Abstract
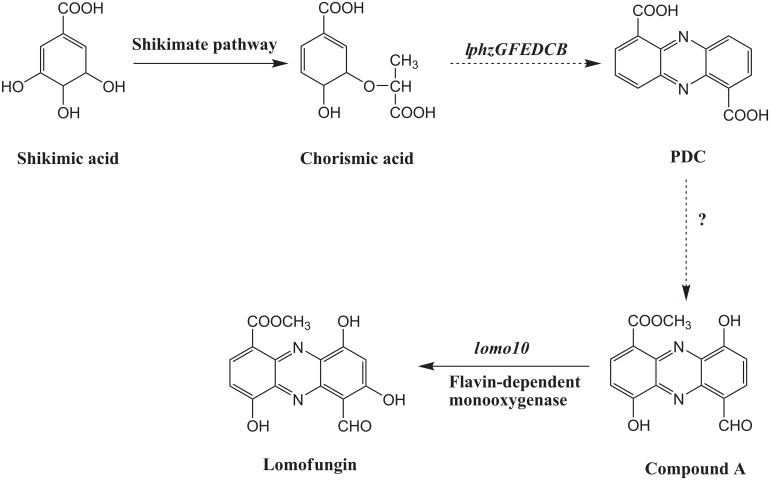
Streptomyces lomondensis S015 synthesizes the broad-spectrum phenazine antibiotic lomofungin. Whole genome sequencing of this strain revealed a genomic locus consisting of 23 open reading frames that includes the core phenazine biosynthesis gene cluster lphzGFEDCB. lomo10, encoding a putative flavin-dependent monooxygenase, was also identified in this locus. Inactivation of lomo10 by in-frame partial deletion resulted in the biosynthesis of a new phenazine metabolite, 1-carbomethoxy-6-formyl-4,9-dihydroxy-phenazine, along with the absence of lomofungin. This result suggests that lomo10 is responsible for the hydroxylation of lomofungin at its C-7 position. This is the first description of a phenazine hydroxylation gene in Streptomyces, and the results of this study lay the foundation for further investigation of phenazine metabolite biosynthesis in Streptomyces.
Introduction
Natural phenazine compounds, a class of secondary metabolites containing a phenazine nucleus, are mainly produced by Streptomyces and Pseudomonas species [1,2]. Phenazines have many biological functions, and demonstrate antimicrobial, antifungal, anti-tumor, antimalarial, and antiparasitic activities. The phenazine compounds produced by Pseudomonas species usually have simple structures, such as phenazine-1-carboxyl acid (PCA) [3], 1-hydroxyphenazine [4], phenazine-1-carboxamide [5], and pyocyanin [6]. In addition to simple phenazines, Streptomyces can biosynthesize phenazine derivatives with more complex structures, such as diphenazines, terpenoidal phenazines, carbohydrate-containing phenazines, and saphenic acid derived phenazines [1]. The biological activity of phenazine derivatives varies with the type and number of functional groups attached to the phenazine nucleus [1,7]. For example, PCA showed higher inhibitory activity than 1-hydroxyphenazine against plant disease pathogens such as Alternaria solani and Fusarium oxysporum [8]. Thus, investigation of side chain modification during the biosynthesis of phenazine derivatives is very important.
In Streptomyces and Pseudomonas, the biosynthetic process of phenazine products begins with shikimic acid. In following steps, chorismic acid is synthesized through the shikimate pathway then converted into intermediates containing a phenazine nucleus [9]. Two intermediates for all phenazine products, PCA and phenazine-1,6-dicarboxyl acid (PDC), are synthesized via this pathway by the highly conserved phz gene cluster [9,10]. The phzABCDEFG gene cluster was first sequenced in Pseudomonas fluorescens 2–79 [11], and has since been sequenced in many other Pseudomonas species, including Pseudomonas aeruginosa strains PAO1 [12] and M18 [13], and Pseudomonas chlororaphis strains PCL 1391 [14], 30–84 [15], GP72 [16], and 2–79 [11]. The first sequenced phenazine biosynthesis gene cluster in Streptomyces was ephzBCDEGA, which contributes to the biosynthesis of endophenazines in Streptomyces cinnamonesis DSM1042 [17]. Until now, six other phz gene clusters have been described in Streptomyces [18].
PCA is the core structure for all phenazine biosynthesis products in Pseudomonas [19], whereas both PCA [20] and PDC [21] can form the core structure in Streptomyces. All phenazine derivatives are further biosynthesized from PCA or PDC by modification of the side chains. The phenazine-modifying genes in Pseudomonas, including the phzM methyltransferase and phzS salicylate hydroxylase genes from P. aeruginosa [22], and the phzH asparagine synthetase gene from P. chlororaphis PCL1391 [14], have been extensively studied. Because of the complicated structure of phenazine derivatives in Streptomyces, only a few genes for phenazine-modifying proteins, such as the prenyltransferase genes, have been examined in Streptomyces [9].
Monooxygenases play an important role in oxidation reactions in a number of secondary metabolite biosynthesis pathways in Streptomyces [23–25]. A FMN-dependent monooxygenase is involved in dihydrokalafungin oxidation catalysis, the last step in the biosynthesis of the natural antibiotic actinorhodin in S. coelicolor [26]. A P450 monooxygenase NysL is responsible for C-10 hydroxylation during biosynthesis of the polyene macrolide antibiotic nystatin in S. noursei [27]. Monooxygenases are also involved in the biosynthesis of phenazine compounds [12,15,16]. A FAD-dependent monooxygenase PhzS from P. aeruginosa PAO1 catalyzes hydroxylative decarboxylation of PCA to yield 1-OH-Phz [12]. phzO, a gene encodes a flavin-dependent aromatic monooxygenase that hydroxylates PCA to produce 2-hydroxyphenazine-1-carboxylic acid (2-OH-PCA), has been found in both P. aureofaciens 30–84 [15] and P. chlororaphis GP72 [16]. Until now, no monooxygenase for the hydroxylation of phenazine compounds in Streptomyces has been reported.
Lomofungin is an olive-yellow phenazine antibiotic that was first discovered in Streptomyces lomondensis sp. n. [28]. This antibiotic has broad-spectrum antibacterial activity against both Gram-positive and Gram-negative bacteria, as well as pathogenic fungi [29–32]. However, despite these advantageous properties, the application of lomofungin has been limited by the low production titer during strain cultivation. S. lomondensis S015, which can biosynthesize lomofungin, was isolated from rhizosphere soil in Shanghai, China, by our group. We have since worked to improve lomofungin production in this strain, both by optimization of fermentation conditions and by overexpression of regulatory genes [33,34].
