Abstract
The first critical stage in salamander or teleost appendage regeneration is creation of a specialized epidermis that instructs growth from underlying stump tissue. Here, we performed a forward genetic screen for mutations that impair this process in amputated zebrafish fins. Positional cloning and complementation assays identified a temperature-sensitive allele of the ECM component laminin beta 1a (lamb1a) that blocks fin regeneration. lamb1a, but not its paralog lamb1b, is sharply induced in a subset of epithelial cells after fin amputation, where it is required to establish and maintain a polarized basal epithelial cell layer. These events facilitate expression of the morphogenetic factors shha and lef1, basolateral positioning of phosphorylated Igf1r, patterning of new osteoblasts, and regeneration of bone. By contrast, lamb1a function is dispensable for juvenile body growth, homeostatic adult tissue maintenance, repair of split fins, or renewal of genetically ablated osteoblasts. fgf20a mutations or transgenic Fgf receptor inhibition disrupt lamb1a expression, linking a central growth factor to epithelial maturation during regeneration. Our findings reveal transient induction of lamb1a in epithelial cells as a key, growth factor-guided step in formation of a signaling-competent regeneration epidermis.
Author Summary
Unlike mammals, adult teleost fish and urodele amphibians can fully regenerate lost appendages. Understanding what initiates regeneration in these vertebrates is of great interest to the scientific community. It has long been known that the epidermis that forms quickly over an amputated limb stump is critical for initiating regenerative programs. Yet, little of understood of the molecular and cellular mechanisms by which a simple adult epithelium transforms into this key signaling source. Here, we performed a large-scale, unbiased genetic screen for epithelial signaling deficiencies during the regeneration of amputated adult zebrafish fins, from which we identified several new mutants. One gene identified from this screen disrupts a specific component of the extracellular matrix material Laminin, Laminin beta 1a, a factor that we find to be dispensable in uninjured adult animals but required for all stages fin regeneration. Transient induction of this component by amputation polarizes the basal layer of the nascent epithelium, and, in turn, facilitates the synthesis of signaling factors, the positioning of ligand receptors, and the patterning of new bone cells. We also find that normal induction of Laminin beta 1a by injury relies on the function of Fibroblast growth factors, secreted polypeptide signals that are released early upon injury. Our results identify key early steps in the endogenous program for vertebrate appendage regeneration.
Introduction
Mammals have a limited ability to regenerate complex structures like limbs, heart or central nervous system tissue. By contrast, teleost fish and urodele amphibians can regenerate major appendages, spinal cord, retina, brain, kidney, and the heart [1–7]. How and why tissue regeneration occurs in non-mammalian vertebrates has fascinated biologists for centuries and is relevant to regenerative medicine strategies.
Previous studies of appendage regeneration have identified three prominent phases: 1) wound healing; 2) blastema formation; and 3) regenerative outgrowth and patterning [8]. Upon amputation injury, the exposed stump tissue is rapidly covered by a sheet of epithelial cells, a process involving little or no cell proliferation [9, 10]. Classical studies in salamanders revealed that if this epithelium is removed, replaced with flank skin, or disrupted by insertion of the limb into the abdominal cavity, limb regeneration does not proceed [11–13]. The wound epithelium becomes multilayered and acquires a layer of cuboidal basal epithelial cells over the next hours to days, maturing into a structure commonly referred to as the wound or regeneration epidermis. Regeneration epidermises of salamander limbs or teleost fins are known to express markers of developmental signaling pathways, including many secreted factors [8, 14]. For instance, after initial epithelialization of an amputated zebrafish fin stump, epithelial and/or mesenchymal cells induce effectors of pathways mediated by Fgfs, Igfs, Wnts, Hhs, Activin-βA/TGFβ, Retinoic acid, Bmps, and Notch [15–27]. Interestingly, fgf20a and igf2b ligand genes are induced within hours of fin amputation in mesenchymal cells, and perturbation of Fgf signaling via a mutation in the fgf20a ligand gene, or of Igf signaling by receptor inhibition, disrupts formation of the regeneration epidermis and subsequent bone regeneration [20, 21]. These findings indicate an important role for early expression of growth factors in structural and functional maturation of epithelial tissue. However, mechanisms by which these pathways define the morphology and signaling activities of the regeneration epidermis have not been addressed.
Here, we used forward genetics to identify a critical role for laminin beta 1a (lamb1a), one of two paralogs encoding a Laminin beta 1 extracellular matrix component, in zebrafish fin regeneration. lamb1a is sharply induced upon fin amputation in the basal layer of the wound epithelium, where its function is required to establish polarity in basal epithelial cells, induce and maintain basal epithelial markers, localize receptors for signaling, and align regenerating osteoblasts. lamb1a induction is dependent on Fgf signaling, both immediately after amputation and throughout regeneration. Thus, Lamb1a is a critical node between growth factor signals and formation of the regeneration epidermis in an amputated vertebrate appendage.
Results
Forward genetic screen for epithelial signaling defects during fin regeneration
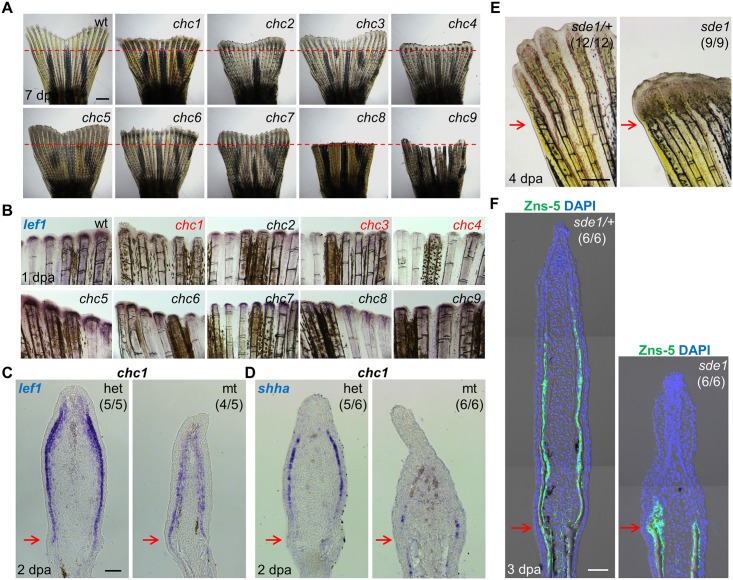
Previous genetic screens for mutants in zebrafish fin regeneration involved parthenogenesis of F1 generation females [1, 28]. This approach saves considerable time and animal facility space, as progeny with homozygous ENU-induced mutations can be screened in the F2 generation. Yet, it also limits animal survival and access to chromosomal regions far from centromeres [29]. For this study, we conducted a three-generation screen, in which we raised 423 F3 families from 108 F2 generation crosses to adulthood at a temperature of 25°C. To identify temperature-sensitive mutants that can be used for toggling gene function, we shifted these adults to 33°C after amputating ~50% of the caudal fins, and then assessed regeneration 7 days later. After several rounds of outcrossing to identify stable phenotypes and dilute unlinked ENU mutations from the genetic background, we found 9 families (chc1-9) with temperature-sensitive defects in fin regeneration inherited as a single recessive determinant (Fig 1A).
Fig 1. Forward genetic screen for signaling defects in the fin regeneration epidermis.
(A) Whole-mount images of wild-type and (chc1-9) mutant regenerates at 7 days post amputation (dpa). Red dashed lines indicate plane of amputation. Scale bars, 1 mm. (B) Whole-mount RNA ISH of lef1 expression in wild-type and mutant regenerates at 1 dpa. chc1, chc3, and chc4 mutant families show reduced or undetectable lef1 expression when compared to their respective heterozygous siblings. (C, D) Longitudinal sections of 2 dpa fin regenerates assessed by RNA ISH, showing reduced lef1 and shha in chc1 mutant regenerates. (E) Whole-mount images of sde1 (formerly, chc1) fin regenerates at 4 dpa. Scale bars, 0.5 mm. (F) Longitudinal sections of 3 dpa fin regenerates show impaired patterning of osteoblasts in sde1 (chc1) mutants, assessed by Zns-5 antibody staining (green). DAPI, blue. Scale bars, 50 μm (unless otherwise indicated). Red arrows indicate plane of amputation.
To identify a subset of mutants that disrupt formation of a functional regeneration epidermis, we examined lef1 expression at 1 dpa by in situ RNA hybridization (ISH; Fig 1B). lef1 is a downstream effector and transcriptional target of Wnt signaling that is induced in wound epithelial cells adjacent to the amputation plane as early as 12 hours post-amputation [30]. Three mutant lines, chc1, chc3, and chc4, consistently displayed reduced lef1 expression at 1 dpa (Fig 1B), from which we initially pursued chc1. chc1 regenerates also had markedly reduced expression of the Hedgehog ligand shha, which, like lef1, is induced in basal epithelial cells (Fig 1C and 1D). shha has been implicated in blastemal cell proliferation and alignment of osteoblasts to areas of prospective bone during zebrafish fin regeneration [15, 31].
