnas.0913482107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman S, Hazra R, Dascher CC, Husson RN. Transcription Regulation by the Mycobacterium tuberculosis Alternative Sigma Factor SigD and Its Role in Virulence. J Bacteriol. 2004;186:6605–6616. doi: 10.1128/JB.186.19.6605-6616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recht MI, Williamson JR. Central domain assembly: thermodynamics and kinetics of S6 and S18 binding to an S15-RNA complex. J Mol Biol. 2001;313:35–48. doi: 10.1006/jmbi.2001.5018. [DOI] [PubMed] [Google Scholar]
- Rowland JL, Niederweis M. Resistance mechanisms of Mycobacterium tuberculosis against phagosomal copper overload. Tuberculosis. 2012;92:202–10. doi: 10.1016/j.tube.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandamurthy VK, Wang X, Chen B, Russell RG, Derrick S, Collins FM, et al. A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med. 2002;8:1171–4. doi: 10.1038/nm765. [DOI] [PubMed] [Google Scholar]
- Sampson SL, Dascher CC, Sambandamurthy VK, Russell RG, Jacobs WR, Bloom BR, Hondalus MK. Protection Elicited by a Double Leucine and Pantothenate Auxotroph of Mycobacterium tuberculosis in Guinea Pigs. Infect Immun. 2004;72:3031–3037. doi: 10.1128/IAI.72.5.3031-3037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya M. a, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Seidel S. a I., Dijkman PM, Lea W. a, Bogaart G. van den, Jerabek-Willemsen M, Lazic A, et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59:301–15. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepelkova G, Pommerenke C, Alberts R, Geffers R, Evstifeev V, Apt A, et al. Analysis of the lung transcriptome in Mycobacterium tuberculosis-infected mice reveals major differences in immune response pathways between TB-susceptible and resistant hosts. Tuberculosis. 2013;93:263–9. doi: 10.1016/j.tube.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Shin J-H, Oh S-Y, Kim S-J, Roe J-H. The zinc-responsive regulator Zur controls a zinc uptake system and some ribosomal proteins in Streptomyces coelicolor A3(2). J Bacteriol. 2007;189:4070–7. doi: 10.1128/JB.01851-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji S, Dambacher CM, Shajani Z, Williamson JR, Schultz PG. Systematic chromosomal deletion of bacterial ribosomal protein genes. J Mol Biol. 2011;413:751–61. doi: 10.1016/j.jmb.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons TJ. Measurement of free Zn2+ ion concentration with the fluorescent probe mag-fura-2 (furaptra). J Biochem Biophys Methods. 1993;27:25–37. doi: 10.1016/0165-022x(93)90065-v. [DOI] [PubMed] [Google Scholar]
- Stover C, la Cruz V. de, Fuerst T, Burlein J, Benson L, Bennett L, et al. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- Trauner A, Bennett MH, Williams HD. Isolation of bacterial ribosomes with monolith chromatography. PLoS One. 2011;6:e16273. doi: 10.1371/journal.pone.0016273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiboli P, Triantafillidou D, Franceschi F, Choli-Papadopoulou T. Studies on the Zn-containing S14 ribosomal protein from Thermus thermophilus. Eur J Biochem. 1998;256:136–41. doi: 10.1046/j.1432-1327.1998.2560136.x. [DOI] [PubMed] [Google Scholar]
- Ueta M, Ohniwa RL, Yoshida H, Maki Y, Wada C, Wada A. Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J Biochem. 2008;143:425–33. doi: 10.1093/jb/mvm243. [DOI] [PubMed] [Google Scholar]
- Ulbrich B, Niearhaus K. Pools of Ribosomal Proteins in Escherichia coli. Eur J Biochem. 1975;57:49–54. doi: 10.1111/j.1432-1033.1975.tb02275.x. [DOI] [PubMed] [Google Scholar]
- Wagner D, Maser J, Lai B, Cai Z, Barry CE, Höner zu Bentrup K, et al. Elemental analysis of Mycobacterium avium, Mycobacterium tuberculosis, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J Immunol. 2005;174:1491–1500. doi: 10.4049/jimmunol.174.3.1491. [DOI] [PubMed] [Google Scholar]
- Walkup GK, Imperiali B. Fluorescent chemosensors for divalent zinc based on zinc finger domains . Enhanced oxidative stability , metal binding affinity , and structural and functional characterization. J Am Chem Soc. 1997;119:3443–3450. [Google Scholar]
- Wang D, Hurst TK, Thompson RB, Fierke C. a. Genetically encoded ratiometric biosensors to measure intracellular exchangeable zinc in Escherichia coli. J Biomed Opt. 2011;16:087011. doi: 10.1117/1.3613926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons Jr WM, Morgan-Warren RJ, Carter AP, Vonrhein C, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Wisniewski J, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–4. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- Zhang YJ, Ioerger TR, Huttenhower C, Long JE, Sassetti CM, Sacchettini JC, Rubin EJ. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 2012;8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.