In this study, the lomofungin biosynthesis genes were examined after the whole genome sequenc of S. lomodensis S105 by comparison with available known sequences in P. chlororaphis GP72 (GenBank: HM594285.1). In addition to the phenazine biosynthesis core gene cluster, a putative flavin-dependent monooxygenase (lomo10), responsible for the hydroxylation of lomofungin, was also identified, and was further characterized by in-frame partial deletion.
Materials and Methods
2.1 Bacterial strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are described in Table 1 [35,36,37]. The primers used for polymerase chain reaction (PCR) assays are described in Table 2 [36].
Table 1. Bacterial strains and plasmids used in this study.
| Strains | Characteristics | Reference/source |
|---|---|---|
| Escherichia coli | ||
| DH5α | Host for general cloning | TranGen Biotech |
| ET12567 (pUZ8002) | Donor strain for intergeneric conjugation KmR, CmR | MacNeil et al. [35] |
| Streptomyces lomondensis | ||
| S015 | Wild-type, lomofungin-producing strain | Our laboratory |
| DCC601 | Δlomo10 mutant | This study |
| DCC602 | lomo10-complemented strain DCC601 (containing pIB139-lomo10), Aprr | This study |
| Plasmids | ||
| pKC1139 | E. coli-Streptomyces shuttle vector temperature-sensitive, Aprr | Bierman et al. [36] |
| pIB139 | E. coli-Streptomyces erythraea integrative shuttle vector containing a strong constitutive PermE* promoter, Aprr | Wilkinson et al. [37] |
| pMD19-T | General cloning plasmid, Amr | Takara Bio |
| pCC601 | Left arm of lomo10 subcloned into pMD19-T, Amr | This study |
| pCC602 | Right arm of lomo10 subcloned into pMD19-T, Amr | This study |
| pCC603 | Left and right arms ligated into pMD19-T, Amr | This study |
| pKC1139-lomo10 | Plasmid for in-frame partial deletion of lomo10, left and right arms ligated into pKC1139 at the HindIII/BglII/XbaI sites, Aprr | This study |
| pCC604 | lomo10 ligated into pMD19-T, Amr | This study |
| pIB139-lomo10 | Vector for lomo10 complementation, Aprr | This study |
Table 2. Primers used in this study.
| Primers | Sequence (5′–3′) | Enzyme sites |
|---|---|---|
| lomo10 left arm-For | AAATTTGAATTCTGGATGATCGCGACGATTTC | HindIII |
| lomo10 left arm-Rev | AAAAAAAAAAAAATTTAGATCTCATCTCCTGCAGACCCCGAGT | BglII |
| lomo10 right arm-For | AAATTTAGATCTGAGTTCGTCAAGGTCAGCTCCC | BglII |
| lomo10 right arm-Rev | AAATTTTCTAGACAGTCGGGGAAGCACTTGAG | XbaI |
| lomo10-For | ACGCATATGGTGGTGCTCGGGGCCAGCATCG | NdeI |
| lomo10-Rev | ACGTCTAGATTCCGATTTCTCAGCGCTGTC | XbaI |
| pIB-F [36] | TTGCGCCCGATGCTAGTCG | |
| pIB-R [36] | GCACGACAGGTTTCCCGACTG |
S. lomondensis S015 (China Center for Type Culture Collection No: M2013140) and its mutants were cultivated at 28°C according to Wang et al. [33]. Seed cultures and fermentations were performed using mannitol soybean (MS) medium (2% mannitol, 2% soybean powder, 2% agar, pH 7.2) and yeast malt (YM) medium (0.4% yeast extract, 1% malt extract, 0.4% glucose, pH 7.2), respectively.
All Escherichia coli strains were grown in Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.2) at 37°C with appropriate antibiotics, as described by Kieser et al. [38].
2.2 DNA isolation, manipulation, and sequencing
Genomic DNA was isolated using the method described by Hopwood et al. [38], and DNA was further manipulated according to Maniatis et al. [39,40]. PCR amplicons were isolated from agarose gel using a DNA Gel Extraction Kit (TranGen Biotech, Beijing, China). PCRs were performed in a 25 μl volume using PrimerSTAR HS DNA polymerase (Takara Bio, Dalian, China) with genomic DNA as template. PCR products were purified using an EasyPure PCR Purification Kit (TranGen Biotech). Primers were synthesized by Invitrogen, Shanghai, China. DNA was sequenced by Huada, Shenzhen, China.
2.3 Genome and protein sequence analysis
The lomofungin biosynthesis gene cluster was identified from the whole genome sequencing results of S. lomondensis S015 and analyzed using the antiSmash program (http://antismash.secondarymetabolites.org, accessed on June 25th, 2013) [41]. The identified sequence was then aligned with the phenazine biosynthesis gene cluster from P. chlororaphis GP72 [3,18] for confirmation. The sequences of the surrounding genes were subjected to similarity comparisons and functional predictions using the BLAST program of the NCBI GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The website was accessed on April 10th, 2013.
2.4 Construction of the lomo10 deletion mutant strain DCC601
The lomo10 gene was disrupted using the pKC1139-lomo10 inactivation plasmid. Two flanking regions or “arms” (1,996 and 1,784 bp), containing the upstream and downstream regions of lomo10, were amplified by PCR (30 cycles, 98°C for 10 s, 55°C for 15 s, and 68°C for 2 min) from S. lomondensis S015 genomic DNA using the primers lomo10 left arm For/Rev and lomo10 right arm For/Rev, respectively. The resulting products were individually ligated into the TA cloning vector pMD19-T to yield pCC601 and pCC602, respectively. Both insertions were verified by DNA sequencing. The downstream fragment from pCC602 was then excised using HindIII and BglII and ligated into the corresponding restriction sites in pCC601 to generate pCC603. The complete 3780 bp fragment from pCC603 was then excised and ligated to the corresponding HindIII and XbaI sites of pKC1139, generating inactivation plasmid pKC1139-lomo10.
pKC1139-lomo10 was first introduced into E. coli ET12567(pUZ8002) via heat shock transformation to generate the donor strain, then introduced into S. lomondensis S015 by conjugation [42]. Following incubation of the transconjugants at 28°C for 18 h, 1 ml of sterile water containing nalidixic acid and apramycin, both at a final concentration of 50 μg/ml, was spread onto the surface of the MS plate. Transconjugants were incubated for a further 2–4 days at 28°C, and resulting colonies were streaked onto solid MS medium containing 50 μg/ml apramycin at 37°C to yield single-crossover homologous recombination mutants. To inactivate lomo10, single crossover mutants were cultured at 37°C with shaking at 220 rpm for 3 days in a 250 ml flask containing 50 ml YM liquid medium. These mutants were cultured for five successive generations without apramycin to generate a double cross-over mutant that was sensitive to apramycin. In-frame deletion of lomo10 in the resulting positive mutant was confirmed by PCR (30 cycles, 98°C for 10 s, 55°C for 15 s, and 68°C for 2 min) with primers lomo10 left arm-For and lomo10 right arm-Rev.
