Abstract
Combinations of molecular signals such as transcription factors and microRNAs in cells are a reliable indicator of multi-gene disorders. A system capable of detecting these conditions in-situ may be used as a tool for diagnosis and monitoring of disease. Here, we engineer genetic circuits that sense endogenous levels of the androgen receptor (AR), the glucocorticoid receptor (GR), and the microRNA hsa-miR-21 (miR-21) in cervical cancer cells (HeLa). Furthermore, using the mediator molecule human chorionic gonadotropin (hCG), we interface the intracellular information to enzyme-linked immunosorbent assay (ELISA) test strips. We demonstrate that this hybrid genetic circuit and test-strip interface can accommodate combinatorial, low-cost, and in-situ reporting, a versatile profiling tool.
Keywords: Synthetic biology, hybrid sensors, microRNAs, diagnostics, profiling
1. Introduction
We report the implementation of a hybrid system engineered to detect combinations of endogenous signals and transduce the information to an external sensor for in-situ and low-cost reporting. The system consists of two autonomous units (sensing and reporting) that are interfaced using a mediator protein molecule. The sensing unit is a genetic circuit that is engineered to perform the endogenous signal sensing in live cells and conditionally produce the mediator molecule which subsequently relays the measurement to the reporting unit. To interface the output of the genetic circuit to the reporting unit we use a mediator that is secreted from the cells and reacts specifically with a paper-based sensor. Specifically, the sensing unit conditionally produces the human chorionic gonadotropin (hCG) β subunit. hCG is a human hormone produced physiologically only when an ovum is fertilized by a sperm. Accordingly, we adopted a variant of an ELISA-based test strip for sensing the hCG protein.
A cell-free paper-based sensor was introduced recently (Pardee et al., 2014), a major advance in the use of synthetic biology for diagnostics. Here, we propose a complementary approach customized for use with human cells that, amongst other properties, bypasses any sample preparation requirements. In sharp contrast to the current paradigm in bioanalytical sensor engineering, where custom microfabricated devices are engineered and optimized to detect a specific input (e.g., a protein), here the sensing is performed by the genetic circuits in vivo. A key advantage of the proposed system is that the endogenous signal sensing unit can be reconfigured as needed and accommodate multiple inputs. The sensing unit can be modified merely by replacing the genetic circuit, notably permitting a range of combinatorial information processing options (Guinn and Bleris, 2014, Benenson, 2012, Rinaudo et al., 2007). Importantly, instead of detecting molecules of interest within a highly complex and heterogeneous cell lysate, the reporting unit will only respond to the presence of a predetermined agent of interest secreted by the cells. This represents a paradigm shift in the area and could significantly diminish the specificity and selectivity issues that generally hamper the reliability of biomolecular sensors.
Robust sensing of combinations of endogenous molecules in cells with genetic circuits is a non-trivial task. A genetic circuit platform was introduced recently for detecting several endogenous microRNAs (Xie et al., 2011). These engineered circuits exploit a miRNA-mediated repression that involves the miRNA and RISC complex (miRISC) directing endonucleolytic mRNA cleavage, known to occur when perfect complementarity between the miRNA target site and the miRISC group exist. On the other hand, reliable sensing of endogenous transcription factors remains a largely unsolved problem. Results include RNA-based (Culler et al., 2010, Kramer et al., 2005) systems that interface with a single endogenous transcription factor.
Here, we developed molecular circuit architectures engineered to sense over-expressed endogenous transcription factors (TFs) and microRNAs (miRNAs). Specifically, we detect the human glucocorticoid receptor (GR), the human androgen receptor (AR), and the human miRNA hsa-miR-21 (miR-21). We engineered genetic circuit architectures that can detect the endogenous TFs separately, in pairs via fan-in architectures, and in finally combinations of TF and miRNA.
Combinations of intracellular molecular signals in cells are an excellent indicator of multi-gene disorders, including cancers and hereditary diseases. Specifically, transcription factors and miRNA are often misregulated simultaneously in disease states (Ribas et al., 2009). Therefore, a system capable of detecting combinations of multiple molecular species may be used in the future as a highly selective tool for diagnosis, prevention, and treatment applications.