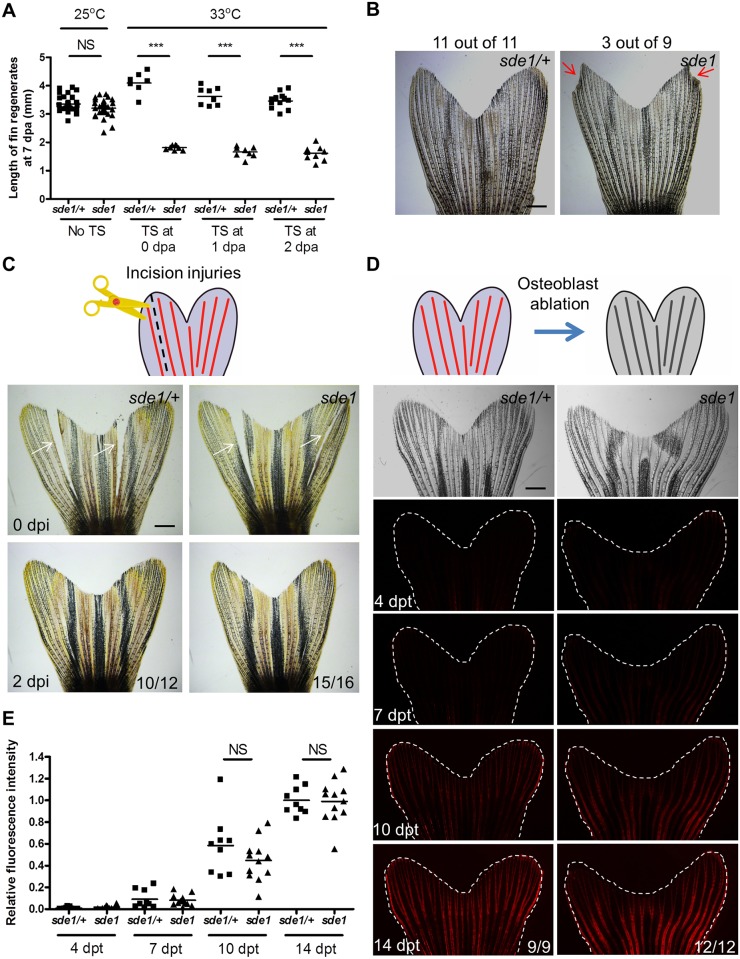
chc1 mutants, renamed signaling deficient epidermis 1 (sde1) mutants, regenerated amputated fins normally at 25°C, indicating a strictly temperature-sensitive effect. Inspection of sde1 fin regenerates showed reduced lengths and no detectable bone at 4 dpa (Fig 1E). Osteoblasts typically begin to align adjacent to the basal epithelial layer by 2 dpa, where they deposit bone minerals that comprise ray hemisegments [1]. Immunofluorescence analysis of 3 dpa sde1 tissue sections revealed the accumulation of osteoblasts in a mass adjacent to the epithelium, or limited presence at all, as opposed to an even distribution to lateral regions (Fig 1F). Analysis of 7 dpa sde1 regenerates indicated reduced but clearly detectable osteoblast-lined bone, with osteoblasts accumulated in small masses at the distal regions of bone (S1A Fig). To determine whether sde1 regenerates eventually reach full length, we assessed fin ray lengths at 14, 21, and 28 dpa. We observed visibly obvious and statistically significant regenerative defects at each of these time points (S1B and S1C Fig). Thus, sde1 mutations have a long-lasting impact on regeneration. To determine temporal requirements for sde1 during fin regeneration, we shifted the animals from 25°C to 33°C at 0, 1, or 2 dpa, and assessed fin lengths at 7 dpa. Each of these procedures resulted in significant defects in sde1 regenerates (Fig 2A), indicative of continued requirements throughout various stages of regeneration. Thus, sde1 is one of a subset of mutations that inhibits induction of morphogenetic signals in the regeneration epidermis. The sde1 gene product is critical for osteoblast patterning, bone formation, and progression of regeneration.
Fig 2. sde1 requirements for tissue regeneration depend on injury context.
(A) Measurement of sde1 fin regenerates at 7 dpa. After amputation, animals were shifted from the permissive temperature (25°C) to the restrictive temperature (33°C) at 0, 1 or 2 dpa. TS, temperature shift. (n = 15, 16, and 21; Student’s t -test, ***P < 0.001; NS, non-significant). (B) Adult sde1/+ and sde1 animals were incubated at 33°C for two months (n = 11 and 9). Red arrows point to a damaged fin edge in sde1. The most severe example of damage in sde1 animals is displayed here (3 of 9 showed damage in the experiment). (C) (Top) Cartoon depicting the model of incision injuries. (Bottom) Whole-mount images were acquired at 0 and 2 days post incision injury (dpi). Images from the same animal are shown before (top) and after repair (bottom). White arrows indicate sites of injury (n = 12 and 16). (D) (Top) Cartoon depicting the model of osteoblast ablation. (Bottom) Fluorescence intensity, indicating recovery of genetically labeled osteoblasts after ablation, was recorded at 4, 7, 10, and 14 days post Mtz treatment (dpt) and quantified using ImageJ software. White dashed lines indicate fin boundaries. Images from the same animal are shown throughout recovery. (E) Quantification of relative fluorescence intensity from individual animals after osteoblasts ablation (n = 9 and 12; mean ± SEM; Student’s t-test; NS, non-significant). Scale bars, 1 mm.
Injury context-specific requirements for sde1
Previous studies of fin regeneration mutants revealed distinct phenotypes when animals are placed at the restrictive temperature for long time periods. sly1 and hsp60 mutants survive poorly beyond 14 days at 33°C, likely reflecting roles in fundamental cell survival or organismal physiology [32, 33]. fgf20a and mps1 mutants, as well as transgenic animals enabling prolonged expression of a dominant-negative Fgf receptor, each survive well at restrictive conditions but display progressive loss of 17–36% of distal fin tissue over a course of 60 days [34]. sde1 mutants showed no adverse effects at 33°C over a 60-day period, displaying higher survival than any other new mutants identified in this study (100%, n = 36; S2 Fig). A minority of sde1 mutants lost small amounts of distal fin tissue (3/9, Fig 2B). These experiments indicate minimal requirements for sde1 in basic cell, tissue, or organismal function, or in homeostatic maintenance of fin structures.
To examine requirements in other forms of regeneration, we performed two additional injury models. First, we made a precise incision within interray tissue that spanned most of the proximodistal length of the caudal fin. This injury, which severed interray mesenchyme and epithelial cells but not bone, is typically healed within 2 days and most likely reflects simple wound-healing [21]. sde1 mutants and heterozygous mutant siblings each rapidly healed these incisions (Fig 2C). Second, we introduced a transgene for visualizing and genetically ablating osteoblasts (osx:mCherry-NTR) into the sde1 background. Adult osx:mCherry-NTR zebrafish quickly repopulate fin rays that have been depleted of virtually all osteoblasts after treatment with the pro-drug metronidazole [35]. Following osteoblast depletion in sde1 mutants and heterozygous clutchmates, we quantified the recovery of osx-driven fluorescence intensity over 14 days (Fig 2D). We detected no defects in the ability of sde1 mutants to regenerate osteoblasts in these experiments (Fig 2E). Thus, sde1 mutations potently affect regeneration of amputated fins, but they have little or no effects on the ability of adult zebrafish to maintain fin tissue or regenerate complex fin injuries that do not require a regeneration epidermis.
sde1 encodes a laminin beta1 paralog
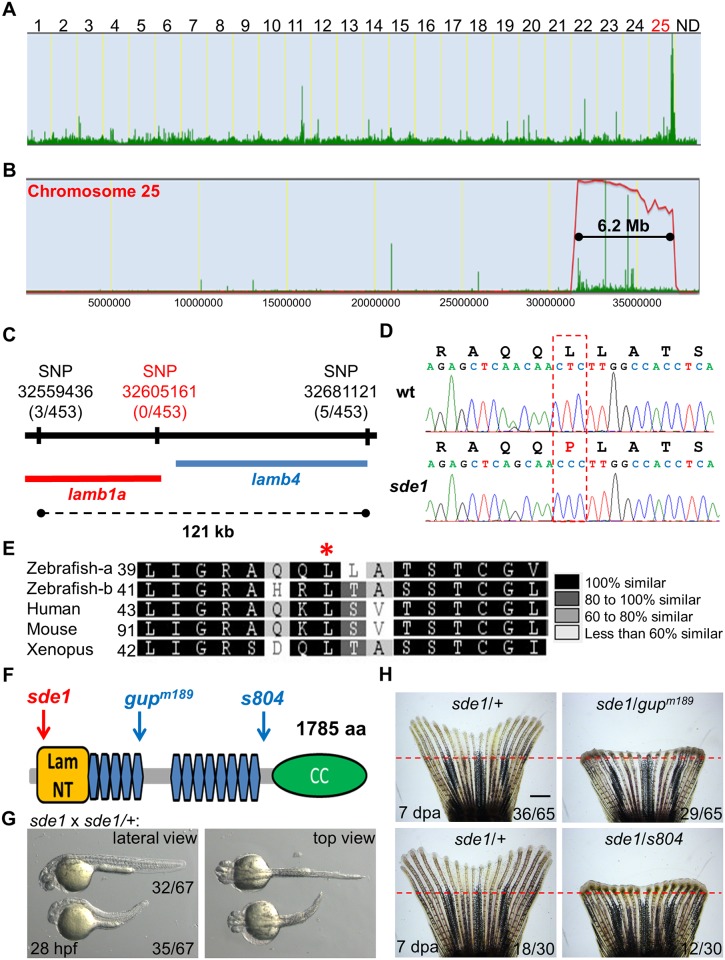
To identify the gene that is disrupted in sde1 mutants, we performed whole-exome sequencing of clutchmate DNA from an sde1 x sde1/+ cross (see Materials and Methods) [36]. Sequencing data were analyzed using the web-based mapping tool SNPtrack, which facilitates linkage analysis based on single nucleotide polymorphisms (SNPs) and regions of homozygosity [37]. Primary analysis from SNPtrack revealed a single 6.2 Mb peak on chromosome 25 containing sde1 (Fig 3A and 3B). We genotyped 453 adults from several sde1 × sde1/+ mapping crosses for polymorphic SNPs in this region. From this linkage analysis, we identified 3 closely linked SNPs that flanked a 121 kb region containing two genes: laminin beta 1a (lamb1a) and laminin beta 4b (lamb4b) (Fig 3C). By filtering against a SNP database established in the Poss lab (see Materials and Methods) and cDNA sequencing, we identified one novel non-synonymous mutation (T to C) in the coding region of the gene lamb1a causing a lysine to proline change at position 46 (Fig 3D). No coding mutations were identified in lamb4b. Zebrafish have two unlinked paralogs encoding laminin beta 1, lamb1a and lamb1b, emerging from a partial genome duplication event estimated at 350 million years ago [38]. Sequence analysis revealed that lysine 46 is conserved between these two zebrafish paralogs, and also among laminin beta 1 genes in other vertebrate species like human and Xenopus laevis (Fig 3E). Additionally, the candidate sde1 mutation is located in a highly conserved Laminin N-terminal (Lam NT) domain (Fig 3F), which has been demonstrated to mediate interactions with other Laminin members [39, 40]. Thus, genetic mapping associates sde1 with a mutation in a conserved residue of the laminin beta 1 paralog, lamb1a.
