SUMMARY
One oscillation of Cyclin-dependent kinase (Cdk) activity, largely driven by periodic synthesis and destruction of cyclins, is tightly coupled to a single complete eukaryotic cell division cycle. Tight linkage of different steps in diverse cell cycle processes to Cdk activity has been proposed to explain this coupling. Here we demonstrate an intrinsically oscillatory module controlling nucleolar release and re-sequestration of the Cdc14 phosphatase, which is essential for mitotic exit in budding yeast. We find that this Cdc14 release oscillator functions at constant and physiological cyclin-Cdk levels, and is therefore independent of the Cdk oscillator. However, the frequency of the release oscillator is regulated by cyclin-Cdk activity. This observation together with its mechanism suggests that the intrinsically autonomous Cdc14 release cycles are locked at once-per-cell-cycle through entrainment by the Cdk oscillator in wild type cells. This concept may have broad implications for the structure and evolution of eukaryotic cell cycle control.
INTRODUCTION
Periodicity of eukaryotic cell cycle events is driven by oscillations of cyclin-dependent kinase (Cdk) activity (Morgan, 2007). Checkpoint surveillance mechanisms enforce correct ordering of events by delaying subsequent events until previous ones are finished; however, checkpoints are not generally essential for ordering the unperturbed cell cycle (Elledge, 1996; Weinert et al., 1994).
Cdk activity oscillation can order cell cycle events by a ‘ratchet’-like mechanism: high cyclin-Cdk activity triggers the initiation of a cell cycle event, but inhibits its completion or re-initiation (Nasmyth, 1996; Stern and Nurse, 1996). Therefore, as a result of having different thresholds for cyclin-Cdk, cell cycle events occur in order, exactly once per cyclin-Cdk cycle. This mechanism is well established in control of DNA replication (Kearsey and Cotterill, 2003), and similar ratchet-like mechanisms may apply to processes such as spindle and bud morphogenesis (Bloom and Cross, 2007).
Independent of molecular mechanisms, ratchet control entails that locking Cdk activity at any constant level should arrest the controlled process at a specific step. However, some cell cycle events may occur cyclically without oscillation of mitotic cyclin-Cdk activity, such as SPB/centrosome duplication cycles, and periodic budding and cell-cycle-regulated transcription in budding yeast (Gard et al., 1990; Haase and Reed, 1999; Haase et al., 2001; McCleland and O'Farrell, 2008; Sluder et al., 1990). Such ‘endocycles’ pose a challenge to the concept of cyclin-Cdk-based ratchet control, but the relevance of these endocycles to the mitotic cell cycle, and what mechanism, if any, entrains them to mitotic cyclin-Cdk cycles, remains unclear
Cdc14 is an essential mitotic phosphatase in budding yeast. Cdc14 is restrained and inhibited in the nucleolus by the constitutively nucleolar Net1p, except in mitosis (Shou et al., 1999; Visintin et al., 1999). The spindle orientation checkpoint (SPOC, regulating the mitotic exit network MEN) and cyclin-Cdk oscillation can regulate Cdc14 localization and activity (Azzam et al., 2004; Bardin et al., 2000; Jaspersen and Morgan, 2000; Pereira et al., 2000; Queralt et al., 2006; Stegmeier and Amon, 2004; Stegmeier et al., 2002). Disrupting SPOC control by removing the Bub2 inhibitor has almost no effect on Cdc14 release in unperturbed cell cycles, suggesting that mitotic cyclin (Clb)-Cdk oscillations may play an important role in regulating Cdc14 release timing. However, it is as yet unclear how Cdc14 localization responses to different Clb levels, and whether these controls constitute a ratchet mechanism sufficient to lock Cdc14 release to once per cell cycle.
To understand whether Clb-Cdk oscillations control Cdc14 localization through a ‘ratchet’-like mechanism, we locked mitotic cyclin Clb2 at stable physiological levels, by titrated pulses of undegradable Clb2, and correlated Cdc14 release and mitotic exit (ME) events to Clb2 levels in individual cells, following a recently developed procedure (Drapkin et al., 2009). High Clb2 blocks ME (Surana et al., 1993); however, the peak level of Clb2-Cdk activity attained in a normal cell cycle was inefficient at restraining ME (Drapkin et al., 2009), posing a problem for the simple cyclin-based ratchet model. Sharpening this contrast, here we show that Cdc14 cycles in and out of the nucleolus multiple times at high but physiological fixed mitotic cyclin levels. These and other results lead us to propose that Cdc14 release, and likely other cell cycle processes, are controlled by intrinsically oscillatory modules, that are entrained to a single occurrence at appropriate cell cycle positions by cyclin-Cdk cycles through a ‘phase-locking’ mechanism.
RESULTS
Blocking mitotic exit with undegradable Clb2kd reveals Cdc14 release endocycles
We determined the response of the Cdc14 release cycle to fixed cyclin-Cdk levels (Drapkin et al., 2009), using a quantitative, single cell measurement for Cdc14 localization based on variation of cellular Cdc14-YFP pixel intensities, standardized to variation of nucleolar Net1-mCherry (Lu and Cross, 2009) (Experimental Procedures; Fig. 1A).
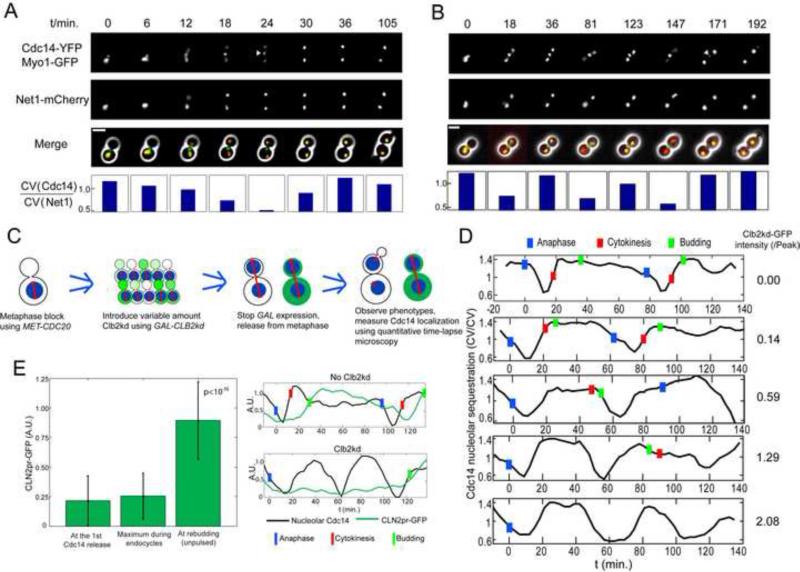
Figure 1. Cyclical Cdc14 release uncoupled from cell cycle progression.
A, B. MET3-CDC20 CDC14-YFP NET1-mCherry MYO1-GFP cells were released from a MET3-CDC20 block (t=0). Bottom: Cdc14 release was quantified at each time point as the following: the coefficient of variation (CV) of Cdc14-YFP signal inside a single cell, computed from fluorescent time-lapse microscopy data, is the standard deviation of YFP pixel intensity across the cell, divided by the mean intensity; CV of Cdc14-YFP is then divided by CV of Net1-mCherry, and this ratio will be high in cells with Cdc14-YFP concentrated in specific regions, and low when Cdc14-YFP is dispersed through the cell. Triangle: disappearing Myo1 ring. Scale bar: 5 microns. A: control. B. Clb2kd was pulsed for 30 min. before release. C. Schematic of procedure for loading cells with undegradable Clb2kd (green) before ME. Nucleus is shown in blue, spindle in red D. Pulsed Clb2kd-GFP was quantified (right column) in units standardized to the peak level of Clb2 attained in a normal cell cycle, and Cdc14 localization quantified (n=170). Blue bars: anaphase (nucleolar separation, marked by Net1-mCherry); Red bars: cytokinesis (Myo1 ring disappearance); Green bars: bud emergence. E. CLN2 promoter expression during Cdc14 endocycles. A CLN2pr-GFP-PEST strain was pulsed with Clb2kd as in (B) for 35 minutes. GFP intensities at the first Cdc14 release, maximum during endocycles (n=40), and at rebudding in unpulsed control cells (P<10−15. Error bars: standard deviation). See also figure S1 and movie S1.