2. Methods
Cell Culture
HeLa cells were maintained at 37°C, 5% CO2 and 100% humidity. The cells were grown in Dulbecco's modified Eagle's medium (DMEM, Life Technologies, #11965-118) supplemented with 10% Fetal Bovine Serum (FBS, Atlanta Biologicals, #S11550), 0.1 mM MEM non-essential amino acids (Life Technologies, #11140-050), 0.045 units/mL of Penicillin and 0.045 units/mL of Streptomycin (Penicillin-Streptomycin liquid, Life Technologies, #15140-122). To pass the cells, the adherent culture was washed with DPBS (Dulbecco's Phosphate-Buffered Saline, 1X with calcium and magnesium, Corning, #21-030-CM) and then trypsinized with Trypsin-EDTA (Trypsin-EDTA (0.25%), phenol red, Life Technologies, #25200-114) and finally diluted in fresh medium upon reaching 50–90% confluence.
Transient Transfection
For transient transfections ~250,000 cells in 1 mL of complete medium were plated into each well of 12-well culture treated plastic plates (Griener Bio-One, #665180) and grown for 16-24 hours. For DNA jetPRIME transfection in 12-well plates, up to 1.85 μg of plasmid was added to 75 μL of jetPRIME buffer and 2.4 μL jetPRIME reagent (Polyplus transfection, #114-15). Transfection solutions were mixed and incubated at room temperature for 10 minutes. The transfection mixture was then applied to the cells and mixed with the medium by gentle shaking. When applicable, hydrocortisone (Alfa Aesar, #A16292), dexamethasone (Enzo Life Sciences, #BML-EI126-0001) and/or ponasterone A (Enzo Life Sciences, #ALX-370-014-M005) were added immediately following transfection.
Fluorescence Microscopy
All microscopy was performed 48 hours post transfection. The live cells were grown on 12-well plates in the complete medium. Cells were imaged using the Olympus IX81 microscope and a Precision Control environmental chamber. The images were captured using a Hamamatsu ORCA-03 Cooled monochrome digital camera. The filter sets (Chroma) are as follows: ET436/20x (excitation) and ET480/40 m (emission) for TagCFP, and ET560/40x (excitation) and ET630/75 m (emission) for mKate2. Data collection and processing was performed in software package Slidebook 5.0. All images within a given experimental set were collected with the same exposure times and underwent identical processing.
Flow Cytometry
48 hours post transfection cells from each well of the 12-well plates were washed with 1.0 mL of DPBS and then trypsinized with 0.3 mL 0.25% Trypsin-EDTA at 37°C for 5 minutes. Trypsin-EDTA was then neutralized by adding 0.7 mL of complete medium. The cell suspension was centrifuged at 1,000 rpm for 5 minutes and after removal of supernatants, the cell pellets were resuspended in 0.5 mL DPBS. The cells were analyzed on a BD LSRFortessa flow analyzer. TagCFP was measured with a 445-nm laser and a 470/20 band-pass filter, and mKate2 with a 561-nm laser, 600 emission filter and 610/20 band-pass filter. For all experiments ~50,000-100,000 events were collected. All wells within a given experimental set had the same number of events collected with the same voltages and underwent identical processing.
The flow cytometry data was processed using FlowJo. A FSC (forward scatter)/SSC (side scatter) gate was generated using an untransfected negative sample with the magnetic gate function in FlowJo and applied to all cell samples. To quantify the response of the sensors within this work, we utilized the integrated mean fluorescence intensity (iMFI) as the sample metric of choice, which is a weighted fluorescence intensity (Guinn and Bleris, 2014, Ausländer et al., 2012). We set a fluorescence gate above the negative control (untransfected) sample, calculated the mean fluorescence above the fluorescence gate, and multiplied the mean by the frequency of cells within the fluorescence gate (cells above the fluorescence gate divided by the total cells in the FSC/SSC gate).