2.5 Construction of the lomo10 self-complementation strain DCC602
The 1,326 bp lomo10 region was amplified (30 cycles, 98°C for 10 s, 55°C for 15 s, and 68°C for 1 min) from S. lomondensis S015 genomic DNA using the primers lomo10-For and lomo10-Rev. The PCR product was gel-purified and ligated into pMD19-T to form plasmid pCC604, which was confirmed by DNA sequencing. The lomo10 fragment was then excised from pCC604 using restriction enzymes NdeI/XbaI, and ligated into the corresponding sites of the integrative vector pIB139 [37] to yield the self-complementary plasmid pIB139-lomo10. pIB139-lomo10 was then introduced into the S. lomondensis S015 lomo10 deletion mutant strain, DCC601, by conjugation from E. coli ET12567(pUZ8002). A positive exconjugant was obtained by apramycin resistance screening and confirmed by PCR amplification and DNA sequencing using the primer pair pIB-F/pIB-R [43] and the thermal cycler parameters mentioned above.
2.6 High performance liquid chromatography (HPLC) analysis
Phenazine products in wild-type S015, lomo10-inactivated mutant DCC601, and lomo10-complemented strain DC602 were analyzed by HPLC as described by Wang et al. [33]. For sampling, up to 5 ml of culture broth were centrifuged at 10,800 × g for 8 min. The supernatant was adjusted to pH 2.0 using an aqueous HCl solution (6 M), and mixed with 5 ml of pure butanone. The resulting mixture was centrifuged at 10,800 × g for 5 min and the upper layer was collected. The water layer (lower layer) was extracted a second time using 5 ml of pure butanone, and the combined extracts were dried using a rotary vacuum dryer (Christ RVC 2–18, Osterode, Germany) at 33°C. The resulting residue was dissolved in 5 ml of HPLC grade solvent, a 1:1 (v/v) mixture of 0.1% formic acid and acetonitrile, and filtered through a 0.22 μm polyvinylidene difluoride syringe filter (Millipore, Shanghai, China). A 20:l sample of the resulting filtrate was analyzed by HPLC using an Agilent 1260 HPLC system (Agilent, Beijing, China) equipped with a DAD detector and an Agilent Eclipse Plus C18 column (250 × 4.6 mm; 5 μm), which was used at 30°C. The mobile phase consisted of solvents A (0.1% formic acid) and B (acetonitrile), which were used with the following gradient profile: 0–4 min, 80–60% A; 4–20 min, 60% A; and 20–30 min, 60–80% A. The HPLC system was operated at a constant flow rate of 1 ml/min.
2.7 Purification and structural analysis of the new phenazine product
Mutant DCC601 was used for large-scale fermentation (10 l) in yeast extract-malt extract broth at 28°C and 180 rpm for 96 h [33]. The liquid culture was centrifuged at 7,104 × g for 30 min, and then adjusted to pH 2.0 using an aqueous HCl solution (6 M). The supernatant was extracted four times with 2.5 l butanone. All four supernatants were combined and then concentrated using a vacuum evaporator at 33°C to remove the organic phase. For further purification, the raw extract was dissolved in 0.1% formic acid/acetonitrile (1:1, v/v) and purified by preparative HPLC using an Agilent 1200 series apparatus with a C18 column (ZOBRAX-C18 column, 5 μm, 10.0 × 250 mm, Elite, Dalian, China). The mobile phase consisted of 0.1% formic acid/acetonitrile (60:40, v/v) with a flow rate of 1 ml/min (detection by absorbance at 270 nm). The peak containing the pure compound was collected and dried under vacuum and freezing.
The dried intermediate metabolite was then used for further liquid chromatography-high resolution mass spectrometery (LC-HRMS) and nuclear magnetic resonance (NMR) analyses. The LC-HRMS was performed on a Waters ACQUITY UPLC system (Waters Corporation, Milford, MA) with an ACQUITY BEH C18 column (100 mm × 2.1 mm, 1.7 μm; Waters). The column was eluted with gradient solvent of 20:80 solutions B:A (0–15 min), 40:60 solutions B:A (15.01–25 min), and 20:80 solutions B:A (25.01–35 min) at a flow rate of 0.40 ml/min, where solution A is 0.1% (v/v) formic acid and solution B is acetonitrile. The system was monitored by measuring UV absorbance at 270 nm. The mass spectrometer was run in positive ion detection mode set to scan between 50 and 1,000 m/z. The NMR assay was performed with a Bruker NMR spectrometer (Avance III 600 MHz; Bruker, Karlsruhe, Germany).
Results
Analysis of lomofungin biosynthesis genes based on whole genome sequence of S. lomondensis S015
Sequencing and assembly of the S. lomondensis S015 genome resulted in a draft genome size of 9,448,526 bp and a GC content of 71.7%. The antiSmash program and alignment of the whole genome sequence with the phenazine biosynthesis gene cluster of P. chlororaphis GP72 [3,18] identified a putative gene cluster for the biosynthesis of lomofungin. This contiguous DNA region contains 23 open reading frames (Fig 1, Table 3). Six of the genes, designated lphzGFEDCB, showed high similarity (53.4–72.2%) at the amino acid level to the phenazine biosynthesis genes phzBCDEFG of P. chlororaphis GP72 [3,18]. Except for the absence of a phzA ortholog, and the order of the genes, the high similarity of these six genes to the corresponding genes in P. chlororaphis GP72 suggested a close evolutionary relationship with this species.
Fig 1. Putative lomofungin biosynthesis gene cluster in Streptomyces lomondensis S015.
