Abstract
Complement pathway activation was found to occur frequently in schizophrenia, and complement 3 (C3) plays a major role in this process. Previous studies have provided evidence for the possible role of C3 in the development of schizophrenia. In this study, we hypothesized that the gene encoding C3 (C3) may confer susceptibility to schizophrenia in Han Chinese. We analyzed 7 common single nucleotide polymorphisms (SNPs) of C3 in 647 schizophrenia patients and 687 healthy controls. Peripheral C3 mRNA expression level was measured in 23 drug-naïve patients with schizophrenia and 24 controls. Two SNPs (rs1047286 and rs2250656) that deviated from Hardy-Weinberg equilibrium were excluded for further analysis. Among the remaining 5 SNPs, there was no significant difference in allele and genotype frequencies between the patient and control groups. Logistic regression analysis showed no significant SNP-gender interaction in either dominant model or recessive model. There was no significant difference in the level of peripheral C3 expression between the drug-naïve schizophrenia patients and healthy controls. In conclusion, the results of this study do not support C3 as a major genetic susceptibility factor in schizophrenia. Other factors in AP may have critical roles in schizophrenia and be worthy of further investigation.
Introduction
Schizophrenia is a chronic, severe and disabling brain disorder that affects approximately 1% of worldwide population. In the past decades, schizophrenia has been regarded as a neurodevelopment disorder. Early literature reported that adverse conditions may result in abnormal brain development during the perinatal period, whilst schizophrenic symptoms appear in later life after the synaptic pruning process [1,2]. However, the pathophysiology of schizophrenia remains unknown [3].
Although heritability estimates for schizophrenia reach 80%, twin concordance is around 50% [4]. Hence, non-genetic factors also play an important role in this disorder [5]. It has been well-documented that maternal virus infection is one of the most consistently identified environmental risk factors for schizophrenia [6]. On the other hand, clinical observations indicated that schizophrenia and certain autoimmune diseases share some key clinical, epidemiological and genetic features [7]. Such findings suggested that immune abnormalities may be implicated with the pathophysiology of schizophrenia [8].
Complement acts as a rapid and efficient immune surveillance system that serves to protect the body against the invasion and proliferation of various microorganisms [9,10]. Complement pathway activation was reported to occur frequently in schizophrenia, in which complement 3 (C3) regulates the process [11]. C3 is a protein of the immune system that plays a central role in the complement cascade and contributes to innate immunity. Recent observations have demonstrated that C3 is a critical mediator for synaptic refinement and plasticity in neurodevelopment [12,13]. In comparison with healthy controls, Hakobyan et al. [14] observed a significant higher level of C3 protein in schizophrenia patients. As such, the above findings provide interesting clues for the potential role of C3 in schizophrenia.
At the molecular level, the gene encoding C3 (C3) is located at chromosome 19, which has been reported to be a genetic schizophrenia susceptibility region [15]. However, few genetic studies have been carried out to investigate the association of C3 with schizophrenia and yielded inconsistent results [16,17,18]. It is known that C3 contains 41 exons and spreads over 41kb. One weakness for the early genetic studies is too few polymorphisms tested. In the present study, we aimed to examine whether the region of C3 is associated with schizophrenia. A total of 7 polymorphisms were selected for a better coverage of this region. As a secondary aim, prior study reported that C3 has a gender-specific effect [19], which may underlie differential susceptibility to schizophrenia [20]. So we attempted to examine whether there was any gender difference in the association of C3 with schizophrenia. Data has shown that C3 polymorphisms result in alternations in its protein function. To validate previous findings, we opted to measure the serum C3 expression level among drug-naïve schizophrenia patients and healthy controls.
Methods
Subjects
All subjects provided written informed consent prior to performing any of the procedures related to this study. All procedures were reviewed and approved by the ethical committees at Tongde Hospital of Zhejiang Province and Hangzhou Seventh People’s Hospital, and performed in strict accordance with the Declaration of Helsinki, and other relevant national and international regulations.
