Abstract
Organotypic 3D cultures of epithelial cells are grown at the air–liquid interface (ALI) and resemble the in vivo counterparts. Although the complexity of in vivo cellular responses could be better manifested in coculture models in which additional cell types such as fibroblasts were incorporated, the presence of another cell type could mask the response of the other. This study reports the impact of whole cigarette smoke (CS) exposure on organotypic mono- and coculture models to evaluate the relevancy of organotypic models for toxicological assessment of aerosols. Two organotypic bronchial models were directly exposed to low and high concentrations of CS of the reference research cigarette 3R4F: monoculture of bronchial epithelial cells without fibroblasts (BR) and coculture with fibroblasts (BRF) models. Adenylate kinase (AK)-based cytotoxicity, cytochrome P450 (CYP) 1A1/1B1 activity, tissue histology, and concentrations of secreted mediators into the basolateral media, as well as transcriptomes were evaluated following the CS exposure. The results demonstrated similar impact of CS on the AK-based cytotoxicity, CYP1A1/1B1 activity, and tissue histology in both models. However, a greater number of secreted mediators was identified in the basolateral media of the monoculture than in the coculture models. Furthermore, annotation analysis and network-based systems biology analysis of the transcriptomic profiles indicated a more prominent cellular stress and tissue damage following CS in the monoculture epithelium model without fibroblasts. Finally, our results indicated that an in vivo smoking-induced xenobiotic metabolism response of bronchial epithelial cells was better reflected from the in vitro CS-exposed coculture model.
Keywords: coculture, air–liquid interface, aerosol, transcriptomic, biological network
Airway epithelial cells are the first point of contact for inhaled agents including cigarette smoke (CS) (Proud and Leigh, 2011). They play a role in defense against airborne pathogens and pollutants (Proud and Leigh, 2011). Growing concern about the toxicity of environmental pollutants has prompted research to investigate mechanisms through which lung injury and repair occur after exposure (BéruBé et al., 2009). In a clinical setting, bronchial brush biopsy samples are frequently used to evaluate changes in pulmonary cells to diagnose respiratory diseases including cancers (NCI, 2014). In a laboratory setting, alternative in vitro test systems have been developed to facilitate inhalation toxicity studies—while minimizing the use of animals—and have become increasingly commercially available (eg, the organotypic 3D airway tissue models EpiAirway (MatTek Corporation, Ashland, Massachusetts) and MucilAir (Epithelix Sàrl, Geneva, Switzerland)) (BéruBé et al., 2009).
In such organotypic culture systems, bronchial epithelial cells are cultured at the air–liquid interface (ALI) and capable of differentiating to form a pseudostratified cell layer containing mucus-secreting goblet cells and ciliated columnar cells (BéruBé et al., 2009). The ALI allows a direct administration of an aerosol onto the apical surface, a situation resembling aerosols exposure of the in vivo respiratory system (BéruBé et al., 2009). Moreover, the epithelium is nourished by a culture medium from the basolateral surface (BéruBé et al., 2009).
Various organotypic epithelium models are available, including mono- and coculture models (Duell et al., 2011). Functionality of epithelial cells can be influenced by their spatiotemporal context such as interactions with neighboring cells and the environment (Fuchs and Watt, 2003). Growing epithelial cells on the top of a fibroblast-seeded collagen layer (eg, in coculture models) increases the differentiation and apoptosis of the basal cells, thus improving the epithelium functionality (BéruBé et al., 2009; Costea et al., 2003). In contrast, without fibroblasts (as in monoculture models), the thickness of the epithelium relies on the proliferation of epithelial cells (Costea et al., 2003).
Moreover, the contribution of fibroblasts to the epithelium functionality can be attributed to the fibroblast-secreted mediators (Costea et al., 2003). Similarly, epithelial cells can also influence the epithelium functionality because they can secrete cytokines and growth factors upon stimulation (Proud and Leigh, 2011). For example, following infection of human rhinovirus (HRV)—a major trigger of respiratory diseases—infected epithelial cells secrete various mediators (eg, vascular endothelial growth factor [VEGF], fibroblast growth factor, amphiregulin, activin A, matrix metalloproteinase [MMP]-9, and tissue inhibitor of metalloproteinase [TIMP]-1) (Proud and Leigh, 2011). Thus, examining the specific epithelial-secreted mediators may increase the understanding of the pro-inflammatory mechanisms elicited by airway epithelial cells in response to insults (eg, environmental airborne and CS exposures). However, the role of the different cell types in the lung epithelia and underlying connective tissues, to secrete mediators has not been fully investigated. In addition, CS exposure can alter the epithelial cell responses by increasing the levels of HRV-induced interleukin (IL)-8 and suppressing the levels of HRV-induced interferon γ-inducible protein (IP)-10 (Hudy et al., 2010). Because measuring mediators in the basolateral media of organotypic epithelium models can be easily done, this approach can be included during the assessment of exposure impact. For example, cell-type specific mediators can be identified by comparing the profiles of secreted medators from various monoculture models that were simultaneously exposed to a stimulus.
In this study, we report the whole CS exposure impact on 2 human organotypic bronchial epithelium models: a monoculture model of bronchial epithelial cells (BR) and a coculture model of bronchial epithelial cells and fibroblasts (BRF). We evaluated the tissue responses to CS exposure using well known in vitro functional assays, such as AK-based cytotoxicity and cytochrome P450s (CYP) activity assays. We measured the CYP1A1 and CYP1B1 activity because they are reported to be the major phase I xenobiotic enzymes induced by CS exposure (Anttila et al., 2011; Port et al., 2004). Moreover, the tissue histology and concentrations of mediators in the basolateral media following CS exposure were analyzed and compared among the different models. Finally, a more comprehensive evaluation of the CS impact was conducted by leveraging the transcriptomics data from the BR mono- and BRF coculture models. We identified the underlying biological processes/mechanisms associated with CS exposure using a gene annotation analysis and a network-based systems biology approach. The latter approach utilizes our published collections of hierarchically structured biological network models (Gebel et al., 2013; Schlage et al., 2011; Westra et al., 2011, 2013), which capture biological processes relevant to the respiratory system, followed by a computation of the degree of network perturbation (Martin et al., 2014). We hope that the study could facilitate the decision making for selecting appropriate epithelium models for toxicological assessment of aerosols.
MATERIALS AND METHODS
Organotypic tissue culture models
Two in vitro models were used and simultaneously exposed to CS:
BR: Human organotypic ‘MucilAir’ consisting of a bronchial epithelial cell monoculture;
BRF: Human organotypic ‘MucilAir-HF’ consisting of a bronchial epithelial cell and fibroblast coculture.
Both models were purchased from Epithelix Sàrl, Geneva, Switzerland. They were cultured at the ALI in 24-well plates with Transwell inserts (6.5-mm diameter, 0.4 µm pore size, Cat No. 3413, Corning Incorporated, Tweksbury, Massachusetts). The bronchial epithelial cells and fibroblasts, which were used to generate the models, were isolated from a healthy 63-year-old female nonsmoker donor who underwent surgical lobectomy. The sample collection was conducted according to the declaration of Helsinki on biomedical research and approved by the local ethics commission. Undifferentiated epithelial cells were used to construct the MucilAir models. The cells were cultured for 45 days while undergoing differentiation. After 45 days of culture, the epithelium forms fully ciliated columnar cells, secretes mucus, and is responsive to proinflammatory stimuli. In the case of the coculture model, the fibroblasts were grown underneath the membrane, thus submerged in the culture medium. The MucilAir model, a commercially available, fully differentiated, and ready-to-use human 3D airway epithelium models, composed of basal cells, ciliated cells, and mucus producing goblet cells (Constant et al., 2014). The proportion of the various cells resembles to what is observed in vivo.
Upon arrival of the tissues in our lab, the MucilAir models were ∼45 days old and maintained at 37°C in 0.7 ml MucilAir culture medium with medium renewal every 48 h according to the supplier’s instructions. Three days after arrival, the tissue cultures were exposed to CS according to the experimental design. Subsequent to the CS exposure, the tissue cultures were maintained in 0.7 ml MucilAir culture medium in 24-well plates for up to 48 h without a medium change. During the study, the tissue cultures were monitored under the microscope (eg, bacterial or fungal contamination, general morphology, and detachment of cells).
Whole CS exposure
3R4F was used as the reference cigarette, which was obtained from the University of Kentucky (www.ca.uky.edu/refcig). The cigarettes were conditioned for 7–21 days at 22 ± 1°C with a relative humidity of 60 ± 3% according to ISO standard 3402 (International Organization for Standardization, 2010). For CS exposure experiments, 10 cigarettes were smoked for 28 min to a standard butt length (∼35 mm) using a 30-port carousel smoking machine (SM2000; Philip Morris, Int.) for a 55-ml puff over 2 s, twice per min with an 8 s pump exhaust time (according to Health Canada smoking regimen).
