Abstract
Cyanobacterial KnowledgeBase (CKB) is a free access database that contains the genomic and proteomic information of 74 fully sequenced cyanobacterial genomes belonging to seven orders. The database also contains tools for sequence analysis. The Species report and the gene report provide details about each species and gene (including sequence features and gene ontology annotations) respectively. The database also includes cyanoBLAST, an advanced tool that facilitates comparative analysis, among cyanobacterial genomes and genomes of E. coli (prokaryote) and Arabidopsis (eukaryote). The database is developed and maintained by the Sub-Distributed Informatics Centre (sponsored by the Department of Biotechnology, Govt. of India) of the National Facility for Marine Cyanobacteria, a facility dedicated to marine cyanobacterial research. CKB is freely available at http://nfmc.res.in/ckb/index.html.
Introduction
Cyanobacteria comprise over 1,600 species with various morphologies and species-specific characteristics, such as cell movement, cell differentiation, and nitrogen fixation [1]. These are the only known oxygenic photosynthetic prokaryotic organisms that inhabit a wide range of ecological habitats (e.g., extreme cold, extreme hot, marine, fresh water, and terrestrial) and exhibit symbiotic associations with other living organisms. These primitive oxygenic Gram negative bacteria are widely used as a valuable model to study the mechanism of carbon fixation and helpful for evolutionary biologists to understand the endosymbiotic theory, as they are considered as the origin of chloroplast. Since these ancient life forms play a major role in many biogeochemical cycles of the global ecological system, they serve as a study material in diverse fields of life-science research [2].
Cyanobacteria are well-known for the formation of toxic cyanobacterial water blooms in freshwater, brackish and coastal marine ecosystems, which are of vital ecological and human health concerns [3]. However, in recent times, these organisms have captured the attention of the researchers worldwide because of their capability of producing prolific bioactive natural products as secondary metabolites, which are of great economic and medical value [4–6].
The National Facility for Marine Cyanobacteria (Sponsored by the Department of Biotechnology, Govt. of India) is dedicated to cyanobacterial research, especially marine cyanobacteria. One of the principal foci of the facility is to build a dedicated knowledge base for cyanobacteria. The increasing number of completely sequenced cyanobacterial genomes provides wide opportunities for understanding the metabolic organization of the cyanobacterial species in diverse environments. Here we introduce the Cyanobacterial KnowldegeBase (CKB), a freely accessible, comprehensive database resource covering information pertaining to 74 completely sequenced cyanobacterial species. The database also includes an informative tool called cyanoBLAST, which helps in comparative analysis between cyanobacterial genomes and the genomes of pro- and eu-karyote, such as E. coli and Arabidopsis.
Results and Discussion
Organisms
Seventy-four fully sequenced genomes of seven orders are currently included in the CKB database. This comprises 12 species of Chroococcales, 1 of Chroococcidiopsidales, 2 of Gloeobacteriales, 12 of Nostocales, 7 of Oscillatoriales, 2 of Pleurocapsales and 38 of Synechococcales. The web user interface of CKB is shown in (Fig 1) and the complete list of the species exists in the CKB is given in Table 1.
Fig 1. CKB web interface.
Table 1. Order wise complete list of species mentioned in CKB.
The table provides information related to order, Morphology (Morph.-U: Unicellular, F: Filamentous and F,H: Filamentous Heterocystous), number of chromosomes (Chr.), number of plasmids (Pla.), genome size (Size, MB), GC %, the number of genes (Genes), number of proteins (Proteins), and Biological Resource Centers (BRCs) from which live specimens can be available for each species.