We blocked cells in metaphase by depleting the essential anaphase-promoting complex (APC) activator Cdc20, by shutoff of MET3-CDC20 (Sullivan et al., 2001). Cdc20 promotes proteolysis of the anaphase inhibitor Pds1, and initial proteolysis of B-type cyclins (Shirayama et al., 1998; Wäsch and Cross, 2002; Yeong et al., 2000). We briefly pulsed cdc20-blocked cells with stable mitotic cyclin Clb2kd-GFP lacking destruction and KEN boxes recognized by the APC activators Cdc20 and Cdh1 (Pfleger and Kirschner, 2000; Schwab et al., 1997; Visintin et al., 1998; Wäsch and Cross, 2002). Stable Clb2kd-GFP accumulates in these cells to a level averaging around the peak Clb2-GFP level in a wild-type cell cycle (‘1X peak’) (Fig. S1A).
Re-inducing MET3-CDC20 induced anaphase, which proceeded on schedule independent of stable Clb2kd-GFP (data not shown). Clb2kd-GFP and associated kinase activity was constant through this protocol (Drapkin et al., 2009). We assayed post-anaphase events as a function of single-cell Clb2kd-GFP levels (Fig. 1C).
>=1X peak Clb2kd-GFP induced dose-dependent delays in cytokinesis and bud emergence (Fig. S1B)(Drapkin et al., 2009). In contrast, Cdc14-YFP was released from the nucleolus and subsequently resequestered, with essentially normal kinetics, up to at least 3X peak Clb2kd-GFP (Fig. 1D) (Drapkin et al., 2009). Strong overexpression of stable Clb2 was shown previously to cause extended Cdc14 release (Stegmeier et al., 2002). We confirmed this observation with continuous galactose induction of GAL1-CLB2kd (yielding >=10-fold peak Clb2kd levels (Fig. S1C)). However, in the following we pursue only results obtained at approximately physiological Clb2kd levels.
Remarkably, cells containing near-1X peak Clb2kd-GFP frequently exhibited multiple ‘endocycles’ of Cdc14-YFP release and resequestration, before finally undergoing cytokinesis and rebudding (Fig. 1B,D; Supplemental Movie 1).
Cdc14 endocycles occurred without budding, cytokinesis, or nuclear or nucleolar division. The G1 cyclin CLN2 is expressed at cell cycle Start (Wittenberg et al., 1990). We observed no significant CLN2pr-GFP expression (Bean et al., 2006; Mateus and Avery, 2000) in cells undergoing Cdc14 endocycles, while a burst of CLN2pr expression occurred when these cells finally budded (Fig. 1E).
In yeast, cyclin-Cdk phosphorylation excludes the MCM helicase complex from the nucleus except in M/G1, helping to restrict pre-replicative complex formation to pre-S phase (Kearsey and Cotterill, 2003). ~1X peak Clb2kd-YFP was sufficient to completely block Mcm2-GFP nuclear re-accumulation (Drapkin et al., 2009), confirming continuous high Clb2kd-associated kinase in this protocol. Consistent with this observation, we observed little or no DNA endoreduplication during Cdc14 endocycles by DNA flow cytometry (Fig. S1D).
Endogenous Clb2-GFP was uniformly degraded after initial Cdc14 release, regardless of exogenously pulsed Clb2kd levels. Endogenous Clb2-GFP remained low during Cdc14 endocycles, only reaccumulating upon cessation of endocycles (Fig. S1E).
Cdc14 endocycles are not driven by oscillations of Sic1, the stoichiometric Clb-Cdk inhibitor (Mendenhall and Hodge, 1998), because Sic1 levels in Clb2kd-pulsed cells were insufficient for Clb2kd inhibition (Drapkin et al., 2009). Consistently, Cdc14 endocycles were independent of Swi5, the major SIC1 transcription factor (Toyn et al., 1997) (Fig. S3A).
Thus, Cdc14 release endocycles are not due to oscillations of Clb2kd-Cdk or of endogenous G1 or mitotic cyclins. Cdc14 endocycles occur in the absence of cytokinesis, rebudding, Mcm complex nuclear reaccumulation and DNA replication.
Frequency control of the Cdc14 release endocycle by Clb2kd levels
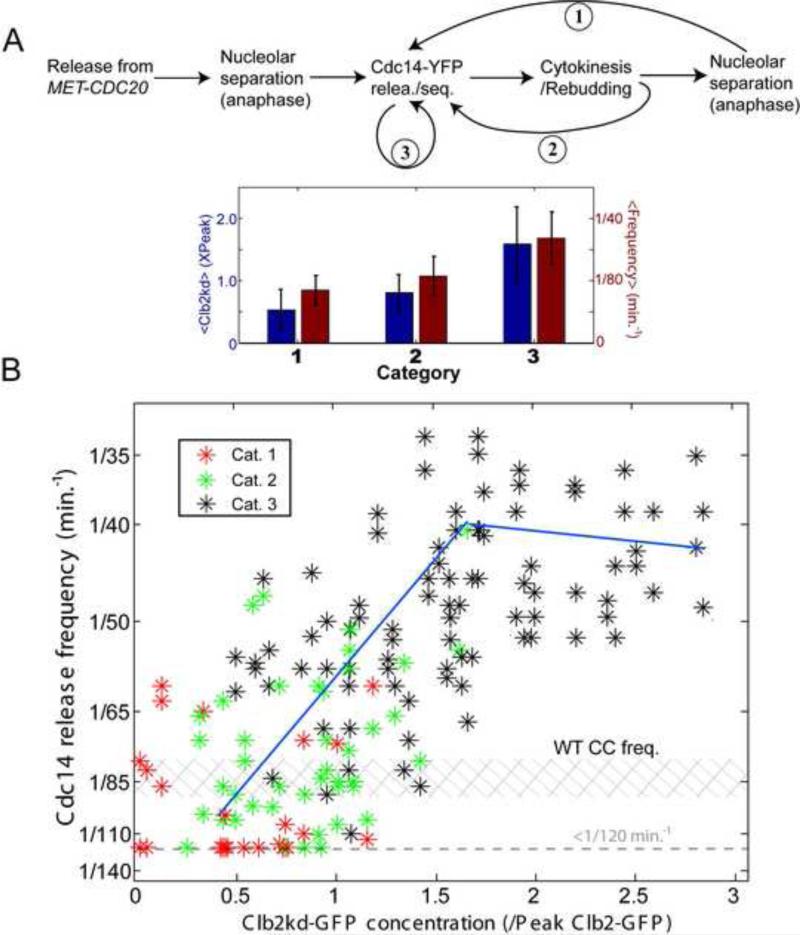
Clb2kd-GFP at less than ~0.5X peak had little effect on cell-cycle progression (Drapkin et al., 2009), and a single normal cycle of Cdc14 release and resequestration followed anaphase (marked by nucleolar Net1 separation) (Fig. 2A, category 1). Cdc14 release in the next cell cycle was frequently delayed, possibly due to 2nd-cycle defects caused by low Clb2kd (Drapkin et al., 2009). At ~0.75X peak levels of Clb2kd-GFP, ME was delayed but not blocked (Drapkin et al., 2009), and a 2nd Cdc14-release event sometimes occurred rapidly after bud emergence, without associated anaphase (Figs. 1D;2A; category 2). At >=1X peak Clb2kd concentrations, we observed Cdc14 endocycles: repeated Cdc14 release and sequestration without evident cell cycle progression (Fig. 2A, category 3).
Figure 2. Clb2kd level controls the Cdc14 endocycle period.
A. Trajectories for Clb2kd-pulsed cells. Category 1: essentially normal cell-cycle progression and Cdc14 release/sequestration; Category 2: a second Cdc14 release occurred between rebudding and nucleolar separation in the next cell cycle; Category 3: Cdc14 endocycles without cytokinesis or rebudding. Below: category means and standard deviations of Clb2kd-GFP concentration (blue) and Cdc14 release frequencies (red). B. Cdc14 release frequencies plotted against Clb2kd-GFP level for cell categories: inverse of intervals between first and second Cdc14 release (categories 1 and 2), or average frequencies of one cell's Cdc14 endocycles (category 3; correlation between the1st and 2nd cycle is shown in Fig. S2B). Shaded: range of cell cycle frequencies for cycling MET3-CDC20 mother cells. 31/170 category 1; 45/170 category 2; 94/170 category 3.. See also figure S2.
Category 2 cells are intermediate between normal tight linkage of Cdc14 release to cell cycle progression, and complete uncoupling as in category 3 endocycling cells, suggesting a continuous transition from normal Cdc14 release cycles to endocycles with increasing Clb2kd.
Below ~1.7× peak Clb2kd, Cdc14 release frequency correlated positively with Clb2kd-GFP level (Fig. 2B; P<2*10−7) using only data from ‘category 3’ cells that undergo endocycles without apparent cell cycle progression. A quantitatively similar, highly significant positive correlation was observed using all three categories of cells, suggesting that Clb2 levels may directly control Cdc14 release frequency, independent of cell cycle progression. At Clb2kd >1.7× peak, endocycle frequency saturated at once per ~45 min. Despite considerable noise in the data, we estimated the slope of the Clb2kd/endocycle frequency relationship (Fig. 2B legend) with a linear function (k=0.013±0.004/PeakClb2−1min−1).