Fig 3. sde1 encodes a laminin beta1 paralog.
(A) Analysis of genomic homozygosity in sde1 mutants. (B) Log likelihood analysis using SNPtrack. (C) High-resolution mapping using linked SNP markers. Three and five recombinants were found for SNP32559436 and SNP32681121, respectively, on each side of a 121 kb region. After genotyping 453 adult animals, no recombinants were identified for a novel SNP at position 32605161. (D) Sanger sequencing readouts from wild-type and sde1 cDNA. SNP32605161 is within the coding region of the gene laminin beta 1a (lamb1a), causing a leucine to proline change. (E) Amino acid alignment across distant species. Red star marks the location of the leucine. Differential gray scale indicates level of conservation across listed species. (F) Cartoon depicting major structural domains in Lamb1a. Blue and red arrows indicate the locations of the sde1 mutation, along with two previously identified alleles gup m189 and s804. Lam NT, Laminin N-terminal domain; Blue hexagons, Laminin-type epidermal growth factor-like domain; CC, uncharacterized coiled-coil domain. (G) sde1 embryos incubated at 31°C have shortened trunks. Representative embryos from an sde1 x sde1/+ cross. Embryos were transferred to 31°C at 3 hours post-fertilization (hpf). Images were acquired at 28 hpf. Approximately 48% of embryos (32 out of 67) showed phenotypes representative of the gup m189 mutation after the temperature shift, consistent with expected Mendelian ratio. The phenotype and the ratio were consistent across three independent crosses. (H) Complementation tests showing both gup m189 and s804 alleles fail to complement the 7 dpa regeneration defects of the sde1 mutation in adult animals, yielding expected ratios (~50%; n = 65 and 30). Red dashed lines indicate plane of amputation. Scale bars, 1 mm.
Two independent loss-of-function mutations in lamb1a have been isolated in zebrafish, gup m189 and s804 (Fig 3F) [41, 42]. We compared phenotypes of sde1 embryos raised at 31°C to those described for the gup m189 embryos, and found trunk shortening reminiscent of the gup m189 phenotype in ~50% of 28 hpf embryos from mapping crosses (Fig 3G). Furthermore, to test whether sde1 complements known lamb1a mutations, we crossed homozygous sde1 mutants with either gup m189 or s804 heterozygous mutant zebrafish. Each of these crosses gave rise to ~50% of progeny with temperature-sensitive defects in adult fin regeneration (Fig 3H). Thus, based on high-resolution genetic mapping, expected embryonic phenotypes, and two independent complementation tests, we conclude that sde1 encodes a conditional allele of lamb1a (lamb1a pd110), most likely acting as a hypomorph at the restrictive temperature.
lamb1a, but not lamb1b, is an induced component of the regeneration epidermis
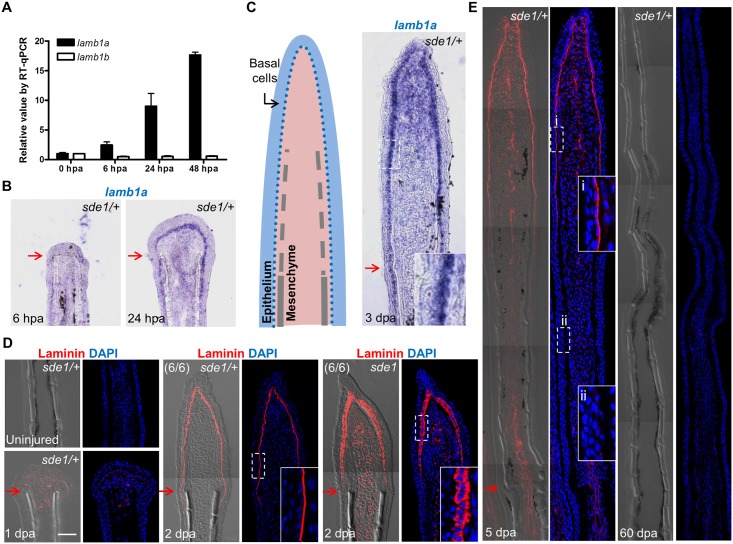
Using specific qPCR probes targeting lamb1a and lamb1b sequences, we found that lamb1a expression, but not lamb1b, was induced during regeneration (Fig 4A). ISH experiments failed to detect lamb1a RNA in uninjured or 6 hpa fins, but visualized lamb1a by 1 dpa mainly in basal epithelial cells, and less prominently in mesenchymal cells (Fig 4B). During regenerative outgrowth, lamb1a expression was maintained in regenerating tissue in a primary basal epithelial cell domain and a secondary mesenchymal domain (Fig 4C). Using an antibody raised against mouse basement membrane Laminin, we found analogous expression domains for Laminin at the protein level. Laminin presence was undetectable in uninjured fins, and evident by 1 dpa (Fig 4D). By 2 dpa, Laminin was primarily localized to the basal side of the basal epithelial cell layer (Fig 4D), ostensibly part of the extracellular basement membrane. Laminin presence gradually waned proximal to the amputation plane from 2 to 5 dpa, and was undetectable in 60 dpa regenerates (Fig 4E). Thus, lamb1a is regulated differently from its lamb1b paralog upon fin amputation, an injury that induces lamb1a transiently in epithelial tissues and maintains its expression during key stages of regeneration.
Fig 4. lamb1a, not lamb1b, is induced during regeneration.
(A) RT-qPCR analysis indicates that lamb1a, but not lamb1b, is induced during regeneration. qPCR results were normalized to rpl13a and to the basal expression of lamb1a/lamb1b at 0 hours post-amputation (hpa). (n = 3; mean ± SEM). (B) Section ISH indicating that lamba1 becomes visually detectable in the basal epithelial layer between 6 and 24 hpa. (C) Left: cartoon depicting basic cellular makeup of the fin regenerate. Right: lamb1a is expressed in basal epithelial cells and some mesenchymal cells at 3 dpa. (D) Antibody staining for Laminin expression in regenerating fins. Laminin protein is restricted to the basal side of the basal epithelial cells layer by 2 dpa in wild-type or sde1/+ regenerates, but mislocalized to all regions of basal cells in sde1 mutants. (E) Laminin expression at 5 dpa and 60 dpa, indicating that Laminin presence is transient during regenerative outgrowth. i: distal, newly regenerated tissue; ii: proximal regenerated tissue. Laminin, red; DAPI, blue. Scale bars, 50 μm. White dashed boxes indicate areas of enlarged view. Red arrows indicate plane of amputation.
lamb1a is required for juvenile fin morphology but not body size
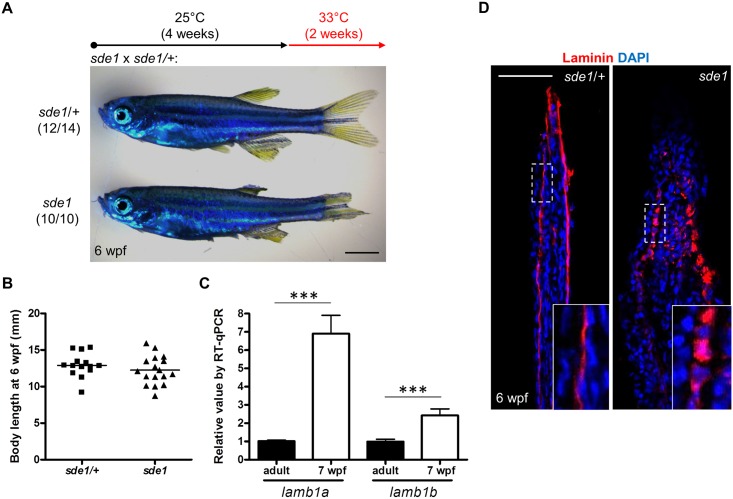
To examine lamb1a functions during juvenile growth, we shifted sde1 animals from 25°C to 33°C at 4 weeks post fertilization (wpf), after zebrafish reach their juvenile stage. After 2 weeks at 33°C, all 6 wpf sde1 animals (10/10) had noticeably degraded fins, whereas majority of sde1/+ siblings displayed no noticeable fin phenotypes (12/14). Interestingly, this temperature shift did not grossly affect the body length of sde1 juvenile animals (Fig 5B). Using specific qPCR probes targeting lamb1a and lamb1b sequences, we found that both lamb1a and lamb1b mRNA levels were higher in juvenile fins than in adult uninjured fins (Fig 5C). These results indicate that lamb1a is required for juvenile fin growth and/or tissue maintenance, but is dispensable for organismal growth at the juvenile stage.
Fig 5. lamb1a is required for juvenile fin growth but not body growth.
