Abstract
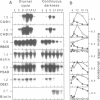
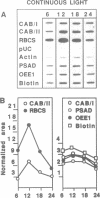
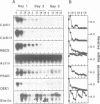
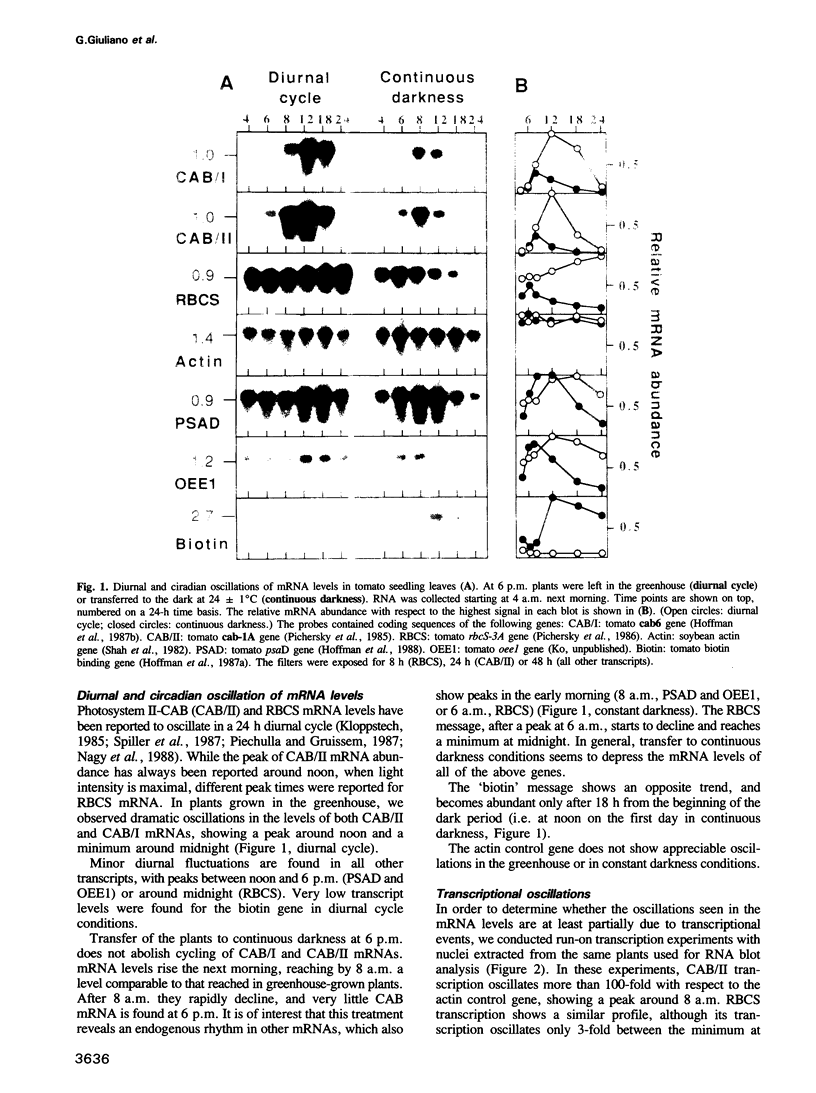
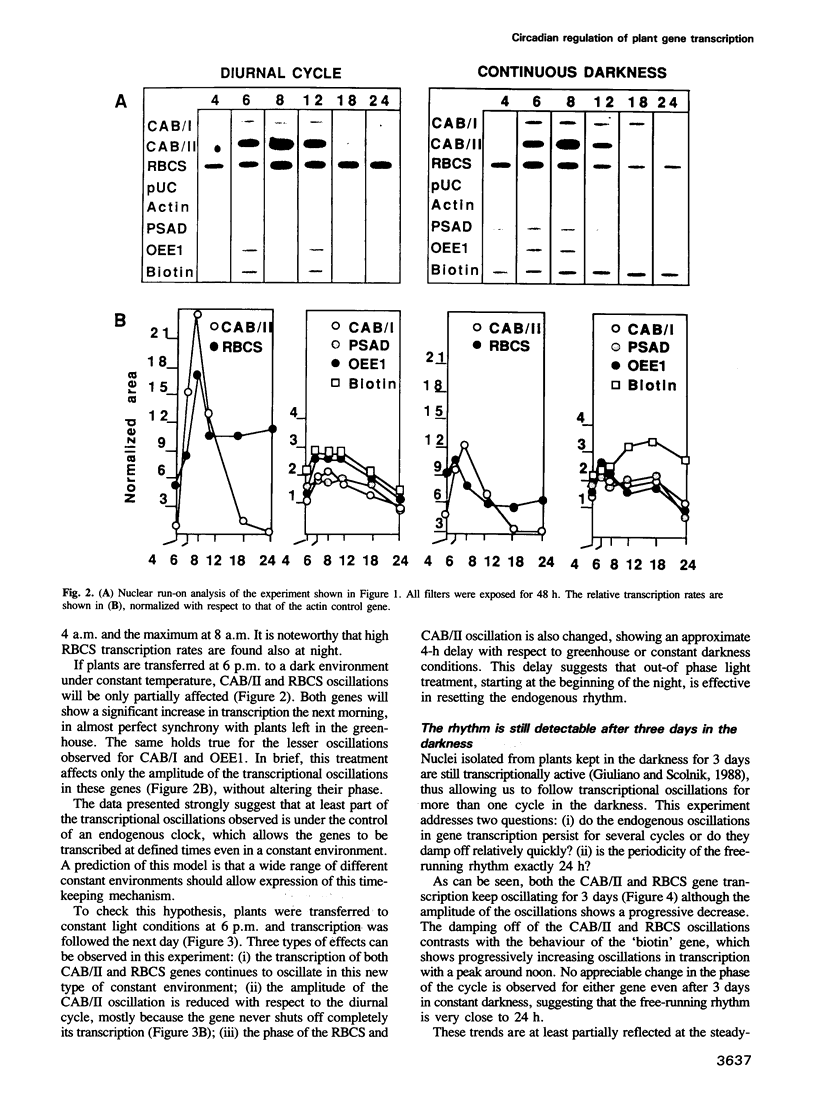
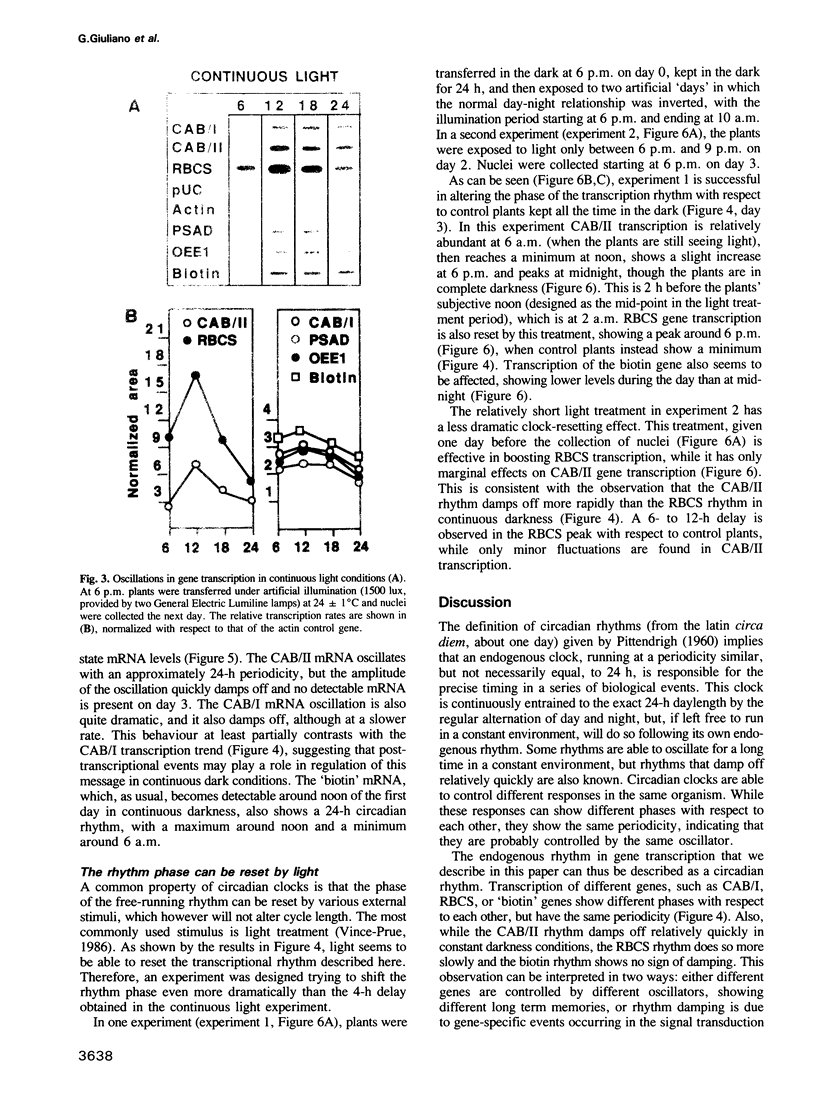
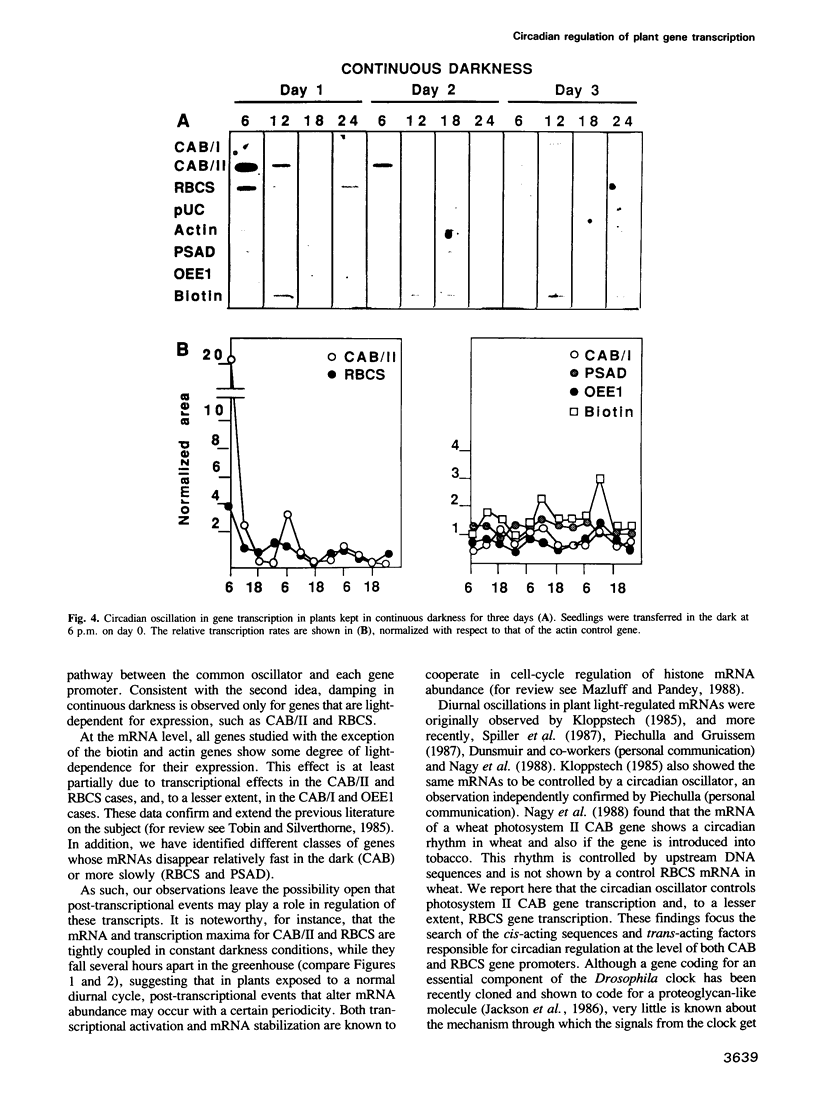
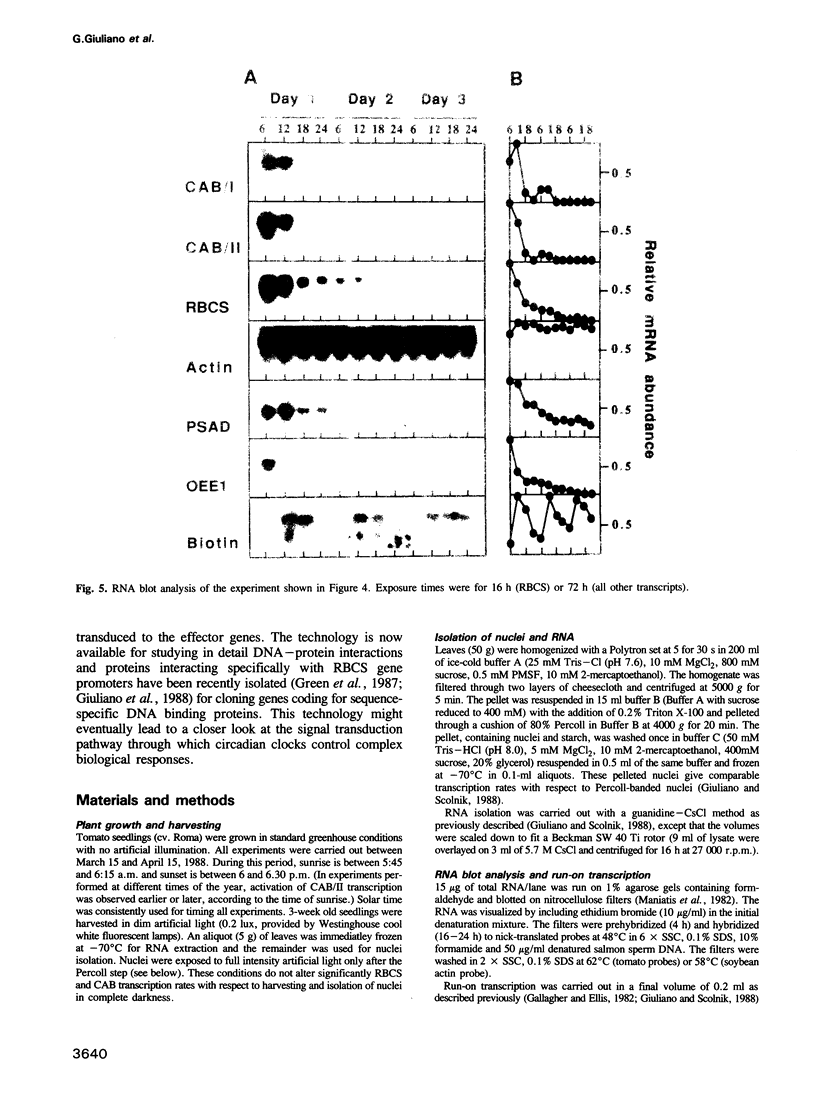
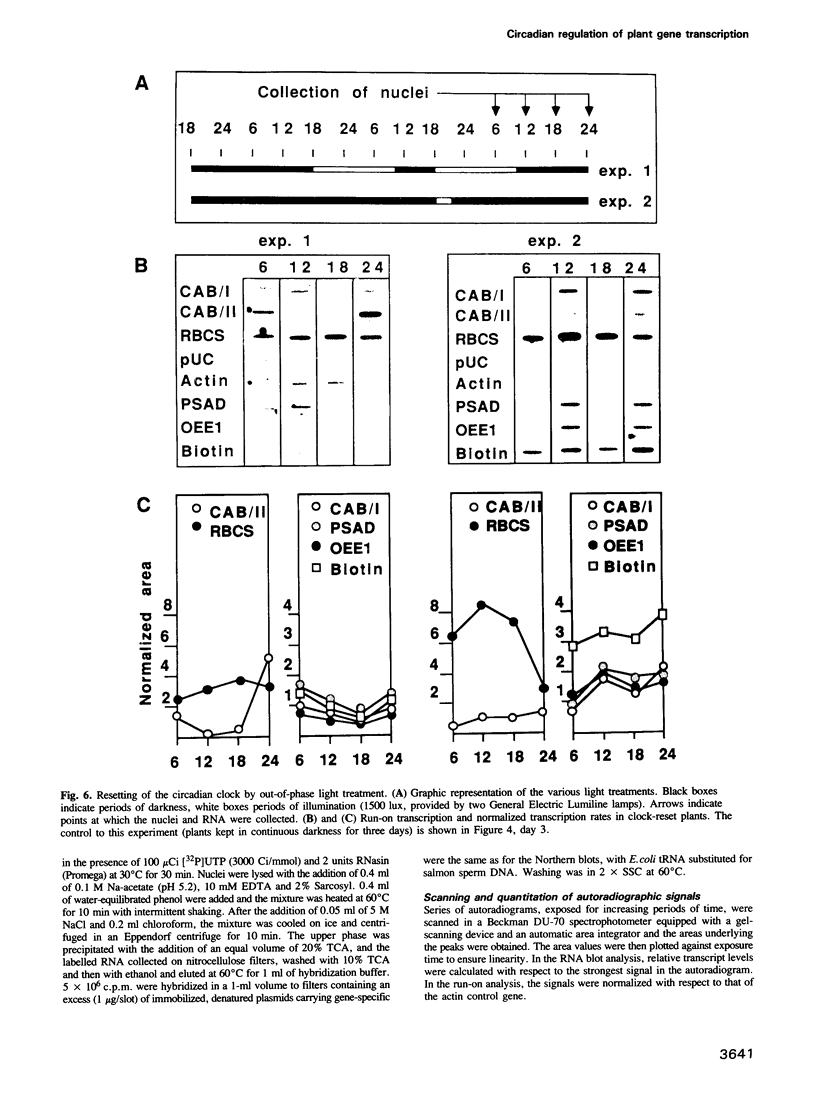
Diurnal oscillations in steady-state mRNA levels and transcription rates were measured for seven transcripts (five of which encode chloroplast-localized proteins) in tomato seedlings: photosystem I and photosystem II chlorophyll a/b binding proteins (CAB/I and CAB/II), small subunit of RuBisCO (RBCS), actin, subunit II of the photosystem I reaction center (PSAD), subunit I of the photosystem II oxygen-evolving enzyme (OEE1), and a biotin-binding protein of unknown function. CAB/II mRNA levels were found to oscillate greater