Abstract
The hyperthermophilic archaeon Thermococcus kodakarensis can utilize sugars or pyruvate for growth. In the absence of elemental sulfur, the electrons via oxidation of these substrates are accepted by protons, generating molecular hydrogen (H2). The hydrogenase responsible for this reaction is a membrane-bound [NiFe]-hydrogenase (Mbh). In this study, we have examined several possibilities to increase the protein levels of Mbh in T. kodakarensis by genetic engineering. Highest levels of intracellular Mbh levels were achieved when the promoter of the entire mbh operon (TK2080-TK2093) was exchanged to a strong constitutive promoter from the glutamate dehydrogenase gene (TK1431) (strain MHG1). When MHG1 was cultivated under continuous culture conditions using pyruvate-based medium, a nearly 25% higher specific hydrogen production rate (SHPR) of 35.3 mmol H2 g-dcw−1 h−1 was observed at a dilution rate of 0.31 h−1. We also combined mbh overexpression using an even stronger constitutive promoter from the cell surface glycoprotein gene (TK0895) with disruption of the genes encoding the cytosolic hydrogenase (Hyh) and an alanine aminotransferase (AlaAT), both of which are involved in hydrogen consumption (strain MAH1). At a dilution rate of 0.30 h−1, the SHPR was 36.2 mmol H2 g-dcw−1 h−1, corresponding to a 28% increase compared to that of the host T. kodakarensis strain. Increasing the dilution rate to 0.83 h−1 or 1.07 h−1 resulted in a SHPR of 120 mmol H2 g-dcw−1 h−1, which is one of the highest production rates observed in microbial fermentation.
Keywords: hydrogen, hydrogenase, hyperthermophile, archaea, genetic engineering, dark fermentation, Thermococcus
Introduction
In view of the high demand for renewable energy resources, biological hydrogen (H2) produced by photosynthetic and anaerobic fermentative microorganisms is a promising biofuel that has attracted research activities during the last decades (Hallenbeck, 2009; Oh et al., 2011; Rittmann et al., 2015). Light-dependent H2 production processes by photosynthetic organisms have been limited by their low cell-specific productivities, and by the requirement of large reactor surface areas for light exposure (Melis et al., 2000; Akkerman et al., 2002; Lo et al., 2010). In contrast, dark fermentation by fermentative anaerobes revealed higher productivities, and studies mostly focused on anaerobic cultures of mesophilic bacteria such as Enterobacter and Clostridium (Taguchi et al., 1995; Kumar and Das, 2001; Rittmann and Herwig, 2012), (hyper-) thermophilic bacteria such as Thermotoga and Caldicellulosiruptor (van Niel et al., 2002; Mars et al., 2010) and hyperthermophilic archaea, especially of the order Thermococcales, such as Pyrococcus and Thermococcus (Schicho et al., 1993; Kanai et al., 2013; Bae et al., 2015).
The hyperthermophilic archaeon T. kodakarensis grows on media with pyruvate or carbohydrates (such as soluble starch or maltodextrin) (Morikawa et al., 1994; Atomi et al., 2004). It displays one of the highest cell-specific H2 production rates when grown in a continuous culture (up to 60 mmol g-dcw−1 h−1) with pyruvate (Kanai et al., 2005). Using similar continuous culture conditions, even higher H2 production rates were reported for Pyrococcus furiosus (up to 102 mmol g-dcw−1 h−1 with maltose) (Schicho et al., 1993). Recently, a maximum cell-specific H2 production rate of 352 mmol g-dcw−1 h−1 with formate was reported in a batch culture of Thermococcus onnurineus (Bae et al., 2015). Bacteria typically exhibit maximum cell-specific H2 production rates below 40 mmol g-dcw−1 h−1 (Rittmann and Herwig, 2012), but have the advantage to reach higher cell densities.