3. Results
The endogenous TFs of interest, AR and GR, are both widely studied mammalian proteins that are misregulated in a variety of human disorders, and have been attributed to the etiology of several diseases (Dehm and Tindall, 2007, Pace et al., 2007, Webster et al., 2001, Sousa et al., 2000). Additionally, miR-21 is misregulated in a wide range of human pathologies (Slaby et al., 2008, Talotta et al., 2008). AR is the mediator for androgens and is known to be involved in prostate cancer; it is therefore a target of therapeutics (Libermann and Zerbini, 2006, Darnell, 2002). GR is the mediator for glucocorticoids, which are involved in many functions including development, metabolism and immune response (Lu et al., 2006, Rhen and Cidlowski, 2005). AR and GR are members of the nuclear receptor subfamily 3, group C. These receptors are localized in the cytoplasm with chaperone proteins until they are bound by a steroid hormone. This ligand binding causes a conformation change in the receptor, allowing dimerization and translocation into the nucleus (Tsai and O'Malley, 1994). Once in the nucleus they regulate target gene transcription by binding to hormone response elements and recruiting coregulators along with the transcriptional machinery (Tsai and O'Malley, 1994).
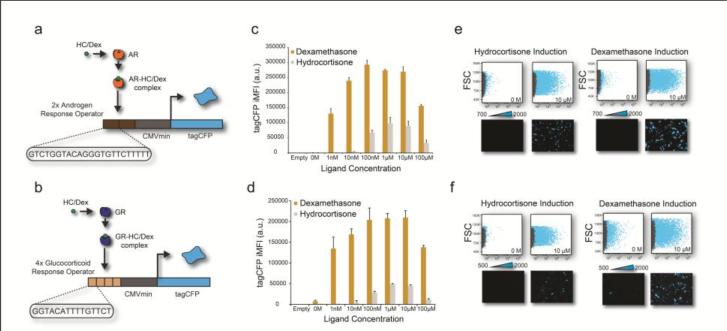
We engineered an AR sensor by inserting two repeats of an androgen response element (ARE) in front of a minimal CMV (minCMV) promoter both upstream of the fluorescent reporter protein TagCFP (Figure 1a). In a similar manner, the GR sensor consists of four repeats of the glucocorticoid response element (GRE) in front of a minCMV promoter both upstream of the fluorescent reporter protein TagCFP (Figure 1b). After sensor construction, we performed a titration of the ligands hydrocortisone and dexamethasone (both of which can act as AR and GR agonists) to induce AR or GR activity in HeLa cells and measured the TagCFP reporter concentrations.
Figure 1. Hydrocortisone and dexamethasone titration of the AR and GR sensor.
(a) Genetic architecture of the AR fluorescent sensor. (b) Genetic architecture of the GR fluorescent sensor. (c) TagCFP iMFI of the AR sensor at different dexamethasone (yellow bars) and hydrocortisone (gray bars) concentrations. Error bars show standard deviation of a triplicate experiment. (d) TagCFP iMFI of the GR sensor at different dexamethasone (yellow bars) and hydrocortisone (gray bars) concentrations. Error bars show standard deviation of a triplicate experiment. (e) Flow cytometry scatter plots (forward scatter versus TagCFP) and fluorescence microscopy showing the low and high concentration of hydrocortisone (HC) and dexamethasone (Dex) on the AR sensor. The gray dots represent the untransfected (control) well, and the blue dots are the transfected cells. (f) Flow cytometry scatter plots (forward scatter versus TagCFP) and fluorescence microscopy showing the low and high concentration of hydrocortisone (HC) and dexamethasone (Dex) on the GR sensor. The gray dots represent the untransfected (control) well, and the blue dots are the transfected cells.
HeLa cells were plated on a 12-well plate, transfected with the sensors 24 hours later, induced immediately post-transfection with hydrocortisone or dexamethasone, and were grown an additional 48 hours, concluding with circuit characterization via fluorescence microscopy and flow cytometry (Supplementary Experimental Table). We performed a titration for both hydrocortisone and dexamethasone (0-100μM), to gauge optimal ligand concentrations for AR or GR induction. We observed maximal TagCFP fluorescence between 100nM-10μM dexamethasone for both the AR and GR sensors (Figure 1c-f). Additionally, we observed non-basal fluorescence signal beginning at 10nM hydrocortisone and 1nM dexamethasone for both the AR and GR sensors. We observe higher TagCFP fluorescence when the cells were induced with dexamethasone compared to hydrocortisone at all concentrations tested (Figure 1). Live cell and fluorescence gating as well as all values of ligand titration for both the AR and GR sensors (for hydrocortisone and dexamethasone) are shown in Supplementary Figures 1-4.