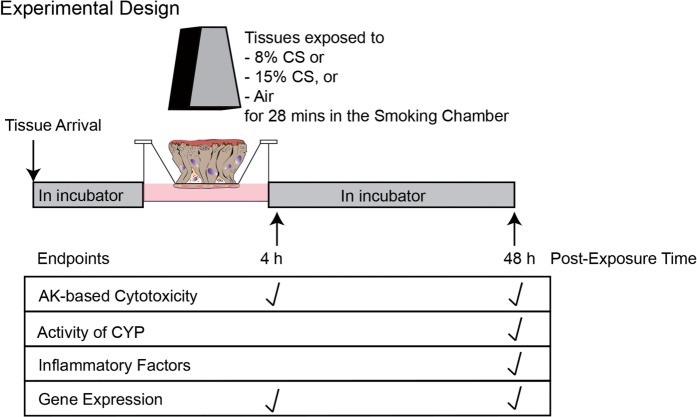
Mono- and coculture tissue inserts were directly exposed to CS for 28 min at the apical side in the climatic chamber of a smoking machine (VITROCELL Systems GmbH, Waldkirch, Germany) at 37°C. For each smoke run (SR), the tissue inserts were exposed to the following concentrations of mainstream CS generated from 3R4F (Fig. 1):
15% CS diluted in filtered air (v/v);
8% CS diluted in filtered air (v/v);
0% CS diluted in filtered air (v/v) (air-exposed control).
FIG. 1.
Experimental design. The concentration of CS 8 and 15% correspond to nicotine concentrations of approximately 0.28 and 0.56 mg/l, respectively, according to analysis performed using the VITROCELL system (Majeed et al., 2014). The concentrations of nicotine in the smoke were determined by a gas chromatography-flame ionization measurement after trapping the aerosol in an Extrelut 3NT column (Merck, Cat. No. 115095) connected to an individual exhaust in the Dilution/Distribution System of the VITROCELL system. Abbreviations: AK, adenylate kinase; CS, cigarette smoke; CYP, cytochrome P450s.
Three to 6 independent SRs were conducted (N = 1 tissue insert, per group and per SR, for each of the tissue models simultaneously in the same row of the exposure base module). The tissue inserts were maintained in an incubator at 37°C after exposure until the tissues and/or basal media were collected for the various endpoint measurements (Fig. 1).
Cytotoxicity assay
Cytotoxicity was assessed by measuring the activity of AK using the basolateral medium of the tissues after exposure. The AK activity was measured using the BioVision Bioluminescence Cytotoxicity Assay kit (Cat. No. 312-500, BioVision, Inc., Milpitas, California). Briefly, 50 µl basolateral medium samples were transferred in to a well in a 96-well plate and mixed with 50 µl Reagent Working Solution. After 5 min of incubation, the luminescence signals were detected using a luminometer. The positive control was the basolateral media from tissue cultures treated with 1% Triton X-100 (Cat. No. 93443 Sigma-Aldrich, Buchs, Switzerland) for 24 h prior to the sample collection. The negative control was the basolateral media from untreated tissues.
Tissue histology
Tissue inserts were collected and processed for histological analysis. The tissue inserts were washed in phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 1 h. Subsequently, the tissue cultures were washed 3 times with PBS. The fixed tissues were embedded in paraffin and used for histology. Standard hematoxylin and eosin (H&E) and Alcian blue staining procedures were conducted.
Measurement of CYP1A1/CYP1B1 activity
Combined activity of CYP1A1/CYP1B1 was determined using the nonlytic P450-Glo assay (Cat. No. V8752; Promega, Madison, Wisconsin) according to the manufacturer’s recommendations. Briefly, the luminogenic CYP-Glo substrate (luciferin-6′ chloroethyl ether), which targets both CYP1A1 and CYP1B1, was added to the basolateral medium 24 h prior to collection. Following the CYP reaction, a luciferin product that generates light was detected in the medium by addition of the Luciferin Detection Reagent. The amount of the luciferin product was measured in the medium using a luminometer. The positive control was media from tissue inserts that were treated for 48 h with 30 nM 2,3,7,8-tetrachlorodibenzo-p-dioxin (Sigma-Aldrich, St. Louis, Missouri) in the basolateral medium. The negative control was media from untreated tissue inserts.
Measurement of secreted mediators
Secreted mediators were measured in the basolateral media of tissue models using a Luminex-based technology (EMD Millipore Corp., Schwalbach, Germany) and Milliplex panels detecting the following mediators: endothelial growth factor (EGF), eotaxin, granulocyte-colony stimulating factor (GCSF), granulocyte macrophage-CSF, growth regulated oncogene (GRO) α/β/γ, IL-1α, IL1β, IL6, IL8, IL10, IL13, interferon-γ IP-10, monocyte chemoattractant protein (MCP) 1, macrophage inflammatory protein (MIP)-3α, MMP-1, MMP9, regulated on activation, normal T cell expressed and secreted (RANTES), soluble intercellular molecule (sICAM)-1, TIMP-1, tumor necrosis factor (TNF)-α, thymic stromal lymphopoietin, and VEGF. Values that were below the detection limit are reported as ‘< LLOD’.
A positive control test to assess the capacity of the tissues to secrete various mediators (basal secretion and after stimulation) was conducted in the following tissues after treatments with PBS or a combination of TNFα and IL-1β (10 ng/ml each), respectively: BR monoculture, BRF coculture, and human fibroblast culture alone. The treatments were added to the basolateral media of the cultures (N = 3 per tissue type) at 24 h before the media were collected for measurements.
RNA isolation and microarray hybridization
The following samples were collected and processed for RNA extraction (Fig. 1): (1) epithelial cells from the BR monoculture; (2) epithelial cells and fibroblasts from the BRF coculture; (3) bronchial epithelial cells that were isolated/scraped from the exposed BRF co-culture (henceforth, they will be referred to as scraped bronchial epithelial samples [sBR]). Briefly, the tissue inserts were washed 3 times with ice-cold PBS and lysed using RLT lysis buffer containing 1% β-mercaptoethanol (Cat. No. 79216; Qiagen, Venlo, The Netherlands) and then stored at −80°C until further processing. Extraction and purification of the mRNAs and microRNAs were performed using a RNA with RNeasy micro kit (Qiagen, Cat. No. 74004) RNA with RNeasy micro kit (Cat. No. 74004 Qiagen). The concentration of the isolated RNAs was measured by a NanoDrop ND1000 Spectrophotometer (Thermo Scientific, Waltham, Massachusetts). RNA quality was verified using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, California). The minimum RNA integrity number (RIN) was 6.3 (the average RIN in this study was 9.4). For mRNA analysis, 5 ng total RNA was processed as described in the User Manual for the Nugen Ovation Pico WTA System V2 (Nugen, Inc., The Netherlands). GeneChip Human Genome U133 Plus 2.0 Arrays were used for hybridization. Samples were randomized prior to RNA extraction and microarray analysis.
Microarray data processing and analysis
Raw CEL files were processed for background correction, normalization, and summarization by the frozen robust multiarray analysis algorithm (McCall et al., 2010). Quality control (QC) for all chips was performed using R packages from Bioconductor (affy, affyPLM) (Bolstad et al., 2003; Brettschneider et al., 2008; Gautier et al., 2004). The following plots and images were generated for QC checks: log-intensities plot, normalized unscaled standard error plot, relative log expression plot, polyA controls boxplot, RNA degradation plot, spike-in controls boxplot, pseudo image, and raw image. A total of 125 CEL files passed the QC process. The gene expression data were submitted to ArrayExpress with the following accession number: E-MTAB-3462. For each postexposure time point, 6 biological replicates per CS concentration of the 3 different samples (ie, BR monoculture, BRF coculture, and sBR cells) were available.
To identify probe sets that were modulated after CS exposure, a linear model was used in Equation (1).
| (1) |
For every dose/type of sample/postexposure time point (DTP), we fitted a model to the samples in the DTP group and the corresponding air-exposed group. The SR was considered as a covariate. The coefficient is equivalent to a paired pair-wise comparison, DTP and Sham (DTP). The doses did not fit in a single model because we expected a strong heteroscedasticity between the dose and postexposure variables. The coefficient β representing the effects of interest described above was estimated using the Limma R package (Smyth, 2004).
Functional annotation analysis of differentially expressed genes
To detect significantly enriched KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) analysis (Huang et al., 2009) version 6.7 (http://david.abcc.ncifcrf.gov). Differentially expressed genes (DEGs) below a false discovery rate (FDR) of <0.01 were uploaded to DAVID. This FDR cut-off was chosen to ensure equal thresholds for each of the contrasts for the annotation analysis in DAVID (limited to 3000 genes). Enrichment was determined at DAVID-calculated p-values of < .05. The KEGG pathways categorized under the ‘human diseases’ category were excluded.
Calculation of the exposure impact on biological processes using a network-based systems biology approach
Transcriptomic data were analyzed in the context of hierarchically structured network models describing the molecular mechanisms underlying essential biological processes in nondiseased lung cells (Hoeng et al., 2012). Table 1 lists all of the network models and their subnetworks that were applied for the analysis.
Table 1.