For the genetic analysis, a total of 647 schizophrenia patients recruited from Tongde Hospital of Zhejiang Province and The Seventh People’s Hospital of Hangzhou. The inclusion criteria for this study were according to our previous ones [20,21,22]. All patients (1) met the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) criteria for schizophrenia; (2) were not first-episode; (3) had no chronic physical disease or other psychiatric disorder aside from schizophrenia. Prior to analysis, all diagnosis and review of psychiatric case records were independently checked and verified by two senior psychiatrists. The control group comprised of 687 Han Chinese enrolled from the local community in Hangzhou. Before sampling, the volunteers self-reported that they were in good physical health and have no family history of psychiatric disorders. Those who have medical illnesses or drug and alcohol abuse/dependence were excluded. Demographic and clinical characteristics were presented in S1 Table.
For the expression analysis, twenty-three drug-naïve patients with first-episode schizophrenia were recruited from Tongde Hospital of Zhejiang Province. The patients were diagnosed according to the DSM-IV criteria for schizophrenia and had no physical disease. Twenty-four healthy subjects from Hangzhou city were also recruited for control group. Basic blood and urine tests were performed prior to recruitment in order to exclude any current physical illness. Patients and controls did not significantly differ for age, gender, BMI and smoking status. Detailed information was presented in S2 Table.
SNP selection
We retrieved CHB data from the HapMap database (http://www.hapmap.org) and defined linkage disequilibrium (LD) blocks using Haploview 4.2 (Broad Institute, Cambridge, MA, USA) to set inclusion criteria for tagging SNPs. Haplotype-tagging single nucleotide polymorphisms (htSNPs) with R 2 cutoff>0.8 and minor allele frequency (MAF)>0.1 were selected. In total, three tag SNPs of C3 were selected for genotyping. Four potential C3 functional SNPs (rs7951, rs2230199, rs2250656 and rs11672613) [23,24,25,26] were also examined in this study (S3 Table).
Genotyping
Genomic DNA of all participants was extracted from peripheral blood using a Tiangen DNA Isolation Kit (Tiangen Biotech, Beijing, China). All 7 SNPs were amplified independently via polymerase chain reaction (PCR) and then genotyped via direct sequencing on an ABI PRISM 3730 Genetic Analyzer (Perkin-Elmer Applied Biosystems). S4 Table detailed the primers information. Genotyping was carried out according to the methods described in our previous studies [27,28]. PCR amplification was performed in a volume of 25 μL containing primer pair for each SNP. PCR primers were also used for sequencing. Sequencing results were handled using DNAStar package (DNA Star Inc., USA), and the original sequencing chromatograms of each sample were then manually checked.
Quantitative real-time polymerase chain reaction (qRT-PCR)
We carried out the C3 mRNA expression analysis using qRT-PCR as previously described [29,30]. Peripheral blood was collected and mononuclear cells were separated by Ficoll-Paque PLUS density gradient centrifugation (GE Healthcare, Amersham, NJ, USA) within 2 hour, placed in TRIzol (Invitrogen, Carlsbad, CA, USA) and stored at -80°C. The total RNA was isolated from peripheral blood mononuclear cells according to the manufacturer’s protocol, and 2 μg total RNA was re-transcribed into complementary DNA with reverse transcription (ReverTra Ace, Toyobo, Osaka, Japan) according to the manufacturer’s instruction. Relative C3 mRNA expression levels were assessed by real-time PCR with commercially available TaqMan gene expression assays for target gene C3 and glyceraldehydes-3-phosphate dehydrogenase (GAPDH) as reference gene (Applied Biosystems, CA, USA). All experiments were conducted in optical 384-well reaction microtiter plates on an ABI Prism 7900HT Sequence Detection System (Applied Biosystems, CA, USA). PCR was performed in a total volume of 10μL containing 1×TaqMan Universal Master Mix with AmpErase UNG, 1×Assay Mix (Applied Biosystems, CA, USA) and complementary DNA template at cycle conditions: 95°C for 15 min, followed by 40 cycles at 95°C for 15s and 60°C for 60s. All reactions were run in triplicate. In each sample, the expression of C3 was normalized to the expression of the reference gene. Results were reported in fold change using 2-ΔΔCt.