| Order | Organism | Morph. | Chr. | Pla. | Size, MB | GC % | Genes | Proteins | BRCs |
|---|---|---|---|---|---|---|---|---|---|
| Chroococcales | Cyanobacterium aponinum PCC 10605 | U | 1 | 0 | 4.72 | 60.5 | 4562 | 4507 | PCC |
| Cyanobacterium stanieri PCC 7202 | U | 1 | 0 | 4.66 | 62 | 4482 | 4430 | PCC | |
| Cyanothece sp. ATCC 51142 | U | 1 | 6 | 7.06 | 38.8 | 6258 | 5838 | ATCC | |
| Cyanothece sp. PCC 7424 | U | 2 | 3 | 5.31 | 38.1 | 4797 | 4511 | PCC | |
| Cyanothece sp. PCC 7425 | U | 2 | 3 | 7.11 | 41.4 | 5813 | 5710 | ATCC; PCC | |
| Cyanothece sp. PCC 7822 | U | 1 | 3 | 6.96 | 39.8 | 5841 | 5535 | PCC | |
| Cyanothece sp. PCC 8801 | U | 1 | 0 | 7.02 | 42.2 | 6250 | 5950 | PCC | |
| Cyanothece sp. PCC 8802 | U | 1 | 3 | 7.61 | 42.2 | 6738 | 6229 | PCC | |
| Dactylococcopsis salina PCC 8305 | U | 1 | 2 | 5.49 | 38.3 | 5380 | 3651 | PCC | |
| Gloeocapsa sp. PCC 7428 | U | 1 | 5 | 9.06 | 41.3 | 7164 | 6689 | ATCC; PCC | |
| Halothece sp. PCC 7418 | U | 1 | 0 | 6.33 | 40.4 | 5538 | 5237 | PCC | |
| Microcystis aeruginosa NIES-843 | U | 1 | 6 | 7.21 | 41.2 | 6213 | 6129 | NIES | |
| Chroococcidiopsidales | Chroococcidiopsis thermalis PCC 7203 | U | 1 | 2 | 6.72 | 41.5 | 5687 | 5449 | ATCC; PCC |
| Gloeobacteriales | Gloeobacter kilaueensis JS1 | U | 1 | 2 | 8.73 | 37.5 | 6946 | 6644 | NA |
| Gloeobacter violaceus PCC 7421 | U | 1 | 9 | 8.36 | 47 | 8571 | 8383 | ATCC; PCC | |
| Nostocales | Anabaena cylindrica PCC 7122 | F, H | 1 | 0 | 6.79 | 44.3 | 6676 | 6630 | ATCC; PCC |
| Anabaena sp. 90 | F, H | 1 | 2 | 6.76 | 45.6 | 6426 | 5945 | NA | |
| Anabaena variabilis ATCC 29413 | F, H | 1 | 8 | 5.62 | 40.2 | 5059 | 4752 | ATCC | |
| Calothrix sp. PCC 630 | F, H | 1 | 1 | 4.18 | 35 | 3614 | 3431 | ATCC; PCC | |
| Calothrix sp. PCC 7507 | F, H | 1 | 0 | 3.16 | 38.7 | 2941 | 2837 | ATCC; PCC | |
| Cylindrospermum stagnale PCC 7417 | F, H | 1 | 0 | 3.34 | 68.7 | 3437 | 3280 | ATCC; PCC | |
| Nostoc azollae 0708 | F, H | 2 | 4 | 5.46 | 38 | 5364 | 5303 | NA | |
| Nostoc punctiforme PCC 73102 | F, H | 1 | 6 | 6.55 | 38.5 | 5942 | 5710 | ATCC; PCC | |
| Nostoc sp. PCC 7107 | F, H | 1 | 3 | 5.79 | 50.7 | 5507 | 5327 | ATCC; PCC | |
| Nostoc sp. PCC 7120 | F, H | 1 | 6 | 7.84 | 39.9 | 7042 | 6642 | ATCC; PCC | |
| Nostoc sp. PCC 7524 | F, H | 1 | 3 | 4.79 | 39.8 | 4619 | 4367 | ATCC; PCC | |
| Rivularia sp. PCC 7116 | F, H | 1 | 4 | 4.8 | 39.8 | 4700 | 4444 | PCC | |
| Oscillatoriales | Arthrospira platensis NIES-39 | F | 1 | 0 | 3.78 | 42.4 | 3684 | 3337 | NIES |
| Crinalium epipsammum PCC 9333 | F | 1 | 0 | 4.68 | 58.5 | 3912 | 3815 | PCC | |
| Geitlerinema sp. PCC 7407 | F | 1 | 4 | 5.88 | 43.4 | 5304 | 5011 | ATCC; PCC | |
| Microcoleus sp. PCC 7113 | F | 1 | 0 | 4.18 | 42.9 | 3920 | 3708 | PCC | |
| Oscillatoria acuminata PCC 6304 | F | 1 | 0 | 5.13 | 43.9 | 4654 | 4228 | ATCC; PCC | |
| Oscillatoria nigro-viridis PCC 7112 | F | 1 | 8 | 7.97 | 46.2 | 6821 | 6441 | PCC | |
| Trichodesmium erythraeum IMS101 | F | 1 | 0 | 5.84 | 42.3 | 6364 | 6312 | NCMA | |
| Pleurocapsales | Pleurocapsa sp. PCC 7327 | U | 1 | 2 | 7.8 | 47.6 | 6100 | 5796 | ATCC; PCC |
| Stanieria cyanosphaera PCC 7437 | U | 1 | 5 | 8.27 | 45.8 | 7006 | 6360 | ATCC; PCC | |
| Synechococcales | Acaryochloris marina MBIC11017 | U | 1 | 1 | 4.89 | 46.2 | 4014 | 3854 | NA |
| Chamaesiphon minutus PCC 6605 | U | 1 | 0 | 2.7 | 55.5 | 2581 | 2522 | PCC | |
| Cyanobium gracile PCC 6307 | U | 1 | 1 | 2.74 | 55.5 | 2715 | 2662 | PCC | |