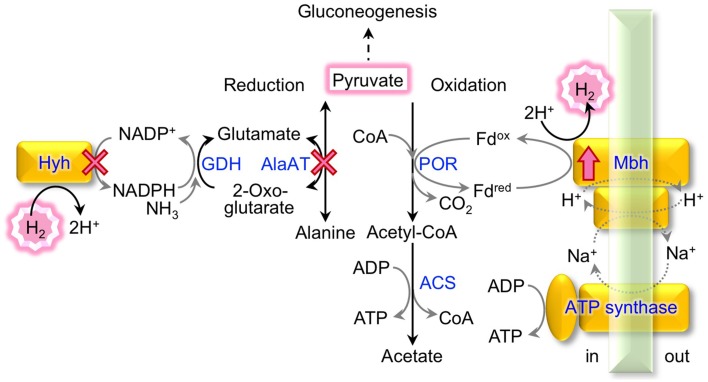
In T. kodakarensis, cultivation on pyruvate was shown to promote a 44% higher cell specific H2 production rate than cultivation on soluble starch (Kanai et al., 2005). Many enzymes involved in pyruvate metabolism and H2 production of Thermococcales were identified in P. furiosus (Verhees et al., 2003; Bräsen et al., 2014) and genome analysis of T. kodakarensis confirmed the presence of equivalent pathways in this organism (Fukui et al., 2005). Besides being used as starting material for gluconeogenesis, pyruvate is mainly either reduced to alanine via alanine aminotransferase (AlaAT) (Ward et al., 2000), or is oxidized to acetate (Figure 1).
Figure 1.
Pyruvate conversion and H2 metabolism in T. kodakarensis. The metabolic pathways of pyruvate reduction to alanine linked to H2 consumption and of pyruvate oxidation to acetate linked to H2 production are indicated. Enzymes marked with a cross were deleted and the Mbh was overproduced in this study; ACS, acetyl-CoA synthetase; AlaAT, alanine aminotransferase; GDH, glutamate dehydrogenase; Hyh, cytosolic [NiFe]-hydrogenase; Mbh, membrane-bound [NiFe]-hydrogenase; POR, pyruvate:ferredoxin oxidoreductase.
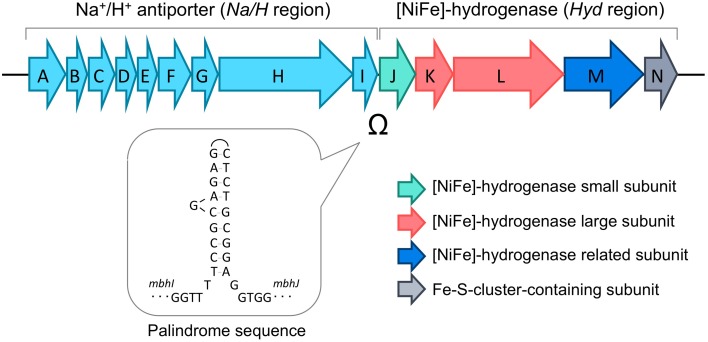
Pyruvate oxidation comprises two steps catalyzed by pyruvate:ferredoxin oxidoreductase (POR) (Blamey and Adams, 1993) and acetyl-CoA synthetases (ACSs), which produce ATP through substrate-level phosphorylation (Mai and Adams, 1996; Glasemacher et al., 1997). The POR reaction produces acetyl-CoA and CO2, and an electron from this reaction is transferred to oxidized ferredoxin (Fdox) to produce reduced ferredoxin (Fdred). A membrane-bound [NiFe]-hydrogenase complex (Mbh; TK2080-TK2093) (Figure 2) utilizes the electrons to produce molecular H2 with protons and regenerates Fdox (Sapra et al., 2000; Silva et al., 2000; Kanai et al., 2011). The metabolism indicates a H2/CO2 gas production ratio of 1 from pyruvate. The Mbh reaction also contributes to energy conservation as it is thought to be coupled to proton export, which via an Na+/H+-antiporter domain, results in a sodium gradient that fuels ATP synthesis by the A1A0-ATP synthase (Sapra et al., 2003; Pisa et al., 2007). Deletion of Mbh abolishes H2production and impairs growth under H2-producing conditions, reflecting that Mbh is the key [NiFe]-hydrogenase that is responsible for H2 production in T. kodakarensis (Kanai et al., 2011; Santangelo et al., 2011) as well as in P. furiosus (Schut et al., 2012).
Figure 2.
Gene structure of the membrane-bound [NiFe]-hydrogenase complex (Mbh). The palindromic sequence found between the genes encoding the Na+/H+ antiporter (Na/H region, mbhA-I) and the genes encoding the catalytic hydrogenase (Hyd region, mbhJ-N) are indicated.