than 20-fold, showing a peak at noon, while only marginal diurnal oscillations are seen in RBCS transcripts. The oscillations are at least partially controlled at the transcriptional level. Transcription rates of both CAB/II and RBCS, measured by nuclear run-on experiments, were found to oscillate, with a peak around 8 a.m. Transcription rates of the 'biotin' clone also oscillated, with a peak around noon. Transfer of plants to constant darkness or constant light conditions alters the amplitude of the transcriptional oscillation, but does not abolish it, suggesting that it is at least partially controlled by a circadian clock. The oscillations are still visible after three days in complete darkness, and have a period very close to 24 h. The oscillator phase can be reset by out-of-phase light treatment.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ehret C. F., Trucco E. Molecular models for the circadian clock. I. The chronon concept. J Theor Biol. 1967 May;15(2):240–262. doi: 10.1016/0022-5193(67)90206-8. [DOI] [PubMed] [Google Scholar]
- Gallagher T. F., Ellis R. J. Light-stimulated transcription of genes for two chloroplast polypeptides in isolated pea leaf nuclei. EMBO J. 1982;1(12):1493–1498. doi: 10.1002/j.1460-2075.1982.tb01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G., Pichersky E., Malik V. S., Timko M. P., Scolnik P. A., Cashmore A. R. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7089–7093. doi: 10.1073/pnas.85.19.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G., Scolnik P. A. Transcription of Two Photosynthesis-Associated Nuclear Gene Families Correlates with the Presence of Chloroplasts in Leaves of the Variegated Tomato ghost Mutant. Plant Physiol. 1988 Jan;86(1):7–9. doi: 10.1104/pp.86.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P. J., Kay S. A., Chua N. H. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 1987 Sep;6(9):2543–2549. doi: 10.1002/j.1460-2075.1987.tb02542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman N. E., Pichersky E., Cashmore A. R. A tomato cDNA encoding a biotin-binding protein. Nucleic Acids Res. 1987 May 11;15(9):3928–3928. doi: 10.1093/nar/15.9.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman N. E., Pichersky E., Malik V. S., Castresana C., Ko K., Darr S. C., Cashmore A. R. A cDNA clone encoding a photosystem I protein with homology to photosystem II chlorophyll a/b-binding polypeptides. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8844–8848. doi: 10.1073/pnas.84.24.8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson F. R., Bargiello T. A., Yun S. H., Young M. W. Product of per locus of Drosophila shares homology with proteoglycans. Nature. 1986 Mar 13;320(6058):185–188. doi: 10.1038/320185a0. [DOI] [PubMed] [Google Scholar]
- Marzluff W. F., Pandey N. B. Multiple regulatory steps control histone mRNA concentrations. Trends Biochem Sci. 1988 Feb;13(2):49–52. doi: 10.1016/0968-0004(88)90027-8. [DOI] [PubMed] [Google Scholar]
- PITTENDRIGH C. S. Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol. 1960;25:159–184. doi: 10.1101/sqb.1960.025.01.015. [DOI] [PubMed] [Google Scholar]
- Pichersky E., Bernatzky R., Tanksley S. D., Breidenbach R. B., Kausch A. P., Cashmore A. R. Molecular characterization and genetic mapping of two clusters of genes encoding chlorophyll a/b-binding proteins in Lycopersicon esculentum (tomato). Gene. 1985;40(2-3):247–258. doi: 10.1016/0378-1119(85)90047-2. [DOI] [PubMed] [Google Scholar]
- Pichersky E., Bernatzky R., Tanksley S. D., Cashmore A. R. Evidence for selection as a mechanism in the concerted evolution of Lycopersicon esculentum (tomato) genes encoding the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3880–3884. doi: 10.1073/pnas.83.11.3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechulla B., Gruissem W. Diurnal mRNA fluctuations of nuclear and plastid genes in developing tomato fruits. EMBO J. 1987 Dec 1;6(12):3593–3599. doi: 10.1002/j.1460-2075.1987.tb02690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D. M., Hightower R. C., Meagher R. B. Complete nucleotide sequence of a soybean actin gene. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1022–1026. doi: 10.1073/pnas.79.4.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller S. C., Kaufman L. S., Thompson W. F., Briggs W. R. Specific mRNA and rRNA Levels in Greening Pea Leaves during Recovery from Iron Stress. Plant Physiol. 1987 Jun;84(2):409–414. doi: 10.1104/pp.84.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]