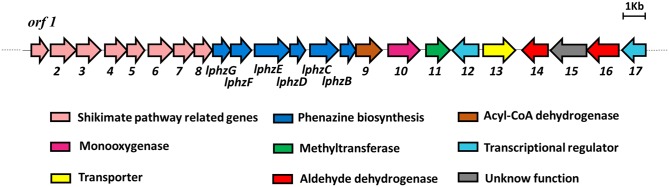
Table 3. Genes in the putative lomofungin biosynthesis cluster.
| ORFs | aa | Proposed function | Identity (%) | Ortholog identified by BLAST search |
|---|---|---|---|---|
| lomo1 | 201 | Shikimate kinase | 88% | Streptomyces fulvoviolaceus |
| lomo2 | 378 | Chorismate synthase | 97% | Streptomyces fulvoviolaceus |
| lomo3 | 272 | 3-phosphoshikimate- 1-carboxyvinyltransferase | 78% | Streptomyces sp. NRRL B-5680 |
| lomo4 | 306 | 5,10-methylenetetrahydrofolate reductase | 90% | Streptomyces sp. NRRL S-646 |
| lomo5 | 203 | Hexokinase | 71% | Streptomyces albaduncus |
| lomo6 | 398 | S-adenosylmethionine synthetase | 90% | Streptomyces sp. NRRL S-646 |
| lomo7 | 201 | Quinone oxidoreductase | 78% | Streptomyces anulatus |
| lomo8 | 162 | Flavin reductase | 85% | Streptomyces sp. NRRL S-646 |
| lphzG | 213 | phzG | 53% | Pseudomonas chlororaphis GP72 |
| lphzF | 278 | phzF | 66% | Pseudomonas chlororaphis GP72 |
| lphzE | 598 | phzE | 63% | Pseudomonas chlororaphis GP72 |
| lphzD | 111 | phzD | 65% | Pseudomonas chlororaphis GP72 |
| lphzC | 368 | phzC | 60% | Pseudomonas chlororaphis GP72 |
| lphzB | 160 | phzB | 72% | Pseudomonas chlororaphis GP72 |
| lomo9 | 354 | Acyl-CoA dehydrogenase | 95% | Streptomyces sp. NRRL S-646 |
| lomo10 | 442 | Flavin-dependent monooxygenase | 48% | Streptomyces viridosporus |
| lomo11 | 273 | Methyltransferase | 39% | Streptomyces auratus |
| lomo12 | 342 | Transcriptional regulator | 94% | Streptomyces sp. NRRL S-646 |
| lomo13 | 484 | Transporter | 91% | Streptomyces sp. NRRL S-646 |
| lomo14 | 314 | Alcohol dehydrogenase | 89% | Streptomyces sp. NRRL S-646 |
| lomo15 | 340 | EsmB1, phenazine antibiotic biosynthesis protein | 72% | Streptomyces antibioticus |
| lomo16 | 435 | Aldehyde dehydrogenase | 86% | Streptomyces sp. NRRL S-646 |
| lomo17 | 337 | Transcriptional regulator | 86% | Streptomyces sp. NRRL S-646 |
Sequences from Streptomyces lomondensis S015 have been deposited in the NCBI GenBank database under accession number KP721214.
A set of eight genes, designated lomo1–8, were most likely to code for enzymes relating to the shikimate pathway [2]. lomo9 showed 95% amino acid similarity to the acyl-CoA dehydrogenase gene from Streptomyces sp. NRRL S-646, which oxidizes branched-chain acyl-CoA fatty acid derivatives and macrolide antibiotics in Streptomyces coelicolor and Streptomyces avermitilis [44]. lomo10 showed 95% amino acid similarity to monooxygenases genes from Streptomyces acidiscabies and Streptomyces noursei, which are responsible for hydroxylation during the biosynthesis of thaxtomin A [9] and the polyene macrolide antibiotic nystatin, respectively [27,45]. lomo11 codes for a protein with moderate similarity (39%) to a putative methyltransferase that has been reported to catalyze the conversion of macrocin to tylosin [46]. Both lomo12 and lomo17 showed high similarity to the transcriptional regulatory genes in Streptomyces sp. NRRL S-646 and they both belong to the AsnC family.
Inactivation and self-complementation of lomo10
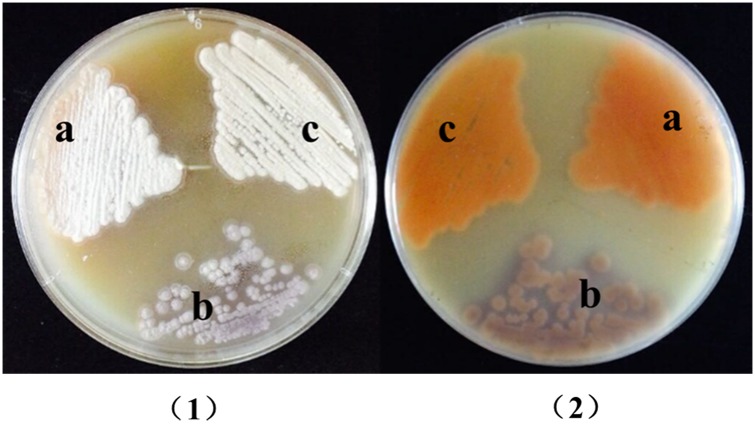
As there are three hydroxyl groups included in the structure of lomofungin, the function of the putative hydroxylation gene lomo10 was investigated by generating an in-frame partial deletion mutant, DCC601, via double-crossover homologous recombination (Fig 2A). Strain DCC601 was obtained by deletion of the 1032 bp in the coding region of lomo10. No promoter was found in this region by using online software (http://www.fruitfly.org/seq_tools/promoter.html) analysis. Thus, the partial deletion of lomo10 might not affect the expression of other genes, such as lomo11. The genotype of the DCC601 lomo10 deletion mutant, as well as that of the lomo10 self-complementation strain DCC602, was confirmed by PCR analysis as shown in Fig 2B and 2C, respectively and DNA sequencing. The phenotypes of wild-type strain S015, DCC601, and DCC602 were examined following culture on solid MS medium (Fig 3). Wild-type strain S015 produced the olive-yellow lomofungin (Fig 3A), whereas the lomo10 deletion mutant (DCC601) produced light purple colored colonies (Fig 3B). Complementation with lomo10 restored lomofungin production (Fig 3C). There was no obvious difference in the phenotype of mycelium between WT, mutant strain DCC601 and complementary strain DCC602.
Fig 2. Inactivation and self-complementation of lomo10 in Streptomyces lomondensis S015.
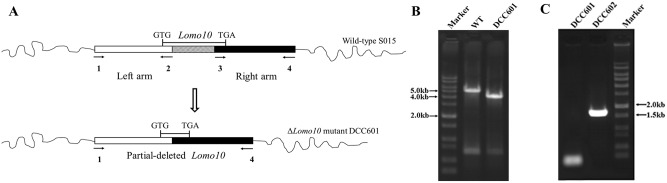
(A) Schematic of the in-frame partial deletion of 1032 bp in lomo10 to generate the Δlomo10 mutant DCC601. Primers 1, 2, 3, and 4 were used to amplify the left and right homology arms. GTG and TGA were the start and termination codons for lomo10, respectively. The expected PCR product from the wild-type (WT) strain was 5,106 bp, and that from DCC601 was 4,074 bp using the primers lomo10 left arm-For and lomo10 right arm-Rev. (B) PCR analysis of WT strain and DCC601. (C) PCR analysis of DCC601 and lomo10 complementation strain DCC602. The amplicon generated from the DCC602 genomic DNA gave the expected 1,726 bp fragment, but no band was amplified from the DCC601 strain.