Pyruvate reduction into alanine potentially competes with H2 production from pyruvate. Glutamate, which is used as an amino donor for pyruvate reduction through AlaAT, is regenerated from 2-oxoglutarate via glutamate dehydrogenase (GDH) coupled with NADPH consumption (Consalvi et al., 1991; Robb et al., 1992; Yokooji et al., 2013). NADPH is partially regenerated by a cytosolic [NiFe]-hydrogenase complex (Hyh; TK2069-2072), which utilizes H2 as an electron donor for NADP+ reduction (Bryant and Adams, 1989; Ma et al., 2000; Kanai et al., 2003, 2011). In a continuous, gas exchange culture of T. kodakarensis with pyruvate as a substrate, the deletion of hyh increases the gas production ratio of H2/CO2 by 8% (Kanai et al., 2011). An increase in cell-specific H2 production of up to three-fold was also reported in a closed batch culture with the same substrates (Santangelo et al., 2011).
Attempts to increase microbial H2 production via genetic engineering revealed two main successful strategies; overexpression of enzymes directly involved in H2 production and the deletion of competing pathways (Yoshida et al., 2005, 2007; Kim et al., 2009; Klein et al., 2010). The effect of homologous overexpression of the H2-evolving hydrogenase on cell-specific H2 production rates depends on the organism and ranges from no effect (Clostridium acetobutylicum) to a 2.8-fold increase (Escherichia coli) (Yoshida et al., 2005; Klein et al., 2010). Heterologous overexpression of the membrane-bound formate hydrogen lyase complex of T. onnurineus in P. furiosus enabled conversion of formate into H2 in addition to its native H2 production from maltose (Lipscomb et al., 2014). In E. coli, overexpression of the hydrogenase from Enterobacter cloacae led to H2 production levels comparable to those observed in Enterobacter species (Chittibabu et al., 2006). The effects of deleting competing pathways (H2-consuming hydrogenases, AlaAT) (Kanai et al., 2011; Santangelo et al., 2011) or pathways generating compounds that inhibit H2 production have been examined (Kim et al., 2009). For example, the disruption of lactate and succinate generating pathways in E. coli, which have a negative effect on H2 production, resulted in an increase in cell-specific H2 production rates by 1.3-fold (Yoshida et al., 2007).
In the present study, we performed homologous overexpression of the Mbh gene in T. kodakarensis via different genetic approaches in combination with the disruption of the genes encoding H2-consuming Hyh and AlaAT (Figure 1). The effects on both cell-specific H2 production rate (SHPR; mmol H2 g-dcw−1 h−1) and media-volume specific H2 evolution rate (HER; mmol H2 L−1 h−1) were analyzed during cultivation with pyruvate under continuous culture conditions.
Materials and methods
Microorganisms and culture conditions
E. coli DH5α was used for general DNA manipulation and sequencing. E. coli strains were cultivated in LB medium (10 g L−1 tryptone, 5 g L−1 yeast extract and 10 g L−1 NaCl) at 37°C. Ampicillin was added to the medium at a concentration of 100 μg mL−1.
T. kodakarensis strains and plasmids used in this study are listed in Table 1. T. kodakarensis strains were routinely grown under anaerobic conditions at 85°C in MA-YT medium with the following composition; 30.4 g L−1 Marine Art SF-1 salt as artificial sea salts (Tomita Pharmaceutical, Tokushima, Japan), 5 g L−1 yeast extract and 5 g L−1 tryptone. In the case of cultivation with S0, sulfur powder was added at a concentration of 2 g L−1 after autoclaving the MA-YT medium. In the case of cultivation with pyruvate, 5 g L−1 sodium pyruvate was added to the MA-YT medium before autoclaving (MA-YT-Pyr).