| Synechococcus elongatus PCC 6301 | U | 1 | 0 | 2.61 | 52.4 | 2944 | 2892 | PCC | |
| Synechococcus elongatus PCC 7942 | U | 1 | 0 | 2.51 | 59.2 | 2756 | 2645 | ATCC; PCC | |
| Synechococcus sp. CC9311 | U | 1 | 0 | 2.23 | 54.2 | 2357 | 2306 | NCMA | |
| Synechococcus sp. CC9605 | U | 1 | 0 | 3.05 | 58.5 | 2942 | 2862 | NCMA | |
| Synechococcus sp. CC9902 | U | 1 | 0 | 2.93 | 60.2 | 2897 | 2760 | NCMA | |
| Synechococcus sp. JA-2-3B'a(2–13) | U | 1 | 1 | 3.72 | 48.5 | 3794 | 3545 | NA | |
| Synechococcus sp. JA-3-3Ab | U | 1 | 6 | 3.41 | 49.2 | 3238 | 3187 | NA | |
| Synechococcus sp. PCC 6312 | U | 1 | 2 | 3.58 | 40.6 | 3666 | 3318 | ATCC; PCC | |
| Synechococcus sp. PCC 7002 | U | 1 | 0 | 2.22 | 60.8 | 2581 | 2533 | ATCC; PCC | |
| Synechococcus sp. PCC 7502 | U | 1 | 0 | 2.37 | 60.2 | 2586 | 2533 | PCC | |
| Synechococcus sp. RCC307 | U | 1 | 0 | 2.43 | 59.4 | 2581 | 2519 | RCC | |
| Synechococcus sp. WH 7803 | U | 1 | 0 | 3.57 | 47.7 | 3219 | 3170 | NCMA | |
| Synechococcus sp. WH 8102 | U | 1 | 4 | 3.95 | 47.3 | 3625 | 3575 | NCMA | |
| Synechocystis sp. PCC 6803 | U | 1 | 7 | 3.95 | 47.3 | 3610 | 3561 | PCC | |
| Synechocystis sp. PCC 6803 | U | 1 | 0 | 3.57 | 47.7 | 3218 | 3169 | PCC | |
| Synechocystis sp. PCC 6803 | U | 1 | 0 | 3.57 | 47.7 | 3217 | 3168 | PCC | |
| Synechocystis sp. PCC 6803 substr. GT-I | U | 1 | 0 | 3.57 | 47.7 | 3217 | 3168 | PCC | |
| Synechocystis sp. PCC 6803 substr. PCC-N | U | 1 | 0 | 2.59 | 53.9 | 2525 | 2476 | PCC | |
| Synechocystis sp. PCC 6803 substr. PCC-P | U | 1 | 0 | 2.52 | 53.8 | 2400 | 2231 | PCC | |
| Thermosynechococcus elongatus BP-1 | U | 1 | 0 | 7.75 | 34.1 | 5126 | 4451 | NA | |
| Thermosynechococcus sp. NK55 | U | 1 | 2 | 6.69 | 44.4 | 6033 | 5752 | NA | |
| Leptolyngbya sp. PCC 7376 | U | 1 | 0 | 4.99 | 45.2 | 4665 | 4268 | ATCC; PCC | |
| Pseudanabaena sp. PCC 7367 | U | 1 | 5 | 5.54 | 36.3 | 5041 | 4781 | PCC | |
| Prochlorococcus marinus str. AS9601 | U | 1 | 0 | 1.67 | 31.3 | 1965 | 1920 | NCMA | |
| Prochlorococcus marinus str. MIT 9211 | U | 1 | 0 | 1.69 | 38 | 1900 | 1854 | NA | |
| Prochlorococcus marinus str. MIT 9215 | U | 1 | 0 | 1.74 | 31.1 | 2054 | 1982 | NCMA | |
| Prochlorococcus marinus str. MIT 9301 | U | 1 | 0 | 1.64 | 31.3 | 1962 | 1906 | NCMA | |
| Prochlorococcus marinus str. MIT 9303 | U | 1 | 0 | 2.68 | 50 | 3136 | 2997 | NCMA | |
| Prochlorococcus marinus str. MIT 9312 | U | 1 | 0 | 1.71 | 31.2 | 1856 | 1810 | NCMA | |
| Prochlorococcus marinus str. MIT 9313 | U | 1 | 0 | 2.41 | 50.7 | 2330 | 2269 | NCMA | |
| Prochlorococcus marinus str. MIT 9515 | U | 1 | 0 | 1.7 | 30.8 | 1964 | 1905 | NCMA | |
| Prochlorococcus marinus str. NATL1A | U | 1 | 0 | 1.86 | 35 | 2250 | 2193 | NCMA | |
| Prochlorococcus marinus str. NATL2A | U | 1 | 0 | 1.84 | 35.1 | 2228 | 2162 | NCMA | |
| Prochlorococcus marinus subsp. marinus str. CCMP1375 | U | 1 | 0 | 1.75 | 36.4 | 1930 | 1882 | NCMA | |
| Prochlorococcus marinus subsp. pastoris str. CCMP1986 | U | 1 | 0 | 1.66 | 30.8 | 1762 | 1717 | NCMA |
The Biological Resource Centers (BRCs) listed (with hyperlinks) includes 1. ATCC (American Type Culture Collection), 2. PCC (Pasteur Culture Collection of Cyanobacteria), 3. NIES (National Institute for Environmental Studies), 4. NCMA (National Center for Marine Algae and Microbiota), 5. RCC (Roscoff Culture Collection) and 6. NA, Not available
Tools
The database analysis portal provides access to the CKB BLAST tool, as well as tools for pattern and fuzzy searches, and restriction digestion.