Although Clb2kd levels controlled Cdc14 release frequency, the amplitude and duration of release was independent of Clb2kd (Fig. S2A). Those observations are important for the theoretical analysis below.
Requirements for Cdc14 endocycles
A B-type cyclin-independent oscillator may drive G1 events (Haase and Reed, 1999; Orlando et al., 2008). To determine if this oscillator might also drive Cdc14 endocycles, we overexpressed the stable Clb-Cdk inhibitor Sic1-4A to inhibit all B-type cyclin-Cdk activity (Verma et al., 1997). Cdc14 remained nucleolar, despite multiple budding without cell division (manifesting the G1 oscillator (Haase and Reed, 1999)) (Fig. S3E). Additionally, we constructed a cdc14-1 temperature-sensitive strain with all B-cyclins deleted. We collected small G1 cells by elutriation, and incubated them at non-permissive temperature for cdc14-1. cdc14-1 and CDC14 B-cyclin-depleted cells budded and rebudded with identical kinetics (Fig. S3F). Therefore, the Cdc14 release endocycle and the G1 endocycle are independent.
Polo kinase Cdc5 is required for Cdc14 release (Visintin et al., 2003), and Cdc14 endocycles required active Cdc5, even after an initial round of Cdc14 release (Fig. 3D). The FEAR network and MEN are required for early and late anaphase Cdc14 release respectively (Azzam et al., 2004; Queralt et al., 2006; Stegmeier and Amon, 2004). The MEN component Cdc15 was required for Cdc14 endocycles, but neither the FEAR network component Spo12, nor the key FEAR pathway event of Net1 phosphorylation by Clb-Cdk, was required (Fig. S3B,C). Thus, Cdc14 endocycles in Clb2kd-blocked cells may involve mechanisms controlling late-anaphase Cdc14 release in the free-running cell cycle, since both require Cdc5 and Cdc15, but not FEAR pathway components. In bub2Δ cells, MEN activity is likely near-constitutive (Alexandru et al., 1999; Pereira et al., 2000; Xu et al., 2000). Cdc14 release endocycles persisted in Clb2kd-arrested bub2Δ cells (Fig. S3D), suggesting that these endocycles do not require cyclical inactivation of the SPOC.
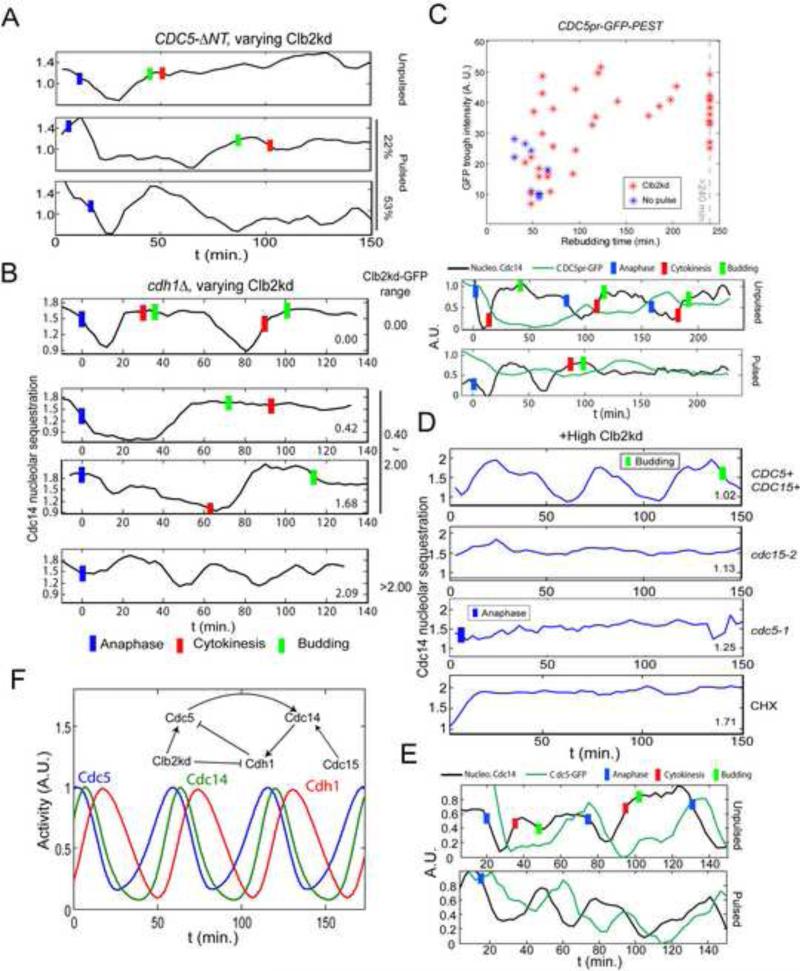
Figure 3. Requirements for Cdc14 endocycles.
A. As in Fig. 1B, but cells also cdc5::3XCDC5-ΔNT. Among cells whose rebudding was delayed for at least 25 minutes (implying >1X peak Clb2kd, Fig. S1B), 19/36 cells showed fast Cdc14 release and resequestration, followed by prolonged Cdc14 release. 8/36 showed only prolonged Cdc14 release. B. As in Fig. 1D, but also cdh1Δ; 30 min Clb2kd-GFP pulse; typical traces for the indicated Clb2kd-GFP ranges. Among cells with >1X peak Clb2kd-GFP (n=86), 41% showed a prolonged Cdc14 release period (middle two traces); 43% (cells with highest Clb2kd-GFP) showed release endocycles with a reduced amplitude (bottom trace); Blue bars: anaphase; Red bars: cytokinesis; Green bars: bud emergence. C. CDC5pr-GFP cells, as in Fig. 1B. Trough GFP intensities before rebudding plotted against rebudding times; rebudding delay indicates Clb2kd levels; sample traces below. D. Cells as in Fig. 1D, but also cdc15-2 or cdc5-1; after 35 min to allow initial Cdc14 release at permissive temperature, cells were plated for time-lapse at 37°C (restrictive temperature) (t=0). CHX: as above, except that time-lapse medium contained 0.2ng/μl cycloheximide (CHX). Among cells with >1X peak Clb2kd-GFP, 18/24 CDC15 CDC5 cells, 0/22 cdc15-2, 0/30 cdc5-1 and 3/64 cells in CHX exhibited Cdc14 release endocycles. Blue bars: anaphase; Green bars: bud emergence. E: As in Fig. 1B, but CDC5-GFP cells; 30 min Clb2kd pulsed; typical traces of Cdc14 release and Cdc5-GFP levels are shown. In 36/45 Cdc14-endocycling cells, Cdc5-GFP signal oscillated out-of-phase with Cdc14 release. F. ODE model simulating Cdc14-Cdh1-Cdc5 negative feedback (Supplemental Methods). See also figure S3.
Cdc5 is down-regulated at the end of mitosis by transcriptional inactivation and by Cdh1-APC-dependent proteolysis (Charles et al., 1998; Shirayama et al., 1998), and Cdc5 proteolysis promotes Cdc14 resequestration (Visintin et al., 2008). To determine whether Cdc14 resequestration in endocycles similarly required Cdc5 proteolysis, we tested the Cdh1-resistant Cdc5-ΔNT mutant (Visintin et al., 2008). In CDC5-ΔNT cells deduced to contain >1X peak Clb2kd, we did not detect multiple Cdc14 release endocycles; 22% of cells showed extended Cdc14 release, and 53% of cells had a short Cdc14 release/re-sequestration cycle prior to a long release (Fig. 3A). These very long release periods in CDC5-ΔNT cells contrasted with the normal duration of Cdc14 release in endocycling CDC5+ cells (see above).
Cdh1 is required for Cdc5 degradation, and cdh1Δ but not CDH1 cells containing 0.5X to 2X peak Clb2kd released Cdc14 for up to 80 minutes, and did not exhibit Cdc14 endocycles (Fig. 3B). Surprisingly, however, low-amplitude Cdc14 endocycles occurred in cdh1Δ cells with > 2X peak Clb2kd (Fig. 3B). This result suggests a parallel Cdh1-independent pathway allowing Cdc14 endocycles (see Discussion); but the primary mechanism likely requires Cdh1-dependent proteolysis of Cdc5, dependent on the Cdc5 N-terminal destruction box.
Cdc14-Cdh1-Cdc5 negative feedback may contribute to Cdc14 endocycles
Cyclical activation and inactivation of Cdc5 may be obligatory for Cdc14 endocycles, since endocycles are prevented by either constitutively high (CDC5-ΔNT) or constitutively low Cdc5 (cdc5-1).