(A) Juvenile sde1 animals, after incubation from 4 to 6 weeks post-fertilization (wpf) at 33°C, acquire degraded fins. Scale bars, 2 mm. (B) sde1 mutations have minimal impact on juvenile outgrowth. Body length was measured from the tip of the snout to the base of caudal fin. (C) RT-qPCR analysis indicates that both lamb1a and lamb1b are induced in fin tissue during juvenile outgrowth. qPCR results were normalized to rpl13a and to the basal expression of lamb1a/lamb1b in adult uninjured fins (n = 4; mean ± SEM). (D) Antibody staining for Laminin expression in juvenile fins. Laminin protein in longitudinal sections of fins is localized to the basal side of the epitheilum in sde1/+ animals, but becomes mislocalized in sde1 mutants.
lamb1a induction is required to induce and maintain polarity and signaling in basal cells of the regeneration epidermis
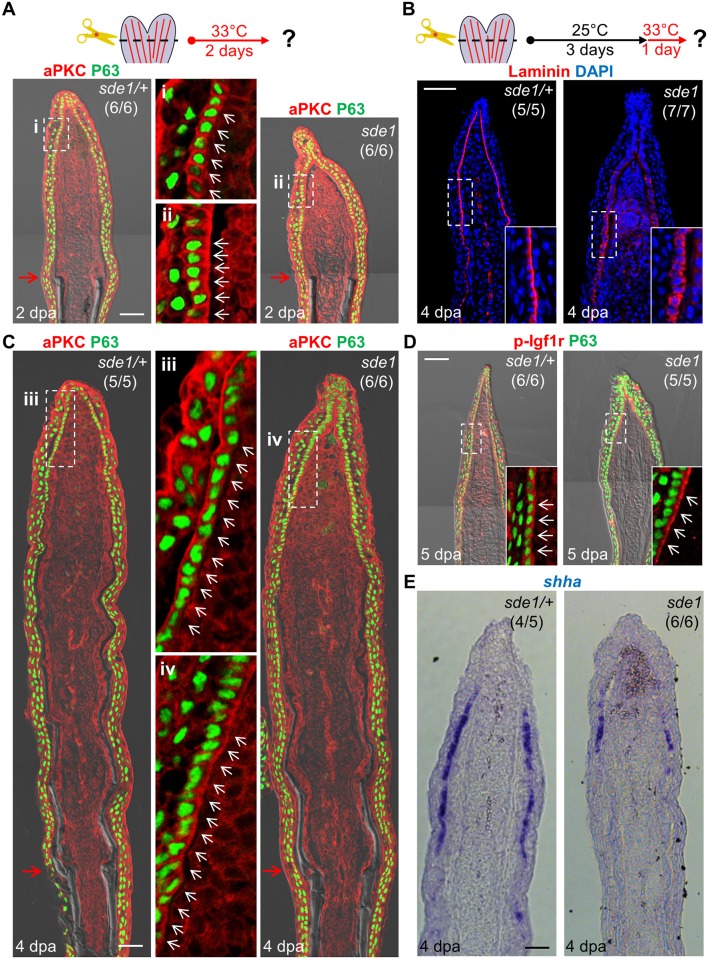
Laminin is widely studied as a component of the basement membrane; thus, its induction in the basal epithelial layer strongly suggested a role in creating this structure. We examined Laminin presence in 2 dpa regenerates of sde1 mutants at 33°C, and found it ectopically localized to all basal epithelial cell regions including the apical and lateral portions (Fig 4D), indicate of intracellular residence. This result supports the idea that Lamb1a interaction with other Laminin members through the N-terminal domain may be important for secretion of Laminin complexes [39, 40]. Laminin was also aberrantly localized in sde1 juvenile fins maintained at 33°C for 14 days (Fig 5D). To examine the polarity of the basal epithelial layer at 2 dpa, we used an antibody raised against atypical Protein kinase C (aPKC), a well-characterized apical marker that is essential to maintain epithelial polarity in many systems, including nematodes, flies, and mammalian cells [43]. Whereas aPKC was localized to apical and lateral regions of basal epithelial cells by 2 dpa in wild-type fins, sde1 regenerates accumulated aPKC expression in basal regions of this cell layer (Fig 6A). This finding is consistent with in vitro [44, 45] and in vivo [46] functional studies indicating Laminin as a polarizing cue for epithelial cells.
Fig 6. lamb1a induction defines cell polarity and signaling competence in basal cells of the regeneration epidermis.
(A) Antibody co-staining for aPKC (red) and P63 (green; an epithelial maker for all basal and some suprabasal epithelial cells) in longitudinal sections of sde1/+ and sde1 fin regenerates at 2 dpa. (B) Antibody staining for Laminin in fin regenerates at 4 dpa after a temperature shift from 25°C to 33°C at 3 dpa, indicating induced mislocalization. Laminin, red; DAPI, blue. White dashed boxes indicate areas of enlarged view. (C) Antibody co-staining for aPKC (red) and P63 (green) in longitudinal sections of sde1/+ and sde1 fin regenerates at 4 dpa after a temperature shift from 25°C to 33°C at 3 dpa, indicating loss of basal cell polarity. iii: distal regenerated tissue (sde1/+); iv: distal regenerated tissue (sde1). White arrows indicate basal cell nuclei. (D) Antibody co-staining for phosphorylated Igf1r (red) and P63 (green) in longitudinal sections of 5 dpa sde1/+ and sde1 fin regenerates, after a temperature shift from 25°C to 33°C at 4 dpa. The basal localization of p-Igf1r is enriched in basal epithelial cells in sde1 mutants. (E) shha RNA expression is reduced in sde1 fin regenerates at 4 dpa after a temperature shift from 25°C to 33°C at 3 dpa. Scale bars, 50 μm. White dashed boxes indicate areas of enlarged view. Red arrows indicate plane of amputation.
To determine whether Lamb1a function actively maintains epithelial cell polarity during regenerative outgrowth, we shifted sde1 animals from 25°C to 33°C at 3 dpa, after the polarity of basal cells was established (Fig 6A). After one day at 33°C, Laminin and aPKC protein became mislocalized in basal epithelial cells (Fig 6B and 6C), indicating that Lamb1a normally maintains this polarity during outgrowth. To examine possible functional consequences of lost basal epithelial polarity, we examined the localization of a receptor of Insulin-like growth factor (Igf) signaling in basal cells of the regeneration epidermis. Igf1r is autophosphorylated in basal epithelial cells during fin regeneration, presumably after engagement by Igf2b and possibly other ligands, and its activity is required for fin regeneration [21]. Whereas phosphorylated Igf1r is located basolaterally in control fin regenerates, one day at 33°C demonstrably enriched the basal expression domain in sde1 regenerates. This rapid change suggests that localization of phosphorylated Igf1r is actively maintained by components of the cell polarity machinery. Additionally, we found that shha expression visualized by ISH was consistently reduced in basal epithelial cells after shifting to 33°C for one day at 3 or 4 dpa (Fig 6E and S3 Fig). The function of Lamb1a in maintaining cell polarity and shha expression is unlikely to be a consequence of a general slowing of regeneration, as 8 hours of 33°C treatment at 3 dpa was sufficient to alter aPKC localization and reduce shha expression (S4 Fig). Additionally, a one-day temperature shift did not grossly decrease indicators of cell proliferation in mesenchymal cells (S5 Fig), suggesting that cell proliferation in one or more subpopulations of blastemal cells is not directly affected. Together, these experiments reveal that lamb1a induction has a central role in formation of a polarized regeneration epidermis after fin amputation. They also suggest that this epithelial polarization is critical for localization of signaling receptors, expression of key morphogenetic signals, osteoblast patterning, and bone regeneration.
Fgf signaling influences lamb1a expression during regeneration
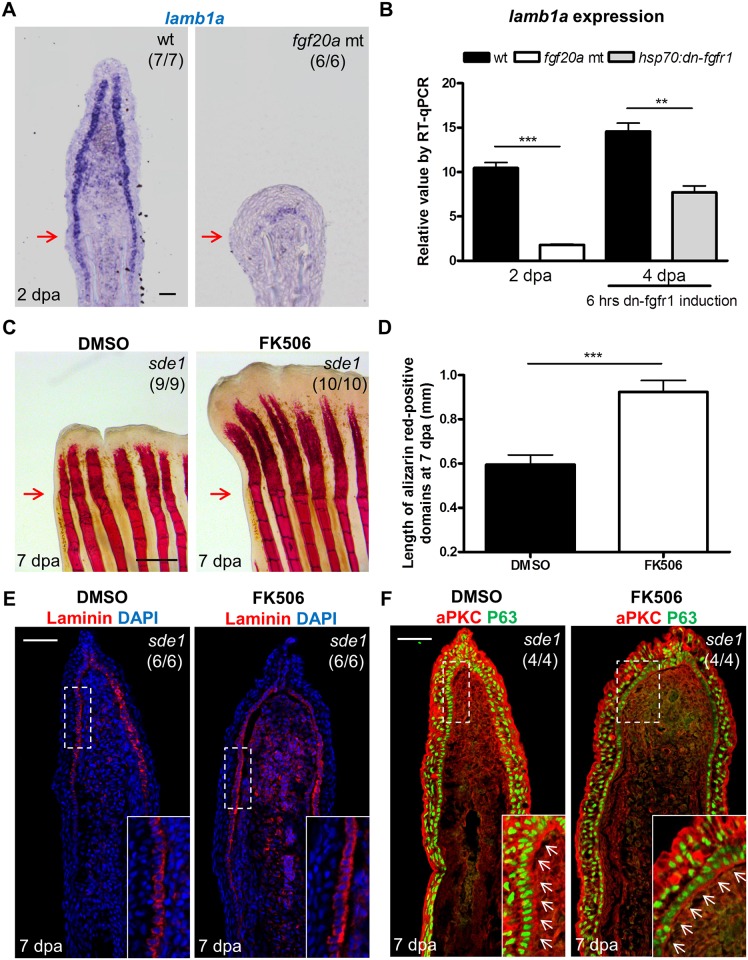
Like lamb1a, the Fgf ligand gene fgf20a is induced during zebrafish fin regeneration. Moreover, fgf20a mutants are also defective in lef1 induction and maturation of the regeneration epidermis [20]. Fgf signaling has been implicated in control of Laminin production in the context of embryoid bodies [47]. Because of these links, we investigated possible expression associations between lamb1a and fgf20a. We found that the induction of fgf20a was not affected in sde1 regenerates (S6A Fig). By contrast, lamb1a induction was severely disrupted in fgf20a mutants at 2 dpa (Fig 7A and 7B), at which point Laminin protein was detectable at very low levels along the epithelial-mesenchymal boundary (S6B Fig). To determine whether Fgf signaling actively sustains lamb1a expression during regeneration, we employed a transgenic line Tg(hsp70:dnfgfr1-EGFP) pd1 that drives a dominant negative Fgfr1 cassette from a heat-shock-inducible promoter [48]. A single heat-shock treatment at 4 dpa to transiently attenuate Fgf signaling during regeneration was sufficient to reduce lamb1a expression by 48% within 6 hours (Fig 7B), suggesting direct control of lamb1a at the transcriptional level by Fgf signaling. By contrast, 24 hours treatment from 3 to 4 dpa with either the Igf receptor antagonist NVP-AEW541 or the Igf signaling agonist NBI-31772 did not significant alter lamb1a expression (S6C Fig). Similarly, the induction of lamb1a was not affected in sde1 regenerates at 2 dpa at the restrictive temperature, as assayed by qPCR (S6D Fig). These results implicate fgf20a upstream of lamb1a in activation of morphogenesis of the regeneration epidermis.