The CKB BLAST tool can be used to compare nucleotide or protein sequences, to identify members of gene families, and to infer functional and evolutionary relationships between sequences.
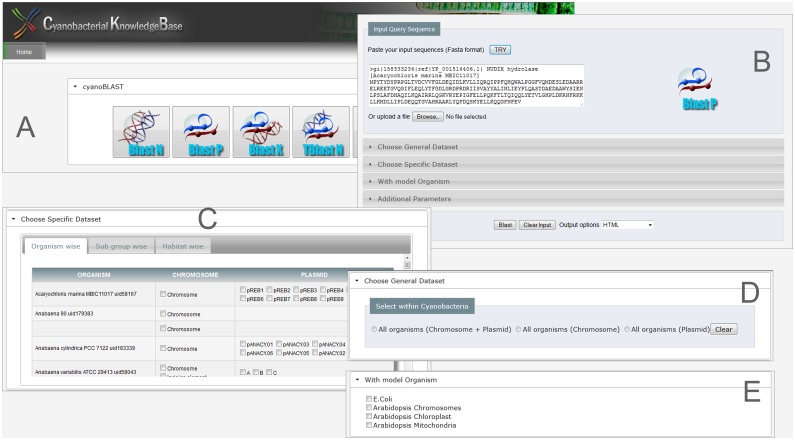
Users are provided with several customized databases for similarity searches within the CKB BLAST analysis tool. This includes a database with information on all cyanobacterial chromosomes and plasmids. The users have the freedom to restrict their analysis to either chromosomes or plasmids. Furthermore, CKB provides databases that allow users to compare individual organisms, multiple organisms and orders also (Fig 2).
Fig 2. CKB BLAST tool.
The BLAST analysis tool compares sequence within cyanobacterial species and with E. coli and Arabidopsis. It consists of A, B, C, D and E divisions where A: denotes different programs of BLAST tool, B: the sequence input page, C: option to select the individual or an order wise cyanobacterial dataset, D: choice to choose genomes of 74 cyanobacterial genomes fully or only chromosomes or only plasmids for analysis and E: option to choose E. coli and /or Arabidopsis.
As cyanobacteria are prokaryotic photosynthetic organisms, a model prokaryotic genome (E. coli) and a photosynthetic eukaryotic genome (Arabidopsis) are included for advancing comparative analysis.
In addition, pattern and fuzzy search tools are available to help in identifying the patterns present in different cyanobacterial genomes. Furthermore, the restriction digestion tool helps to identify restriction sites within the sequences.
Searching and browsing through the database
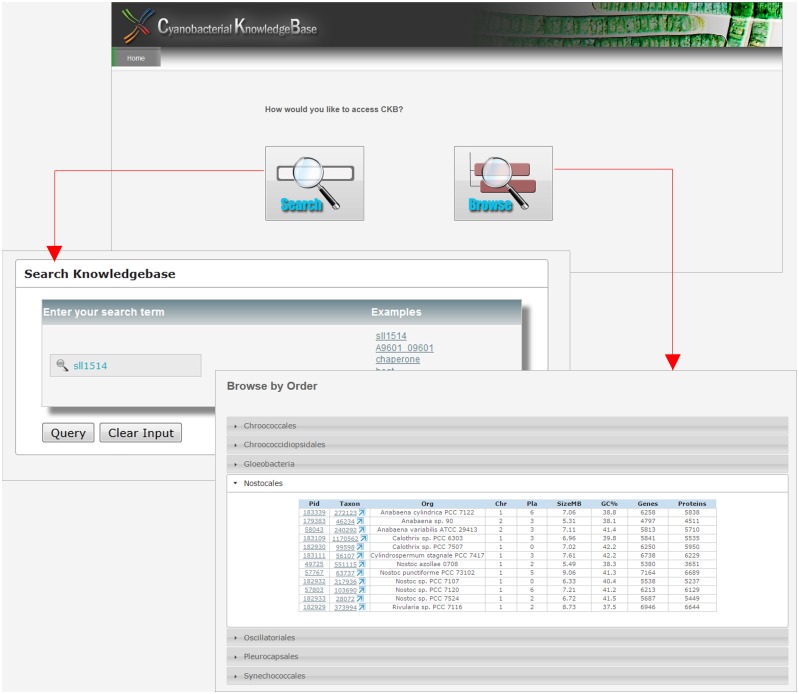
The Cyanobacterial KnowledgeBase consists of information related to 74 fully sequenced cyanobacterial species of seven orders, namely Chroococcales, Chroococcidiopsidales, Gloeobacteriales, Nostocales, Oscillatoriales, Pleurocapsales and Synechococcales. The browse option helps with orientation and navigation through the species under each order (Fig 3). The species report can be reached from the “Browse by Order” option, which provides brief information about the species, taxonomy, morphological features, genome status, and its genome details.
Fig 3. Search and browse tool.