We hypothesized a negative feedback loop to explain Cdc14 endocycles: Cdc5 promoting Cdc14 release, Cdc14 activating Cdh1 leading to Cdc5 proteolysis, and consequent Cdc14 resequestration. In the wild-type cell cycle, Cdc5 cannot reaccumulate until the next M-phase: CDC5 is co-transcribed with CLB2 under Clb2-dependent positive feedback control (Wittenberg and Reed, 2005), so Clb2 removal at anaphase blocks further CDC5 transcription. In Clb2kd-blocked cells, persistent Clb2kd could both maintain CDC5 transcription, and inactivate Cdh1 by phosphorylation (in the absence of counterbalancing dephosphorylation by Cdc14), allowing rapid reaccumulation of Cdc5 and endocyclic Cdc14 release and resequestration. This negative feedback hypothesis could explain absence of endocycles in cdc5-1, CDC5-ΔNT and cdh1Δ cells.
We used a cell-cycle-regulated CDC5pr-GFP reporter (J. Skotheim, pers. comm.) to measure CDC5 transcription. Consistent with the negative feedback hypothesis, we observed higher trough levels and rapid rebound of CDC5 transcription oscillation in Clb2kd-pulsed cells compared to unpulsed controls (Fig. 3C). Also consistent with the hypothesis, Cdc14 endocycles required new protein synthesis, since endocycles were blocked by addition of cycloheximide after an initial Cdc14 release (Fig. 3D).
Finally, in cells undergoing Cdc14 endocycles, we observed cyclical accumulation and degradation of Cdc5-GFP fusion protein expressed from the endogenous promoter, out of phase with Cdc14 release (Fig. 3E; the actual peak of Cdc5-GFP protein is likely to be ~15 min. earlier than the observed peak due to the time required for fluorescent maturation of newly synthesized Cdc5-GFP).
A qualitative ODE model (Fig. 3F;S3G) of a Cdc5-Cdc14-Cdh1 negative feedback loop reproduced our major results, including dependence of Cdc14 endocycle frequency on Clb2kd concentrations (Fig. 2B).
The experiments above suggest that Cdc14 localization could be controlled by an oscillatory module. Cdc14 release endocycles could come about because Clb2kd accelerates the Cdc14-release-control module, by driving rapid rebound of the Cdc5-Cdc14-Cdh1 negative feedback oscillator (Fig. 2B;4A), and blocks Clb-Cdk oscillation, resulting in a temporal separation of the two oscillators.
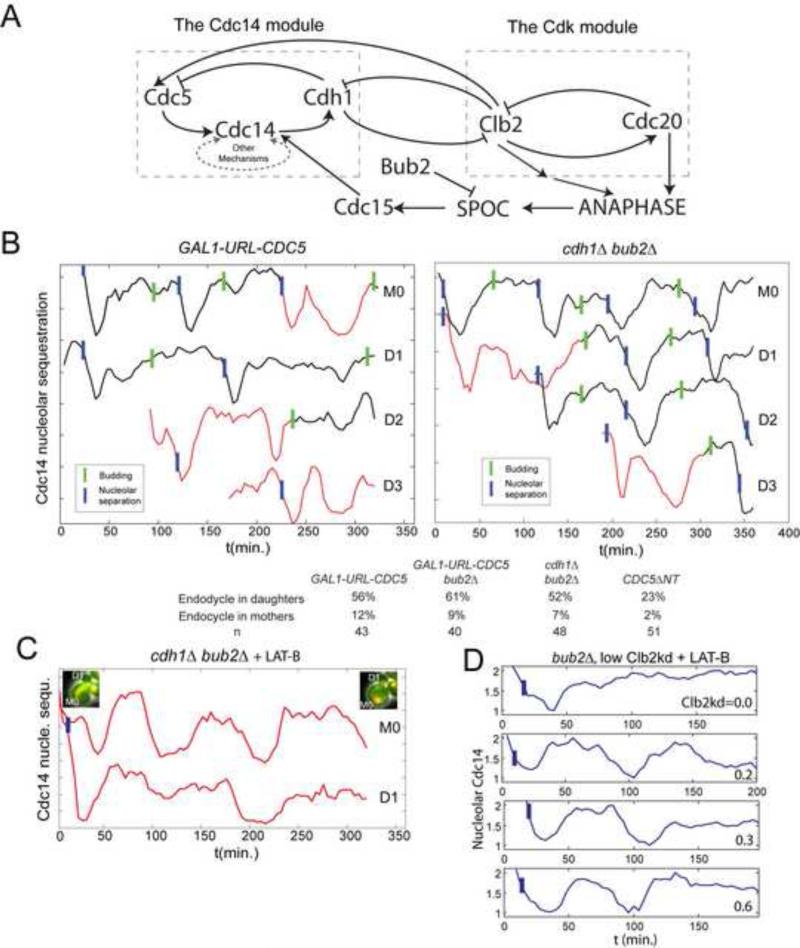
Figure 4. Cdc14 release endocycles in cycling cells or with low Clb2kd.
A. Cdc14 release and Clb-Cdk control mechanisms. Left: potential autonomous Cdc14 release oscillator; right: Clb-Cdk negative-feedback oscillator. SPOC: spindle orientation checkpoint. B. Cdc14 release analyzed as in Fig. 1, in bub2Δ cdh1Δ and GAL1-URL-CDC5 cells (cycling cells, without cdc20 block-release or Clb2kd pulse). A representative lineage (M0: mother, D1, D2...sequential daughters) exhibiting ectopic Cdc14 release endocycles (red) before bud emergence. Cdc14 release curves shifted for visualization. Below: probabilities of G1 Cdc14 endocycles. C. 67μM Latrunculin-B (LAT-B) was added to the medium (t=0) to inhibit budding of a cycling bub2Δ cdh1Δ strain. 32/44 cells demonstrated G1 Cdc14 endocycles. 14/19 daughter cells and 13/19 mother cells exhibited endocycles (maximum 4 endocycles; average=2.8). Cell images at the beginning/end of the time-course indicates no cell cycle progression. D. MET-CDC20 bub2Δ cdc15-2 cells were arrested in metaphase at 34°C and pulsed with Clb2kd-GFP for 20 minutes (using cdc15-2 to achieve a stable block), then released into cell cycle at 28°C with 67μM LAT-B. First anaphase (nucleolar separation) happened with normal kinetics. LAT-B effectively blocked budding, cytokinesis, and subsequent anaphase in ~90% cells. 39/46 cells containing 0.2~1.0x peak Clb2kd-GFP demonstrated Cdc14 endocycles. Four traces are shown. Blue bars: anaphase. See also figure S4.
Elucidating the mechanism of Cdc14 endocycles does not address whether the potential for Cdc14 endocycles is physiologically relevant in the normal cell cycle. To address this issue required additional experimental and analytical studies.
Oscillator uncoupling reveals Cdc14 endocycles in free-running cell cycles
Does the oscillation of this Cdc14 module control timing and execution of normal Cdc14 release in unperturbed cells cycles? Any manipulation trying to study the autonomous behavior of Cdc14-control module will inevitably perturb cell cycle progression. So, we analyzed this question in two steps: first, we asked whether the Cdc14-control module could oscillate autonomously in free-running cell cycles; second, we studied whether oscillatory behavior of the intact module might be involved in controlling normal Cdc14 release events.
First, we tried to uncouple the Cdc14 module from the Clb-Cdk oscillator, without blocking the cell cycle. Genetic manipulations providing sufficient uncoupling could allow detection of Cdc14 release endocycles in free-running cell cycles. Plausible coupling components include: 1. CDC5 transcription, activated by Clb-Cdk; 2. Cdh1, activated by Cdc14 and degrading Clbs and Cdc5; 3. the MEN, indirectly activated by mitotic cyclin-Cdk, since anaphase (promoted by cyclin-Cdk (Fitch et al., 1992)) activates the MEN (Fig. 4A).
We made CDC5 transcription independent of Clb-Cdk, using GAL1-URL-CDC5 ((Visintin et al., 2008); the destabilizing URL tag kept Cdc5 at a non-lethal level). Consistent with the oscillatory module hypothesis, in freely cycling GAL1-URL-CDC5 cells, we observed G1-specific Cdc14 endocycles in ~70% daughter cells (Fig. 4B). G1 cells stably contain almost no cyclin-Cdk activity (Mendenhall and Hodge, 1998), so events directly controlled by a cyclin-Cdk ratchet have no basis for repeated activity in G1 cells.