List of networks used for computation of network perturbation
| No | Network | Subnetwork | References | |
|---|---|---|---|---|
| 1 | Cell proliferation | Calcium | (Schlage et al., 2011; Thomson et al., 2013) | |
| 2 | Cell proliferation | Cell cycle | ||
| 3 | Cell proliferation | Cell interaction | ||
| 4 | Cell proliferation | Clock | ||
| 5 | Cell proliferation | Epigenetics | ||
| 6 | Cell proliferation | Growth factor | ||
| 7 | Cell proliferation | Hedgehog | ||
| 8 | Cell proliferation | Jak Stat | ||
| 9 | Cell proliferation | MAPK | ||
| 10 | Cell proliferation | Notch | ||
| 11 | Cell proliferation | Nuclear receptors | ||
| 12 | Cell proliferation | PGE2 | ||
| 13 | Cell proliferation | Wnt | ||
| 14 | Cell stress | Xenobiotic metabolism response | (Schlage et al., 2011) | |
| 15 | Cell stress | Endoplasmic reticulum stress | ||
| 16 | Cell stress | Hypoxic stress | ||
| 17 | Cell stress | NFE2L2 signaling | ||
| 18 | Cell stress | Osmotic stress | ||
| 19 | Cell stress | Oxidative stress | ||
| 20 | Inflammatory process | Mucus hypersecretion | (Westra et al., 2013) | |
| 21 | Inflammatory process | Epithelial cell barrier defense | ||
| 22 | Inflammatory process | Epithelial proinflammatory signaling | ||
| 23 | Inflammatory process | Tissue damage | ||
| 24 | Apoptosis | Caspase cascade | (Gebel et al., 2013) | |
| 25 | Apoptosis | ER stress-induced apoptosis | ||
| 26 | Apoptosis | MAPK signaling | ||
| 27 | Apoptosis | NFκB signaling | ||
| 28 | Apoptosis | PKC signaling | ||
| 29 | Apoptosis | Prosurvival mitochondrial signaling | ||
| 30 | Apoptosis | TNFR1 | ||
| 31 | DNA damage | Components affecting TP53 activity | ||
| 32 | DNA damage | Components affecting TP73 activity | ||
| 33 | DNA damage | Components affecting TP63 Activity | ||
| 34 | DNA damage | DNA Damage to G1 | ||
| 35 | DNA damage | DNA Damage to G2 | ||
| 36 | DNA damage | Double strand break response | ||
| 37 | DNA damage | Inhibition of DNA Repair | ||
| 38 | DNA damage | TP53 TS | ||
| 39 | Necroptosis | Fas activation | ||
| 40 | Autophagy | ATG Induction of Autophagy | ||
| 41 | Autophagy | mTOR signaling | ||
| 42 | Senescence | Oncogene induced senescence | ||
| 43 | Senescence | Regulation by tumor suppressors | ||
| 44 | Senescence | Replicative senescence | ||
| 45 | Senescence | Stress-induced premature senescence | ||
| 46 | Senescence | Transcriptional regulation of SASP | ||
| 45 | Senescence | Stress induced premature senescence | ||
| 46 | Senescence | Transcriptional regulation of SASP | ||
Abbreviations: ATG, anti-thymocyte globulin; ER, endoplasmic reticulum; MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin; NFE2L2, nuclear factor (erythroid-derived 2)-like 2; NFκB, nuclear factor kappa-light-chain-enhancer of activated B-cells; SASP, senescence-associated secretory phenotype; TNFR1, tumor necrosis factor receptor 1; TP53 TS, tumor protein 53 transcriptional signature.
The effects of exposure were quantified by scoring the impact on each subnetwork (referred to as ‘network perturbation amplitude’ [NPA]) (Martin et al., 2014). NPA aims to reduce the high-dimensional transcriptomics data by combining the gene expression log2-fold changes into fewer differential backbone values (between a few dozen and 200). By definition of the 2-layer structure, no measurements corresponding to nodes in the functional layer are available. The differential backbone values are therefore determined by a fitting procedure that infers values that best satisfy the directional and signed relationships contained in the backbone model, while being constrained by the experimental data (the gene log2-fold changes). Once the perturbation is quantified, 3 statistics, the confidence intervals, ‘O’ statistic, and ‘K’ statistic, are computed to assess the significance of the NPA with respect to the experimental error and its specificity to the given 2-layer subnetwork structure. The confidence intervals test whether the current NPA score is significantly more than 0 at a 5% type I error. The ‘O’ statistic tests whether the current NPA score is larger than the 95th percentile of the estimated probability density function (PDF) of random NPA scores based on 500 random permutations of downstream genes. The ‘K’ statistic tests whether the current NPA score is larger than the 95th percentile of the estimated PDF of random NPA scores based on 500 random permutations of edges of the backbone subnetwork (function layer). A network score can be significant without all of its subnetworks being significant. Finally, the network is considered to be specifically perturbed (*) if both ‘O’ and ‘K’ statistics are significant (p < .05) (Martin et al., 2014).
Comparability of in vivo/in vitro responses to CS exposure
To determine correlations between the effects of CS exposure in vitro and the effects of smoking in vivo, the network perturbation scores of the Xenobiotic Metabolism Response network (Iskandar et al., 2013) generated from the in vitro datasets (CS-exposed vs air-exposed organotypic tissues) were compared with those generated from publicly available in vivo datasets (smokers vs nonsmokers in large airway brushing samples): GGSE7895 (Beane et al., 2007); GSE160058 (Zhang et al., 2010); and GSE14633 (Schembri et al., 2009).
RESULTS
Cytotoxicity Following CS Exposure
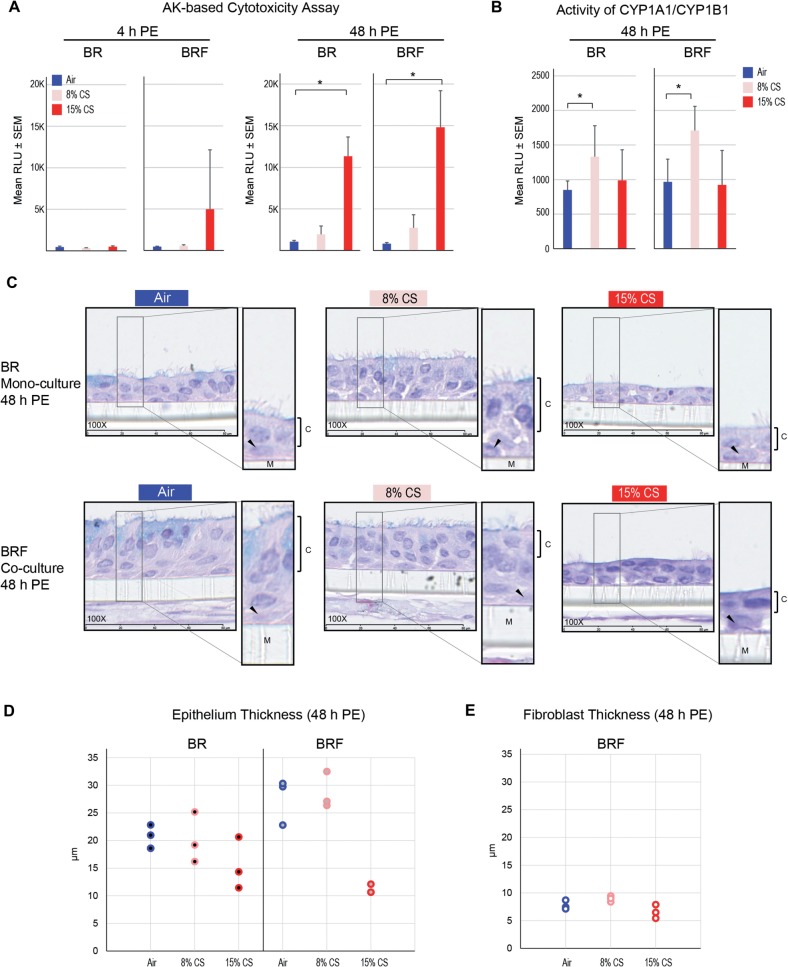
Cytotoxicity of the tissue models after CS exposure was assessed at 4-h and 48-h postexposure. At 4-h postexposure, the levels of AK activity of the CS-exposed BR monoculture were not significantly different from those of the CS-exposed BRF coculture (Fig. 2A), despite a slight increase of AK in the 15% CS-exposed BRF. However, at 48-h postexposure, a significant concentration-dependent increase of AK activity levels was observed in both models (p < .05, Fig. 2A). Overall, CS induced similar cytotoxicity in both models and the cytotoxicity was increasing with the duration of postexposure (Fig. 2A).
FIG. 2.
Impact of CS exposure on cytotoxicity, CYP1A1/1B1 activity, and tissue morphology. A, cytotoxicity-based AK activity was assessed in the BR monoculture and BRF coculture at 4 h and 48 h after CS exposure (N= 6 independent SRs, one insert/tissue type for each run). *p < .05 as compared with the air-exposed. B, Combined activity of CYP1A1/CYP1B1 was measured at 48 h after CS exposure (N = 6 independent SRs, one insert/tissue type for each run). *p < .05 as compared with the air-exposed. C, H&E-stained tissue sections observed at 48 h after CS exposure (N = 3, 100 × magnification). Insets show ciliated columnar epithelial cells (C), basal cells (black arrow), and the membrane (M). D, Thickness of the epithelium in the BR monoculture and BRF coculture at 48 h after exposure (N = 3, each data point represents an average of 5 measurements from one H&E-stained tissue cross-section). E, Thickness of the fibroblast layer in the BRF coculture at 48 h after exposure (N = 3, each data point represents an average of 5 measurements from one H&E-stained tissue cross-section). Abbreviations: BR, bronchial epithelial cell monoculture; BRF, bronchial epithelial cells cocultured with fibroblasts, PE, postexposure.