Statistical analysis
The Hardy-Weinberg equilibrium testing and individual SNP association analyses were conducted using SHEsis (http://analysis.bio-x.cn). The odds ratio (OR) and corresponding 95% confidence interval (CI) were calculated with the major allele as reference. Pairwise linkage disequilibrium of all pairs of htSNPs was performed using HaploView 4.2 (Broad Institute, Cambridge, MA, USA), and the extent of linkage disequilibrium (LD) was measured by the standardized D’ and R 2. Referring to the previous report [19], logistic regression was performed with SNP-gender interaction to adjust the effect of gender on SNPs. For the expression analysis, ANCOVA was carried out with age, gender, smoking status and BMI as covariates controlled in the model, to minimize the potential effect of these factors on the expression levels of C3 mRNA. The ANCOVA analysis was performed using SPSS 17.0 (SPSS Inc., Chicago, IL, USA). To adjust for multiple testing, the level of significance was corrected via Bonferroni correction. Power calculations were carried out using Quanto 1.2.3 (http://hydra.usc.edu/GxE).
Results
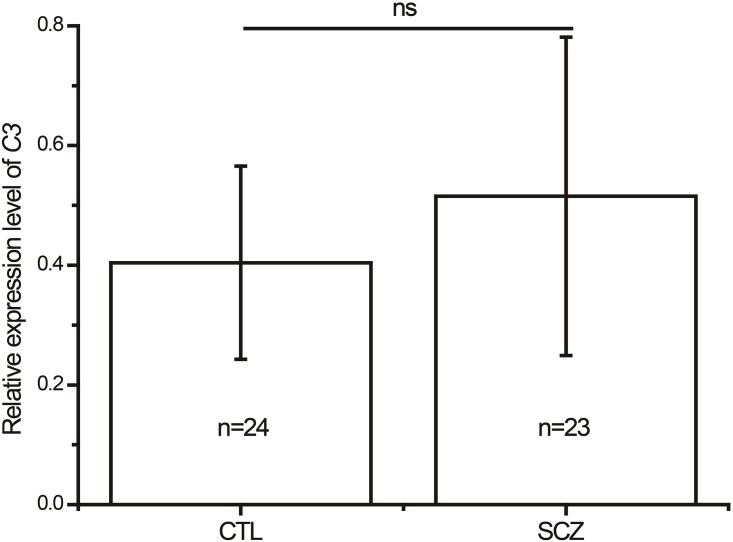
For the genetic analysis, there was no significant difference between the schizophrenia and control groups in term of age and gender. Seven SNPs were genotyped to investigate the association of C3 with schizophrenia. Two SNPs (rs1047286 and rs2250656) that deviated from Hardy-Weinberg equilibrium were excluded for the further analysis. Among the remaining 5 SNPs, no deviation from the Hardy-Weinberg was observed in genotype distribution. Table 1 showed that there was no significant difference in allele and genotype frequencies between the patient and control groups. After calculating LD for all pairs of SNPs, we found a low R 2 in C3 (S1 Fig), indicating that no specific haplotype block could be identified. We further investigate whether there was any gender difference in the association of C3 with schizophrenia. Logistic regression analysis showed no significant SNP-gender interaction in either dominant model or recessive model (Tables 2 and 3). A total of 23 drug-naïve patients with schizophrenia and 24 well-matched healthy controls were recruited for the C3 expression study. As shown in Fig 1, there was no significant difference in the level of peripheral C3 mRNA expression between the drug-naïve schizophrenia patients and healthy controls. On the basis of the genotype data, the statistical power of the 5 SNPs within C3 was more than 85% (α = 0.05) for schizophrenia samples under the assumption of a moderate effect size (OR = 1.5), a log additive model, and the prevalence of schizophrenia (≈1%).