We did not observe Cdc14 endocycles or Cdc14 release anomalies in cycling cdh1Δ cells (data not shown), perhaps because Cdh1 contributes to the Cdc14 intrinsic oscillatory mechanism, as well as to its coupling with Cdk oscillations (Fig. 3B). More complete decoupling could reveal Cdc14 endocycles in the absence of Cdh1, assuming parallel coupled pathways within the Cdc14 oscillatory module (see above). Indeed, additionally deletion of BUB2, eliminating dependence of MEN activation on mitotic cyclin-induced anaphase (Fesquet et al., 1999; Fitch et al., 1992; Pereira et al., 2000), revealed G1-specific Cdc14 endocycles in cycling cdh1Δ bub2Δ cells (Fig. 4B). BUB2 deletion had little or no effect on its own or in GAL1-URL-CDC5 cells (Fig. S4A,B). CDC5-ΔNT in a CDH1 BUB2 background also induced G1 Cdc14 endocycles (Fig. S4C).
Daughter preference of endocycles could be due to G1-cyclin-dependent relocalization of MEN activators such as Lte1 to the incipient bud (Jensen et al., 2002), sequestering these activators until spindle pole entry into the bud in the next anaphase. Mother cells bud and express G1 cyclins much earlier than daughters (Di Talia et al., 2007), which could forestall mother G1 Cdc14 endocycles. Actin depolymerization with latrunculin B (LAT-B) prevents Lte1 sequestration by inhibiting budding; LAT-B treatment also inhibits all overt cell cycle progression (Jensen et al., 2002). cdh1Δ bub2Δ cells incubated with LAT-B exhibited Cdc14 endocycles in unbudded mothers and daughters with equal probability (Fig. 4C) (up to four endocycles, usually without associated anaphase), supporting the idea that rapid bud emergence and sequestration of MEN activators could prevent Cdc14 endocycles in G1 mother cells.
Therefore we conclude that the Cdc14-release module can oscillate autonomously in free-running cell cycles. These observations also emphasize the importance of oscillator coupling for maintaining timely, once-per-cell-cycle Cdc14 release in the normal cell cycle.
The Cdc14-release module is intrinsically oscillatory at mid-cell-cycle Clb-Cdk levels
The minimal Clb2 level allowing oscillation of the Cdc14 module determines whether this oscillatory behavior could control normal Cdc14 release in unperturbed cell cycles. If this level is low, oscillation of the module will last for most of the cell cycle, implying a likely role in controlling normal Cdc14 release timing and execution.
To determine this critical level, we used LAT-B to inhibit cytokinesis, budding and anaphase, to study the autonomous behavior of the Cdc14 module at low Clb2kd levels. We also deleted BUB2 to avoid the interference of SPOC. BUB2 deletion alone caused no difference in Cdc14 endocycles (Fig. S4A). In LAT-B-treated bub2 cells, 90-min period Cdc14 release cycles emerged at 0.2X peak Clb2kd-GFP (Fig. 4D). With <1X peak Clb2kd, 39/46 cells exhibited Cdc14 endocycles. The frequency-Clb2kd relationship had a similar slope as was obtained in Fig. 2B at higher Clb2kd levels (Fig. S5). Therefore, the Cdc14-release module oscillates even at Clb2 levels characteristic of early-mid cell cycle, with a frequency very close to normal Cdc14 release cycles (~90 min.), consistent with the oscillator functioning to control Cdc14 release timing in normal cell cycles. The apparently continuous transition from normal Cdc14 release cycles to endocycles also support this conclusion (Fig. 2A).
Control of Cdc14 release by an autonomous oscillator must be compatible with the well-established restriction of Cdc14 release to late mitosis, at the end of the cyclin-Cdk cycle. We show that coupling of the autonomous oscillation of the Cdc14 module to Clb-Cdk is sufficient to yield appropriate regulation and timing of Cdc14 release in the unperturbed cell cycle; this, coupled with the likelihood that the Cdc14 oscillator is functioning during much of the normal cell cycle (see above), makes the Cdc14 oscillator an important candidate for regulating Cdc14 release timing in the normal cell cycle. We extend this concept to oscillatory modules controlling other cell cycle events.
Cyclin-Cdk oscillations could order cell cycle events by phase-locking
Other cell cycle events besides Cdc14 release can occur ‘endocyclically’, driven by intrinsic oscillating modules without mitotic cyclin-Cdk oscillation. These observations are difficult to fit into Cdk-ratchet models (see Introduction). We considered alternative, non-ratchet-like ways that cyclin-Cdk oscillations could order cell cycle events.
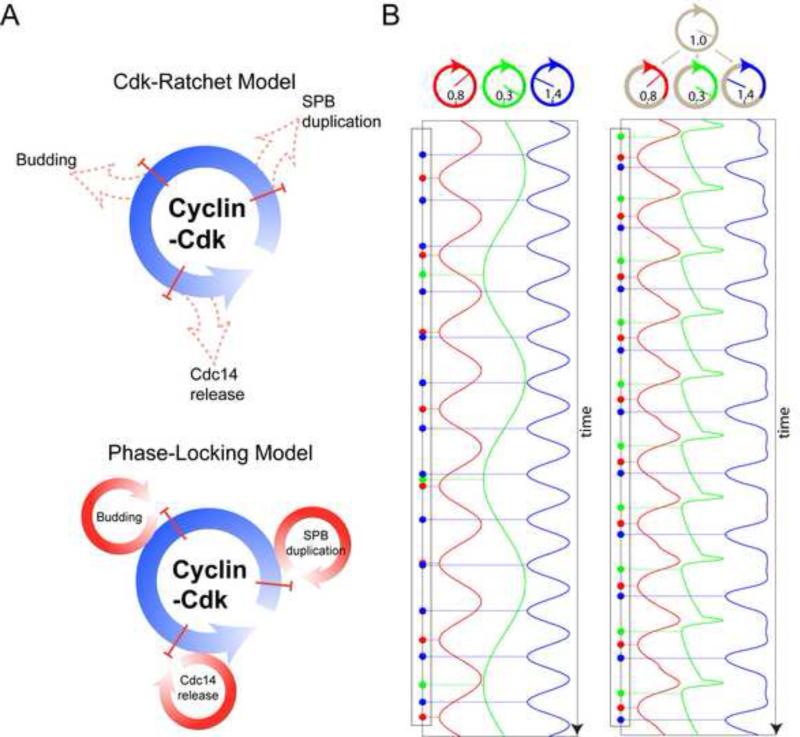
Physiological Clb2kd concentrations linearly controlled Cdc14 release endocycle frequency (Fig. 2B). Oscillators with different intrinsic frequencies can be synchronized to oscillate at one collective frequency if the frequency of one oscillator is sufficiently controlled by the activity of another, a phenomenon called ‘phase-locking (PL)’ (Glass, 2001; Winfree, 1967). A well-known biological example of PL is the entrainment of the circadian clock to diurnal cycles. Cyclin-Cdk oscillations could similarly entrain or phase-lock Cdc14 release, as well as other autonomous oscillatory modules, promoting orderly and coherent cell cycle progression (Fig. 5A).
Figure 5. Cell cycle control through phase-locking.
A. Schematic of ratchet (above) and phase-locking (below). B. Conceptual model: three simply oscillators ((Kuramoto, 1975), Supplemental Methods) with different intrinsic frequencies (indicated) control different cell cycle events. Assuming a single event is generated as the oscillator's phase evolves passing n*2π or ‘12 o’clock’. Without entrainment by cyclin-Cdk oscillator, each peripheral oscillator evolves uniformly, and there is no fixed order (left) among events they generate. Allowing a cyclin-Cdk oscillator to advance or delay part of the peripheral oscillators’ cycles leads to phase-locking and a stable order of events (Right). See also figure S5.
To demonstrate how PL could order cell cycle events, we built a conceptual model containing four oscillating modules: the cyclin-Cdk oscillator, and three autonomous ‘peripheral’ oscillators controlling specific cell cycle events (e.g., budding, SPB duplication, Cdc14 release). Without coupling, different intrinsic frequencies of the peripheral oscillators results in a disordered and irregular sequence of cell cycle events (Fig. 5B, left). In simulation, allowing cyclin-Cdk master oscillator to affect the frequencies of the peripheral ones resulted in all oscillators cycling exactly once per cyclin-Cdk cycle, in a fixed order (Fig. 5B, right).
This result of a distinct, dynamically stable PL solution for each oscillating module, provided they are coupled to the central Cdk oscillator, is derived from a very simple model, but may be robust to biological complexity in real systems. This is because PL is largely independent of detailed mechanism. Stability is determined by the local dynamical structure near the fixed point (Winfree, 1967), given sufficiently strong linkage, and reasonably close intrinsic frequencies. These features may be highly evolvable (see Discussion).