CYP1A1/1B1 Activity Following CS Exposure
The xenobiotic metabolizing enzymes CYP1A1 and CYP1B1 are involved in the metabolism of tobacco smoke constituents, (Anttila et al., 2011; Port et al., 2004). The CYPs-mediated reactions contribute to the generation of toxic metabolites, which can bind to DNA thereby forming DNA adducts (Anttila et al., 2011; Port et al., 2004). Therefore, we measured the activity of CYP1A1 and CYP1B1 in the tissues following CS exposure.
The activity of CYP1A1 and CYP1B1 (combined) was assessed 48-h postexposure to CS. Increased levels of CYP1A1/CYP1B1 activity were observed in 8% CS-exposed tissues but not in the 15% CS-exposed tissues compared with their respective air-exposed controls (Fig. 2B). The CS-induced activities of CYP1A1/CYP1B1 in the BR monoculture were not significantly different from those in the BR monoculture (Fig. 2B).
Tissue Histology Following CS Exposure
Histological assessment was performed on tissues 48-h postexposure. A reduced thickness of the epithelial cell layer and decreased occurrence of columnar ciliated cells were similarly observed in CS-exposed BR monoculture and BRF coculture (Fig. 2C). The reduced epithelium thickness was more pronounced in the tissue inserts exposed to 15% CS than those exposed to 8% CS (Fig. 2D). Similarly, the fibroblast layer of the BRF coculture was considerably thinner following 15% CS exposure (Fig. 2C and 2E). Overall, comparable changes were observed in both models.
Effects of CS on the Secretion of Various Mediators
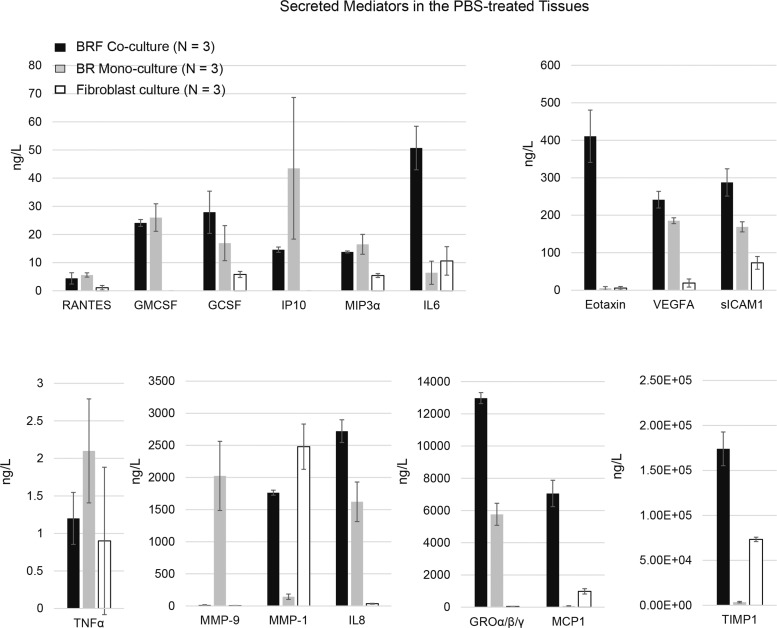
To determine the effects of CS on the secretion of various mediators, the concentrations of cytokines, chemokines, and growth factors were measured in the basolateral media of the tissue models 48-h after exposure. Initially, the capacity of the tissue models to secrete mediators was examined by treating the tissue inserts with a combination of known proinflammatory stimuli (TNFα + IL1β for 24 h) (as a positive control, data not shown). Here, we first report the basal capacity of the different cell types (ie, bronchial epithelial cells and fibroblasts, alone or combined) to secrete mediators. Following a treatment with PBS for 24 h (negative control), the concentration of various mediators was measured in the basolateral media (Fig. 3). Among the tissue models, higher concentrations of GCSF, IL6, eotaxin, VEGFA, sICAM1, IL8, GROα/β/γ, MCP1, and TIMP1 were found in the basolateral medium of the BRF coculture (Fig. 3). Moreover, MMP1, MCP1, and TIMP1 were predominantly observed in the media of the BRF coculture and fibroblast monoculture, but not in the BR monoculture (Fig. 3). Finally, MMP9 was detectable primarity in the BR monoculture (Fig. 3). These findings were similar to those in the tissues exposed to fresh air at the ALI (Supplementary Figure S1).
FIG. 3.
Secreted mediators in the basolateral medium of PBS-treated tissues. Measurements were conducted in the basolateral media of the BR monoculture, BRF coculture, and fibroblast culture after PBS treatment for 48 h (N = 3 per tissue type).
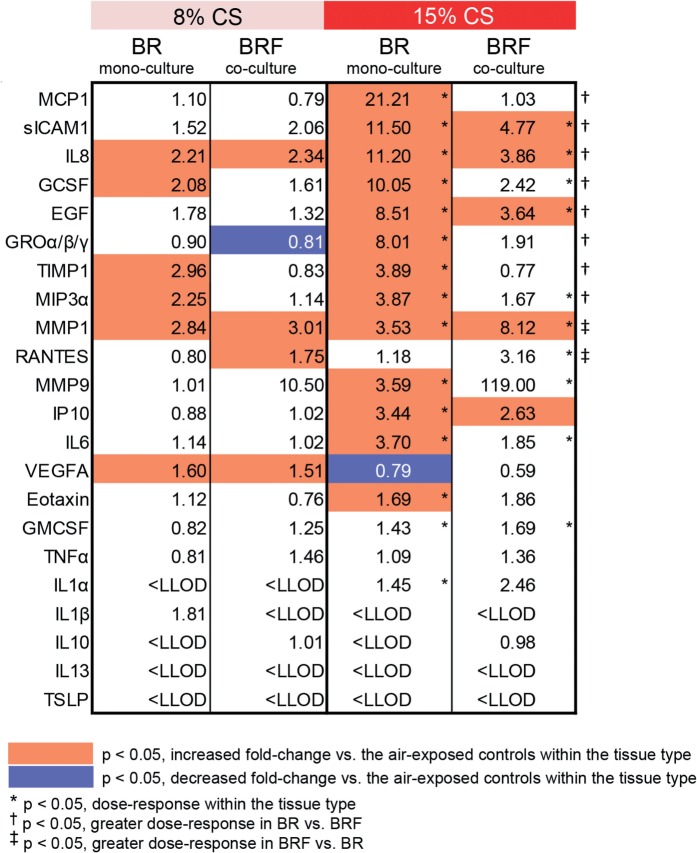
Furthermore, the impact of CS on the secretion of mediators was determined 48-h postexposure. Compared with the air-exposed tissues, more mediators were altered following CS exposure in the BR monoculture than in the BRF coculture (orange and blue indicate fold increase and fold decrease, respectively, as compared with the air-exposed samples, Fig. 4). In addition, a higher concentration dependent response of CS exposure on MCP1, sICAM1, IL8, GCSF, EGF, GROα/β/γ, TIMP1, and MIP3α was found in the BR monoculture as compared with BRF co-culture (indicated with ‘†’, p < .05, Fig. 4). Whereas, for MMP1 and RANTES, a higher concentration dependent response of CS exposure was found in the BRF co-culture as compared to BR mono-culture (indicated with ‘‡’, p < .05, Fig. 4).
FIG. 4.
Fold changes of the secreted mediators in basolateral media following CS exposure. Values indicate the fold changes as compared with the air-exposed controls for a given tissue model. Measurements were conducted in the media of the BR monoculture and BRF coculture at 48 h after CS exposure (N = 6 independent SRs, one insert/tissue type for each run). Abbreviations: BR, bronchial epithelial cell monoculture; BRF, bronchial epithelial cells cocultured with fibroblasts; LLOD, lower limit of detection.
Impact of CS on the Transcriptomes of Monoculture and Coculture Models
Canonical pathway annotation
In addition to the gene expression data from the BR monoculture and BRF coculture, data were also obtained from sBR samples (ie, bronchial epithelial cells that were isolated/scraped from the exposed BRF coculture and referred to as scraped bronchial epithelial samples (sBR), as described in Materials and methods). Biological interpretation of the gene expression data was done using DAVID annotation tool (Huang et al., 2009) from the DEGs (FDR of ≤ 0.01). We found more pathways annotated from the gene expression changes at the 48-h postexposure time point.