Table 1. Comparison of genotype and allele frequencies of 5 C3 SNPs in 647 schizophrenia patients and 687 healthy controls.
| Genotype, n (%) | Allele, n (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2277984 | N | G/G | G/A | A/A | P a | P b | N | G | A | OR (95%CI) | P a | P c |
| Case | 647 | 132 (20.4) | 336 (51.9) | 179 (27.7) | 0.95 | 0.26 | 1294 | 600 (46.4) | 694 (53.6) | 1.02 (0.88–1.19) | 0.76 | |
| Control | 687 | 137 (19.9) | 355 (51.7) | 195 (28.4) | 0.28 | 1374 | 629 (45.8) | 745 (54.2) | ||||
| rs7951 | T/T | T/C | C/C | T | C | |||||||
| Case | 647 | 7 (1.1) | 122 (18.9) | 518 (80.1) | 0.42 | 0.95 | 1294 | 136 (10.5) | 1158 (89.5) | 0.91 (0.71–1.16) | 0.45 | |
| Control | 687 | 5 (0.7) | 147 (21.4) | 535 (77.9) | 0.13 | 1374 | 157 (11.4) | 1217 (88.6) | ||||
| rs11672613 | C/C | C/T | T/T | C | T | |||||||
| Case | 647 | 98 (15.1) | 307 (47.4) | 242 (37.4) | 0.22 | 0.97 | 1294 | 503 (38.9) | 791 (61.1) | 0.90 (0.77–1.05) | 0.17 | |
| Control | 687 | 109 (15.9) | 352 (51.2) | 226 (32.9) | 0.15 | 1374 | 570 (41.5) | 804 (58.5) | ||||
| rs2230205 | A/A | A/G | G/G | A | G | |||||||
| Case | 647 | 141 (21.8) | 346 (53.5) | 160 (24.7) | 0.09 | 0.07 | 1294 | 628 (48.5) | 666 (51.5) | 1.18 (1.01–1.37) | 0.035 | 0.175 |
| Control | 687 | 127 (18.5) | 357 (52.0) | 203 (29.5) | 0.17 | 1374 | 611 (44.5) | 763 (55.5) | ||||
| rs2230199 | G/G | G/C | C/C | G | C | |||||||
| Case | 647 | 0 (0.0) | 10 (1.5) | 637 (98.5) | 0.50 | 0.84 | 1294 | 10 (0.8) | 1284 (99.2) | 0.76 (0.33–1.71) | 0.50 | |
| Control | 687 | 0 (0.0) | 14 (2.0) | 673 (98.0) | 0.79 | 1374 | 14 (1.0) | 1360 (99.0) | ||||
a P values were not adjusted by Bonferroni correction
b P values were calculated for Hardy-Weinberg equilibrium
c P values were adjusted by Bonferroni correction.
Table 2. Logistic regression analysis of C3 SNPs×gender interaction in dominant model.
| Variables | B | S.E | Wals | OR (95%CI) | P a | P b |
|---|---|---|---|---|---|---|
| rs2277984×gender | 0.54 | 0.22 | 5.87 | 1.71 (1.11–2.65) | 0.015 | 0.075 |
| rs7951×gender | -0.40 | 0.24 | 2.78 | 0.67 (0.42–1.07) | 0.10 | |
| rs11672613×gender | -0.23 | 0.21 | 1.26 | 0.79 (0.53–1.19) | 0.26 | |
| rs2230205×gender | 0.21 | 0.21 | 0.95 | 1.23 (0.81–1.87) | 0.33 | |
| rs2230199×gender | -0.21 | 0.27 | 0.62 | 0.81 (0.47–1.38) | 0.43 |
a P values were not adjusted by Bonferroni correction
b P values were adjusted by Bonferroni correction.
Table 3. Logistic regression analysis of C3 SNPs×gender interaction in recessive model.
| Variables | B | S.E | Wals | OR (95%CI) | P a |
|---|---|---|---|---|---|
| rs2277984×gender | 0.04 | 0.24 | 0.02 | 1.04 (0.65–1.64) | 0.88 |
| rs7951×gender | 0.35 | 1.25 | 0.08 | 1.42 (0.12–16.26) | 0.78 |
| rs11672613×gender | -0.17 | 0.26 | 0.43 | 0.84 (0.60–1.41) | 0.79 |
| rs2230205×gender | -0.14 | 0.23 | 0.37 | 0.87 (0.55–1.37) | 0.55 |
| rs2230199×gender | NA | NA | NA | NA | NA |
a P values were not adjusted by Bonferroni correction
NA, Not applicable.