Fig 3. Photographs of the different strains grown on MS medium.
(A) Wild-type strain. (B) lomo10 inactivated mutant DCC601. (C) lomo10 complementation strain DCC602.
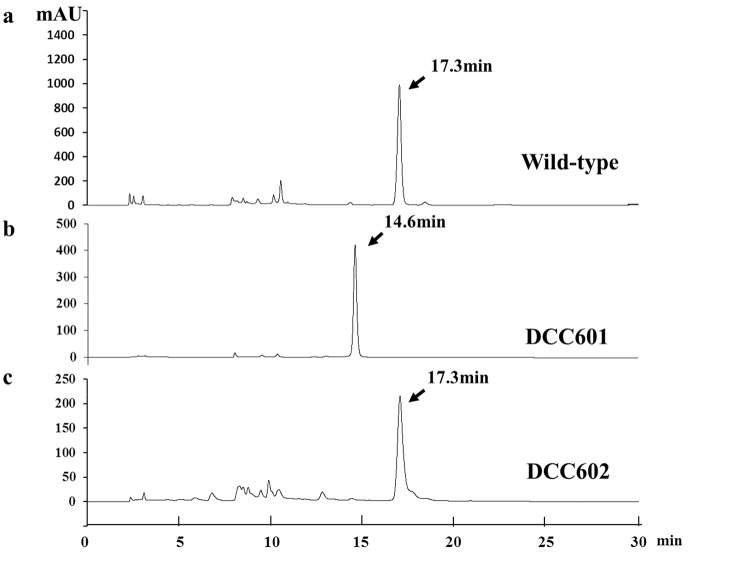
The HPLC profiles of the fermentation products of wild-type S015, DCC601, and DCC602 are illustrated in Fig 4. The peak corresponding to lomofungin appeared at a retention time of 17.3 min in the wild-type strain, but was eliminated in the knockout mutant, and a new peak with a retention time of 14.6 min appeared. Comparison of the HPLC profiles of the fermentation products of extracts from complementation strain DCC602 showed that lomofungin production had been restored in this strain. The new compound produced by the mutant strain, compound A, was purified, and its structure was characterized by LC-HRMS and NMR analyses.
Fig 4. HPLC profiles of fermentation products from different strains.
(A) Wild-type strain. (B) lomo10 inactivated mutant DCC601. (C) lomo10 complementation strain DCC602. lomofungin (Rt = 17.3 min), compound A (Rt = 14.6 min).
The methods and results of whole-cell biotransformation of compound A by Lomo10 were shown in S1 File and S1 Fig, respectively. After 2 h reaction, lomofungin was synthesized in the reaction system of E.coli DH5μ/pMD-18T-lomo10 mixed with compound A. The results further verified that Lomo10 could hydroxylates compound A to produce lomofungin.
Purification and structural analysis of compound A
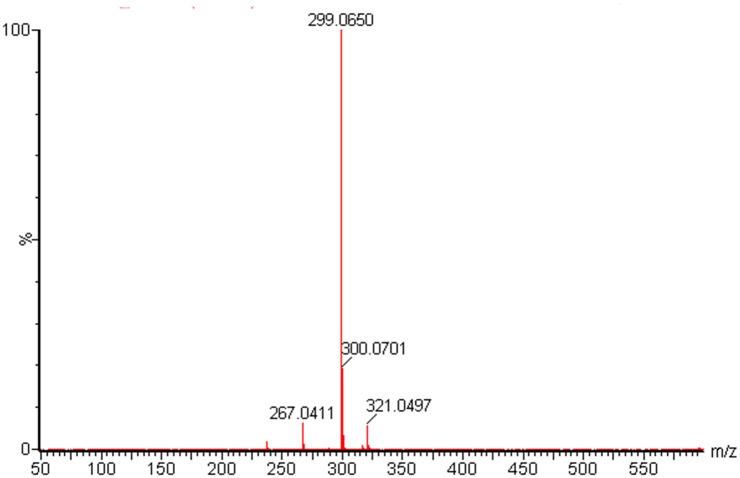
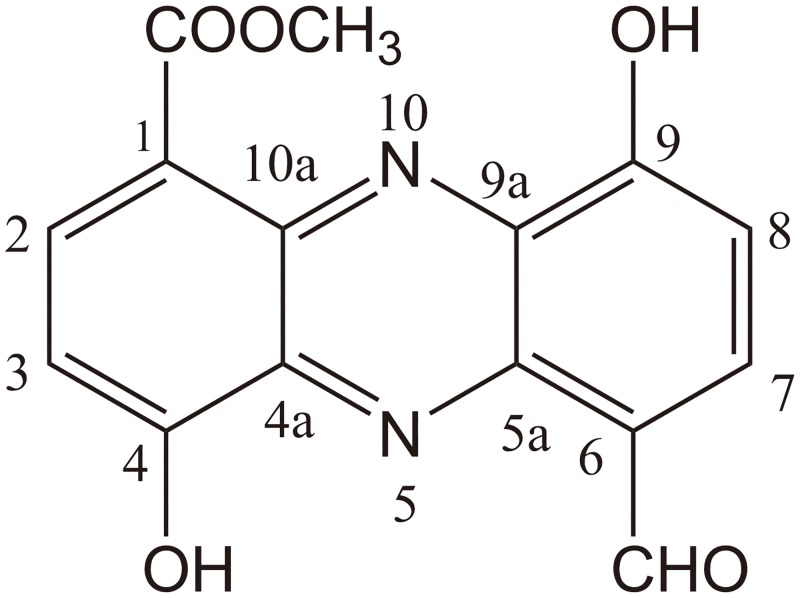
To confirm the structure of the new compound, compound A was purified from 10 l of fermentation broth. In total, 80 mg of compound A was obtained following purification. The exact mass of compound A obtained from LC-HRMS was 299.07 for [C15H10N2O5]+ (calculated mass, 298.25; Fig 5). This calculated mass was 16 Da smaller than the known mass of lomofungin, suggesting that compound A contained one less hydroxyl than lomofungin (C15H10N2O6, MW 314 Da) [47]. Structural analysis was performed by 1H NMR, 13C NMR, distortionless enhancement by polarization transfer-90, 1H-1H correlation spectroscopy, heteronuclear multiple bond correlation, and heteronuclear multiple quantum coherence analyses, and the proton and carbon chemical shifts of compound A are shown in Table 4 and in S2 Fig, respectively. The 1H-NMR spectra contained four proton signals (δH, 7.39–8.41 ppm) that are typical of double bonds (ring hydrogen), along with corresponding 13C-NMR spectra of carbon atoms (δC, 110.1–134.5 ppm), suggesting that compound A has a phenazine ring in the core of its structure. Compared with the reported NMR data for lomofungin [47], the main differences were the absence of a hydroxyl hydrogen at δH 11.22, and the presence of hydrogen ring at δH 8.27. Therefore, compound A was predicted to be 1-carbomethoxy-6-formy-4,9-dihydroxy phenazine, a new chemical compound missing the C-7 hydroxyl of lomofungin (Fig 6).