The CKB search tools allow to search by keywords or accession and browse tool, through which users can peruse to individual organism.
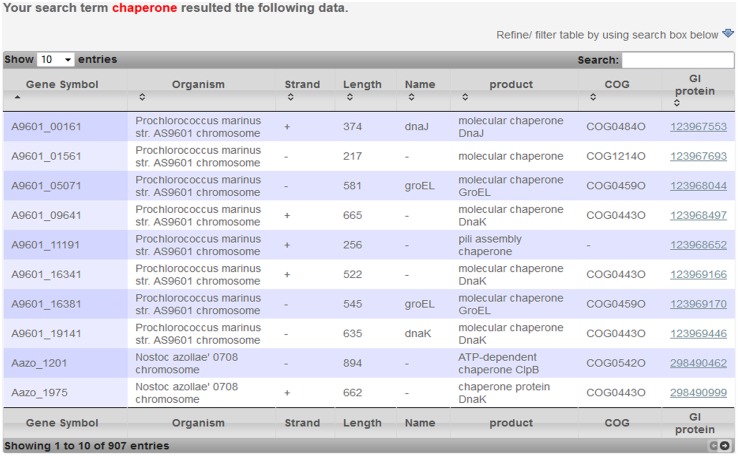
The search tool can also be used to retrieve information related to specific genes, functions, or keywords, etc. An example search result for a query keyword "Chaperone" returned 907 entries (Fig 4)
Fig 4. Search results.
Results for keyword "Chaperone" showing 907 entries.
Proteome profiling
The complete gene set of each genome can be accessed under the proteome profiling from the “Species reporter” tool. The table provides a complete gene list with PID gene name (locus_tag), synonym, product name, strand, start and end, length, COG (Clusters of Orthologous Groups) id and GI (Genbank) accession number. Furthermore, the search tool within the table provides an option to search and retrieve the results by specific keyword.
Gene report
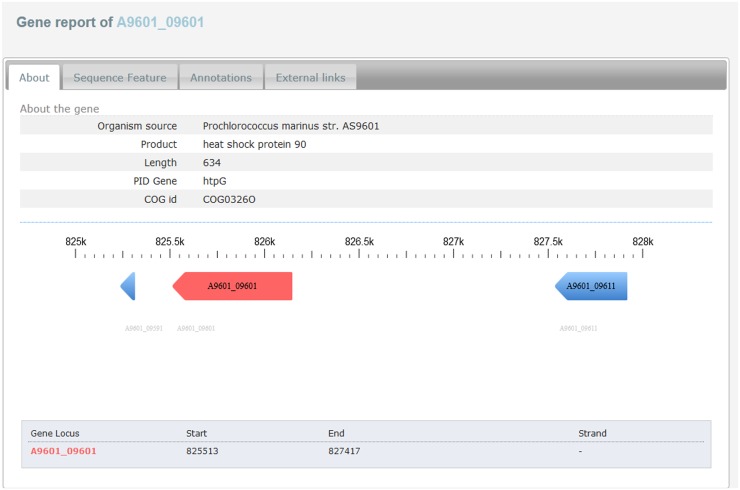
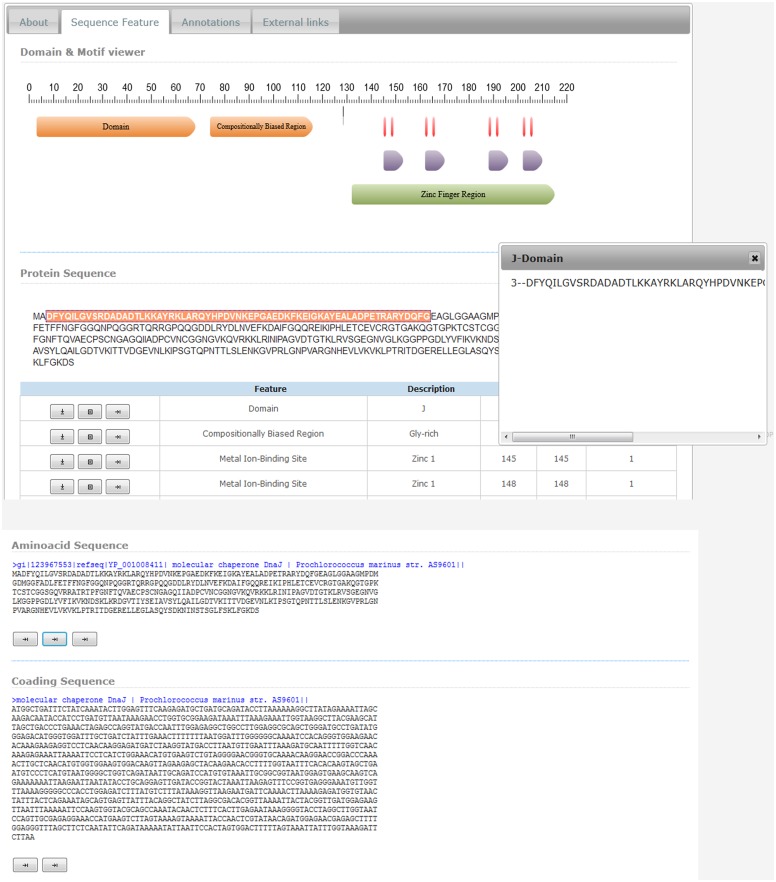
Information related to each gene is displayed under five sections. The ‘details section’ provides brief information related to the gene, and allows the user to navigate to the nearest genes present on either side of the gene of interest (Fig 5). The ‘sequence feature section’ provides domains, repeats, motif, and binding site information in both graphical and tabular form (Fig 6). The FASTA format of protein and nucleotide sequences are provided at the bottom section with direct links for BLAST analysis. The ‘annotation section’ displays the functions of the gene with gene ontology and UniProt keywords. The last two sections provide links to other external databases and list of homologous proteins respectively.