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics | Sources or references | |
|---|---|---|---|
| Strains | KU216 | KOD1 ΔpyrF | Sato et al., 2005 |
| MHG1 | KU216 mbh::Pmbh−2μ-Pgdh-mbhA | This study | |
| MHC1 | KU216 ΔchiA::Pcsg-mbhJKLMN-2μ | This study | |
| MPD1 | KU216 mbh::ΔΩ | This study | |
| DPHA1 | KU216 ΔhyhBGSL::2μ′Δaat::2μ′ | Kanai et al., 2011 | |
| MAH1 | DPHA1 mbh::Pmbh-Pcsg-mbhA | This study | |
| Plasmids | pUC118 | Ampr general cloning vector | Takara Bio (Otsu, Japan) |
| pUD | pUC118 derivative; pyrF marker cassette | Sato et al., 2003 | |
| pUD2 | pUC118 derivative; pyrF marker cassette | Sato et al., 2005 | |
| pUP1 | pUC118 derivative; 2μ −pyrF−2μ | This study | |
| pMHG1 | pUC118 derivative; Pmbh−2μ-pyrF−2μ- Pgdh-mbhA | This study | |
| pMHC1 | pUC118 derivative; chiAN-Pcsg-mbhJKLMN−2μ-pyrF−2μ-chiAC | This study | |
| pMPD1 | pUD2 derivative; ΔΩ | This study | |
| pMAH1 | pUD2 derivative; Pmbh-Pcsg-mbhA | This study |
Construction of T. kodakarensis mutant strains
Disruption of specific genes by double-crossover homologous recombination (for MHG1 and MHC1) or single-crossover homologous recombination followed by pop-out deletion of region containing pyrF marker (for MPD1 and MAH1) in T. kodakarensis was performed as described previously (Sato et al., 2003, 2005; Hirata et al., 2008). The sequences of all PCR primers used for this study are listed in Table 2. For Mbh overexpression in T. kodakarensis, four vectors (pMHG1, pMHC1, pMPD1, and pMAH1) were constructed as follows. Schemes of the cloning strategies are shown in the Supplementary Materials, Figure S1 for construction of pMHG1, Figure S2 for pMHC1, Figure S3 for pMPD1 and Figure S4 for pMAH1.
Table 2.
Sequences of primers used in this study.
| Plasmid used for | Name | Sequence (from 3′ to 5′) |
|---|---|---|
| pUP1 | PyrF-N-SP | AAAAACTAGTCCGCAACGCGCATTTTGCTCACCC |
| pUP1 | M13RV | CAGGAAACAGCTATGAC |
| pUP1 | 2μm-Sp | AAAAACTAGTGATAAGCTGTCAAAGATGAG |
| pUP1 | 2μm-Xb | AAAATCTAGAATGCGACGTGCAAGATTACC |
| pMHG1 | gdh-Nd | AAAACATATGTACCACCTCATTTCGGTAATCTGCGAGG |
| pMHG1 | gdh-Xb | AAAATCTAGATATCCCACCTCCGATTCCGTTGG |
| pMHG1 | mhp1 | AAAAGAATTCGGCTGGAGCGTTCATCGCCTTCG |
| pMHG1 | mhp2 | AAAATCTAGAGCTTAAAACGCTTTTCCCAAGC |
| pMHG1 | mhp3-3 | AAAATCTAGAAAAAACATATGTTGCCGTTCATAGTGGCGTTCCTC |
| pMHG1 | mhp4 | AAAAGTCGACCCTCGTAGGCATCAACAACCGC |
| pMHC1 | Tk-mbhJ-Nh | AAAAGCTAGCATGGCGATAACAGTTCCCGCCAAC |
| pMHC1 | Tk-mbhN-Bm | AAAAGGATCCACCTACGGTGAAGAACCGAAAAAA |
| pMHP1 | mhpd1 | AAAGGATCCAACCCTCATAGTAGGCAACGCGA |
| pMHP1 | mhpd4 | AAAGAATTCAGGCGGAGCGGGTAGATGCCCTC |
| pMHP1 | mhpd2-2 | AAACCCTTCATCCCCATATCA |
| pMHP1 | mhpd3-2 | CAAAAACACACTCTGCGGAGGTGGTAGCTGATG |
| pMAH1 | csgx | AAAATCTAGACGGCAAAAGGCGAATTATGTG |
| pMAH1 | csgn | AAAACATATGACAACACCTCCTTGGGTTG |
Construction of pMHG1
pUP1 is a plasmid that contains the pyrF marker gene of T. kodakarensis flanked by identical sequences (2 μm), necessary for marker removal via homologous recombination after cloning. pUP1 was constructed by amplification of the pyrF region from the pUD plasmid (Sato et al., 2003) with the primer set, PyrF-N-SP/M13RV and inserting the fragment into the SpeI and XbaI sites of pUC19-Sp. pUC19-Sp is a modified pUC19 plasmid containing an SpeI recognition site instead of the SmaI recognition site. Next, the primer set 2 μm-Sp/2 μm-Xb was used to amplify the 2 μm region from the yeast expression vector pYES (Life Technologies, Carlsbad, CA), and the fragment was inserted into the SpeI site upstream of the pyrF gene, and again into the XbaI site downstream of the pyrF gene. To enable further cloning via NdeI, an NdeI site (CATATG) inside of the pyrF gene of pUP1 was changed to CACATG by point mutation (underline indicates the position of the changed nucleotide), resulting in plasmid pUP1m. The promoter region of the glutamate dehydrogenase gene (TK1431) (Pgdh) was amplified from the genomic DNA of