After validation of the AR and GR sensors, we coupled the AR sensor with an endogenous miRNA. miRNAs are a class of short, 17-22 nt, noncoding RNAs that play roles in regulating the expression of genes involved in various functions by binding to sequences in an expressed messenger RNA (mRNA), resulting in decreased protein expression. miRNAs have been shown to control the activity of nearly 30% of all protein-coding genes and appear to play a regulatory role in nearly every cellular process (Baek et al., 2008, Tang et al., 2008, Bartel, 2004, He and Hannon, 2004). Studies over the past decade have demonstrated that differential expression of miRNAs in a given cell type play an important role in the pathogenesis of various cancers (Du and Pertsemlidis, 2010, Pang et al., 2010, Garzon et al., 2009). We selected hsamiR-21 (miR-21), which is upregulated in multiple cancer types and was shown to be activated by AR in prostate cancer (Jazbutyte and Thum, 2010, Ribas et al., 2009).
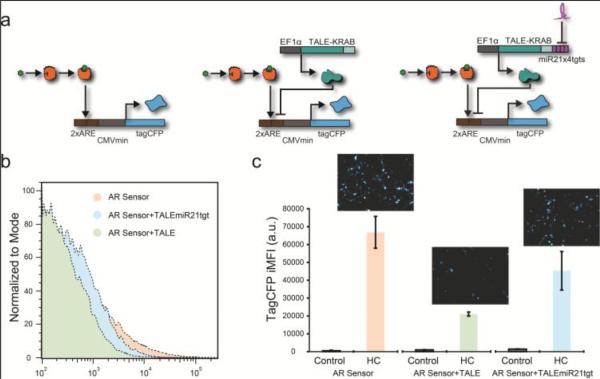
Our sensors for endogenous miRNA use four fully complementary target sites located in the 3'UTR of a target protein (Xie et al., 2011). As illustrated in Figure 2a, the miR-21 target sequence was incorporated into the 3'UTR of a transcriptional activator-like effector (TALE) repressor gene, which in turn inhibits the reporter protein tagCFP. TALEs are DNA binding proteins from the pathogenic plant bacteria, Xanthomonas, which can be engineered to function in mammalian cells as transcription factors (Li et al., 2014, Moore et al., 2014, Lienert et al., 2013, Li et al., 2012). Accordingly, increased levels of miR-21 will relieve the TALE based repression of tagCFP. We observe that adding a TALE repressor module without the microRNA targets, decreases the expression of the sensor in the ON state (Figure 2b-c). When we add the TALE with the miRNA targets we partially recover the system original ON state (Figure 2b-c). For the TF and miRNA combined sensing, we found optimal expression of the system with hydrocortisone at a concentration of 100 nM.
Figure 2. TF and miRNA coupled sensor.
(a) Biological schematic and genetic architecture of the AR sensor, the AR sensor with TALE module, and the AR sensor with TALE module and miRNA module. The AR sensor is targeted for repression by the TALE module, which is in turn regulated by the endogenous miR-21 (the miRNA module). (b) Histograms of HeLa cells transfected with the AR sensor and additional modules. The red population is the AR sensor alone, the green population is the AR sensor with the TALE module, and the blue population is the AR sensor with the TALE and miRNA module. (c) Fluorescence microscopy of the AR sensor alone and with the different modules and bar graph showing TagCFP iMFI calculated from the flow cytometry data. Error bars show standard deviation of a triplicate experiment.