Fig 1. Expression levels of C3 mRNA in peripheral blood in drug-naïve schizophrenia patients and healthy subjects.
C3 mRNA was normalized to that of GAPDH. CTL, control subjects (n = 24); SCZ, schizophrenia patients (n = 23); ns, no significance.
Discussion
As a key component of innate immunity, accumulating evidence has indicated that abnormalities in the complement system are implicated in the etiology of schizophrenia [23]. We have examined the association of schizophrenia with the gene encoding C4-binding protein (C4BPB/C4BPA), a potent circulating soluble inhibitor of the classical and lectin pathways of complement. However, our results did not support the involvement of C4BPB/C4BPA in schizophrenia [22]. Here, we aimed to investigate the association of C3, another critical factor in complement system, with schizophrenia in Chinese Han population.
In this study, we did not observe any significant difference of allele and genotype frequencies between the schizophrenia patients and healthy controls. The statistical power of our study was also enough to detect an association between the variants and schizophrenia. Although Liu et al. [19] reported a SNP-gender interaction in C3, we did not have such findings in our sample. These results demonstrated that there is no genetic association between C3 and schizophrenia, at least in Han Chinese. However, a recent study showed that increased levels of C3, acting as activation of complement system, can be found in schizophrenia patients when compared with healthy controls [31]. In contrast, Wong et al. [32] found a lower level of C3 in schizophrenia patients than that in controls. We noticed that the patients in both studies were those with chronic schizophrenia [31,32]. The aforementioned inconsistent results prompted us to determine the expression of C3 in drug-naïve patients with first-episode schizophrenia. Our results showed no significant difference in the level of C3 expression between schizophrenia patients and healthy controls. Therefore, our findings suggested that C3 may not confer susceptibility to schizophrenia in Han Chinese.
Recently, Li et al. [33] performed a label-free quantitative proteomics analysis to identify 27 proteins as being schizophrenia related proteins, and found dysregulation of the alternative complement pathway in schizophrenia patients. The alternative pathway (AP) is one of three complement pathways, which is initiated by the spontaneous hydrolysis of C3. A number of molecules are involved in the occurrence of AP. Even though no association of C3 with schizophrenia was found in this study, we could not exclude possible role of AP in the development of schizophrenia.
On the other side, cytokines are believed to play a vital role in coordinating immunologic and inflammatory responses in physiological and pathological conditions [34]. Therefore, cytokines may be critical mediators of the cross-talk between immune system and neuropsychiatric disorders [35]. Miller et al. [36] meta-analyzed 40 studies on cytokines and schizophrenia, and observed significant alternations of cytokine network in schizophrenia. Therefore, imbalance of cytokine network may be involved in the pathophysiology of schizophrenia. Prior literature indicated that complement activation products, such as C3a and C3a desArg, may enhance cytokine synthesis and inhibit the systemic synthesis of proinflammatory cytokines [37]. It is known that schizophrenia results from the cumulative impact of multiple common small-effect genetic variants and interactions between genes with small effect may contribute a larger heritable proportion to the overall risk of this disorder [38]. Therefore, we assumed that interaction of C3 with genes encoding cytokines may be more sensitive to account for its susceptibility to schizophrenia. There is a need for further investigations to validate this hypothesis.