Fig 7. Association of lamb1a expression and function with key regeneration effector pathways.
(A) Longitudinal sections of 2 dpa fin regenerates stained for lamb1a by ISH, indicating sparse expression in fgf20a mutants (dob). Scale bars, 50 μm. (B) RT-qPCR analysis indicating depleted levels of lamb1a RNA in fgf20a mutants (left). When Fgf signaling is blocked by induced expression of a dominant–negative Fgf receptor for just 6 hours at 4 dpa, lamb1a levels drop by nearly 50%. qPCR results were normalized to rpl13a and to the basal expression of lamb1a at 0 hpa. (n = 4; mean ± SEM; Student’s t -test, ***P < 0.001, **P < 0.01). (C) Whole-mount images of fin regenerates stained by alizarin red staining for calcium deposition, after treatment of sde1 animals with DMSO or FK506. Scale bars, 0.5 mm. (D) Measurement of the length of alizarin red-positive domains at 7 dpa (n = 9 and 10; mean ± SEM; Student’s t -test, ***P < 0.001). (E) Antibody staining for Laminin protein in vehicle- or FK506-treated sde1 fin regenerates at 7 dpa. Laminin, red; DAPI, blue. (F) Antibody co-staining for aPKC (red) and P63 (green) expression in vehicle or FK506-treated sde1 fin regenerates at 7 dpa. FK506 treatment partially rescued bone regeneration in sde1 mutants, with no detectable impact on Laminin localization or basal epithelial cell polarity. Scale bars, 50 μm. White dashed boxes indicate areas of enlarged view. White arrows indicate basal cell nuclei. Red arrows indicate plane of amputation.
To test whether bone growth programs potentially downstream of epithelial lamb1a function could rescue regeneration, we treated sde1 animals with the Calcineurin inhibitor FK506 during fin regeneration. A recent study reported that increases in Calcineurin activity reduce ray growth as regeneration progresses, a model supported by the finding that extended FK506 treatment causes gross lengthening of regenerated fin rays [27]. Interestingly, we found that FK506 treatment for 7 days increased the length of sde1 regenerates (from 39% of untreated sde1/+ regenerates at 7 dpa to 54%; S7A, S7B and S7C Fig), partially rescuing the length of regenerated bone (Fig 7C and 7D). By contrast, treatment of Smoothened Agonist (SAG) to activate the Hedgehog signaling pathway did not increase the lengths of sde1 regenerates (S7D and S7E Fig). Notably, FK506 treatment had little or no impact on mislocalized Laminin and aPKC in sde1 basal epithelial cells (Fig 7E and 7F). We were also unable to detect increased expression of shha in FK506-treated regenerates (S8 Fig). While partial rescue of length and bone formation occurred, FK506-treated sde1 regenerates were clearly dysmorphic compared to clutchmate control regenerates (S7A Fig). Although it is possible that FK506 treatment acts independently of lamb1a-mediated functions, our findings indicate that restoration of some mesenchymal compartment osteogenesis may occur in the presence of epithelial defects.
Discussion
Here, we carried out a forward genetic screen for zebrafish mutants defective in creation of a signaling-competent regeneration epidermis. The sde1 product is not required to repopulate genetically ablated osteoblasts or heal incision wounds, and appears largely dispensable in uninjured adult animals. However, after fin amputation, sde1 lesions disrupt epidermal maturation and signaling, and impair osteoblast patterning, and they remain inhibitory throughout the process of tissue replacement. Genetic analyses, including gene product assessment and complementation with known mutants, define sde1 as a temperature-sensitive allele of lamb1a, an ECM component that is transiently induced by injury in a subset of epithelial cells. Thus, Lamb1a is a key component of the regeneration epidermis with context-specific roles in appendage regeneration.
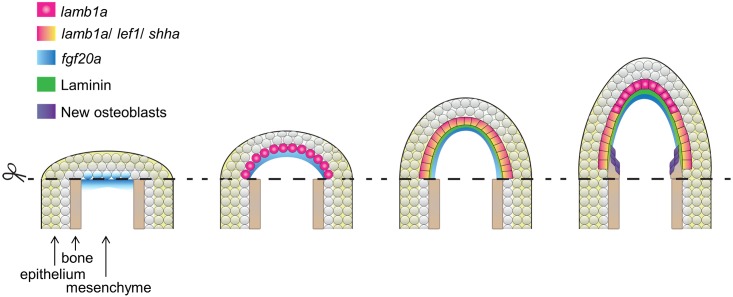
Our findings, combined with those of past studies, suggest a regulatory model for construction and maintenance of the regeneration epidermis (Fig 8). After an amputation injury, the wound closes within the next several hours by epithelial cell migration, ostensibly controlled by tension changes within the epithelial sheet. fgf20a is induced at the epithelial-mesenchymal boundary by 6 hours post-amputation. One direct or indirect function of fgf20a signaling is to induce expression of the lamb1a paralog in the adjacent epithelial cell layer, which establishes a basement membrane and polarizes the basal cell layer. This polarization is essential for positioning signaling receptors, such as phosphorylated Igf1r, and induction of factors involved in Hh signaling and Wnt/β-catenin signaling (via indirect influences [17, 49]) that guide osteoblast patterning and bone formation. During regenerative outgrowth, Fgf signaling retains lamb1a expression in the basal cell layer, maintaining polarization in basal epithelial cells and the competence to signal to osteoblasts. Our results with FK506 indicate that Lamb1a function is predominantly epithelial, as activation of the mesenchymal component is sufficient to promote bone regeneration to a certain degree even in the presence of a disrupted regeneration epidermis. Fgf ligand presence and Lamb1a expression wane after completion of regeneration [48].
Fig 8. Model for morphogenesis of the regeneration epidermis.
After initial wound closure, fgf20a is induced at the epithelial-mesenchymal boundary by 6 hours post-amputation. fgf20a signaling then contributes to induction of expression of the lamb1a paralog in the adjacent epithelial cell layer to establish a basement membrane and to polarize the basal cell layer. This polarization is essential for positioning signaling receptors and induction of morphogenetic factors that guide osteoblast patterning and bone formation.
ECM components have been implicated in tissue regeneration in several recent studies, including tadpole tail bud regeneration, zebrafish heart regeneration, and newt heart regeneration [50–52]. It is generally challenging to dissect their biological functions during regeneration, as Laminins and other ECM molecules are often required during embryogenesis. For example, mice with null mutations in Lama1, Lamb1, or Lamc1, and zebrafish with mutations in lamb1a, lama5, or several other ECM components [41, 42, 53–55], each die at embryonic stages. However, as we reveal here, the regulation of Laminin is also critical in the context of regenerating large regions of bone, in a way we expect is both structurally to create an epidermal scaffold and chemically as a source of signaling factors. While our data implicate fgf20a as an inducer of lamb1a during fin regeneration, there may exist other regulatory inputs for this and distinct ECM factors. For example, Nagendran and colleagues recently reported in a morpholino-based study that lama5 expression is under control of canonical Wnt signaling in the embryonic zebrafish fin epithelium [56]. Thus, it is possible that the regeneration epidermis is shaped by a network of growth factor-ECM regulatory interactions.
Interestingly, the estimated timing of teleost genome duplication precedes the dramatic rise of biological diversification in teleosts, according to paleontological evidence [38]. This is consistent with the hypothesis that gene duplication offers surplus genetic materials as origins of new biological functions. Recently, Rohner and colleagues reported that either of the two fibroblast growth factor receptor 1 paralogs (fgfr1a and fgfr1b) is sufficient for embryogenesis in zebrafish, whereas loss-of-function in both paralogs is lethal [57]. Only one of the paralogs, fgfr1a, is specifically expressed in the skin of 30 day-old juveniles, where its function is required for normal adult scale development in adult zebrafish and carp. Here, we found that lamb1a, but not lamb1b, is induced during fin regeneration, where it is essential. We speculate that maintenance of two lamb1 paralogs preserved viability in the setting of new mutations, permitting selection events that were favorable to adult regenerative potential. Comparison of the regulatory sequences of the lamb1 paralogs, and other sets of paralogs with similar divergent expression upon injury, could help decode genetic modifications that have preserved or dampened regenerative capacity in the evolution of vertebrate species.