Reducing amplitude of the cyclin-Cdk oscillator results in disordered cell cycle events
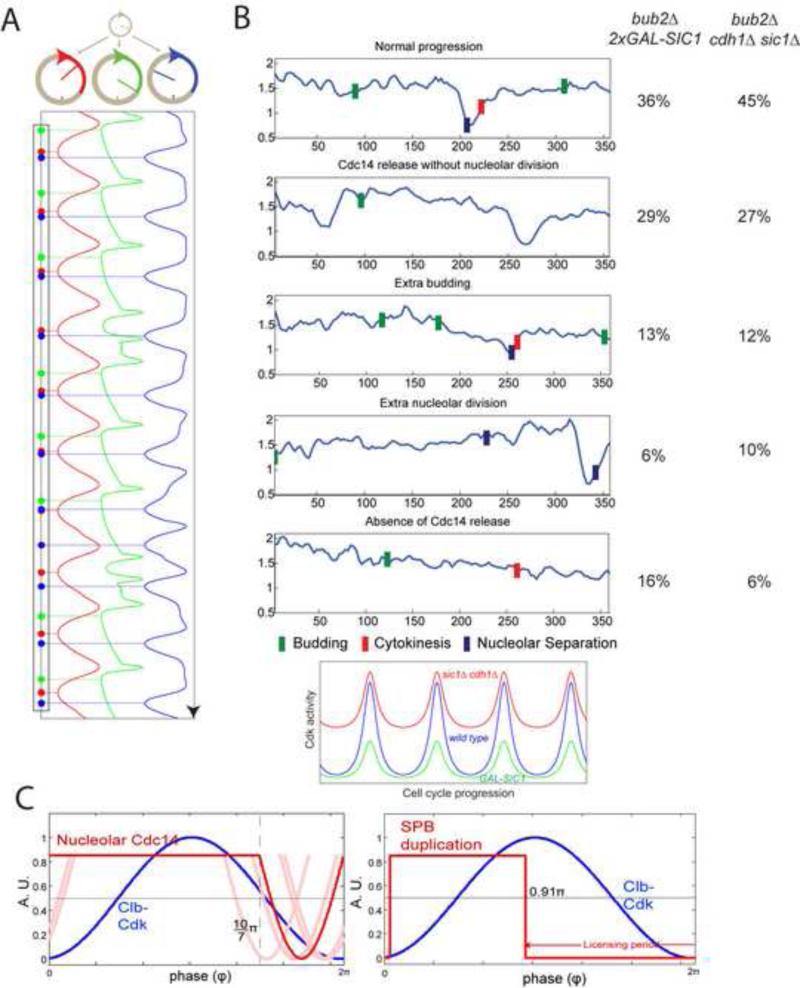
PL models (Fig. 5B) lead to a key prediction: lowering the amplitude of Clb-Cdk oscillation should weaken entrainment between the Clb cycle and other oscillators and perturb order of PL-controlled cell-cycle events (Fig. 6A). Cell cycle control through ratchet-like mechanisms yields entirely different predictions: lowering amplitude should block or delay events, but should not alter event order. In both cases these considerations are largely independent of model details.
Figure 6. Experimental test of phase-locking predictions.
A. Model as in Fig. 5B, but the amplitude of cyclin-Cdk oscillation is reduced by 15%, resulting in event disorder (compare Fig. 5B, right). B. 2XGAL1-SIC1 bub2Δ or cdh1Δ sic1Δ bub2Δ strains may reduce the amplitude of cyclin-Cdk oscillation by lowering the peak Cdk activity or raising the trough (cartoon below). 2XGAL1-SIC1 cells (n=37) were grown in raffinose medium prior to time-lapse analysis on galactose medium to induce Sic1 (t=0). cdh1Δ sic1Δ GALL-CDC20 cells (n=35) were imaged on glucose to turn off GALL-CDC20. Cells were classified according the cell cycle disorder that appeared first, shown with representative traces (right). Blue bars: nucleolar separation; Green bars: budding; Red bars: cytokinesis. C. Cdc14 release and SPB duplication timing in free-running cell cycles predicted by a quantitative phase-locking model. Red: best estimation of parameters; blue: sine wave simulation of Clb-Cdk cycle. Light-red: solutions with parameters altered +/− one standard deviation for one or both of the two parameters (for two combinations no solution could be obtained due to mathematical restrictions of the model; Supplemental Methods). Right panel: phase-locking could entrain SPB duplication oscillator (red) to the correct position early in the Clb-Cdk cycle (blue) (Supplemental Methods). See also figure S6.
We tried two experimental strategies to lower the amplitude of cyclin-Cdk oscillation. We first used strong constitutive overexpression of the Sic1 inhibitor of Clb-Cdk activity (Mendenhall and Hodge, 1998). The SIC1 overexpression cassette we used (‘2X GAL-SIC1’ ) was capable of significantly reducing Clb-Cdk levels in cells previously blocked with approximately peak Clb-Cdk activity, consistent with the idea that this cassette will similarly lower the peak in cycling cells (Fig. S6A; Fig. 6B). 2X GAL-SIC1 cells are fully viable. As a contrasting means to lower amplitude, we deleted CDH1 and SIC1, which results in a high trough level of Clb2-Cdk activity but probably little reduction in the peak (Wäsch and Cross, 2002; Yeong et al., 2000) (Fig. 6B). Although cdh1Δ sic1Δ cells are ultimately inviable (Schwab et al., 1997; Visintin et al., 1997), they undergo multiple cell cycles (Wäsch and Cross, 2002), allowing analysis of order of events by time-lapse microscopy. Thus, in GAL1-SIC1 cells, Clb-Cdk levels are extremely low during an extended G1, then probably rise to a reduced peak compared to wild-type, while in cdh1Δ sic1Δ cells there is probably no period with low Clb-Cdk levels, but likely a relatively normal peak level.
These two mechanisms for reducing the amplitude do so by opposite means: lowering the peak, vs. raising the trough. Ratchet control, in which specific Clb-Cdk levels control sequential steps in a process, predicts very different results from these manipulations, since a ratchet mechanism demands distinct responses to different Clb-Cdk activity levels. In contrast, the PL perspective can predict qualitatively similar outcomes of event disorder in these two cases.
To focus specifically on reduction of amplitude of the Clb-Cdk oscillation, we deleted the spindle orientation checkpoint component BUB2, to relieve dependence of Cdc14 release on spindle position (Bardin et al., 2000) (Qualitatively similar although lesser effects on budding, cytokinesis, and anaphase were observed in a BUB2 background; bub2 deletion alone had no effect (Fig. S4A)).
GAL1-SIC1 expression in bub2Δ cells resulted in 64% abnormal first cell cycles after Sic1 expression on galactose medium (supernumerary Cdc14 release without nucleolar separation; extra budding without Cdc14 release or cell division; or cytokinesis without Cdc14 release or nucleolar separation (Fig. 6B)). Remarkably, similar cell cycle anomalies with close frequencies were also observed in cycling cdh1Δ sic1Δ bub2Δ cells. Those anomalies are not likely due to nonspecific growth defects since GAL1-SIC1 bub2 cells grow well both in bulk culture and when monitored as single cells. cdh1Δ sic1Δ occasionally causes pre-anaphase block, probably due to replication problems (Wäsch and Cross, 2002); such cells were excluded from analysis.
The conceptual PL model yielded loss-of-order phenotypes as a result of lowering the amplitude of Cdk oscillation (Fig. 6A). In contrast, any Cdk-ratchet model would predict that under amplitude changes, the normal orders of cell cycle events would remain the same, excepting delays or blockages of some events. Further, near-opposite phenotypes would be predicted for lowering the peak vs. raising the trough. Thus, the result of out-of-order phenotypes, and the surprising similarity of the phenotypes of two entirely distinct ways of reducing amplitude, support a PL model over a ratchet model.
The PL model describes how Cdk oscillation entrains autonomous peripheral oscillators to form a coherent cell cycle, associating once-per-cell-cycle occurrence of an event (e.g. Cdc14 release, SPB duplication) in normal cell cycles with its capacity to undergo endocycles in blocked cells In contrast, in any ratchet model, endocycles must be pathological consequences of cell cycle blockage. Therefore, an empirical test of the PL vs. the ratchet perspectives is to ask whether the response of the oscillatory module to fixed Clb-Cdk levels is consistent with timing in the normal cell cycle. We calculated the stable phase offset (the difference between peak Clb2 and peak Cdc14 release), based on a simple quantitative PL model, containing only three parameters extracted directly from empirical observations on Clb2kd-blocked cells (Fig. 2B; Supplemental Methods). (For example, one key parameter, strength of coupling of Clb2 to Cdc14 cycle frequency, is measured from the slope of the straight-line fit in Fig. 2B.) An analytical solution yielded a phase-offset with Cdc14 release occurring as Clb-Cdk levels decreased (Supplemental Methods; Fig. 6C), consistent with experiment (Stegmeier and Amon, 2004). Uncertainties in parameter estimation do not significantly affect this conclusion, since increasing or decreasing the parameter estimates by one standard deviation of measurement error, alone or in combination, had little effect (Fig. 6C). This fit would be unlikely if endocycles are pathological consequences of cell cycle blockage.