Figure 5 showed that at the 48-h postexposure time point, 8% CS exposure impacted ‘tryptophan metabolism’, ‘steroid hormone biosynthesis’, and ‘metabolism of xenobiotic by CYP’, all of which were linked to only 2 genes (CYP1A1 and CYP1B1) (Supplementary Table S1). Furthermore, for the higher concentration of CS (15%), more pathway annotations were identified, revealing alterations in stress-related metabolism (‘metabolism of xenobiotics by CYP’, ‘tryptophan metabolism’, and ‘drug metabolism’), and inflammatory responses (‘complement and coagulation cascade’, ‘chemokine signaling pathway’, ‘cytokine-cytokine receptor interaction’, and ‘natural killer cell-mediated cytotoxicity’), as well as various signaling pathways.
FIG. 5.
Pathway annotations associated with CS exposure. The DEGs of the CS-exposed versus air-exposed samples at a FDR of ≤ 0.01 were uploaded to DAVID. Canonical pathway of the KEGG database was annotated in the datasets using the DAVID annotation tool at a p-value of < .05. The KEGG pathways that were categorized under the ‘human diseases’ category were excluded. The different colors designate the p-value annotations. Abbreviations: BR, bronchial epithelial cell monoculture; BRF, bronchial epithelial cell and fibroblast coculture; sBR, isolated/scraped bronchial epithelial cells from the coculture; DEG, differentially expressed genes; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Biological network approach
A more comprehensive analysis of the tissue responses among the various sample types following CS exposure was conducted using a threshold-free, network-based systems biology approach. The network-based analysis leverages the transcriptomics data to computationally infer the biological processes perturbed following CS exposure. The approach employs our published collection of hierarchically structured network models that are specific to the respiratory and cardiovascular system and capture biological processes and mechanisms (specified as subnetworks), such as Cell Proliferation (Schlage et al., 2011), Cell tress (Schlage et al., 2011), DNA Damage, Autophagy, Cell Death, and Senescence (Gebel et al., 2013), and Pulmonary Inflammation Processes (Gebel et al., 2013) networks. These biological network models were constructed from cause-and-effect relationships among biological entities derived from published literature within specific boundaries, ie, within the context of nondiseased mammalian pulmonary tissue. Because the hierarchical network models capture mechanisms at the levels of biological processes, pathways, and specific molecular entities, the network models have been useful to evaluate mechanisms/pathways affected by CS exposure (Gebel et al., 2013; Hoeng et al., 2012). Using the network models and systems biology approach (Martin et al., 2014), we assessed the impact of CS exposure (ie, perturbation of the networks) using a set of networks that are relevant to bronchial tissue (Table 1), as used in a previous publication (Talikka et al., 2014).
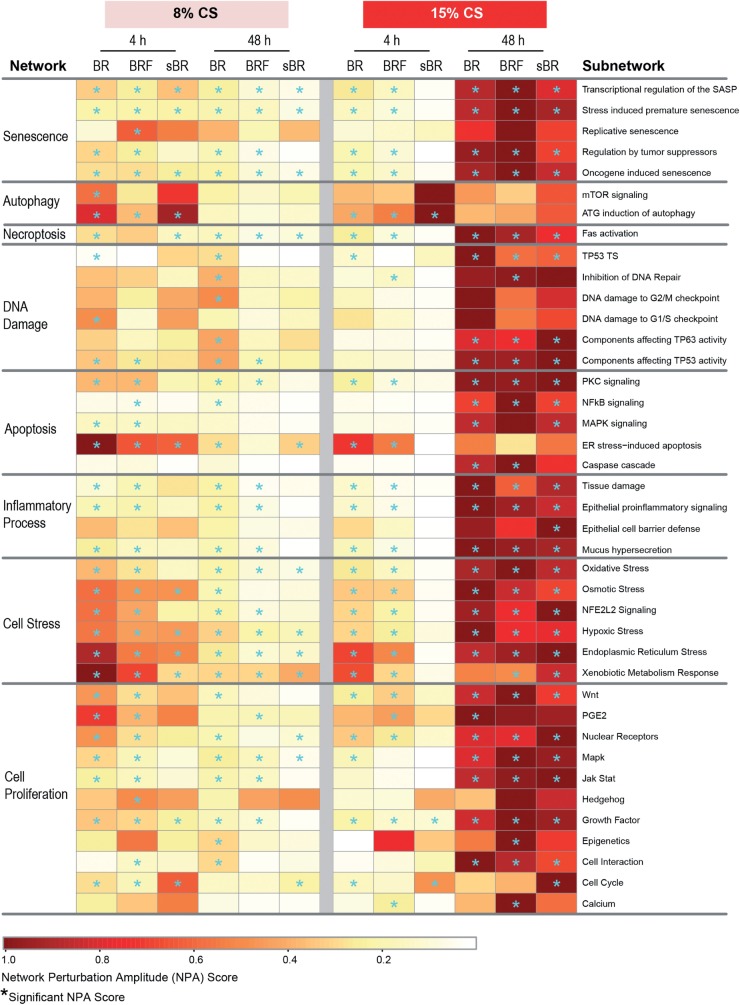
The network-based analysis showed that the tissues exposed to the lower concentration of CS (8%) were impacted at a greater degree at the earlier postexposure time point (4 h) than at 48 h (Fig. 6). Among the various sample types, a greater impact on the BR monoculture was observed most notably in the following networks: Autophagy, Apoptosis, and Cell Stress networks. Nonetheless, at the 48-h postexposure time point, a predominant impact was observed for the DNA Damage network in the BR monoculture.
FIG. 6.
Impact of CS on various biological processes analyzed using a network-based systems biology approach analyzed at 4 and 48-h postexposure. The network is considered to be specifically perturbed (*) if both ‘O’ and ‘K’ statistics are significant (p < .05) and confidence interval is more than 0 (Martin et al., 2014). Abbreviations: ATG, antithymocyte globulin; BR, bronchial epithelial cell monoculture; BRF, bronchial epithelial cell and fibroblast co-culture; MAPK, mitogen activated protein kinase; mTOR, mammalian target of rapamycin; NFE2L2, nuclear factor (erythroid-derived 2)-like 2; NFκB, nuclear factor kappa-light-chain-enhancer of activated B-cells; PGE2, prostaglandin E2; SASP, senescence-associated secretory phenotype; sBR, isolated/scraped bronchial epithelial cells from the co-culture; TP53 TS, tumor protein 53 transcriptional signature.
Following the higher concentration of CS exposure (15%), the BR monoculture and BRF coculture were impacted at a similar degree in the majority of the networks at 4 h time point after exposure, whereas sBR sample were minimally impacted, with the exception of Autophagy network (Fig. 6). Moreover, at the later postexposure time point (48 h), the networks were strongly perturbed in all samples. The degree of impact on the Senescence and Cell Proliferation networks was higher in the BRF as compared to the BR monocultures. In contrast, a stronger impact on the Inflammatory Process and Cell Stress networks was observed in the BR monoculture than in the BRF
Comparison of in vitro Versus in vivo Bronchial Epithelial Signatures of Exposure to CS
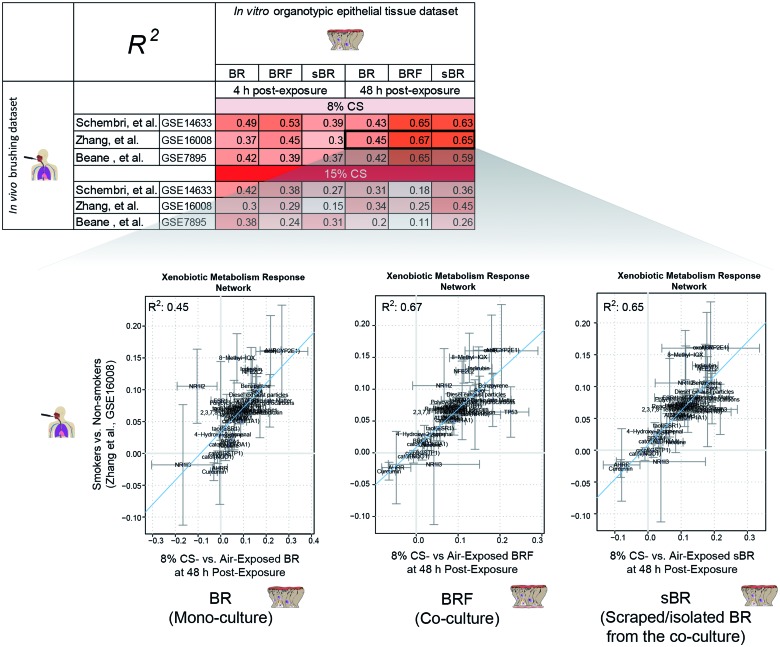
Activation of xenobiotic metabolism could indicate tissue responses to foreign compounds, including CS exposure (Courcot et al., 2012). Using the published Xenobiotic Metabolism Response network model (Iskandar et al., 2013), we compared the in vitro tissue response following CS exposure to that of the in vivo after smoking conventional cigarettes. The in vivo tissue response to smoking was probed from transcriptomics data of the bronchial epithelial cells obtained from smoker and nonsmoker volunteers by brushing (from 3 public datasets GSE14633, GSE16008, and GSE7895). The in vitro tissue response to CS was probed from the transcriptomics data of the organotypic models. The backbone-node value of the Xenobiotic Metabolism Response network model, a computable value derived using backward reasoning from the altered transcript levels (exposed vs nonexposed) of a set of gene (Martin et al., 2014), would indicate smoking-induced perturbation/impact of a specific entity that makes up the xenobiotic metabolism.