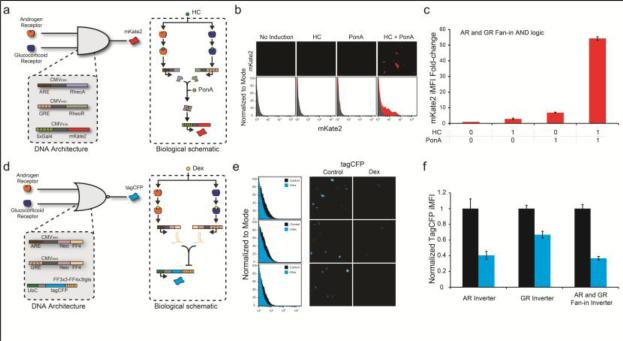
We next engineered sensors that interface the two transcription factors. The first fan-in circuit produces the mKate2 output signal only when both TFs are active. The second fan-in circuit produces the output only when both TFs are absent. The fan-in AND circuit utilizes the Rheo system which consists of a heterodimer pair, the RheoActivator (RheoA) and the RheoReceptor (RheoR) (New England Biolabs), which binds to the Gal4 enhancer and activates transcription in the presence of a small molecule, ponasterone A (Kumar et al., 2002). The first plasmid contains the ARE in front of the minCMV promoter, and transcribes RheoA in response to AR. The second plasmid contains the GRE in front of the minCMV promoter, and transcribes RheoR in response to GR. The third plasmid contains five copies of the Gal4 enhancer in front of the minCMV promoter, and transcribes the fluorescent reporter protein mKate2 in response to the Rheo dimer (Figure 3a). This architecture produces mKate2 fluorescence only when RheoA and RheoR are produced and dimerize in the presence of Ponasterone A (Figure 3a). We induced the system with 5μM hydrocortisone and 10μM ponasterone. All possible states of this sensor are shown in Supplementary Figure 5.
Figure 3. Fan-In Architectures.
(a) Biological schematic and genetic architecture of the fan-in AND gene circuit. (b) Fluorescence microscopy and histograms (flow cytometry) of the fan-in AND gene circuit under different induction states. (c) mKate2 iMFI fold change for the fan-in AND gene circuit under different induction states. (d-f) Same as a-c for the fan-in NOR gene circuit. HC: induced with hydrocortisone; PonA: induced with ponasterone A. Dex: induced with dexamethasone. Error bars show standard deviation of a triplicate experiment.
Qualitative analysis using fluorescence microscopy shows mKate2 fluorescence only when the fan-in AND sensor plasmids are used and induced with both hydrocortisone and ponasterone A (Figure 3b). Analyzing the system quantitatively using flow cytometry confirms the microscopy measurements (Figure 3c). We observe high mKate2 signal with the fan-in AND sensor plasmids and both hydrocortisone and ponasterone A, and basal level mKate2 signal in all other cases (Supplementary Figure 5).
The fan-in NOR circuit produces a TagCFP output signal when both AR and GR are absent. The fan-in NOR sensor architecture makes use of the synthetic miRNA-FF4 (Kashyap et al., 2013, Bleris et al., 2011, Leisner et al., 2010). The first plasmid contains the ARE minCMV promoter controlling the transcription of the synthetic miRNA-FF4 after the neomycin resistance gene. The second plasmid contains the GRE minCMV promoter controlling the transcription of the synthetic miRNA-FF4 (Figure 3d). The third plasmid contains the UbC promoter, which constitutively transcribes the fluorescent protein TagCFP which has three perfectly complementary miRNA-FF4 target sites in its 3’ untranslated region (Guinn and Bleris, 2014). This architecture only produces TagCFP fluorescence when miRNA-FF4 is absent, which occurs when AR and GR are absent or at low levels within the nucleus. We induced the system with 1μM dexamethasone.
Fluorescence microscopy and flow cytometry show that the AR and GR are able to repress the output as single inputs or combined in the fan-in architecture (Figure 3e-f). In agreement with Figures 1 and Supplementary Figures 1-4, which show that the AR effect is higher than GR's effect on the sensors, we observe that AR is able to suppress 60% of the OFF state while the GR is able to suppress approximately 35%. The cumulative effect of both AR and GR shows a combined inhibition of approximately 65%. The optimization data and control states of the system are shown in Supplementary Figure 6.