Fig 5. Gene report-About section.
About section of the gene provides brief information about the gene of interest and genes located either side of the gene.
Fig 6. Sequence features.
Graphical and tabular display of sequence features.
Review of other related databases and web-resources
The rapidly increasing genomics and proteomics data due to advancements in high throughput data generation has created a need for enhanced data management to empower basic and applied research in cyanobacteria. Many web-based databases and community resources have been created specifically for cyanobacteria to facilitate systems biology analysis using these large data. Table 2 provides the list of databases summarized by Hernández-Prieto et al. [7] which has analytical tools along with the additional web resources and databases that are currently available.
Table 2. List of other databases or web-resources available as at May-2015.
| Name | Database/Web resource | Details | Tools | Link to website |
|---|---|---|---|---|
| CyanoBase | Database | Database of genetic data of about 39 species | BLAST2, KazusaMart | http://genome.microbedb.jp/cyanobase/ |
| CYORF | Database | Contains ORF list of about 33 species | BLAST, FASTA | http://cyano.genome.ad.jp/ |
| CyanoBIKE | Database | A web-based programmable knowledge base for genomic, metabolic, and experimental data | In built and custom tools | http://biobike.csbc.vcu.edu/ |
| cTFbase | Database | Database of transcription factors in the cyanobacterial genomes | BLAST, Multiple sequence alignment tool | http://bioinformatics.zj.cn/cTFbase/DatabaseLink.php |
| CyanoPhyChe | Database | Contains data related to physico-chemical properties, structure and biochemical pathway information of cyanobacterial proteins | NA | http://bif.uohyd.ac.in/cpc/ |
| CyanoClust | Database | Database of homologous proteins in cyanobacteria and plastids | BLAST | http://cyanoclust.c.u-tokyo.ac.jp/ |
| CyanoEXpress | Database | Used for examination and visualisation of gene expression changes for various experimental or genetic manipulations from numerous transcriptome studies | NA | http://cyanoexpress.sysbiolab.eu/ |
| CyanoLyase | Database | A manually curated sequence and motif database of phycobilin lyases and related proteins | BLAST, Pattern matching tool (Protomatch) | http://cyanolyase.genouest.org/ |
| Cyanorak | Database | Information system of clusters of orthologous sequences from marine picocyanobacteria | Blast | http://www.sb-roscoff.fr/cyanorak |
| SynechoNET | Database | Integrated protein-protein interaction database of Synechocystis | NA | http://bioportal.kobic.kr/SynechoNET/ |
| ProPortal | Database | A resource for integrated systems biology of isolates of Prochlorococcus | NA | http://proportal.mit.edu/ |
| CyanoNews | Web resource | Newsletter intended for cyanobacteriologists | NA | http://www.vcu.edu/cyanonews/ |
| Cyanosite | Web resource | A general webserver for cyanobacterial research | NA | http://www-cyanosite.bio.purdue.edu/ |
| CyanoData | Web resource | A database for methods for cyanobacterial bloom management | NA | http://www.cyanodata.net/faq.php |
| CyanoDB | Web resource | A taxonomic database of cyanobacterial genera. | NA | http://www.cyanodb.cz/ |
The most comprehensive and widely used web based database is CyanoBase [8], which contains currently sequenced and annotated genome sequences, along with gene annotations and information related to various mutations involved in 39 species of cyanobacteria. It also includes tools such as BLAST for genes and genome similarity searches and KazusaMart which can be used to convert identifiers from one format to different formats. CYORF is another community annotated database that provides the open reading frame (ORF) list for approximately 33 genomes along with data from KEGG and DBGET at the GenomeNet, Pfam and Prosite motifs, predicted localization sites and protein 3D structures and tools to search for similar sequences [9]. CyanoBIKE is an instance of BioBike which provides web-based programmable knowledge base for genomic, metabolic and experimental data specifically for cyanobacteria. It has the collection of different datasets along with built-in tools for analysis, which require some basic programming skills for its application [10].
Apart from the above three generalized cyanobacterial databases, there are a few more databases which are developed specifically for a particular species or a group of cyanobacteria, which includes Cyanorak [11], SynechoNET [12] and ProPortal [13]. These are dedicated resources with annotations for orthologous sequences of marine picocyanobacteria, protein-protein interaction data for Synechocystis, and information related Prochlorococcus isolates respectively.