Materials and Methods
Animals
Adult zebrafish 3–5 months of age were used for most experiments. Animal density was maintained at 3–4 per liter in all experiments. gup m189 and s804 mutants were used to test complementation [41, 42]. fgf20a mutants (dob), Tg(hsp70:dn-fgfr1) pd1, and Tg(osterix:mCherry-NTRo) pd46 fish were described previously [20, 35, 48]. sde1 mutants are referred to as lamb1a pd110. For NVP-AEW541and NBI-31772 treatments, 16 wild-type animals (each) with 3 dpa fin regenerates were treated for 24 hours at 25°C in fish water containing either 2 μM NVP-AEW541 (Cayman Chemical) or 10 μM NBI-31772 (Calbiochem, 479830-5MG). Stocks were prepared in DMSO (5 mM and 20 mM) and control animals were treated with 0.04% DMSO. For FK506 treatments, after amputation, 12–15 fish were maintained at 33°C in 1L fish water containing 0.1 μg/ml FK506 (Sigma, F4679-5MG). Animals were fed every other day followed by a water change with fresh drug. FK506 was dissolved in DMSO for a stock solution of 5 mg/ml and control animals were treated with 0.004% DMSO. For SAG treatment, 8–9 animals were treated with either DMSO (0.05%) or SAG (5 μM; Cayman Chemical, CAS 912545-86-9) for two hours at room temperature every other day as described [31]. For juvenile experiments, 4 wpf sde1 and sde1/+ animals were transferred from 25°C to 33°C for a two-week treatment. All animal procedures were performed in accordance with Duke University guidelines, under protocol #A100-12-04.
Mutagenesis and screen for fin regeneration mutants
EK or WIK strain males were mutagenized with ENU (3 mM) using published protocols (i.e. 1 hr/treatment/week for 4 weeks) [58] and mated to females of the WIK or EK strains to generate F1 families. A total of 108 F2 families and 423 F3 families were generated for identifying temperature-sensitive (TS) fin regeneration mutants. After fin amputation, F3 adult fish at 2–3 months of age were screened individually at 7 dpa at the restrictive temperature (33°C) for regeneration defects, using a dissecting microscope. Putative mutant families were out-crossed again to either EK or WIK strains to generate F4 and F5 families. A total of 9 mutants were subsequently selected from 25 putative F5 families for heritability, robustness of the defects, and survival rate at the restrictive temperature. For genetic mapping, homozygous mutant males of the F5 generation were crossed to F4 heterozygous females to generate pools of homozygous mutants and heterozygous siblings.
Exome sequencing and genetic mapping
Genomic DNA was isolated from pools of 64 homozygous mutants and 51 heterozygous siblings using the Puregene Core Kit from Qiagen. For library preparation, 3 μg genomic DNA was sheared to 150–250 bp fragments using a Covaris sonicator. The sheared DNA was assessed on an Agilent chip to verify the size range. Library construction and exome capture were performed as described [36], using an Agilent early access SureSelect XT Zebrafish exome kit. Sequencing was performed using an Illumina HiSeq2000 with 100 bp paired-end (PE) runs. For the mutant pool, 47,116,257 paired-end reads were collected. For the het sibling pool, 44,813,293 paired-end reads were collected.
Fastq files for each mutant and heterozygous pool were concatenated and compressed before being uploaded to SNPTrack (http://genetics.bwh.harvard.edu/snptrack/). The appropriate pool sizes (64 vs. 51) were entered before starting analysis. We used 4 data sources to generate our “SNP universe”. The first is the Ensembl Release 71 VCF file containing 1,352,592 SNPs. The second data source is the Megason "Universe" containing 16,075,952 SNPs in a VCF file. The third data source is 6 whole genomes of various Zebrafish strains (AB, TLF, TUB, TUG, WKB, WKG) sequenced by the Harris lab. The final source of SNPs is sequencing data generated in the Poss lab by exome sequencing of 12 zebrafish pools and one whole-genome sequencing pool.
Preliminary analysis by SNPTrack revealed a 6.2 Mb region on chromosome 25 with a high homozygosity score. We identified 67,365 SNPs in the region. After filtering against our SNP universe, only 2 novel non-synonymous SNPs were found at the locations 32605161 and 34060320. Within this region, the percent usable on-target bases was 42%, and the mean target coverage was 51X for the mutant pool, while the percent usable on-target bases was 41%, and the mean target coverage was 47X for the het sibling pool.
For genetic mapping, we designed primers using Primer 3 (http://bioinfo.ut.ee/primer3-0.4.0/) to genotype individual SNPs. A high Resolution Melting (HRM)-based assay was used for genotyping. The assay was performed using the Roche LightCycler 480 and LightCycler 480 High Resolution Melting Master (Cat. No: 04909631001), follow the manufacturer's instructions. The primer sequences for SNP 32559436, SNP 32605161, and SNP 32681121 were listed in the S1 Table. All progeny from sde1 x sde1/+ mapping crosses were raised to 2–3 months old at 25°C, before phenotyping for regeneration defects at 33°C at 7 dpa.
Histological assays
Whole-mount ISH with caudal fins was performed as described previously [22]. To generate digoxigenin-labeled probes for this study, we used lef1 and shha cDNA fragments [30], and a partial 1 kb lamb1a cDNA fragment as templates (see S1 Table). Immunohistochemistry on sectioned fins was performed as described [30], using antibodies against aPKC (Santa Cruz Biotechnology, C-20-sc-216, 1:200), P63 (Santa Cruz Biotechnology, 4A4, 1:200), Zns-5 (ZIRC, 1:200), phosphorylated-Igf1r (Santa Cruz Biotechnology, sc-101703, 1:100), and Laminin (Sigma, L9393, 1:200). For EdU incorporation assays, EdU solution (10 mM) was injected intraperitoneally 60 minutes prior to collection of fin regenerates, which were then fixed and processed as described [59]. Frozen blocks were sectioned at 16 μm, mounted using Vectashield with DAPI, and examined by laser confocal microscopy (Zeiss 700).
For whole-mount Alizarin red staining, 4% PFA-fixed fins were rehydrated in 50% ethanol for 30 minutes. Fin tissues were then incubated in a solution with 0.5% KOH and 0.01% alizarin red S (Sigma A5533) for overnight staining. Next, fin tissues were bleached for 20 minutes at room temperature in a freshly made solution containing 1.5% H2O2 and 1% KOH. After three washes with water, tissues were transferred to 80% glycerol for storage and imaging.
RT-qPCR
For each sample, fin tissues were collected from 4 individuals and homogenized in 1 ml Trizol (Invitrogen). cDNA was synthesized from 1 μg of RNA using Transcriptor First Strand cDNA Synthesis Kit (Cat. No: 04379012001). qPCR was performed using the Roche LightCycler 480 and LightCycler 480 Probes Master (Cat. No: 04887301001). Primer sequences and probes are listed in S1 Table. All samples were analyzed in biological and technical triplicate. Analysis was performed using the ΔΔCT method [60] against the level of ribosomal protein L13a (rpl13a) cDNA, which was found to be constant during fin regeneration.
Supporting Information
(A) sde1 regenerates (right) are severely shortened at 7 dpa, comparable to a 3 dpa clutchmate sample (left). Osteoblasts (green) line the bone in proximal areas but accumulate in masses in distal regions. Scale bars, 50 μm. Red arrows indicate plane of amputation. (B) Whole-mount images of sde1/+ and sde1 regenerates at 7, 14, 21, and 28 dpa. Images from the same animal are shown here across different time points. Red arrows and dashed lines indicate plane of amputation. Scale bars, 1 mm. (C) Measurement of fin regenerates at different time points (n = 14 vs. 11; Student’s t -test, ***P < 0.001).
(TIF)
Adult homozygous mutants (mt) and their heterozygous siblings (het) were incubated at the restrictive temperature (33°C) for two months. Animals were examined on a daily basis. sde1 (chc1) mutation has no apparent impact on adult animals over a two-month period (n = 18 each).
(TIF)
(A) shha RNA expression is reduced in sde1 regenerates at 4 dpa after a temperature shift from 25°C to 33°C at 3 dpa. (B) Similar results were observed in sde1 regenerates at 5 dpa after a temperature shift at 4 dpa. Scale bars, 100 μm. Representative images from different samples are shown here.
(TIF)
(A) Antibody co-staining for aPKC (red) and P63 (green) in longitudinal sections of sde1/+ and sde1 fin regenerates after 8 hours of 33°C treatment at 3 dpa, indicating rapid loss of basal cell polarity. Scale bars, 50 μm. (B) shha RNA expression is also reduced in sde1 regenerates after 8 hours of 33°C treatment (n = 5 each). Scale bars, 100 μm.
(TIF)
(A) Longitudinal sections of 5 dpa fin regenerates collected after 60 minutes of EdU incorporation. (B) Counting of EdU+ mesenchymal cells in distal fin ray area. Mesenchymal cell proliferation was grossly similar between sde1/+ and sde1 regenerates at 5 dpa after a temperature shift from 25°C to 33°C at 4 dpa. Scale bars, 50 μm. (n = 4, counts from three sections were averaged for each sample; Student’s t -test, NS, non-significant).
(TIF)
(A) RT-qPCR analysis for levels of fgf20a in sde1 fin regenerates. qPCR results were normalized to rpl13a and to the basal expression of fgf20a at 0 hpa. (n = 3; mean ± SEM; Student’s t -test, NS, non-significant). (B) Antibody co-staining for Laminin (red) and P63 (green) in longitudinal sections of wild-type and dob fin regenerates at 2 dpa. Scale bars, 50 μm. White arrows indicate where (low) levels of Laminin are detectable. Red arrows indicate plane of amputation. (C) RT-qPCR analysis lamb1a levels in 4 dpa wild-type fin regenerates after 24 hours of treatment with either Igf receptor antagonist NVP-AEW541 (2 μM), or Igf signaling agonist NBI-31772 (10 μM). (D) RT-qPCR analysis of lamb1a levels in 2 dpa sde1/+ and sde1 fin regenerates at the restrictive temperature. qPCR results were normalized to rpl13a and to the basal expression of lamb1a at 0 hpa. (n = 4; mean ± SEM; Student’s t -test, NS, non-significant).