This study has some limitations that should be noted. First, the lack of a significant association may be caused by the modest sample size, possibly resulting in a type II error. Second, we did not psychiatrically screen the control subjects. Third, the principal hypothesis underlying this study is that common SNPs within C3 may confer susceptibility to schizophrenia. Therefore, we did not sequence the C3 to assess the influence of rare variants on schizophrenia, and this prevented us to detect their active role in the development of this disorder. Fourth, the case-control association analyses have the potential for population stratification, although all participants were ethnically matched in our sample. Finally, Wong et al. [32] reported lower level of C3 protein in schizophrenia patients in comparison to healthy controls only in male subjects, suggesting that there might be interesting to test C3 expression separately in male and female subjects. However, we recruited only 47 individuals with or without schizophrenia in the C3 expression study. The small sample size limited us to further analyze the gender-specific effect of C3 expression on schizophrenia. Meanwhile, this also limited us to detect the association of studied C3 SNPs with C3 mRNA.
In conclusion, the results of this study do not support C3 as a major genetic susceptibility factor in schizophrenia. Other factors in AP may have critical roles in schizophrenia and be worthy of further investigation.
Supporting Information
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Acknowledgments
We are deeply grateful to all participants. We thank the anonymous reviewers for their insightful comments.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Natural Science Foundation of China (81000581 and 81471358), the Shanghai Science and Technology Commission (14411969000), the Shanghai Mental Health Center Foundation (2014-FX-03), and the Medical Health Technique Project of Zhejiang Province (2010KYA047). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Weinberger DR (1996) On the plausibility of "the neurodevelopmental hypothesis" of schizophrenia. Neuropsychopharmacology 14: 1S–11S. [DOI] [PubMed] [Google Scholar]
- 2. Schmitt A, Malchow B, Hasan A, Falkai P (2014) The impact of environmental factors in severe psychiatric disorders. Front Neurosci 8: 19 10.3389/fnins.2014.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang C, Fang Y, Xie B, Cheng W, Du Y, Wang D, et al. (2009) DNA methyltransferase 3B gene increases risk of early onset schizophrenia. Neurosci Lett 462: 308–311. 10.1016/j.neulet.2009.06.085 [DOI] [PubMed] [Google Scholar]
- 4. Cannon TD, Kaprio J, Lonnqvist J, Huttunen M, Koskenvuo M (1998) The genetic epidemiology of schizophrenia in a Finnish twin cohort. A population-based modeling study. Arch Gen Psychiatry 55: 67–74. [DOI] [PubMed] [Google Scholar]
- 5. Avramopoulos D, Pearce BD, McGrath J, Wolyniec P, Wang R, Eckart N, et al. (2015) Infection and inflammation in schizophrenia and bipolar disorder: a genome wide study for interactions with genetic variation. PLoS One 10: e0116696 10.1371/journal.pone.0116696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmitt A, Malchow B, Hasan A, Falkai P (2014) The impact of environmental factors in severe psychiatric disorders. Frontiers in Neuroscience 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benros ME, Nielsen PR, Nordentoft M, Eaton WW, Dalton SO, Mortensen PB. (2011) Autoimmune Diseases and Severe Infections as Risk Factors for Schizophrenia: A 30-Year Population-Based Register Study. Am J Psychiatry 168: 1303–1310. 10.1176/appi.ajp.2011.11030516 [DOI] [PubMed] [Google Scholar]