Fig 5. Spectrum of compound A following LC-HRMS analysis.
The caculated mass of compound 1 was determined to be 298.25 (m/z 299.07).
Table 4. Chemical shift summarized from 1H (DMSO-d6) and 13C (MeOD-d6) analyses recorded by 600 MHz NMR spectrometry.
| 1H NMR | Chemical shift δ (ppm) | 13C NMR | Chemical shift δ (ppm) |
|---|---|---|---|
| -COOCH 3 | 3.95, s | C-1 | 120.9 |
| C-2 | 8.41, d | C-2 | 134.4 |
| C-3 | 7.30, d | C-3 | 111.2 |
| C-7 | 8.28, d | C-4 | 156.5 |
| C-8 | 7.39, d | C-4a | 141.3 |
| -CHO | 11.40,s | C-5a | 135.2 |
| C-6 | 123.0 | ||
| C-7 | 134.5 | ||
| C-8 | 110.1 | ||
| C-9 | 160.2 | ||
| C-9a | 133.4 | ||
| C-10a | 139.8 | ||
| -CHO | 190.7 | ||
| -COOCH 3 | 52.1 | ||
| -COOCH3 | 166.2 |
Fig 6. Chemical structure of compound A.
Discussion
Phenazine biosynthesis gene clusters are normally composed of seven genes arranged in order (e.g. phzABCDEFG), and have been located downstream of shikimate pathway-related genes in Pseudomonas species including P. fluorescens 2–79, P. aureofaciens 30–84, and P. chlororaphis PCL1391 [11,14,15]. Two nearly identical core phz gene clusters, called phzA1-G1 and phzA2-G2, with different promoters and flanking regions, have also been found in Pseudomonas sp. M18 [48] and P. aeruginosa PAO1 [12].
Compared with Pseudomonas, the number and order of genes within the phenazine biosynthetic gene cluster in Streptomyces are more varied. Streptomyces anulatus 9663 also has a seven-gene cluster for the biosynthesis of endophenazine A and endophenazine B, but unlike the corresponding region in Pseudomonas species, phzA is located at the end of the cluster: ppzBCDEFGA [9,49]. A six-gene cluster for prenylated phenazine biosynthesis in Streptomyces cinnamonensis DSM 1042 is ordered ephzBCDEGA, and completely lacks the gene encoding the PhzF protein [17]. Another gene cluster consisting of just epzAGFC was identified in S. cinnamonensis DSM 1042, and is also involved in prenylated phenazine biosynthesis [50]. The whole genome sequencing performed in the current study showed that the phenazine biosynthetic gene cluster in S. lomondensis S015 is structured lphzGFEDCB, and while still located downstream of the shikimate pathway-related genes, the gene order is completely reversed compared with the cluster in Pseudomonas, and lacks phzA. In addition, we found another two phenazine biosynthesis-related genes, phzC2 and phzE2, in another scaffold of the whole genome of S. lomondensis S015. Further study is needed to confirm whether they are involved in the biosynthesis of lomofungin.
Until now, monooxygenases for the hydroxylation of phenazine compounds have been identified only in Pseudomonas. Most of these monooxygenases use PCA as their substrate [12,15,16]. In the current study, alignment of the lomo10 amino acid sequence showed that lomo10 encodes a putative flavin-dependent monooxygenase. Inactivation of lomo10 in S. lomondensis S015 resulted in the production of a novel phenazine product containing a deletion of the hydroxyl at the C-7 position of lomofungin. The four monooxygenases that catalyzed the hydroxylation of phenazines were compared and analyzed using DNASTAR Lasergene.v7.1 software and the results were shown in S3 Fig The monooxygenases in P. aureofaciens 30–84 and P. chlororaphis GP72 showed very high similarity and they both catalyze the hydroxylation of PCA at its C-2 position. The monooxygenase in S. lomondensis S015 showed low similarity to others might due to the differences in substrate structure and hydroxylation position because it hydroxylates compound A at its C-7 position.
There are three hydroxyl groups in lomofungin, located at the C-4, C-7, and C-9 positions. Only the C-7 hydroxyl was deleted in the lomo10 mutant strain DCC601, suggesting that there might be other hydroxylation genes in S. lomondensis S015. Two P450 monooxygenases, named lomo56 and lomo57, were located downstream of the lomofungin biosynthesis gene cluster in S. lomondensis S015. These two genes may be involved in the transfer of the C-4 and C-7 hydroxyl groups of lomofungin.
Based on the alignment of the S. lomondensis S015 genes and the other results obtained in this study, a putative lomofungin biosynthesis pathway is shown in Fig 7. Buckland et al. [51] confirmed that lomofungin is biosynthesized from PDC. Thus, here we propose that the lphzGFDECB gene cluster can biosynthesize PDC. The new compound obtained in this study, 1-carbomethoxy-6-formyl-4,9-dihydroxy-phenazine (compound A), might be synthesized from PDC in several steps catalyzed by Lomo9 and Lomo11 et al., and then converted into lomofungin by lomo10. Further validation of this pathway is required.
Fig 7. Proposed lomofungin biosynthetic pathway in Streptomyces lomondensis S015.
Supporting Information
(A) Lomofungin standard. (B) Biotransformation system with Lomo10. (C) Control without Lomo10.
(TIF)
(A) 1H NMR spectrum in DMSO-d6. (B) 13C NMR spectrum in MeOD. (C) COSY spectrum in DMSO-d6. (D) HMBC spectrum in DMSO-d6.
(TIF)
(TIF)
(PDF)
Acknowledgments
The authors express their appreciation to the Instrumental Analysis Center of Shanghai Jiao Tong University, particularly Dr. Lei Feng, for help with LC-HRMS analysis. We also thank Prof. Shuangjun Lin for help analyzing the NMR results.
