Abstract
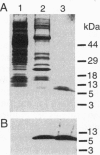
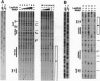
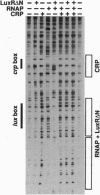
LuxR, the Vibrio fischeri luminescence gene (lux) activator, is the best-studied member of a family of bacterial transcription factors required for cell density-dependent expression of specific genes involved in associations with eukaryotic hosts. Neither LuxR nor any other LuxR homolog has been shown to bind DNA directly. We have purified the LuxR C-terminal transcriptional activator domain from extracts of recombinant Escherichia coli in which this polypeptide was expressed. The purified polypeptide by itself binds to lux regulatory DNA upstream of the lux box, a 20-bp palindrome that is required for LuxR activity in vivo, but it does not bind to the lux box. However, the LuxR C-terminal domain together with RNA polymerase protects a region including the lux box and the lux operon promoter from DNase I cleavage. There is very little protection of the lux operon promoter region from DNase I digestion in the presence of RNA polymerase alone. Apparently, there is a synergistic binding of the LuxR C-terminal domain and RNA polymerase to the promoter region. The upstream binding region for the purified polypeptide encompasses a binding site for cAMP receptor protein (CRP). Under some conditions, CRP binding can block the binding of the LuxR C-terminal domain to the upstream binding region, and it can also block the synergistic binding of the LuxR C-terminal domain and RNA polymerase to the lux box and luminescence gene promoter region. This description of DNA binding by the LuxR C-terminal domain should lead to an understanding of the molecular interactions of the LuxR family of transcriptional activators with regulatory DNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adar Y. Y., Simaan M., Ulitzur S. Formation of the LuxR protein in the Vibrio fischeri lux system is controlled by HtpR through the GroESL proteins. J Bacteriol. 1992 Nov;174(22):7138–7143. doi: 10.1128/jb.174.22.7138-7143.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adar Y. Y., Ulitzur S. GroESL proteins facilitate binding of externally added inducer by LuxR protein-containing E. coli cells. J Biolumin Chemilumin. 1993 Sep-Oct;8(5):261–266. doi: 10.1002/bio.1170080506. [DOI] [PubMed] [Google Scholar]
- Choi S. H., Greenberg E. P. Genetic dissection of DNA binding and luminescence gene activation by the Vibrio fischeri LuxR protein. J Bacteriol. 1992 Jun;174(12):4064–4069. doi: 10.1128/jb.174.12.4064-4069.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. H., Greenberg E. P. The C-terminal region of the Vibrio fischeri LuxR protein contains an inducer-independent lux gene activating domain. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11115–11119. doi: 10.1073/pnas.88.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine J. H., Shadel G. S., Baldwin T. O. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5688–5692. doi: 10.1073/pnas.86.15.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan K. M., Greenberg E. P. Evidence that GroEL, not sigma 32, is involved in transcriptional regulation of the Vibrio fischeri luminescence genes in Escherichia coli. J Bacteriol. 1992 Aug;174(15):5132–5135. doi: 10.1128/jb.174.15.5132-5135.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap P. V., Greenberg E. P. Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein-luxR protein regulatory circuit. J Bacteriol. 1988 Sep;170(9):4040–4046. doi: 10.1128/jb.170.9.4040-4046.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard A., Burlingame A. L., Eberhard C., Kenyon G. L., Nealson K. H., Oppenheimer N. J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981 Apr 28;20(9):2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Nealson K., Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983 Mar;32(3):773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- Engebrecht J., Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J., Silverman M. Nucleotide sequence of the regulatory locus controlling expression of bacterial genes for bioluminescence. Nucleic Acids Res. 1987 Dec 23;15(24):10455–10467. doi: 10.1093/nar/15.24.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua W. C., Winans S. C., Greenberg E. P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994 Jan;176(2):269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray K. M., Passador L., Iglewski B. H., Greenberg E. P. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J Bacteriol. 1994 May;176(10):3076–3080. doi: 10.1128/jb.176.10.3076-3080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J. J., Gussin G. N. Interactions between Escherichia coli RNA polymerase and lambda repressor. Mutations in PRM affect repression of PR. J Mol Biol. 1988 Apr 20;200(4):735–739. doi: 10.1016/0022-2836(88)90484-6. [DOI] [PubMed] [Google Scholar]
- Kaplan H. B., Greenberg E. P. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985 Sep;163(3):1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan H. B., Greenberg E. P. Overproduction and purification of the luxR gene product: Transcriptional activator of the Vibrio fischeri luminescence system. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6639–6643. doi: 10.1073/pnas.84.19.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb A., Igarashi K., Ishihama A., Lavigne M., Buckle M., Buc H. E. coli RNA polymerase, deleted in the C-terminal part of its alpha-subunit, interacts differently with the cAMP-CRP complex at the lacP1 and at the galP1 promoter. Nucleic Acids Res. 1993 Jan 25;21(2):319–326. doi: 10.1093/nar/21.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolibachuk D., Greenberg E. P. The Vibrio fischeri luminescence gene activator LuxR is a membrane-associated protein. J Bacteriol. 1993 Nov;175(22):7307–7312. doi: 10.1128/jb.175.22.7307-7312.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski A. M., Parr T. R., Jr, Angus B. L., Hancock R. E., Ghiorse W. C., Greenberg E. P. Isolation of the outer membrane and characterization of the major outer membrane protein from Spirochaeta aurantia. J Bacteriol. 1987 Jan;169(1):172–179. doi: 10.1128/jb.169.1.172-179.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lane D., Prentki P., Chandler M. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol Rev. 1992 Dec;56(4):509–528. doi: 10.1128/mr.56.4.509-528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meighen E. A. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991 Mar;55(1):123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O. Nucleoprotein structures at positively regulated bacterial promoters: homology with replication origins and some hypotheses on the quaternary structure of the activator proteins in these complexes. Mol Microbiol. 1989 Mar;3(3):455–458. doi: 10.1111/j.1365-2958.1989.tb00191.x. [DOI] [PubMed] [Google Scholar]
- Ren Y. L., Garges S., Adhya S., Krakow J. S. Cooperative DNA binding of heterologous proteins: evidence for contact between the cyclic AMP receptor protein and RNA polymerase. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4138–4142. doi: 10.1073/pnas.85.12.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel G. S., Devine J. H., Baldwin T. O. Control of the lux regulon of Vibrio fischeri. J Biolumin Chemilumin. 1990 Apr-Jun;5(2):99–106. doi: 10.1002/bio.1170050205. [DOI] [PubMed] [Google Scholar]
- Shadel G. S., Young R., Baldwin T. O. Use of regulated cell lysis in a lethal genetic selection in Escherichia coli: identification of the autoinducer-binding region of the LuxR protein from Vibrio fischeri ATCC 7744. J Bacteriol. 1990 Jul;172(7):3980–3987. doi: 10.1128/jb.172.7.3980-3987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slock J., VanRiet D., Kolibachuk D., Greenberg E. P. Critical regions of the Vibrio fischeri luxR protein defined by mutational analysis. J Bacteriol. 1990 Jul;172(7):3974–3979. doi: 10.1128/jb.172.7.3974-3979.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]