Incorporating the inhibitory effects of Cdc14 release on Clb-Cdk activity is likely to further stabilize the PL between Cdc14 and Cdk oscillators. We lack sufficient information to determine all parameters in the ‘bidirectional’ model, but the same general conclusion is reached (Supplemental Methods).
Any oscillating module whose frequency is controlled by Cdk can potentially be phase-locked. We constructed a similar analytical model for PL of a hypothesized SPB duplication module. Published data are insufficient to obtain empirical estimates of parameters. Still, we could estimate predicted timing using our model and plausible parameter choices (Supplemental Methods). SPB duplication was predicted to occur early in the cell cycle, before the rise in Clb-Cdk, as observed experimentally (Fig. 6C), showing consistency of timing of SPB duplication with a PL model.
DISCUSSION
Cdc14 endocycles, oscillatory modules, and phase-locking
The canonical ratchet model for once-per-cell-cycle regulation by cyclin-Cdk oscillations proposes that high and low cyclin-Cdk levels promote alternating steps of a process. In this case, any fixed level of cyclin-Cdk should result in a terminal arrest with one or the other steps of each cell cycle process completed and the next never occurring. Cdc14 endocycles at fixed Clb2 levels challenge this idea, as do other autonomous cell cycle oscillators (see introduction). The regular oscillation of Cdc14 localization, with frequency controlled by Clb2kd levels, suggests a regulated process rather than noisy or sporadic failure of normal control. As an alternative to ratchet models, to account for existence and entrainment of these endocycles at a common frequency, we propose that the Cdk cycle phase-locks other independent oscillators to order cell cycle events.
Such a PL model is consistent with mechanistic regulatory details that can also support ratchet models; for example, the ability of high mitotic cyclin-Cdk activity to prevent SPB ‘licensing’ for duplication is readily interpreted as a mechanism of oscillator coupling. The PL model may provide deeper insight into cell cycle regulation since it integrates endocycle phenotypes and normal cell cycle regulation into a coherent framework. Above, we presented experiments supporting PL over ratchet control in the free-running cell cycle, as well as in blocked cells.
Cdc5 proteolysis, controlled by a Cdc14-Cdh1-Cdc5 negative feedback loop, probably contributes to Cdc14 endocycles at high mitotic cyclin levels. Similar conclusions have been reached by others, by different methods (R. Visintin, pers. comm.). Nevertheless, Cdh1-dependent proteolysis of Cdc5 is not the only mechanism causing Cdc14 re-sequestration, since re-sequestration still happened after a delay in the absence of Cdh1; we observed G1 Cdc14 endocycles in bub2Δ cdh1Δ cells; and the Cdh1 requirement for endocycles in Clb2kd-blocked cells was lost at very high (>2X peak) Clb2kd levels (Fig. 3B). The oscillatory Cdc14 module may include parallel mechanisms, ensuring robust oscillation. Interestingly, we found that Mob1-GFP localized to SPB periodically during Cdc14 endocycles in phase with Cdc14 release (Fig. S6B). Since Mob1's SPB localization may signal MEN activation (Stegmeier and Amon, 2004; Yoshida and Toh-e, 2001), MEN activity may oscillate to drive Cdc14 endocycle even in the absence of Cdc14-Cdh1-Cdc5 negative feedback.
The severe disruptions in normal Cdc14 release by manipulations specifically predicted to perturb the autonomous Cdc14 release module, even in free-running cell cycles, imply that oscillatory function of this module is required for normal Cdc14 release timing. A PL control system could transit evolutionarily to a pure ratchet-control system; indeed, this is formally the result of increasing coupling strength to sufficiently high values that the peripheral oscillator will not move within a physiological timescale without a ‘kick’ from the entraining oscillator. Different cell cycle systems, even if initially independently oscillatory, may vary in the degree to which they are at present under ratchet control; for example, DNA replication may be entirely under cyclin-Cdk ratchet control in modern eukaryotes, despite its intrinsically oscillatory character. This may represent a tradeoff between the robustness and simplicity of the PL mechanism, and the cost in some systems of occasional uncoupling (aneuploidy in the case of DNA replication).
Modular design and evolution of the cell cycle
Modularity may create functional robustness and evolvability (Bhattacharyya et al., 2006; Hartwell et al., 1999). Our study suggests that some cell cycle modules are intrinsically oscillatory; coupling of these oscillators to the central Cdk oscillator can nevertheless readily ensure once-per-cell-cycle control (Fig. 5A,B).
The eukaryotic cell cycle evolved from a Cdk-free precursor, since Cdk is a eukaryotic-specific kinase (Nasmyth, 1995). Further, Cdks may have evolved late, after other eukaryotic cell-cycle-regulatory protein kinases (Krylov et al., 2003). If processes such as DNA replication and cell division were intrinsically oscillatory before the emergence of Cdk, then Cdk would only need to gain the ability to modulate these oscillators to gradually become a master regulator. This satisfies the evolutionary requirement of utility of intermediate forms (Darwin, 1859).
We speculate that in primordial eukaryotes, multiple autonomous biochemical oscillators entrained each other to an approximate aggregate rhythm. Cyclin-Cdk oscillation could then have evolved, yielding a stable PL structure. Selection for entrainment in large populations of simulated oscillators selects central pacemaker oscillators (Brede, 2008; Sendina-Nadal et al., 2008), which provided stability to weakening of oscillator coupling as an unselected ‘phenotype’. Further, unevolved random networks of coupled oscillators, constructed computationally in a standard manner (Kuramoto, 1975), showed increased stability of spontaneous entrainment with increasing centralization of control in network generation (Fig. S6C).
The predominant model for cell cycle control involves direct cyclin-Cdk control of cell cycle events, with surveillance checkpoints to ensure order (Morgan, 2007). An earlier model (Morgan, 2007; Pringle and Hartwell, 1981), considered cell cycle control to be due to parallel, interlocking but unidirectional morphogenetic pathways arranged in a functional sequence map. Our PL model combines aspects of both: a central cyclin-Cdk oscillator phase-locks independent oscillatory modules controlling specific cell cycle events. Such independent cyclical processes are analogous to ‘independent functional sequences’ in the cdc formulation – processes that can operate freely in parallel, such as DNA replication and spindle morphogenesis (Hartwell, 1974). Phase-locking of these autonomous oscillators by a central Cdk pacemaker could have been important in early cell cycle evolution, and this mechanism may control multiple autonomous cell cycle oscillators, even in modern eukaryotes.
EXPERIMENTAL PROCEDURES
Time-lapse microscopy
Pulsed expression and release of GAL1-CLB2kd(-GFP) in MET3-CDC20 blocked cells and time-lapse microscopy were previously described Drapkin et al. (in press) (Bean et al., 2006).
Image analysis
Image segmentation and quantification were described(Bean et al., 2006; Charvin et al., 2008). Cdc14 release was quantified as the coefficient of variation (CV, standard deviation divided by mean) of Cdc14-YFP pixel intensities (mother and bud treated separately), divided by the Net1-mCherry CV. A narrow band-width GFP filter (Chroma, 41020) and the contribution of pure GFP to the YFP channel allows computational recovery of pure YFP signal. The small amount of bud-neck-localized Cdc14 was excluded from quantification. Smoothing of time-lapse series was by 3-neighbour averaging method.
Model calculations and computer simulations
Basic forms of the ODE model in Fig.3F and parameters are adopted from Chen's 2004 cell cycle model. Simulation is performed using MATLAB, as is in Fig.5B. Simulation of Kuramoto oscillator population in Fig.S6C was programmed in C language with Numerical Recipe 2.0 package.