Data Availability
All gene sequence files are available from the NCBI database (accession number KP721214.1).
Funding Statement
This study was supported by grants from the 863 Program of China (Grant No. 2012AA022107), the National Natural Science Foundation of China (NSFC project No. 20706037), and the 973 Project of China (Grant No. 2012CB721005).
References
- 1. Laursen JB, Nielsen J. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chemical Rev 2004; 104: 1663–1686. [DOI] [PubMed] [Google Scholar]
- 2. Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas Spp. biosynthesis and regulation. Annu Rev Phytopathol 2006; 44: 417–445. [DOI] [PubMed] [Google Scholar]
- 3. Liu H, He Y, Jiang H, Peng H, Huang X, Zhang X, et al. Characterization of a phenazine-producing strain Pseudomonas chlororaphis GP72 with broad-spectrum antifungal activity from green pepper rhizosphere. Current Microbiol 2007; 54: 302–306. [DOI] [PubMed] [Google Scholar]
- 4. Kerr J, Taylor G, Rutman A, Høiby N, Cole P, Wilson R. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J Clinical Pathology 1999; 52: 385–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chin-A-Woeng TF, Bloemberg GV, van der Bij AJ, van der Drift KM, Schripsema J, Kroon B, et al. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Molecular Plant-Microbe Interactions 1998; 11: 1069–1077. [Google Scholar]
- 6. Lau GW, Ran H, Kong F, Hassett DJ, Mavrodi D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infection Mmmunity 2004; 72: 4275–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turner JM, Messenger AJ. Occurrence, biochemistry and physiology of phenazine pigment production. Advances in Microbial Physiology 1986; 27: 211–275. [DOI] [PubMed] [Google Scholar]
- 8. Luo Q, Hu H, Peng H, Zhang X, Wang W. Isolation and structural identification of two bioactive phenazines from Streptomyces griseoluteus P510. Chinese J Chemical Engineering. 2015; 23: 699–703. [Google Scholar]
- 9. Mavrodi DV, Peever TL, Mavrodi OV, Parejko JA, Raaijmakers JM, Lemanceau P, et al. Diversity and evolution of the phenazine biosynthesis pathway. Appl Environmental Microbiol 2010; 76: 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blankenfeldt W, Kuzin AP, Skarina T, Korniyenko Y, Tong L, Bayer P, et al. Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens . PNAS 2004; 101: 16431–16436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mavrodi DV, Ksenzenko VN, Bonsall RF, Cook RJ, Boronin AM, Thomashow LS. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens2-79. J Bacteriol 1998; 180: 2541–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol 2001; 183: 6454–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu DQ, Ye J, Ou HY, Wei X, Huang X, He YW, et al. Genomic analysis and temperature-dependent transcriptome profiles of the rhizosphere originating strain Pseudomonas aeruginosa M18. BMC Genomics 2011; 12: 438 Available: http://www.biomedcentral.com/1471-2164/12/438. 10.1186/1471-2164-12-438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chin-A-Woeng TF, Thomas-Oates JE, Lugtenberg BJ, Bloemberg GV. Introduction of the phzH gene of Pseudomonas chlororaphis PCL1391 extends the range of biocontrol ability of phenazine-1-carboxylic acid-producing Pseudomonas spp. strains. Molecular Plant-Microbe Interactions 2001; 14: 1006–1015. [DOI] [PubMed] [Google Scholar]
- 15. Delaney SM, Mavrodi DV, Bonsall RF, Thomashow LS. phzO, a gene for biosynthesis of 2-hydroxylated phenazine compounds in Pseudomonas aureofaciens 30–84. J Bacteriol 2001; 183: 318–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang L, Chen MM, Wang W, Hu HB, Peng HS, Zhang XH. Enhanced production of 2-hydroxyphenazine in Pseudomonas chlororaphis GP72. Appl Microbiol Biotechnol 2011; 89: 169–177. 10.1007/s00253-010-2863-1 [DOI] [PubMed] [Google Scholar]
- 17. Haagen Y, Glück K, Fay K, Kammerer B, Gust B, Heide L. A gene cluster for prenylated naphthoquinone and prenylated phenazine biosynthesis in Streptomyces cinnamonensis DSM 1042. ChemBioChem 2006; 7: 2016–2027. [DOI] [PubMed] [Google Scholar]
- 18. Shen X, Chen M, Hu H, Wang W, Peng H, Zhang XH. Genome sequence of Pseudomonas chlororaphis GP72, a root-colonizing biocontrol strain. J Bacteriol 2012; 194: 1269–1270. 10.1128/JB.06713-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mentel M, Ahuja EG, Mavrodi DV, Breinbauer R, Thomashow LS, Blankenfeldt W. Of two make one: the biosynthesis of phenazines. ChemBioChem 2009; 10: 2295–2304. 10.1002/cbic.200900323 [DOI] [PubMed] [Google Scholar]
- 20. Saleh O, Flinspach K, Westrich L, Kulik A, Gust B, Fiedler HP, et al. Mutational analysis of a phenazine biosynthetic gene cluster in Streptomyces anulatus 9663. Beilstein J Organic Chemistry 2012; 8: 501–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zeyhle P, Bauer JS, Steimle M, Leipoldt F, Rösch M, Kalinowski J, et al. A membrane-Bound prenyltransferase catalyzes the O-prenylation of 1, 6-dihydroxyphenazine in the marine bacterium Streptomyces sp. CNQ-509. ChemBioChem 2014; 15: 2385–2392. 10.1002/cbic.201402394 [DOI] [PubMed] [Google Scholar]
- 22. Parsons JF, Greenhagen BT, Shi K, Calabrese K, Robinson H, Ladner JE. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa . Biochemistry 2007; 46: 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu Y, Mortimer MW, Fisher TS, Kahn ML, Brockman FJ, Xun L. Cloning, sequencing, and analysis of a gene cluster from Chelatobacter heintzii ATCC 29600 encoding nitrilotriacetate monooxygenase and NADH: flavin mononucleotide oxidoreductase. J Bacteriol 1997; 179: 1112–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rix U, Remsing LL, Hoffmeister D, Bechthold A, Rohr J. Urdamycin L: a novel metabolic shunt product that provides evidence for the role of the urdM gene in the urdamycin A biosynthetic pathway of Streptomyces fradiae TÜ 2717. ChemBioChem 2003; 4: 109–111. [DOI] [PubMed] [Google Scholar]
- 25. Valton J, Filisetti L, Fontecave M, Nivière V. A two-component flavin-dependent monooxygenase involved in actinorhodin biosynthesis in Streptomyces coelicolor . J Biological Chemistry 2004; 279: 44362–44369. [DOI] [PubMed] [Google Scholar]