(TIF)
(A) Whole-mount images of sde1/+ and sde1 fin regenerates at 7 dpa after treatment of DMSO (0.04%) or FK506 (0.1 μg/ml). Scale bars, 1 mm. (B) Measurement of the fin length at 7 dpa (Student’s t -test, ***P < 0.001). (C) Fold change of fin regenerates at 7 dpa. Relative values are normalized to the length of sde1/+ regenerates after DMSO treatment (mean ± SEM). (D) Whole-mount images of sde1/+ and sde1 fin regenerates at 6 dpa after treatment of DMSO (0.05%) or SAG (5 μM). (E) Measurement of the fin lengths at 6 dpa (Student’s t -test, NS, non-significant).
(TIF)
shha RNA expression was grossly similar between DMSO- and FK506-treated sde1 regenerates at 7 dpa. Consistent with S3 Fig, sde1 regenerates have a reduced level of shha expression. Scale bars, 100 μm. Representative images from different samples are shown here.
(TIF)
(DOCX)
Acknowledgments
We thank J. Burris, N. Blake, A. Dunlap, and S. Davies for zebrafish care; A. Dickson for artwork; Tatiana Hochgreb-Hägele and Didier Stainier for s804 zebrafish; Michel Bagnat for discussions; and Poss laboratory members for comments on the manuscript.
Data Availability
Exome sequencing data for sde1 mutants are accessible at http://genetics.bwh.harvard.edu/ggi/snptrack/ca9abb392566337de5785547bb796323415c1775/2/viewresults.html. All other relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Howard Hughes Medical Institute (hhmi.org), Early Career Scientist award to KDP; and the NIH (nih.gov), grant R01 GM074057 to KDP. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Johnson SL, Weston JA. Temperature-sensitive mutations that cause stage-specific defects in Zebrafish fin regeneration. Genetics. 1995;141(4):1583–95. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Becker T, Wullimann MF, Becker CG, Bernhardt RR, Schachner M. Axonal regrowth after spinal cord transection in adult zebrafish. J Comp Neurol. 1997;377(4):577–95. . [DOI] [PubMed] [Google Scholar]
- 3. Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 2000;44(3):289–307. . [DOI] [PubMed] [Google Scholar]
- 4. Diep CQ, Ma D, Deo RC, Holm TM, Naylor RW, Arora N, et al. Identification of adult nephron progenitors capable of kidney regeneration in zebrafish. Nature. 2011;470(7332):95–100. 10.1038/nature09669 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goldshmit Y, Sztal TE, Jusuf PR, Hall TE, Nguyen-Chi M, Currie PD. Fgf-dependent glial cell bridges facilitate spinal cord regeneration in zebrafish. J Neurosci. 2012;32(22):7477–92. 10.1523/JNEUROSCI.0758-12.2012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kizil C, Kyritsis N, Dudczig S, Kroehne V, Freudenreich D, Kaslin J, et al. Regenerative neurogenesis from neural progenitor cells requires injury-induced expression of Gata3. Dev Cell. 2012;23(6):1230–7. 10.1016/j.devcel.2012.10.014 . [DOI] [PubMed] [Google Scholar]
- 7. Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298(5601):2188–90. 10.1126/science.1077857 . [DOI] [PubMed] [Google Scholar]
- 8. Poss KD. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet. 2010;11(10):710–22. 10.1038/nrg2879 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poleo G, Brown CW, Laforest L, Akimenko MA. Cell proliferation and movement during early fin regeneration in zebrafish. Dev Dyn. 2001;221(4):380–90. 10.1002/dvdy.1152 . [DOI] [PubMed] [Google Scholar]
- 10. Santos-Ruiz L, Santamaria JA, Ruiz-Sanchez J, Becerra J. Cell proliferation during blastema formation in the regenerating teleost fin. Dev Dyn. 2002;223(2):262–72. 10.1002/dvdy.10055 . [DOI] [PubMed] [Google Scholar]
- 11. Mescher AL. Effects on adult newt limb regeneration of partial and complete skin flaps over the amputation surface. J Exp Zool. 1976;195(1):117–28. 10.1002/jez.1401950111 . [DOI] [PubMed] [Google Scholar]
- 12. Thornton CS. The effect of apical cap removal on limb regeneration in Amblystoma larvae. J Exp Zool. 1957;134(2):357–81. . [DOI] [PubMed] [Google Scholar]
- 13. Tassava RA, Garling DJ. Regenerative responses in larval axolotl limbs with skin grafts over the amputation surface. J Exp Zool. 1979;208(1):97–110. 10.1002/jez.1402080111 . [DOI] [PubMed] [Google Scholar]
- 14. Yoshinari N, Kawakami A. Mature and juvenile tissue models of regeneration in small fish species. Biol Bull. 2011;221(1):62–78. . [DOI] [PubMed] [Google Scholar]
- 15. Quint E, Smith A, Avaron F, Laforest L, Miles J, Gaffield W, et al. Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc Natl Acad Sci U S A. 2002;99(13):8713–8. 10.1073/pnas.122571799 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stewart S, Gomez AW, Armstrong BE, Henner A, Stankunas K. Sequential and opposing activities of Wnt and BMP coordinate zebrafish bone regeneration. Cell Rep. 2014;6(3):482–98. 10.1016/j.celrep.2014.01.010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wehner D, Cizelsky W, Vasudevaro MD, Ozhan G, Haase C, Kagermeier-Schenk B, et al. Wnt/beta-catenin signaling defines organizing centers that orchestrate growth and differentiation of the regenerating zebrafish caudal fin. Cell Rep. 2014;6(3):467–81. 10.1016/j.celrep.2013.12.036 . [DOI] [PubMed] [Google Scholar]
- 18. Stoick-Cooper CL, Weidinger G, Riehle KJ, Hubbert C, Major MB, Fausto N, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134(3):479–89. 10.1242/dev.001123 . [DOI] [PubMed] [Google Scholar]
- 19. Kawakami Y, Rodriguez Esteban C, Raya M, Kawakami H, Marti M, Dubova I, et al. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20(23):3232–7. 10.1101/gad.1475106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310(5756):1957–60. 10.1126/science.1117637 . [DOI] [PubMed] [Google Scholar]
- 21. Chablais F, Jazwinska A. IGF signaling between blastema and wound epidermis is required for fin regeneration. Development. 2010;137(6):871–9. 10.1242/dev.043885 . [DOI] [PubMed] [Google Scholar]
- 22. Poss KD, Shen J, Nechiporuk A, McMahon G, Thisse B, Thisse C, et al. Roles for Fgf signaling during zebrafish fin regeneration. Dev Biol. 2000;222(2):347–58. 10.1006/dbio.2000.9722 . [DOI] [PubMed] [Google Scholar]
- 23. Jazwinska A, Badakov R, Keating MT. Activin-betaA signaling is required for zebrafish fin regeneration. Curr Biol. 2007;17(16):1390–5. 10.1016/j.cub.2007.07.019 . [DOI] [PubMed] [Google Scholar]
- 24. Grotek B, Wehner D, Weidinger G. Notch signaling coordinates cellular proliferation with differentiation during zebrafish fin regeneration. Development. 2013;140(7):1412–23. 10.1242/dev.087452 . [DOI] [PubMed] [Google Scholar]
- 25. Munch J, Gonzalez-Rajal A, de la Pompa JL. Notch regulates blastema proliferation and prevents differentiation during adult zebrafish fin regeneration. Development. 2013;140(7):1402–11. 10.1242/dev.087346 . [DOI] [PubMed] [Google Scholar]
- 26. Blum N, Begemann G. Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration. Development. 2012;139(1):107–16. 10.1242/dev.065391 . [DOI] [PubMed] [Google Scholar]
- 27. Kujawski S, Lin W, Kitte F, Bormel M, Fuchs S, Arulmozhivarman G, et al. Calcineurin regulates coordinated outgrowth of zebrafish regenerating fins. Dev Cell. 2014;28(5):573–87. 10.1016/j.devcel.2014.01.019 . [DOI] [PubMed] [Google Scholar]
- 28. Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129(22):5141–9. . [DOI] [PubMed] [Google Scholar]
- 29. Streisinger G, Singer F, Walker C, Knauber D, Dower N. Segregation analyses and gene-centromere distances in zebrafish. Genetics. 1986;112(2):311–9. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Poss KD, Shen J, Keating MT. Induction of lef1 during zebrafish fin regeneration. Dev Dyn. 2000;219(2):282–6. . [DOI] [PubMed] [Google Scholar]
- 31. Lee Y, Hami D, De Val S, Kagermeier-Schenk B, Wills AA, Black BL, et al. Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev Biol. 2009;331(2):270–80. 10.1016/j.ydbio.2009.05.545 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nechiporuk A, Poss KD, Johnson SL, Keating MT. Positional cloning of a temperature-sensitive mutant emmental reveals a role for sly1 during cell proliferation in zebrafish fin regeneration. Dev Biol. 2003;258(2):291–306. . [DOI] [PubMed] [Google Scholar]
- 33. Makino S, Whitehead GG, Lien CL, Kim S, Jhawar P, Kono A, et al. Heat-shock protein 60 is required for blastema formation and maintenance during regeneration. Proc Natl Acad Sci U S A. 2005;102(41):14599–604. 10.1073/pnas.0507408102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wills AA, Kidd AR 3rd, Lepilina A, Poss KD. Fgfs control homeostatic regeneration in adult zebrafish fins. Development. 2008;135(18):3063–70. 10.1242/dev.024588 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell. 2012;22(4):879–86. 10.1016/j.devcel.2012.03.006 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ryan S, Willer J, Marjoram L, Bagwell J, Mankiewicz J, Leshchiner I, et al. Rapid identification of kidney cyst mutations by whole exome sequencing in zebrafish. Development. 2013;140(21):4445–51. 10.1242/dev.101170 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leshchiner I, Alexa K, Kelsey P, Adzhubei I, Austin-Tse CA, Cooney JD, et al. Mutation mapping and identification by whole-genome sequencing. Genome Res. 2012;22(8):1541–8. 10.1101/gr.135541.111 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays. 2005;27(9):937–45. 10.1002/bies.20293 . [DOI] [PubMed] [Google Scholar]