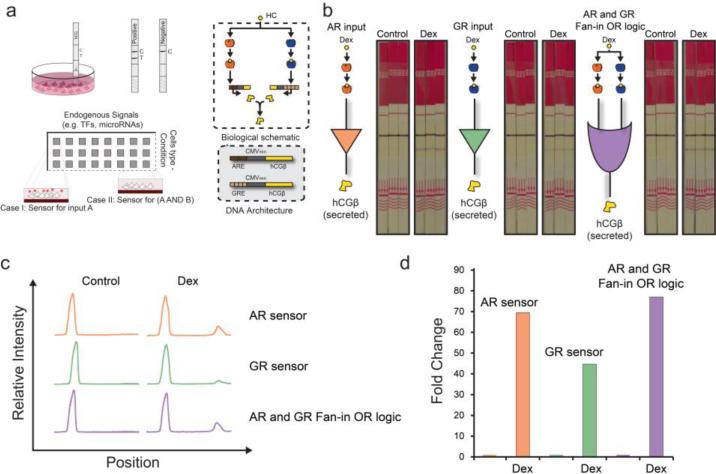
Lastly, we established the interface between the output of the genetic circuits and a colorimetric reporter. We selected ELISA-based test strips used for detecting the pregnancy marker, human chorionic gonadotropin. Physiologically, the hCG protein complex consists of two subunits, α and β, but the β subunit is the molecular marker that leads to a positive or negative test result on the test strip. Receiving two bands on the test strip (the control band which is the top band and the test band which is the bottom band) indicates that hCGβ is present in the sample while one band on the test strip (the control band only) indicates that hCGβ is absent (Figure 4a).
Figure 4. Interface with ELISA test strips.
(a) Example of the proposed system in a multi-well implementation, biological schematic and genetic architecture of hCG system. (b) Experimental schematic of the AR, GR and fan-in OR gene circuit interfaced with commercially available ELISA test strips. Images of the ELISA test strips used for the measurements. (c) Histograms of the signal intensity obtained from test strips. (d) Fold change of the test peak between induced and uninduced states of the OR gene circuit.
In principle, the reporting unit can be replaced with any desired biosensing modality (Zhou et al., 2014). For example, using a functionalized sensor with a biotin surface (Duan et al., 2012) would allow streptavidin to be used as the mediator molecule produced by the genetic circuits. A simple modification on the mediator molecule (e.g. introducing the BM40 signal peptide) will result to secretion of the selected protein. In our case, the mediator molecule hCGβ, is excreted from the cell naturally. We envision high-throughput implementations where live cells are introduced in a device together with the transfection reagents that carry genetic circuits (Figure 4a). The delivered DNA will begin expressing the active components which will interact with the intracellular milieu and respond by releasing only when necessary, an agent that reacts with the selected functionalized reporting device.
Accordingly, we replaced the fluorescent reporters of our sensors with hCGβ, allowing colorimetric detection of the output. To validate the sensors, approximately HeLa cells were plated on a culture dish, transfected 24 hours later, induced immediately post-transfection with 1μM dexamethasone, and were grown an additional 72 hours. At 72 hours post-transfection and induction, we immersed a test strip in the media and quantified the bands 10 minutes later.
As shown in the first panel of Figure 4b we first test the AR sensor. The first three test strips are triplicate measurements of the AR sensor transfected, in the absence of dexamethasone. There is a single band which is the control band produced when a sample is added. The second set of test strips is the AR sensor transfected in the presence of dexamethasone. This induces the AR to translocate to the nucleus, activate the AR sensor to produce hCGβ, which subsequently is exported in the media. In this case, we obtain two bands for each strip; the control band and test band. The bottom bands are only present when the marker of interest is present in the sample. This same set-up was performed for the GR sensor shown in the next two sets of tests strips, showing proper signal production in the case of active TFs. Lastly, we coupled both the AR and GR sensor into a fan-in OR gate. Again as with the two sensors separately, we obtain only the control bands only when the TFs are inactive and both control and test bands when the TFs are active.
Finally, we quantified the test-strip bands using ImageJ (Figure 4c-d, Supplementary Figure 6 & 7). The histograms correspond to the relative intensity of the triplicate test strips bands of Figure 4b. The orange bands are the AR sensor, the green bands are the GR sensor, and the purple bands are the fan-in OR gate sensor. Comparing the test bands among the AR sensor, the GR sensor, and the fan-in OR gate, we show between 45 and 77 fold change for the ON-OFF states, with the strongest band observed when both AR and GR sensors were introduced into the cells as an AR and GR fan-in OR logic gate (Figure 4b-d).
4. Conclusion
Here, we present a novel hybrid system engineered to detect combinations of intracellular signals and transduce the information to a paper-based sensor for in-situ colorimetric reporting. We detect combinations of over-expressed endogenous transcription factors and a miRNA. We demonstrate that TFs that translocate to the nucleus in response to signaling mechanisms are excellent candidates for interfacing with genetic sensors. Additionally, we show for the first time a coupling of endogenous TFs and miRNAs at the sensor that may provide superior specificity for cellular classification.