The in vitro CS exposure-induced modulation of the backbone-node values (from the BRF, BR, and sBR samples) were correlated to those of the in vivo samples (Fig. 7). The correlation plots illustrates that the in vitro xenobiotic responses following exposure of 8% CS (at 48-h postexposure) to the BRF coculture and sBR cells were more comparable with the in vivo situation of apparently healthy smokers (R2 = 0.59–0.67). The same outcomes were observed consistently when the in vitro data were compared with the 3 different in vivo datasets.
FIG. 7.
Correlations between the network backbone values of the nodes in the Xenobiotic Metabolism Response network. The table shows the R2 values of the different correlations. The correlation plots are representative plots between the in vivo-derived backbone-node values (Zhang et al. GSE16008, y-axis) and the in vitro-derived backbone node values (x-axis). Abbreviations: BR, bronchial epithelial cell monoculture; BRF, bronchial epithelial cell and fibroblast coculture; sBR, isolated/scraped bronchial epithelial cells from the coculture.
DISCUSSION
Here, we demonstrated the impact of whole mainstream CS exposure on 2 bronchial organotypic models: BR monoculture and BRF coculture. We aim to advance the understanding of the models for toxicological assessment of aerosols. The impact of 2 doses of CS exposure on various endpoints was investigated not only using well-known toxicity assays but also a systems biology approach.
Xenobiotic Metabolism Response Following CS-Induced Cellular Stress
Our results demonstrated that 8% CS exposure led to cellular stress; the network-based analysis showed that this concentration of CS was linked to a significant impact on many of the subnetworks within the Cell Stress network. The impact was more pronounced at the earlier time point after exposure (4 h) than at the later time (48 h), suggesting that the tissues were progressively recovering from stress. Independent of the postexposure time points, the degree of impact on the Cell Stress network was more pronounced in the BR monoculture as compared with the BRF coculture or sBR samples, suggesting that fibroblasts suppressed the CS-induced stress. One possible explanation could be the differing antioxidant capacities between epithelial cells and fibroblasts, as reported before (Kartvelishvili et al., 2004).
Moreover, the stress response of the tissue following 8% CS exposure can be linked to the activation of xenobiotic metabolism. This finding was supported by our CYP activity assay, pathway annotation analysis, and network-based systems biology analysis. First, we observed increased enzymatic activity of the major phase I xenobiotic metabolizing enzymes CYP1A1/CYP1B1 48 h after 8% CS exposure in both models. Second, from the pathway annotations, we found that the DEGs (8% CS-exposed vs air-exposed tissues at 48-h postexposure) were significantly linked to the following pathways: ‘tryptophan metabolism’, ‘steroid hormone biosynthesis’, and ‘metabolism of xenobiotics by CYP’. Regardless of the annotation names (according to the KEGG database), these annotations were linked to the upregulation of CYP1A1 and CYP1B1 genes (Supplementary Table S1). Third, we observed higher correlation coefficients for xenobiotic response for the comparison between the 8% CS-exposed models and the bronchial brushing of smokers, than for the comparison to the 15% CS-exposed models (ie, the R2 in the context of Xenobiotic Metabolism Response network model, see Fig. 7). The relationship between xenobiotic metabolism activation and tissue stress response can be further supported by other studies, in which CS toxicants (eg, polycyclic aromatic hydrocarbons, nitrosamines) bind and activate the transcription factor aromatic hydrocarbon receptor, leading to the transcription of CYP1A1 and CYP1B1 genes (Cheng et al., 2012; Zhang et al., 2006). This tissue stress response may further point to the attempt to restore tissue homeostasis following 8% CS exposure.
In the context of Xenobiotic Metabolism Response network model, high correlation coefficients were generally observed when the in vivo bronchial brushing samples (smokes vs nonsmokers) were compared to the in vitro BRF coculture or sBR (8% CS-exposed vs air-exposed) (R2 = 0.59–0.67). However, the correlation coefficients were reduced when the in vivo brushing samples were compared to the BR monoculture (R2 = 0.43–0.45) at the 48-h postexposure time. This observation can be explained by the feature of the in vivo bronchial brushing samples, which we thought to be more comparable to the isolated epithelial cells from the in vitro coculture. During the in vitro CS exposure experiment, the isolated epithelial cells (sBR samples) were a part of the intact in vitro BRF coculture model, which then separately isolated for the transcription analysis. Similarly, in reality during smoking situation, the brushed epithelial cells were part of the intact smoker’s bronchial epithelial tissue in which epithelial—fibroblasts interaction occurs. A previous study has reported that a brushing procedure collects mostly the epithelial cells (≥90% of the collected samples) (Spira et al., 2007). Thus, our results support that the presence of fibroblasts may induce a more robust xenobiotic metabolism response. Furthermore, we obtained higher correlation coefficients of the Xenobiotic Metabolism Network response when the in vivo brushing samples were compared to the in vitro coculture samples collected at 48-h postexposure time point, than to those collected at the 4 h time point. This could be attributed to the standard practice of bronchial brushing procedure, for which smokers need to refrain from smoking at least 24 h before undergoing the procedure (American Thoracic Society, 2004).
Tissue Response Against CS-Induced Tissue Damage
The results suggest that tissue damage occurred following the exposure of the higher dose of CS (15%). Several factors support this conclusion. First, gross histology examination of the tissues 48 h after CS exposure demonstrated alteration of the tissue morphology in both BR monoculture and BRF coculture. The thickness of the epithelial layer and height of the columnar cells were reduced. Additionally, fewer ciliated cells were observed in the 15% CS-exposed tissues as compared with the air-exposed tissues. These changes were analogous to those observed in the respiratory tissue of asthmatic patients (Persson et al., 1996; Shebani et al., 2005) and also in a recent in vitro study using human bronchial epithelial cells exposed to CS extract (Schamberger et al., 2015). Second, the increased AK activity in the 15% CS-exposed tissues suggests a disruption of the epithelial integrity. An increase of AK activity may signify cell death or damage of the cellular membrane (Jacobs et al., 2013).
Furthermore, the changes in the transcriptome profiles following 15% CS exposure were linked to tissue damage. The KEGG pathway ‘apoptosis’ was significantly annotated in the DEGs generated from the BR monoculture and BRF coculture 48-h postexposure to 15% CS (as compared with their respective air-exposed controls). Moreover, the network-based analysis demonstrated a significant impact of 15% CS exposure on the Tissue Damage subnetwork (one of the subnetwork within the Inflammatory Process network) at 48-h postexposure. Finally, Necroptosis, DNA Damage, and Apoptosis networks were also impacted, all of which reflect various modes of cell death.
A more comprehensive evaluation using the network-based systems biology analysis showed that the impact of 15% CS on the Tissue Damage subnetwork of the BRF coculture was the smallest than the other samples. This suggests that the presence of fibroblasts may suppress the CS-induced epithelium damage. The degree of the CS-induced secretion of MCP1 levels can further support this argument. A previous study reported that injured bronchial epithelial cells release MCP1 (Lundien et al., 2002). Here, we found higher levels of MCP1 in the CS-exposed BR monoculture without fibroblast, suggesting that the presence of fibroblasts may suppress the epithelial cells injury.
The ability of fibroblast to minimize the epithelium damage may also be attributed to their capacity to become senescent. The levels of MMP1, which has been linked to senescent fibroblast (Cho et al., 2014; Dasgupta et al., 2010), were higher in the basolateral media of the BRF coculture than the BR monoculture. Furthermore, our network-based analysis showed that greater score of the Senescence NPA was observed in the BRF coculture than in the BR monoculture. Moreover, senescent fibroblasts can stimulate the proliferation of their surrounding cells (Bavik et al., 2006; Dean and Nelson, 2008; Eyman et al., 2009; Liu and Hornsby, 2007). Consistent with this notion, the perturbation amplitude scores of the various Cell Proliferation subnetworks were generally smaller in the BR monoculture without fibroblast, as compared with those in the BRF coculture and sBR (ie, the isolated epithelial cells from the BRF coculture). This observation may further suggest that cellular repair was taking place (Rodier and Campisi, 2011). Overall, the data suggested that fibroblasts contribute to the endurance of the epithelial cells following insults.
Secretion of Mediators
Because the fibroblasts were cultured underneath the membrane (submerged in the basolateral medium), the culture of fibroblast alone was not exposed to aerosols (CS or fresh air). However, by measuring secreted mediators in the tissues treated with PBS or exposed to fresh air, we could evaluate the basal capacity of the different cell types to secrete mediators. In the basolateral media of the PBS-treated cultures, the levels of MMP1, MCP1, and TIMP1 were detected in both BRF coculture and fibroblast culture, suggesting that the fibroblasts regulate the secretion of these mediators. A previous study has reported that fibroblasts secrete a higher level of MMP1 compared with other lung cell types (O’Kane et al., 2010) and also secrete TIMP1 (Gueders et al., 2006). In contrast, GMCSF and IP10 were only detected in the BRF coculture and BR monoculture, implying that their secretions were likely to have originated from the epithelial cells, whereas the contribution of fibroblasts was miniscule. Secretions of GMCSF and IP10 from airway epithelial cells have been reported before (Cox et al., 1992; Kelsen et al., 2004). This observation also excluded the argument that coculturing with fibroblast underneath the membrane may hamper the secretion of mediators to the basolateral media. Moreover, MMP9 could only be detected in the media of the BR monoculture, suggesting that its basal secretion may be regulated (inhibited) by coculturing with fibroblasts.