- 39. Yurchenco PD, Cheng YS. Self-assembly and calcium-binding sites in laminin. A three-arm interaction model. J Biol Chem. 1993;268(23):17286–99. . [PubMed] [Google Scholar]
- 40. Odenthal U, Haehn S, Tunggal P, Merkl B, Schomburg D, Frie C, et al. Molecular analysis of laminin N-terminal domains mediating self-interactions. J Biol Chem. 2004;279(43):44504–12. 10.1074/jbc.M402455200 . [DOI] [PubMed] [Google Scholar]
- 41. Parsons MJ, Pollard SM, Saude L, Feldman B, Coutinho P, Hirst EM, et al. Zebrafish mutants identify an essential role for laminins in notochord formation. Development. 2002;129(13):3137–46. . [DOI] [PubMed] [Google Scholar]
- 42. Hochgreb-Hagele T, Yin C, Koo DE, Bronner ME, Stainier DY. Laminin beta1a controls distinct steps during the establishment of digestive organ laterality. Development. 2013;140(13):2734–45. 10.1242/dev.097618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McCaffrey LM, Macara IG. Signaling pathways in cell polarity. Cold Spring Harb Perspect Biol. 2012;4(6). 10.1101/cshperspect.a009654 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. O'Brien LE, Jou TS, Pollack AL, Zhang Q, Hansen SH, Yurchenco P, et al. Rac1 orientates epithelial apical polarity through effects on basolateral laminin assembly. Nat Cell Biol. 2001;3(9):831–8. 10.1038/ncb0901-831 . [DOI] [PubMed] [Google Scholar]
- 45. Klein G, Langegger M, Timpl R, Ekblom P. Role of laminin A chain in the development of epithelial cell polarity. Cell. 1988;55(2):331–41. . [DOI] [PubMed] [Google Scholar]
- 46. Rasmussen JP, Reddy SS, Priess JR. Laminin is required to orient epithelial polarity in the C. elegans pharynx. Development. 2012;139(11):2050–60. 10.1242/dev.078360 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li X, Chen Y, Scheele S, Arman E, Haffner-Krausz R, Ekblom P, et al. Fibroblast growth factor signaling and basement membrane assembly are connected during epithelial morphogenesis of the embryoid body. J Cell Biol. 2001;153(4):811–22. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee Y, Grill S, Sanchez A, Murphy-Ryan M, Poss KD. Fgf signaling instructs position-dependent growth rate during zebrafish fin regeneration. Development. 2005;132(23):5173–83. 10.1242/dev.02101 . [DOI] [PubMed] [Google Scholar]
- 49. Wehner D, Weidinger G. Signaling networks organizing regenerative growth of the zebrafish fin. Trends Genet. 2015. 10.1016/j.tig.2015.03.012 . [DOI] [PubMed] [Google Scholar]
- 50. Contreras EG, Gaete M, Sanchez N, Carrasco H, Larrain J. Early requirement of Hyaluronan for tail regeneration in Xenopus tadpoles. Development. 2009;136(17):2987–96. 10.1242/dev.035501 . [DOI] [PubMed] [Google Scholar]
- 51. Mercer SE, Odelberg SJ, Simon HG. A dynamic spatiotemporal extracellular matrix facilitates epicardial-mediated vertebrate heart regeneration. Dev Biol. 2013;382(2):457–69. 10.1016/j.ydbio.2013.08.002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang J, Karra R, Dickson AL, Poss KD. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev Biol. 2013;382(2):427–35. 10.1016/j.ydbio.2013.08.012 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Webb AE, Sanderford J, Frank D, Talbot WS, Driever W, Kimelman D. Laminin alpha5 is essential for the formation of the zebrafish fins. Dev Biol. 2007;311(2):369–82. 10.1016/j.ydbio.2007.08.034 . [DOI] [PubMed] [Google Scholar]
- 54. Carney TJ, Feitosa NM, Sonntag C, Slanchev K, Kluger J, Kiyozumi D, et al. Genetic analysis of fin development in zebrafish identifies furin and hemicentin1 as potential novel fraser syndrome disease genes. PLoS Genet. 2010;6(4):e1000907 10.1371/journal.pgen.1000907 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Domogatskaya A, Rodin S, Tryggvason K. Functional diversity of laminins. Annu Rev Cell Dev Biol. 2012;28:523–53. 10.1146/annurev-cellbio-101011-155750 . [DOI] [PubMed] [Google Scholar]
- 56. Nagendran M, Arora P, Gori P, Mulay A, Ray S, Jacob T, et al. Canonical Wnt signalling regulates epithelial patterning by modulating levels of laminins in zebrafish appendages. Development. 2014. 10.1242/dev.118703 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rohner N, Bercsenyi M, Orban L, Kolanczyk ME, Linke D, Brand M, et al. Duplication of fgfr1 permits Fgf signaling to serve as a target for selection during domestication. Curr Biol. 2009;19(19):1642–7. 10.1016/j.cub.2009.07.065 [DOI] [PubMed] [Google Scholar]
- 58. Trevarrow B. Techniques for optimizing the creation of mutations in zebrafish using N-ethyl-N-nitrosourea. Lab Anim (NY). 2011;40(11):353–61. 10.1038/laban1111-353 . [DOI] [PubMed] [Google Scholar]
- 59. Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci U S A. 2008;105(7):2415–20. 10.1073/pnas.0712168105 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) sde1 regenerates (right) are severely shortened at 7 dpa, comparable to a 3 dpa clutchmate sample (left). Osteoblasts (green) line the bone in proximal areas but accumulate in masses in distal regions. Scale bars, 50 μm. Red arrows indicate plane of amputation. (B) Whole-mount images of sde1/+ and sde1 regenerates at 7, 14, 21, and 28 dpa. Images from the same animal are shown here across different time points. Red arrows and dashed lines indicate plane of amputation. Scale bars, 1 mm. (C) Measurement of fin regenerates at different time points (n = 14 vs. 11; Student’s t -test, ***P < 0.001).
(TIF)
Adult homozygous mutants (mt) and their heterozygous siblings (het) were incubated at the restrictive temperature (33°C) for two months. Animals were examined on a daily basis. sde1 (chc1) mutation has no apparent impact on adult animals over a two-month period (n = 18 each).
(TIF)
(A) shha RNA expression is reduced in sde1 regenerates at 4 dpa after a temperature shift from 25°C to 33°C at 3 dpa. (B) Similar results were observed in sde1 regenerates at 5 dpa after a temperature shift at 4 dpa. Scale bars, 100 μm. Representative images from different samples are shown here.
(TIF)
(A) Antibody co-staining for aPKC (red) and P63 (green) in longitudinal sections of sde1/+ and sde1 fin regenerates after 8 hours of 33°C treatment at 3 dpa, indicating rapid loss of basal cell polarity. Scale bars, 50 μm. (B) shha RNA expression is also reduced in sde1 regenerates after 8 hours of 33°C treatment (n = 5 each). Scale bars, 100 μm.
(TIF)
(A) Longitudinal sections of 5 dpa fin regenerates collected after 60 minutes of EdU incorporation. (B) Counting of EdU+ mesenchymal cells in distal fin ray area. Mesenchymal cell proliferation was grossly similar between sde1/+ and sde1 regenerates at 5 dpa after a temperature shift from 25°C to 33°C at 4 dpa. Scale bars, 50 μm. (n = 4, counts from three sections were averaged for each sample; Student’s t -test, NS, non-significant).
(TIF)
(A) RT-qPCR analysis for levels of fgf20a in sde1 fin regenerates. qPCR results were normalized to rpl13a and to the basal expression of fgf20a at 0 hpa. (n = 3; mean ± SEM; Student’s t -test, NS, non-significant). (B) Antibody co-staining for Laminin (red) and P63 (green) in longitudinal sections of wild-type and dob fin regenerates at 2 dpa. Scale bars, 50 μm. White arrows indicate where (low) levels of Laminin are detectable. Red arrows indicate plane of amputation. (C) RT-qPCR analysis lamb1a levels in 4 dpa wild-type fin regenerates after 24 hours of treatment with either Igf receptor antagonist NVP-AEW541 (2 μM), or Igf signaling agonist NBI-31772 (10 μM). (D) RT-qPCR analysis of lamb1a levels in 2 dpa sde1/+ and sde1 fin regenerates at the restrictive temperature. qPCR results were normalized to rpl13a and to the basal expression of lamb1a at 0 hpa. (n = 4; mean ± SEM; Student’s t -test, NS, non-significant).
(TIF)
(A) Whole-mount images of sde1/+ and sde1 fin regenerates at 7 dpa after treatment of DMSO (0.04%) or FK506 (0.1 μg/ml). Scale bars, 1 mm. (B) Measurement of the fin length at 7 dpa (Student’s t -test, ***P < 0.001). (C) Fold change of fin regenerates at 7 dpa. Relative values are normalized to the length of sde1/+ regenerates after DMSO treatment (mean ± SEM). (D) Whole-mount images of sde1/+ and sde1 fin regenerates at 6 dpa after treatment of DMSO (0.05%) or SAG (5 μM). (E) Measurement of the fin lengths at 6 dpa (Student’s t -test, NS, non-significant).
(TIF)
shha RNA expression was grossly similar between DMSO- and FK506-treated sde1 regenerates at 7 dpa. Consistent with S3 Fig, sde1 regenerates have a reduced level of shha expression. Scale bars, 100 μm. Representative images from different samples are shown here.
(TIF)
(DOCX)
Data Availability Statement
Exome sequencing data for sde1 mutants are accessible at http://genetics.bwh.harvard.edu/ggi/snptrack/ca9abb392566337de5785547bb796323415c1775/2/viewresults.html. All other relevant data are within the paper and its Supporting Information files.