- 8. Leza JC, Bueno B, Bioque M, Arango C, Parellada M, Do K, et al. (2015) Inflammation in schizophrenia: A question of balance. Neurosci Biobehav Rev 55: 612–626. 10.1016/j.neubiorev.2015.05.014 [DOI] [PubMed] [Google Scholar]
- 9. Ricklin D, Hajishengallis G, Yang K, Lambris JD (2010) Complement: a key system for immune surveillance and homeostasis. Nat Immunol 11: 785–797. 10.1038/ni.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tichaczek-Goska D (2012) Deficiencies and excessive human complement system activation in disorders of multifarious etiology. Adv Clin Exp Med 21: 105–114. [PubMed] [Google Scholar]
- 11. Severance EG, Gressitt KL, Halling M, Stallings CR, Origoni AE, Vaughan C, et al. (2012) Complement C1q formation of immune complexes with milk caseins and wheat glutens in schizophrenia. Neurobiol Dis 48: 447–453. 10.1016/j.nbd.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fourgeaud L, Boulanger LM (2007) Synapse remodeling, compliments of the complement system. Cell 131: 1034–1036. [DOI] [PubMed] [Google Scholar]
- 13. Michailidou I, Willems JG, Kooi EJ, van Eden C, Gold SM, Geurts JJ, et al. (2015) Complement C1q-C3 associated synaptic changes in multiple sclerosis hippocampus. Ann Neurol 77: 1007–1026. 10.1002/ana.24398 [DOI] [PubMed] [Google Scholar]
- 14. Hakobyan S, Boyajyan A, Sim RB (2005) Classical pathway complement activity in schizophrenia. Neurosci Lett 374: 35–37. [DOI] [PubMed] [Google Scholar]
- 15. Francks C, Tozzi F, Farmer A, Vincent JB, Rujescu D, St Clair D, et al. (2010) Population-based linkage analysis of schizophrenia and bipolar case-control cohorts identifies a potential susceptibility locus on 19q13. Mol Psychiatry 15: 319–325. 10.1038/mp.2008.100 [DOI] [PubMed] [Google Scholar]
- 16. Rudduck C, Beckman L, Franzen G, Lindstrom L (1985) C3 and C6 complement types in schizophrenia. Hum Hered 35: 255–258. [DOI] [PubMed] [Google Scholar]
- 17. Fananas L, Moral P, Panadero MA, Bertranpetit J (1992) Complement genetic markers in schizophrenia: C3, BF and C6 polymorphisms. Hum Hered 42: 162–167. [DOI] [PubMed] [Google Scholar]
- 18. Blackwood DHR, Muir WJ, Stephenson A, Wentzel J, Adhiah A, Walker MJ, et al. (1996) Reduced expression of HLA-B35 in schizophrenia. Psychiatr Genet 6: 51–59. [DOI] [PubMed] [Google Scholar]
- 19. Liu K, Lai TYY, Chiang SWY, Chan VCK, Young AL, Tam PO, et al. (2014) Gender specific association of a complement component 3 polymorphism with polypoidal choroidal vasculopathy. Sci Rep 4: 7018 10.1038/srep07018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tang WX, Cai J, Yi ZH, Zhang Y, Lu WH, Zhang C. (2014) Association study of common variants within the G protein-coupled receptor kinase 6 gene and schizophrenia susceptibility in Han Chinese. Hum Psychopharmacol 29: 100–103. 10.1002/hup.2375 [DOI] [PubMed] [Google Scholar]
- 21. Zhu YL, Wang ZL, Ni JL, Zhang Y, Chen MJ, Cai J, et al. (2015) Genetic variant in NDUFS1 gene is associated with schizophrenia and negative symptoms in Han Chinese. J Hum Genet 60: 11–16. 10.1038/jhg.2014.94 [DOI] [PubMed] [Google Scholar]
- 22. Wang SH, Lu HQ, Ni JL, Zhang JT, Tang WX, Lu WH, et al. (2015) An evaluation of association between common variants in C4BPB/C4BPA genes and schizophrenia. Neurosci Lett 590: 189–192. 10.1016/j.neulet.2015.02.005 [DOI] [PubMed] [Google Scholar]
- 23. Miyagawa H, Yamai M, Sakaguchi D, Kiyohara C, Tsukamoto H, Kimoto Y, et al. (2008) Association of polymorphisms in complement component C3 gene with susceptibility to systemic lupus erythematosus. Rheumatology 47: 158–164. 10.1093/rheumatology/kem321 [DOI] [PubMed] [Google Scholar]