In contrast to the current paradigm in biosensor engineering, where custom fabricated devices are engineered and optimized to detect a specific input, here we propose a hybrid system where the sensing is performed by reconfigurable genetic circuits. The following properties differentiate it from available bioanalytical sensor and profiling platforms: (a) It is low-cost, (b) there is no need for preprocessing (e.g., RNA extraction), (c) it has the capacity to perform medium-sized profiling, and finally (d) it can report on combinations of intracellular signals.
Looking ahead, there are several challenges and opportunities for this family of hybrid biosensors. Firstly, the circuit components can be updated and optimized depending on the sensing application and desired specifications. For example, here we utilized the Rheo switch system, which requires ponasterone A, because it is well-characterized heterodimerization system. To eliminate the ponasterone A requirement, instead of using the Rheo system, a dimerization domain could be added to a split TALE, with one side having the DNA binding domain and the other the activation domain (Truong et al., 2015, Reinke et al., 2010). Secondly, the delay in detecting hCGβ remains a limitation for in-situ sensing applications. We predict that the time duration needed for signal detection can be significantly reduced through circuit as well as assay modifications. For example, increasing the number of response elements within the circuit could increase the recruitment of the transcription factors to the sensor and increase the sensitivity to the endogenous inputs. Custom genetic architectures can be incorporated to accelerate the response time of the circuit (e.g. negative feedback (Shimoga et al., 2013, Rosenfeld et al., 2002)). Finally, another option for increasing the sensitivity could be adopting commercially available ELISA amplification methods or alternative reporters (Luk et al., 2009).
Supplementary Material
Highlights.
We engineer genetic circuits that sense endogenous levels of the androgen receptor, the glucocorticoid receptor, and the microRNA hsa-miR-21.
Using the mediator molecule human chorionic gonadotropin (hCG), we interface intracellular information to enzyme-linked immunosorbent assay test strips.
We demonstrate that our hybrid genetic circuit and test-strip interface can accommodate combinatorial, low-cost, and in-situ reporting.
Acknowledgements
This work was funded by the US National Institutes of Health (NIH) grant R15GM09627, TxACE SRC grant P12095, and National Science Foundation (NSF) grant 1105524.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
K.E. and M.G. designed and performed the experiments. T.Q. assisted with cloning and characterization experiments, K.E., M.G. M.Z and L.B. analyzed the data and prepared the manuscript. M.G. conceived the hCG reporting system. L.B. conceived and supervised the project.
Competing financial interests
No competing financial interests.
Additional information
Supplementary information is available in the online version of the paper.
References
- Ausländer S, Ausländer D, Müller M, Wieland M, Fussenegger M. Nature. 2012;487:123–127. doi: 10.1038/nature11149. [DOI] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Benenson Y. Nature Reviews Genetics. 2012;13:455–468. doi: 10.1038/nrg3197. [DOI] [PubMed] [Google Scholar]
- Bleris L, Xie Z, Glass D, Adadey A, Sontag E, Benenson Y. Molecular Systems Biology. 2011;7:519. doi: 10.1038/msb.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culler SJ, Hoff KG, Smolke CD. Science. 2010;330:1251. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE. Nature Reviews Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- Dehm SM, Tindall DJ. Molecular Endocrinology. 2007;21:2855. doi: 10.1210/me.2007-0223. [DOI] [PubMed] [Google Scholar]
- Du L, Pertsemlidis A. Cancer and Metastasis Reviews. 2010;29:109–122. doi: 10.1007/s10555-010-9204-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Li Y, Rajan NK, Routenberg DA, Modis Y, Reed MA. Nature nanotechnology. 2012;7:401–407. doi: 10.1038/nnano.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. Annual Review of Medicine. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Guinn M, Bleris L. ACS synthetic biology. 2014;3:627–633. doi: 10.1021/sb4001596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. Nature Reviews Genetics. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Jazbutyte V, Thum T. Current Drug Targets. 2010;11:926–935. doi: 10.2174/138945010791591403. [DOI] [PubMed] [Google Scholar]