- 26. Valton J, Fontecave M, Douki T, Kendrew SG, Nivière V. An aromatic hydroxylation reaction catalyzed by a two-component FMN-dependent monooxygenase the ActVA-ActVB system from Streptomyces coelicolor . J Biological Chemistry 2006; 281: 27–35. [DOI] [PubMed] [Google Scholar]
- 27. Volokhan O, Sletta H, Ellingsen TE, Zotchev SB. Characterization of the P450 monooxygenase NysL, responsible for C-10 hydroxylation during biosynthesis of the polyene macrolide antibiotic nystatin in Streptomyces noursei . Appl Environmental Microbiol 2006; 72: 2514–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson LE, Dietz A. Lomofungin, a new antibiotic produced by Streptomyces lomondensis sp. n. Appl Microbiol 1969; 17: 755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kopecká M, Gabriel M. Staining the nuclei in cells and protoplasts of living yeasts, moulds and green algae with the antibiotic lomofungin. Archives Microbiol 119: 305–311. [Google Scholar]
- 30. Gottlieb D, Nicolas G. Mode of action of lomofungin. Appl Microbiol 1969; 18: 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Manerba M, Vettraino M, Fiume L, Di Stefano G, Sartini A, Giacomini E, et al. Galloflavin (CAS 568-80-9): a novel inhibitor of lactate dehydrogenase. ChemMedChem 2012; 7: 311–317. 10.1002/cmdc.201100471 [DOI] [PubMed] [Google Scholar]
- 32. Kaneko Y, Fukazawa H, Ohno H, Miyazaki Y. Combinatory effect of fluconazole and FDA-approved drugs against Candida albicans . J Infection Chemotherapy 2013; 19: 1141–1145. [DOI] [PubMed] [Google Scholar]
- 33. Wang W, Wang H, Hu H, Peng H, Zhang X. Overexpression of afsR and optimization of metal chloride to improve lomofungin production in Streptomyces lomondensis S015. J Microbiol Biotechnol 2015; 25(5): 672–680. [DOI] [PubMed] [Google Scholar]
- 34. Wang H, Bei X, Hu H, Peng H, Zhang X, Wang W. Isolation, identification and medium optimization of modified phenazine derivative lomofungin from Streptomyces lomondensis S015. Shanghai Jiaotong University: Agricul Sci 2014; 32(2): 48–54. [Google Scholar]
- 35. MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 1992; 111: 61–68.52. [DOI] [PubMed] [Google Scholar]
- 36. Bierman M, Logan R, O'Brien K, Seno E, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 1992; 116: 43–49. [DOI] [PubMed] [Google Scholar]
- 37. Wilkinson CJ, Hughes-Thomas ZA, Martin CJ, Bohm I, Mironenko T, Deacon M, et al. Increasing the efficiency of heterologous promoters in actinomycetes. J Molecular Microbiol Biotechnol 2002; 4: 417–426. [PubMed] [Google Scholar]
- 38. Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, et al. Genetic manipulation of Streptomyces: A laboratory manual. 1st ed. The John Innes Foundation, Norwich, UK and Cold Spring Harbour Laboratory; 1985. [Google Scholar]
- 39. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical streptomyces genetics. 1st ed. The John Innes Foundation; 2000. [Google Scholar]
- 40. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. 2nd ed. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41. Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 2011; 39: 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mazodier P, Petter R, Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. J Bacteriol 1989; 171: 3583–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huang D, Xia M, Li S, Wen J, Jia X. Enhancement of FK506 production by engineering secondary pathways of Streptomyces tsukubaensis and exogenous feeding strategies. J Industrial Microbiol Biotechnol 2013; 40: 1023–1037. [DOI] [PubMed] [Google Scholar]
- 44. Zhang YX, Denoya CD, Skinner DD, Fedechko RW, McArthur HA, Morgenstern MR, et al. Genes encoding acyl-CoA dehydrogenase (AcdH) homologues from Streptomyces coelicolor and Streptomyces avermitilis provide insights into the metabolism of small branched-chain fatty acids and macrolide antibiotic production. Microbiology 1999; 145: 2323–2334. [DOI] [PubMed] [Google Scholar]
- 45. Healy FG, Krasnoff SB, Wach M, Gibson DM, Loria R. Involvement of a cytochrome P450 monooxygenase in thaxtomin A biosynthesis by Streptomyces acidiscabies . J Bacteriol 2002; 184: 2019–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bauer NJ, Kreuzman AJ, Dotzlaf JE, Yeh WK. Purification, characterization, and kinetic mechanism of S-adenosyl-L-methionine: macrocin O-methyltransferase from Streptomyces fradiae . J Biological Chemistry 1988; 263: 15619–15625. [PubMed] [Google Scholar]
- 47. Bergy M. Lomofungin, a new broad spectrum antibiotic: isolation and characterization. J Antibiotics 1969; 22: 126–128. [DOI] [PubMed] [Google Scholar]
- 48. Li Y, Du X, Lu ZJ, Wu D, Zhao Y, Ren B, et al. Regulatory feedback loop of two phz gene clusters through 5′-untranslated regions in Pseudomonas sp. M18. PLoS One 2011; 6: e19413 10.1371/journal.pone.0019413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saleh O, Gust B, Boll B, Fiedler HP, Heide L. Aromatic prenylation in phenazine biosynthesis: dihydrophenazine-1-carboxylate dimethylallyl transferase from Streptomyces anulatus . J Biological Chemistry 2009; 284: 14439–14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Seeger K, Flinspach K, Haug-Schifferdecker E, Kulik A, Gust B, Fiedler HP, et al. The biosynthetic genes for prenylated phenazines are located at two different chromosomal loci of Streptomyces cinnamonensis DSM 1042. Microbiol Biotechnol 2011; 4: 252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Buckland P, Gulliford S, Herbert R, Holliman F. The biosynthesis of phenazines: biosynthesis of lomofungin via phenazine-1,6-dicarboxylic acid. J Chemical Res 1981; 12: 362–362. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Lomofungin standard. (B) Biotransformation system with Lomo10. (C) Control without Lomo10.
(TIF)
(A) 1H NMR spectrum in DMSO-d6. (B) 13C NMR spectrum in MeOD. (C) COSY spectrum in DMSO-d6. (D) HMBC spectrum in DMSO-d6.
(TIF)
(TIF)
(PDF)
Data Availability Statement
All gene sequence files are available from the NCBI database (accession number KP721214.1).