Additionally, many specialized databases that are available focusing on specific protein class or property exclusively for cyanobacterial species. It includes cTFbase, a database containing transcription factors [14], CyanoPhyChe, which contains physico-chemical properties of cyanobacterial proteins [15], CyanoClust, which includes homolog groups in cyanobacteria and plastids produced by the program Gclust [16], CyanoEXpress, with curated genome-wide expression data [17] and CyanoLyase, a database of phycobilin lyase sequences, motifs and functions [18].
Along with these online databases, CyanoNews [19], Cyanosite [20], CyanoData [21] and CyanoDB [22] are the major web resources that provide the basic information about cyanobacteria, current happenings in cyanobacterial research, the methods used in cyanobacteriology, bibliography archive, research groups involved in cyanobacterial research, etc. that are extensively referred by cyanobacteriologists.
CKB, the present available database has incorporated all 74 currently fully sequenced genomes of cyanobacteria, including customized tools for inclusive analysis of these genomes. The tool also helps in interpreting newly sequenced genomes by comparing them with the previously annotated cyanobacterial and/or other model organism genomes. The flexibility of defining datasets by either organism or order, or as whole genome or plasmids, helps the user to segregate their search and its results according to their specific needs. An additional significant characteristic is the inclusion of the model prokaryotic genome (E. coli) and presence of a photosynthetic eukaryotic genome (Arabidopsis), which further assists in comparative sequence analysis thereby making CKB a unique and beneficial resource for cyanobacterial genome analysis.
Future Prospects
It is planned to improve and update the content of the database of CKB in the following aspects. First, gene information will be enriched by adding experimentally proven results related to biological functions, expression, and protein-protein interactions by manually curating the data from peer reviewed literature. In addition, we intend to include or develop further analysis tools to support the analysis of cyanobacterial genomes. The necessary efforts will also be made to ensure the database as user-friendly and efficient as possible, using the reflection and feedback from users of the first version of CKB to guide our efforts.
Conclusions
Here we present CKB as a knowledge database for the cyanobacteriologists. CKB provides access to information related to fully sequenced genomes and can be utilized for analysis and retrieving information. The CKB database website is freely accessible as a web application at: http://nfmc.res.in/ckb.
Materials and Methods
Data Collection and Organization
The complete genomes of 74 cyanobacteria were downloaded from the NCBI ftp site and their accession numbers are listed in S1 File [23]. Sequence features, annotations, and external links were downloaded from the UniProt database in xml format for each gene [24]. All the downloaded data from NCBI and UniProt databases were converted into csv format and uploaded into a SQL database. The full schema of the database is included as the S1 Fig.
Web Interface and Application
CKB is built on a 64 bit CentOS (version 5) server running WAMPSERVER (V2.2d), which integrates the Apache HTTP Server (V2.2.21) with PHP (V5.3.10) and the MySQL Server (V5.5.20). Complete data related to the sequence and annotations are stored in a MySQL database. The database is designed using PHP, with jQuery JavaScript Library (V1.10), and Cascading Style Sheets (CSS) for the web interface. In addition, a simple gene browser in HTML5 is incorporated into the gene report page, which is provided by Chase Miller [25]. The BLAST 2.2.29+ tool is downloaded from NCBI ftp and pattern and fuzzy search tool and the restriction digestion tools are downloaded from Sequence Manipulation Suite [26–27]. The web server and all information parts of the database are hosted at NFMC portal www.nfmc.res.in.
Supporting Information
(TIF)
(DOCX)
Acknowledgments
The authors sincerely thank the Department of Biotechnology, Government of India, New Delhi for funding the Sub-Distributed Bioinformatics Centre (Grant No. BT/BI/04/038/98)
Data Availability
The data used for this database is retrieved from the NCBI (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/) and UniProt (http://www.uniprot.org/) repositories for the listed organisms and there are no legal or ethical restrictions on usage of this data. The raw data downloaded from NCBI and UniProt repositories is formatted as per the CKB database requirement. List of the accession numbers used is uploaded as Supporting Information.
Funding Statement
The authors thank the facility BT/03/040/89 and the Sub-DIC (BT/BI/)4/038/02 for their fiscal support.