- Kashyap N, Pham B, Xie Z, Bleris L. Molecular Biosystems. 2013;9:1919–1925. doi: 10.1039/c3mb70043g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer BP, Fischer M, Fussenegger M. Metabolic Engineering. 2005;7:241–250. doi: 10.1016/j.ymben.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Kumar MB, Fujimoto T, Potter DW, Deng Q, Palli SR. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14710–14715. doi: 10.1073/pnas.222278999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner M, Bleris L, Lohmueller J, Xie Z, Benenson Y. Nature nanotechnology. 2010;813:169–186. doi: 10.1007/978-1-61779-412-4_10. [DOI] [PubMed] [Google Scholar]
- Li Y, Ehrhardt K, Zhang MQ, Bleris L. Scientific reports. 2014;4 doi: 10.1038/srep04857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Moore R, Guinn M, Bleris L. Scientific reports. 2012;2 doi: 10.1038/srep00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libermann TA, Zerbini LF. Current gene therapy. 2006;6:17–33. doi: 10.2174/156652306775515501. [DOI] [PubMed] [Google Scholar]
- Lienert F, Torella JP, Chen JH, Norsworthy M, Richardson RR, Silver PA. Nucleic Acids Research. 2013;41:9967–9975. doi: 10.1093/nar/gkt758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu NZ, Wardell SE, Burnstein KL, Defranco D, Fuller PJ, Giguere V, Hochberg RB, McKay L, Renoir JM, Weigel NL. Pharmacological Reviews. 2006;58:782. doi: 10.1124/pr.58.4.9. [DOI] [PubMed] [Google Scholar]
- Luk C, Giovannoni G, Williams DR, Lees AJ, de Silva R. J. Neurosci. Methods. 2009;180:34–42. doi: 10.1016/j.jneumeth.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Moore R, Chandrahas A, Bleris L. ACS Synthetic Biology. 2014;3:707–716. doi: 10.1021/sb400137b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace TW, Hu F, Miller AH. Brain, Behavior, and Immunity. 2007;21:9–19. doi: 10.1016/j.bbi.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Young CY, Yuan H. Acta biochimica et biophysica Sinica. 2010;42:363–369. doi: 10.1093/abbs/gmq038. [DOI] [PubMed] [Google Scholar]
- Pardee K, Green AA, Ferrante T, Cameron DE, DaleyKeyser A, Yin P, Collins JJ. Cell. 2014;159:940–954. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke AW, Grant RA, Keating AE. Journal of the American Chemical Society. 2010;132:6025–6031. doi: 10.1021/ja907617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhen T, Cidlowski JA. New England Journal of Medicine. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, Kudrolli TA, Yegnasubramanian S, Luo J, Rodriguez R, Mendell JT, Lupold SE. Cancer Research. 2009;69:7165–7169. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. Nature Biotechnology. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Elowitz MB, Alon U. Journal of Molecular Biology. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- Shimoga V, White JT, Li Y, Sontag E, Bleris L. Molecular systems biology. 2013;9 doi: 10.1038/msb.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Oncology. 2008;72:397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- Sousa AR, Lane SJ, Cidlowski JA, Staynov DZ, Lee TH. Journal of Allergy and Clinical Immunology. 2000;105:943–950. doi: 10.1067/mai.2000.106486. [DOI] [PubMed] [Google Scholar]
- Talotta F, Cimmino A, Matarazzo M, Casalino L, De Vita G, D'Esposito M, Di Lauro R, Verde P. Oncogene. 2008;28:73–84. doi: 10.1038/onc.2008.370. [DOI] [PubMed] [Google Scholar]
- Tang F, Hajkova P, O'Carroll D, Lee C, Tarakhovsky A, Lao K, Surani MA. Biochemical and Biophysical Research Communications. 2008;372:24–29. doi: 10.1016/j.bbrc.2008.04.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong DJ, Kuhner K, Kuhn R, Werfel S, Engelhardt S, Wurst W, Ortiz O. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M, O'Malley BW. Annual Review of Biochemistry. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- Webster JC, Oakley RH, Jewell CM, Cidlowski JA. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6865–6870. doi: 10.1073/pnas.121455098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Hu W, Peng B, Liu Y. The Journal of Physical Chemistry C. 2014;118:14586–14594. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.