CONCLUSION AND OUTLOOK
Based on the AK-based toxicity assays, CYP activity assays, and histological evaluation, this study demonstrated that the impact of CS exposure in the 2 culture models were closely analogous. However, based on the transcriptomics data and systems biology approach, a more detailed molecular investigation showed that coculturing with fibroblasts contributed to a stronger tissue capacity to defend against CS-induced cellular stress and damage. One of the limitations of the study comes from our inability to obtain satisfactory RNA from the isolated fibroblast only (from the BRF coculture) that hinders the confirmation of the fibroblast-specific mechanisms. Although various techniques of obtaining fibroblast samples were tested, the particularly thin layer of the fibroblasts was challenging for the isolation of satisfactory RNA and the subsequent generation of the transcriptomic data. Ultimately, we could only obtain a small number of transcriptomic samples (experimental replicates) from the scraped fibroblast group. Thus, an accurate and meaningful biological interpretation could not be derived. Future study may consider using a larger insert size or increasing the number of replicate per group to overcome these challenges.
Nevertheless, our results demonstrated the manifestation of cell–cell crosstalk within the MucilAir models, ie, cell–cell communication via the secretion of inflammatory cytokines, chemokines, and growth factors. As we have eluded earlier, this phenomenon may reflect an early repair process and airway remodeling following 15% CS exposure. However, we acknowledge that the later manifestations of airway remodeling, such as cell spreading and migration (Crosby and Waters, 2010) could not be demonstrated in this study. One possible explanation could be related to the material of the insert that is not optimal for studying migration of cells. The MucilAir cultures used in this study were constructed using a corning Transwell polycarbonate membrane-insert that has pores with a relatively small size (0.4 µm), which is suitable for small molecule transport and permeability studies but inappropriate for cell invasion and transepithelial migration studies (Corning, 2013). Moreover, because epithelial mesenchymal transition (EMT) contributes to airway remodeling and participates in tissue repair (Crosby and Water, 2010), we compared the gene expression levels of eminent EMT-related markers (Kalluri and Weinberg, 2009) among the samples following exposure (Supplementary Figure S2). Our assessment illustrated no observable difference of the EMT-related markers among the different samples (Supplementary Figure S2), suggesting that the tissue models—within the context of the current study design—cannot sufficiently display mesenchymal remodeling. Because cell spreading and migration may take several weeks (Crosby and Water, 2010), future studies could consider monitoring the tissues for extended periods after exposure to better explore tissue remodeling. Finally, a more complex culture system that integrates additional cell types (eg, immune cells) and extracellular matrix (eg, collagen, fibronectin, and laminin) may offer a better recapitulation of the in vivo airway tissue to study tissue remodeling and EMT-related processes.
Despite the limitations of the tissue models used in this study, our results further highlight that the use of a particular tissue model should be aligned with the primary research hypothesis. For example, based on the network-based analysis, we found that the in vivo xenobiotic metabolism response against cellular stress was better assessed in vitro using a coculture model. Moreover, the contribution of fibroblasts to tissue repair (eg, senescence and proliferation) seemed to be better reflected in coculture models. On the other hand, the impact of exposures on a specific cell type could be more straightforwardly identified from the monoculture models. Nevertheless, the functional endpoints, such as cytotoxicity, CYP activity, and histological changes following CS exposure were less influenced by the cell-type compositions.
FUNDING
This study was supported by Philip Morris International R&D, Neuchatel, Switzerland.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
ACKNOWLEDGMENTS
The authors acknowledge the technical expertise of Shoaib Majeed in conducting the smoke exposure experiment. The authors also acknowledge the technical expertise of Keyur Trivedi and Abdelkader Benyagoub on the histological processing; Celine Merg on the Luminex-based detection of mediators; as well as Karine Baumer, Remi Dulize, Dariusz Peric, and Sophie Scheuner on the microarray experiment. The authors thank Stephanie Boue for performing the sample randomization, as well as Sam Ansari and Vincenzo Belcastro for their supports on the dataset submission to ArrayExpress.
REFERENCES
- American Thoracic Society. (2004). Fiberobtic Bronchoscopy. Am. J Respir Crit Care Med.169, P1–P2. Available at http://www.thoracic.org/patients/patient-resources/resources/fiberoptic-bronchoscopy.pdf. Accessed February 2, 2015. [Google Scholar]
- Anttila S., Raunio H., Hakkola J. (2011). Cytochrome P450-mediated pulmonary metabolism of carcinogens: Regulation and cross-talk in lung carcinogenesis. Am. J. Respir., Cell Mol. Biol. 44, 583–590. [DOI] [PubMed] [Google Scholar]
- Bavik C., Coleman I., Dean J. P., Knudsen B., Plymate S., Nelson P. S. (2006). The gene expression program of prostate fibroblast senescence modulates neoplastic epithelial cell proliferation through paracrine mechanisms. Cancer Res. 66, 794–802. [DOI] [PubMed] [Google Scholar]
- Beane J., Sebastiani P., Liu G., Brody J. S., Lenburg M. E., Spira A. (2007). Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 8, R201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BéruBé K., Aufderheide M., Breheny D., Clothier R., Combes R., Duffin R., Forbes B., Gaça M., Gray A., Hall I. (2009). In vitro models of inhalation toxicity and disease. Altern. Lab. Anim. 37, 89–141. [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193. [DOI] [PubMed] [Google Scholar]
- Brettschneider J., Collin F., Bolstad B. M., Speed T. P. (2008). Quality assessment for short oligonucleotide microarray data. Technometrics 50(3). [Google Scholar]
- Cheng Y. H., Huang S. C., Lin C. J., Cheng L. C., Li L. A. (2012). Aryl hydrocarbon receptor protects lung adenocarcinoma cells against cigarette sidestream smoke particulates-induced oxidative stress. Toxicol. Appl. Pharmacol. 259, 293–301. [DOI] [PubMed] [Google Scholar]
- Cho S. Y., Seo D.-B., Kim W. G., Lee S.-J. (2014). Mild mitochondrial uncoupling prevents premature senescence in human dermal fibroblasts. J. Invest. Dermatol. 134, 540–543. [DOI] [PubMed] [Google Scholar]
- Constant S., Wiszniewski L., Huang S. (2014). The use of in vitro 3D cell models of human airway epithelia (MucilAirTM) in inhalation toxicity. In Cellular In Vitro Testing: Methods and Protocols (J. Haycock, A. Whluwalia, and M. Wilkinson, Eds.), pp. 15–33. CRC Press, London, UK. [Google Scholar]
- Corning (2013). Transwell(R) Permeable Supports Selection and Use Guide. Available at: http://csmedia2.corning.com/LifeSciences/Media/pdf/transwell_guide.pdf doi. Accessed June 5, 2015. [Google Scholar]
- Costea D. E., Loro L. L., Dimba E. A. O., Vintermyr O. K., Johannessen A. C. (2003). Crucial effects of fibroblasts and keratinocyte growth factor on morphogenesis of reconstituted human oral epithelium. J. Invest. Dermatol. 121, 1479–1486. [DOI] [PubMed] [Google Scholar]
- Courcot E., Leclerc J., Lafitte J.-J., Mensier E., Jaillard S., Gosset P., Shirali P., Pottier N., Broly F., Lo-Guidice J.-M. (2012). Xenobiotic metabolism and disposition in human lung cell models: Comparison with in vivo expression profiles. Drug Metab. Dispos. 40, 1953–1965. [DOI] [PubMed] [Google Scholar]
- Cox G., Gauldie J., Jordana M. (1992). Bronchial epithelial cell-derived cytokines (G-CSF and GM-CSF) promote the survival of peripheral blood neutrophils in vitro. Am. J. Respir. Cell Mol. Biol. 7, 507–513. [DOI] [PubMed] [Google Scholar]
- Crosby L. M., Waters C. M. (2010). Epithelial repair mechanisms in the lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 298, L715–L731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta J., Kar S., Liu R., Joseph J., Kalyanaraman B., Remington S. J., Chen C., Melendez J. A. (2010). Reactive oxygen species control senescence-associated matrix metalloproteinase-1 through c-Jun-N-terminal kinase. J. Cell. Physiol. 225, 52–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean J. P., Nelson P. S. (2008). Profiling influences of senescent and aged fibroblasts on prostate carcinogenesis. Br. J. Cancer 98, 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duell B. L., Cripps A. W., Schembri M. A., Ulett G. C. (2011). Epithelial cell coculture models for studying infectious diseases: benefits and limitations. J. BioMed Biotechnol. (2011), 852419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyman D., Damodarasamy M., Plymate S. R., Reed M. J. (2009). CCL5 secreted by senescent aged fibroblasts induces proliferation of prostate epithelial cells and expression of genes that modulate angiogenesis. J. Cell. Physiol. 220,376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E., Watt F. M. (2003). Cell differentiation: Focus on epithelia. Curr. Opin. Cell Biol. 15, 738–739. [Google Scholar]