T. kodakarensis using the primer set gdh-Xb/gdh-Nd. The amplified fragment was inserted into the SpeI/NdeI site of pUP1m to yield plasmid pUPG1. Two genomic regions (1.0–1.1 kb each) including the promoter of the mbh operon (Pmbh) and a part of the mbh structural genes (mbhA) were amplified with the primer sets mhp1/mhp2 and mhp3-3/mhp4, respectively. The resulting fragments were cut by EcoRI/XbaI and XbaI/SalI, respectively, and were fused and inserted into the EcoRI/SalI sites of pUC118, resulting in plasmid pMHGa. A point mutation (T to C) was introduced to the 204th nucleotide of mbhA, to change an existing NdeI site (CATATG) to CATACG (underline indicates the position of the changed nucleotide), yielding plasmid pMHGam. The point mutation resulted in a change of the respective (68th) codon from TAT to TAC, both encoding the same amino acid (tyrosine). Next, a fragment containing 2 μm-pyrF−2 μm-Pgdh was cut from pUPG1 by NdeI/XbaI and introduced to the respective sites of pMHGam, to obtain plasmid pMHG1.
Construction of pMHC1
First, the mbhJKLMN genes as well as its terminator region and the cell surface glycoprotein gene (TK0895) promoter (Pcsg) were amplified from the genomic DNA of T. kodakarensis by PCR using the primer sets Tk-mbhJ-Nh/Tk-mbhN-Bm and Pcsg-Sp/Pcsg-Nh, respectively. Via the introduced SpeI, NheI, and BamHI cleavage sites of these fragments, Pcsg and mbhJKLMN were fused and inserted into the SpeI/BamHI sites of pUC19-Sp, yielding the plasmid pMH1. Second, SpeI and XbaI were used to clone the promoter gene cassette into the XbaI site of plasmid pUP1 containing the 2 μm-pyrF−2 μm cassette, resulting in plasmid pMHUP1. In the third step, the cassette including Pcsg, mbhJKLMN and 2 μm-pyrF−2 μm was excised via SpeI and inserted into the SpeI site of the plasmid pchiA-NC, to yield plasmid pMHC1. pchiA-NC is a pUC118 derivative with 0.9–1.0 kb homologous sequences of the 5′-flanking region of the T. kodakarensis chitinase gene (chiA,TK1765) and of the 3′-portion of the gene itself. After amplification of the 5′-flanking region and the 3′-portion of chiA from the genome using primer sets ChiA-1/ChiA-2 and ChiA-3/ChiA-4, respectively, both fragments contained overlapping regions upstream of the introduced SpeI sites and were fused in a second fusion PCR reaction using the primer set ChiA-1/ChiA-4. The resulting fragment was inserted into the EcoRI and SalI sites of the multi-cloning site of pUC118 upon digestion and blunt ending to yield pchiA-NC. The final plasmid pMHC1 carries the mbhJKLMN genes with its terminator region (Tmbh) and the 2 μm-pyrF−2 μm cassette, flanked by the chiA sequences for homologous recombination.
Construction of pMPD1
A palindrome sequence (5′-TCCGCGAGAGCTCTGCGGA-3′) is located within a non-coding region (37 bp) between the Mbh subunit structure genes mbhI and mbhJ. The non-coding region was amplified together with its adjacent mbh genes from the genomic DNA of T. kodakarensis using the primer set mhpd1/mhpd4. The fragment was cut by BamHI/EcoRI, and ligated into the respective sites of plasmid pUD2 (Sato et al., 2005), to yield plasmid pMPDa. In order to disrupt the palindrome sequence on pMPDa via nucleotide substitution (5′-CAAAAACACACTCTGCGGA-3′; underline indicates the positions of mutated nucleotides), inverse PCR was performed using the primer set mhpd2-2/mhpd3-2, and the amplified fragment was self-ligated to obtain plasmid pMPD1.
Construction of pMAH1
Using genomic DNA of T. kodakarensis, Pcsg was amplified with the primer set csgx/csgn, and the fragment cut by XbaI/NdeI was introduced to the respective sites of pMHGam, to yield plasmid pMAHa. A fragment containing the Pmbh region, Pcsg and a part of the mbhA structure gene was excised from this plasmid by sequentially applying SalI, DNA blunting and EcoRI digestion. The fragment was introduced into the EcoRI/SmaI site of pUD2, to obtain plasmid pMAH1.