References
- 1. Bryant DA (1994) The Molecular Biology of Cyanobacteria. Dordrecht: Kluwer Academic Publishers; 613–639. 10.1007/978-94-011-0227-8 [DOI] [Google Scholar]
- 2. Whitton BA, Potts M (2002) The Ecology of Cyanobacteria, Their Diversity in Time and Space. Kluwer Academic Publisher; Dordrecht: 563–589. 10.1007/0-306-46855-7 [DOI] [Google Scholar]
- 3. Blaha L, Babica P, Marsalek B (2009) Toxins produced in cyanobacterial water blooms—toxicity and risks. Inter discip Toxicol 2:36–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tan LT (2007) Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry 68: 954–979. [DOI] [PubMed] [Google Scholar]
- 5. Burja AM, Banaigs B, Abou-Mansour E, Grant Burgess J, Phillip C. Wright (2001) Marine cyanobacteria-a prolific source of natural products. Tetrahedron 57:9347–9377. 10.1016/S0040-4020(01)00931-0 [DOI] [Google Scholar]
- 6. Kehr JC, GattePicchi D, Dittmann E (2011) Natural product biosyntheses in cyanobacteria: A treasure trove of unique enzymes. Beilstein J Org Chem 1622–1635. 10.3762/bjoc.7.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hernández-Prieto MA, Semeniuk TA, Futschik ME (2014) Toward a systems-level understanding of gene regulatory, protein interaction, and metabolic networks in cyanobacteria. Front Genet. 2014 July 2;5:191 10.3389/fgene.2014.00191. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fujisawa T, Okamoto S, Katayama T, Nakao M, Yoshimura H, Kajiya-Kanegae H et al. (2014) CyanoBase and RhizoBase: databases of manually curated annotations for cyanobacterial and rhizobial genomes. Nucleic Acids Res 42: 666–670. 10.1093/nar/gkt1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Furumichi M, Sato Y, Omata T, Masahiko Ikeuchi, Minoru Kanehisa (2002) CYORF: Community Annotation of Cyanobacteria Genes. Genome Informatics 13: 402–403. [Google Scholar]
- 10. Elhai J, Taton A, Massar JP, Myers JK, Travers M, Casey J et al. (2009) BioBIKE: a Web-based, programmable, integrated biological knowledge base. Nucleic Acids Res. 2009 July;37(Web Server issue):W28–32. 10.1093/nar/gkp354. Epub 2009 May 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cyanorak Database. Accessed: http://www.sb-roscoff.fr/cyanorak
- 12. Kim WY, Kang S, Kim BC, Oh J, Cho S, Bhak J et al. (2008) SynechoNET: integrated protein-protein interaction database of a model cyanobacterium Synechocystis sp. PCC 6803. BMC Bioinformatics. 2008;9 Suppl 1:S20 10.1186/1471-2105-9-S1-S20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kelly L1, Huang KH, Ding H, Chisholm SW. ProPortal: a resource for integrated systems biology of Prochlorococcus and its phage. Nucleic Acids Res. 2012. January;40(Database issue):D632–40. 10.1093/nar/gkr1022. Epub 2011 Nov 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu J, Zhao F, Wang S, Deng G, Wang J, Bai J et al. (2007) cTFbase: a database for comparative genomics of transcription factors in cyanobacteria. BMC Genomics 18:104 10.1186/1471-2164-8-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arun PV, Bakku RK, Subhashini M, Singh P, Prabhu NP, Suzuki I et al. (2012) CyanoPhyChe: a database for physico-chemical properties, structure and biochemical pathway information of cyanobacterial proteins. PLoS One 7:e49425 10.1371/journal.pone.0049425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sasaki NV, Sato N. (2010) CyanoClust: comparative genome resources of cyanobacteria and plastids. Database (Oxford). 2010;2010:bap025 10.1093/database/bap025. Epub 2010 Jan 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hernandez-Prieto MA, Futschik ME. CyanoEXpress: A web database for exploration and visualisation of the integrated transcriptome of cyanobacterium Synechocystis sp. PCC6803. Bioinformation. 2012;8(13):634–8. 10.6026/97320630008634. Epub 2012 Jul 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bretaudeau A, Coste F, Humily F, Garczarek L, Le Corguillé G, Six C et al. (2013) CyanoLyase: a database of phycobilin lyase sequences, motifs and functions. Nucleic Acids Res. 2013 January;41(Database issue):D396–401. 10.1093/nar/gks1091. Epub 2012 Nov 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CyanoNews. Accessed: http://www.vcu.edu/cyanonews/
- 20.Cyanosite. Accessed: http://www-cyanosite.bio.purdue.edu/
- 21.CyanoData. Accessed: http://www.cyanodata.net/faq.php
- 22.CyanoDB. Accessed: http://www.cyanodb.cz/
- 23.FTP site for genome sequence. Accessed: ftp://ftp.ncbi.nlm.nih.gov/genomes/
- 24.UniProt database. Accessed: http://www.uniprot.org/
- 25. Miller CA, Anthony J, Meyer MM, Marth G (2013) Scribl: an HTML5 Canvas-based graphics library for visualizing genomic data over the web. Bioinformatics. 29:381–383. 10.1093/bioinformatics/bts677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.NCBI standalone blast. Accessed: ftp://ftp.ncbi.nlm.nih.gov/blast/
- 27.Sequence Manipulation Suite. Accessed: http://www.bioinformatics.org/sms2/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(DOCX)
Data Availability Statement
The data used for this database is retrieved from the NCBI (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/) and UniProt (http://www.uniprot.org/) repositories for the listed organisms and there are no legal or ethical restrictions on usage of this data. The raw data downloaded from NCBI and UniProt repositories is formatted as per the CKB database requirement. List of the accession numbers used is uploaded as Supporting Information.