Supplementary Material
Acknowledgments
Special thanks go to B. Drapkin for developing and characterizing the quantitative Clb2kd-GFP loading method, and for communicating results before publication. We thank A. Amon, B. Drapkin, A. Geoghegan, K. Lee, D. Morgan, K. Nasmyth, J. Robbins, E. Schiebel, J. Skotheim and R. Visintin for providing strains and plasmids. We thank L. Bai, N. Buchler, B. Drapkin, H. Funabiki, J. Robbins, W. Shou, E. Siggia and J. Skotheim for comments on the manuscript. We thank R. Visintin for sharing unpublished data. We also thank E. Siggia for extended discussion. This work was supported by grants from the National Institutes of Health GM078153 and GM47238 to F. C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alexandru G, Zachariae W, Schleiffer A, Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. The EMBO journal. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam R, Chen SL, Shou W, Mah AS, Alexandru G, Nasmyth K, Annan RS, Carr SA, Deshaies RJ. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science (New York, NY. 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Bean JM, Siggia ED, Cross FR. Coherence and timing of cell cycle start examined at single-cell resolution. Molecular cell. 2006;21:3–14. doi: 10.1016/j.molcel.2005.10.035. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annual review of biochemistry. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nature reviews. 2007;8:149–160. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- Brede M. Construction principles for highly synchronizable sparse directed networks. Physics Letters A. 2008;372:5305–5308. [Google Scholar]
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS ONE. 2008;3:e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin CR. The origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. EZreads Publications, LLC; 1859. March 4, 2009. [PMC free article] [PubMed] [Google Scholar]
- Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- Drapkin BJ, Lu Y, Procko AL, Timney BL, Cross FR. Analysis of the mitotic exit control system using locked levels of stable mitotic cyclin. Molecular systems biology. 2009;5:328. doi: 10.1038/msb.2009.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science (New York, NY. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Fesquet D, Fitzpatrick PJ, Johnson AL, Kramer KM, Toyn JH, Johnston LH. A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. The EMBO journal. 1999;18:2424–2434. doi: 10.1093/emboj/18.9.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch I, Dahmann C, Surana U, Amon A, Nasmyth K, Goetsch L, Byers B, Futcher B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Molecular biology of the cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Hafezi S, Zhang T, Doxsey SJ. Centrosome duplication continues in cycloheximide-treated Xenopus blastulae in the absence of a detectable cell cycle. The Journal of cell biology. 1990;110:2033–2042. doi: 10.1083/jcb.110.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass L. Synchronization and rhythmic processes in physiology. Nature. 2001;410:277–284. doi: 10.1038/35065745. [DOI] [PubMed] [Google Scholar]
- Haase SB, Reed SI. Evidence that a free-running oscillator drives G1 events in the budding yeast cell cycle. Nature. 1999;401:394–397. doi: 10.1038/43927. [DOI] [PubMed] [Google Scholar]
- Haase SB, Winey M, Reed SI. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nature cell biology. 2001;3:38–42. doi: 10.1038/35050543. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Saccharomyces cerevisiae cell cycle. Bacteriological reviews. 1974;38:164–198. doi: 10.1128/br.38.2.164-198.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402:C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Morgan DO. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr Biol. 2000;10:615–618. doi: 10.1016/s0960-9822(00)00491-7. [DOI] [PubMed] [Google Scholar]
- Jensen S, Geymonat M, Johnson AL, Segal M, Johnston LH. Spatial regulation of the guanine nucleotide exchange factor Lte1 in Saccharomyces cerevisiae. Journal of cell science. 2002;115:4977–4991. doi: 10.1242/jcs.00189. [DOI] [PubMed] [Google Scholar]
- Kearsey SE, Cotterill S. Enigmatic variations: divergent modes of regulating eukaryotic DNA replication. Molecular cell. 2003;12:1067–1075. doi: 10.1016/s1097-2765(03)00441-6. [DOI] [PubMed] [Google Scholar]
- Krylov DM, Nasmyth K, Koonin EV. Evolution of eukaryotic cell cycle regulation: stepwise addition of regulatory kinases and late advent of the CDKs. Curr Biol. 2003;13:173–177. doi: 10.1016/s0960-9822(03)00008-3. [DOI] [PubMed] [Google Scholar]
- Kuramoto Y. Int. Symp. on Mathematical problems in theoretical physics. In: Araki H, editor. Lect N Phys. Springer; New York: 1975. pp. 420–422. [Google Scholar]
- Lu Y, Cross F. Mitotic exit in the absence of separase activity. Molecular biology of the cell. 2009;20:1576–1591. doi: 10.1091/mbc.E08-10-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus C, Avery SV. Yeast. Vol. 16. Chichester, England: 2000. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. pp. 1313–1323. [DOI] [PubMed] [Google Scholar]
- McCleland ML, O'Farrell PH. RNAi of mitotic cyclins in Drosophila uncouples the nuclear and centrosome cycle. Curr Biol. 2008;18:245–254. doi: 10.1016/j.cub.2008.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall MD, Hodge AE. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1191–1243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO. The cell cycle : principles of control. Published by New Science Press in association with Oxford University Press; London Sunderland, MA: 2007. [Google Scholar]
- Nasmyth K. Evolution of the cell cycle. Philosophical transactions of the Royal Society of London. 1995;349:271–281. doi: 10.1098/rstb.1995.0113. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. At the heart of the budding yeast cell cycle. Trends Genet. 1996;12:405–412. doi: 10.1016/0168-9525(96)10041-x. [DOI] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Wang JY, Socolar JE, Iversen ES, Hartemink AJ, Haase SB. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature. 2008;453:944–947. doi: 10.1038/nature06955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Molecular cell. 2000;6:1–10. [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes & development. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Hartwell LH. The Molecular biology of the yeast Saccharomyces Life cycle and inheritance. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y.: 1981. The Saccharomyces cerevisiae cell cycle. pp. 97–142. [Google Scholar]
- Queralt E, Lehane C, Novak B, Uhlmann F. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast. Cell. 2006;125:719–732. doi: 10.1016/j.cell.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Sendina-Nadal I, Buldu JM, Leyva I, Boccaletti S. Phase locking induces scale-free topologies in networks of coupled oscillators. PLoS ONE. 2008;3:e2644. doi: 10.1371/journal.pone.0002644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. The EMBO journal. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Sluder G, Miller FJ, Cole R, Rieder CL. Protein synthesis and the cell cycle: centrosome reproduction in sea urchin eggs is not under translational control. The Journal of cell biology. 1990;110:2025–2032. doi: 10.1083/jcb.110.6.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annual review of genetics. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Visintin R, Amon A. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell. 2002;108:207–220. doi: 10.1016/s0092-8674(02)00618-9. [DOI] [PubMed] [Google Scholar]
- Stern B, Nurse P. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 1996;12:345–350. [PubMed] [Google Scholar]
- Sullivan M, Lehane C, Uhlmann F. Orchestrating anaphase and mitotic exit: separase cleavage and localization of Slk19. Nature cell biology. 2001;3:771–777. doi: 10.1038/ncb0901-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. The EMBO journal. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyn JH, Johnson AL, Donovan JD, Toone WM, Johnston LH. The Swi5 transcription factor of Saccharomyces cerevisiae has a role in exit from mitosis through induction of the cdk-inhibitor Sic1 in telophase. Genetics. 1997;145:85–96. doi: 10.1093/genetics/145.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science (New York, NY. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- Visintin C, Tomson BN, Rahal R, Paulson J, Cohen M, Taunton J, Amon A, Visintin R. APC/C-Cdh1-mediated degradation of the Polo kinase Cdc5 promotes the return of Cdc14 into the nucleolus. Genes & development. 2008;22:79–90. doi: 10.1101/gad.1601308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Molecular cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science (New York, NY. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Visintin R, Stegmeier F, Amon A. The role of the polo kinase Cdc5 in controlling Cdc14 localization. Molecular biology of the cell. 2003;14:4486–4498. doi: 10.1091/mbc.E03-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wäsch R, Cross FR. APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature. 2002;418:556–562. doi: 10.1038/nature00856. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes & development. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Winfree AT. Biological rhythms and the behavior of populations of coupled oscillators. Journal of theoretical biology. 1967;16:15–42. doi: 10.1016/0022-5193(67)90051-3. [DOI] [PubMed] [Google Scholar]
- Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–2755. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- Wittenberg C, Sugimoto K, Reed SI. G1-specific cyclins of S. cerevisiae: cell cycle periodicity, regulation by mating pheromone, and association with the p34CDC28 protein kinase. Cell. 1990;62:225–237. doi: 10.1016/0092-8674(90)90361-h. [DOI] [PubMed] [Google Scholar]
- Xu S, Huang HK, Kaiser P, Latterich M, Hunter T. Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr Biol. 2000;10:329–332. doi: 10.1016/s0960-9822(00)00382-1. [DOI] [PubMed] [Google Scholar]
- Yeong FM, Lim HH, Padmashree CG, Surana U. Exit from mitosis in budding yeast: biphasic inactivation of the Cdc28-Clb2 mitotic kinase and the role of Cdc20. Molecular cell. 2000;5:501–511. doi: 10.1016/s1097-2765(00)80444-x. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Toh-e A. Regulation of the localization of Dbf2 and mob1 during cell division of saccharomyces cerevisiae. Genes & genetic systems. 2001;76:141–147. doi: 10.1266/ggs.76.141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.