- Gautier L., Cope L., Bolstad B. M., Irizarry R. A. (2004). affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315. [DOI] [PubMed] [Google Scholar]
- Gebel S., Lichtner R. B., Frushour B., Schlage W. K., Hoang V., Talikka M., Hengstermann A., Mathis C., Veljkovic E., Peck M., et al. (2013). Construction of a computable network model for DNA damage, autophagy, cell death, and senescence. Bioinform. Biol. Insights 7, 97–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueders M. M., Foidart J. M., Noel A., Cataldo D. D. (2006). Matrix metalloproteinases (MMPs) and tissue inhibitors of MMPs in the respiratory tract: Potential implications in asthma and other lung diseases. Eur. J. Pharmacol. 533,133–144. [DOI] [PubMed] [Google Scholar]
- Hoeng J., Deehan R., Pratt D., Martin F., Sewer A., Thomson T. M., Drubin D. A., Waters C. A., de Graaf D., Peitsch M. C. (2012). A network-based approach to quantifying the impact of biologically active substances. Drug Discov. Today 17, 413–418. [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Hudy M. H., Traves S. L., Wiehler S., Proud D. (2010). Cigarette smoke modulates rhinovirus-induced airway epithelial cell chemokine production. Eur. Respir. J. 35, 1256–1263. [DOI] [PubMed] [Google Scholar]
- International Organization for Standardization. (1999). ISO 3402: 1999—Tobacco and tobacco products—Atmosphere for conditioning and testing. In 4th Edition. Available at: http://www.iso.org/iso/home/store/catalogue_tc/catalogue_detail.htm?csnumber=28324. Accessed June 5, 2015. [Google Scholar]
- Iskandar A. R., Martin F., Talikka M., Schlage W. K., Kostadinova R., Mathis C., Hoeng J., Peitsch M. C. (2013). Systems approaches evaluating the perturbation of xenobiotic metabolism in response to cigarette smoke exposure in nasal and bronchial tissues. Biomed. Res. Int. (2013), 512086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A. C., Didone L., Jobson J., Sofia M. K., Krysan D., Dunman P. M. (2013). Adenylate kinase release as a high-throughput-screening-compatible reporter of bacterial lysis for identification of antibacterial agents. Antimicrob. Agents Chemother. 57, 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R., Weinberg R. A. (2009). The basics of epithelial-mesenchymal transition. The J. Clin. Investig. 119, 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartvelishvili T., Abuladze M., Asatiani N., Akhvlediani J., Asanishvili L., Holman H. Y., Sapojnikova N. (2004). Antioxidant capacity of cultured mammalian cells estimated by ESR method. ScientificWorldJournal 4, 490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsen S. G., Aksoy M. O., Yang Y., Shahabuddin S., Litvin J., Safadi F., Rogers T. J. (2004). The chemokine receptor CXCR3 and its splice variant are expressed in human airway epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 287, L584–L591. [DOI] [PubMed] [Google Scholar]
- Liu D., Hornsby P. J. (2007). Senescent human fibroblasts increase the early growth of xenograft tumors via matrix metalloproteinase secretion. Cancer Res. 67, 3117–3126. [DOI] [PubMed] [Google Scholar]
- Lundien M. C., Mohammed K. A., Nasreen N., Tepper R. S., Hardwick J. A., Sanders K. L., Van Horn R. D., Antony V. B. (2002). Induction of MCP-1 expression in airway epithelial cells: role of CCR2 receptor in airway epithelial injury. J. Clin. Immunol. 22, 144–152. [DOI] [PubMed] [Google Scholar]
- Majeed S., Frentzel S., Wagner S., Kuehn D., Leroy P., Guy P. A., Knorr A., Hoeng J., Peitsch M. C. (2014). Characterization of the Vitrocell(R) 24/48 aerosol exposure system using mainstream cigarette smoke. Chem. Cent. J. 8, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F., Sewer A., Talikka M., Xiang Y., Hoeng J., Peitsch M. C. (2014). Quantification of biological network perturbations for mechanistic insight and diagnostics using two-layer causal models. BMC Bioinform. 15, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall M. N., Bolstad B. M., Irizarry R. A. (2010). Frozen robust multiarray analysis (fRMA). Biostatistics 11, 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCI, N. C. I. NCI Dictionary of Cancer Terms: bronchial brush biopsy. Available at: http://www.cancer.gov/dictionary?cdrid=661165. Accessed October 20, 2014.
- O’Kane C. M., Elkington P. T., Jones M. D., Caviedes L., Tovar M., Gilman R. H., Stamp G., Friedland J. S. (2010). STAT3, p38 MAPK, and NF-kappaB drive unopposed monocyte-dependent fibroblast MMP-1 secretion in tuberculosis. Am. J. Respir. Cell. Mol. Biol. 43, 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C. G., Erjefalt J. S., Erjefalt I., Korsgren M. C., Nilsson M. C., Sundler F. (1996). Epithelial shedding—Restitution as a causative process in airway inflammation. Clin. Exp. Allergy 26, 746–755. [PubMed] [Google Scholar]
- Port J. L., Yamaguchi K., Du B., De Lorenzo M., Chang M., Heerdt P. M., Kopelovich L., Marcus C. B., Altorki N. K., Subbaramaiah K., et al. (2004). Tobacco smoke induces CYP1B1 in the aerodigestive tract. Carcinogenesis 25, 2275–2281. [DOI] [PubMed] [Google Scholar]
- Proud D., Leigh R. (2011). Epithelial cells and airway diseases. Immunol. Rev. 242, 186–204. [DOI] [PubMed] [Google Scholar]
- Rodier F., Campisi J. (2011). Four faces of cellular senescence. J. Cell Biol. 192, 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schamberger A. C., Staab-Weijnitz C. A., Mise-Racek N., Eickelberg O. (2015). Cigarette smoke alters primary human bronchial epithelial cell differentiation at the air-liquid interface. Sci. Rep. 5, 8163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schembri F., Sridhar S., Perdomo C., Gustafson A. M., Zhang X., Ergun A., Lu J., Liu G., Zhang X., Bowers J., et al. (2009). MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl. Acad. Sci. U.S.A. 106, 2319–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlage W. K., Westra J. W., Gebel S., Catlett N. L., Mathis C., Frushour B. P., Hengstermann A., Van Hooser A., Poussin C., Wong B. (2011). A computable cellular stress network model for non-diseased pulmonary and cardiovascular tissue. BMC Syst. Biol. 5, 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shebani E., Shahana S., Janson C., Roomans G. M. (2005). Attachment of columnar airway epithelial cells in asthma. Tissue Cell 37, 145–152. [DOI] [PubMed] [Google Scholar]
- Smyth G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3. [DOI] [PubMed] [Google Scholar]
- Spira A., Beane J. E., Shah V., Steiling K., Liu G., Schembri F., Gilman S., Dumas Y. M., Calner P., Sebastiani P., et al. (2007). Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat. Med. 13, 361–366. [DOI] [PubMed] [Google Scholar]
- Talikka M., Kostadinova R., Xiang Y., Mathis C., Sewer A., Majeed S., Kuehn D., Frentzel S., Merg C., Geertz M., et al. (2014). The response of human nasal and bronchial organotypic tissue cultures to repeated whole cigarette smoke exposure. Int. J. Toxicol. 33, 506–517. [DOI] [PubMed] [Google Scholar]
- Thomson T. M., Sewer A., Martin F., et al. (2013). Quantitative assessment of biological impact using transcriptomic data and mechanistic network models. Toxicol Appl Pharmacol. 273, 863–878. [DOI] [PubMed] [Google Scholar]
- Westra J. W., Schlage W. K., Frushour B. P., Gebel S., Catlett N. L., Han W., Eddy S. F., Hengstermann A., Matthews A. L., Mathis C. (2011). Construction of a computable cell proliferation network focused on non-diseased lung cells. BMC Syst. Biol. 5, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra J. W., Schlage W. K., Hengstermann A., Gebel S., Mathis C., Thomson T., Wong B., Hoang V., Veljkovic E., Peck M. (2013). A modular cell-type focused inflammatory process network model for non-diseased pulmonary tissue. Bioinform. Biol. Insights 7, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Y., Wang Y., Prakash C. (2006). Xenobiotic-metabolizing enzymes in human lung. Curr. Drug Metab. 7, 939–948. [DOI] [PubMed] [Google Scholar]
- Zhang X., Sebastiani P., Liu G., Schembri F., Zhang X., Dumas Y. M., Langer E. M., Alekseyev Y., O’Connor G. T., Brooks D. R., et al. (2010). Similarities and differences between smoking-related gene expression in nasal and bronchial epithelium. Physiol. Genomics 41, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.