- 24. Yu QQ, Yao Y, Zhu J, Bao X, Xie TH, Sun C, et al. (2015) Nonsynonymous single nucleotide polymorphisms in the complement component 3 gene are associated with risk of age-related macular degeneration: A meta-analysis. Gene 561: 249–255. 10.1016/j.gene.2015.02.039 [DOI] [PubMed] [Google Scholar]
- 25. Phillips CM, Goumidi L, Bertrais S, Ferguson JF, Field MR, Kelly ED, et al. (2009) Complement component 3 polymorphisms interact with polyunsaturated fatty acids to modulate risk of metabolic syndrome. Am J Clin Nutr 90: 1665–1673. 10.3945/ajcn.2009.28101 [DOI] [PubMed] [Google Scholar]
- 26. Rhodes B, Hunnangkul S, Morris DL, Hsaio LC, Graham DSC, Nitsch D, et al. (2009) The heritability and genetics of complement C3 expression in UK SLE families. Genes Immun 10: 525–530. 10.1038/gene.2009.23 [DOI] [PubMed] [Google Scholar]
- 27. Zhang Y, Chen MJ, Wu ZG, Chen J, Yu SY, Fang Y, et al. (2013) Association Study of Val66Met Polymorphism in Brain-Derived Neurotrophic Factor Gene with Clozapine-Induced Metabolic Syndrome: Preliminary Results. PLoS One 8: e72652 10.1371/journal.pone.0072652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y, Chen M, Chen J, Wu Z, Yu S, Fang Y, et al. (2014) Metabolic syndrome in patients taking clozapine: prevalence and influence of catechol-O-methyltransferase genotype. Psychopharmacology (Berl) 231: 2211–2218. [DOI] [PubMed] [Google Scholar]
- 29. Zhang C, Wang Z, Hong W, Wu Z, Peng D, Fang Y. (2015) ZNF804A Genetic Variation Confers Risk to Bipolar Disorder. Mol Neurobiol. 10.1007/s12035-015-9193-3 [DOI] [PubMed] [Google Scholar]
- 30. Zhang C, Wu Z, Hong W, Wang Z, Peng D, Chen J, et al. (2014) Influence of BCL2 gene in major depression susceptibility and antidepressant treatment outcome. J Affect Disord 155: 288–294. 10.1016/j.jad.2013.11.010 [DOI] [PubMed] [Google Scholar]
- 31. Soria LD, Gubert CD, Cereser KM, Gama CS, Kapczinski F (2012) Increased serum levels of C3 and C4 in patients with schizophrenia compared to eutymic patients with bipolar disorder and healthy. Revista Brasileira De Psiquiatria 34: 119–120. [PubMed] [Google Scholar]
- 32. Wong CT, Tsoi WF, Saha N (1996) Acute phase proteins in male Chinese schizophrenic patients in Singapore. Schizophrenia Research 22: 165–171. [DOI] [PubMed] [Google Scholar]
- 33. Li Y, Zhou KJ, Zhang Z, Sun LY, Yang JL, Zhang M, et al. (2012) Label-free quantitative proteomic analysis reveals dysfunction of complement pathway in peripheral blood of schizophrenia patients: evidence for the immune hypothesis of schizophrenia. Mol Biosyst 8: 2664–2671. 10.1039/c2mb25158b [DOI] [PubMed] [Google Scholar]
- 34. Deleidi M, Jaggle M, Rubino G (2015) Immune aging, dysmetabolism, and inflammation in neurological diseases. Front Neurosci 9: 172 10.3389/fnins.2015.00172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Potvin S, Stip E, Sepehry AA, Gendron A, Bah R, Kouassi E. (2008) Inflammatory cytokine alterations in schizophrenia: a systematic quantitative review. Biol Psychiatry 63: 801–808. [DOI] [PubMed] [Google Scholar]
- 36. Miller BJ, Buckley P, Seabolt W, Mellor A, Kirkpatrick B (2011) Meta-analysis of cytokine alterations in schizophrenia: clinical status and antipsychotic effects. Biol Psychiatry 70: 663–671. 10.1016/j.biopsych.2011.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Takabayashi T, Vannier E, Clark BD, Margolis NH, Dinarello CA, Burke JF, et al. (1996) A new biologic role for C3a and C3a desArg: regulation of TNF-alpha and IL-1 beta synthesis. J Immunol 156: 3455–3460. [PubMed] [Google Scholar]
- 38. McClellan JM, Susser E, King MC (2007) Schizophrenia: a common disease caused by multiple rare alleles. Br J Psychiatry 190: 194–199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.