DNA restriction and modification enzymes as well as general cloning plasmids were purchased from TaKaRa (Otsu, Japan) or Toyobo (Osaka, Japan). The KOD plus NEO DNA polymerase (Toyobo) was used for amplification, and DNA fragments separated via agarose gel electrophoresis were isolated using the MinElute gel extraction kit (Qiagen, Hilden, Germany). Plasmids were isolated with the Plasmid Mini kit (Qiagen). The cloning products were confirmed via sequencing with the BigDye Terminator cycle sequencing kit, version 3.1 and a model 3130 capillary DNA sequencer (Applied Biosystems, Foster City, CA).
For transformation, the T. kodakarensis uracil-auxotroph strains KU216 (Sato et al., 2005) (for MHG1, MHC1, and MPD1) and DPHA1 (Kanai et al., 2011) (for MAH1) were used as host strains. The transformation procedures included selection of pyrF+ strains with uracil-prototrophy and positive selection of pyrF-eliminated strains with 5-fluoroorotic acid and was performed as described elsewhere (Sato et al., 2005; Hirata et al., 2008; Kanai et al., 2011). Recombinant strains carrying the desired genetic modifications on the genome were identified by colony PCR and sequencing.
Western blot analysis
To determine intracellular protein levels of MbhL, Western blot analysis was performed. T. kodakarensis strains (KU216, DPHA1, MHG1, MHC1, MPD1, and MAH1) were cultivated in MA-YT medium supplemented with 0.5% (w/v) sodium pyruvate. After 11 h of cultivation at 85°C, cells were harvested by centrifugation under 5000 g for 10 min at 4°C. Cell pellets were resuspended in 25 mM Tris-HCl (pH 8.0) buffer containing 0.1% (v/v) Triton-X100, and disrupted by vortex for 30 min at 4°C. After removing the insoluble fraction by centrifugation under 5000 g for 10 min, the resulting cell extracts were used for Western blot analysis. Protein concentrations were measured using the Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA), with bovine serum albumin as the standard. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed in a 12.5% gel. Western blot analysis was performed as described previously (Endoh et al., 2006) using rabbit polyclonal antibodies against the MbhL protein.
Continuous culture experiments
Continuous culture experiments of the host strains KU216 and DPHA1 and the engineered strains MHG1, MHC1, MPD1, and MAH1 were performed as described previously (Kanai et al., 2011) using a gas-lift fermenter designed for cultivation of hyperthermophiles (Taiyo Nippon Sanso Corporation, Tokyo, Japan). In a 1 L cultivation vessel, 500 mL of MA-YT-Pyr medium was introduced and cultivation was performed at 85°C with continuous agitation using a rotor at 50 rpm. The evolved gas metabolites were flushed out by nitrogen gas, which was introduced continuously into the vessel at a rate of 100 mL min−1. Fresh medium was supplied into the vessel using a peristaltic pump and the volume of the culture was monitored with a water level sensor (B.E. Marubishi, Tokyo, Japan), which was connected to a pump for culture discharging. Cell densities were monitored by measuring the turbidity at 660 nm (OD660) and according biomasses (dcw) were calculated from OD660 via calibration information determined beforehand. The pH of the culture broth was maintained at 7.4 and the amounts of H2 gas and CO2 gas in the exhaust gas were measured periodically using gas chromatography (provided by Taiyo Nippon Sanso Corporation) as described previously (Kanai et al., 2005).
Results
Construction of T. kodakarensis strains that overexpress the Mbh genes
In T. kodakarensis, the membrane-bound hydrogenase, Mbh, is the key enzyme that is responsible for the evolution of H2 (Kanai et al., 2011). The mbh operon can be divided into two regions; the former region containing genes presumed to encode Na+/H+ antiporter subunits (Na/H region; mbhA-I; TK2080-TK2088), and the latter region containing genes for the catalytic [NiFe]-hydrogenase subunits (Hyd region; mbhJ-N; TK2089-TK2093) (Figure 2). These two regions are separated by a palindrome sequence (5′-TCCGCGAGAGCTCTGCGGA-3′) that can form a remarkably long stem-loop structure and may potentially inhibit transcription and/or translation.
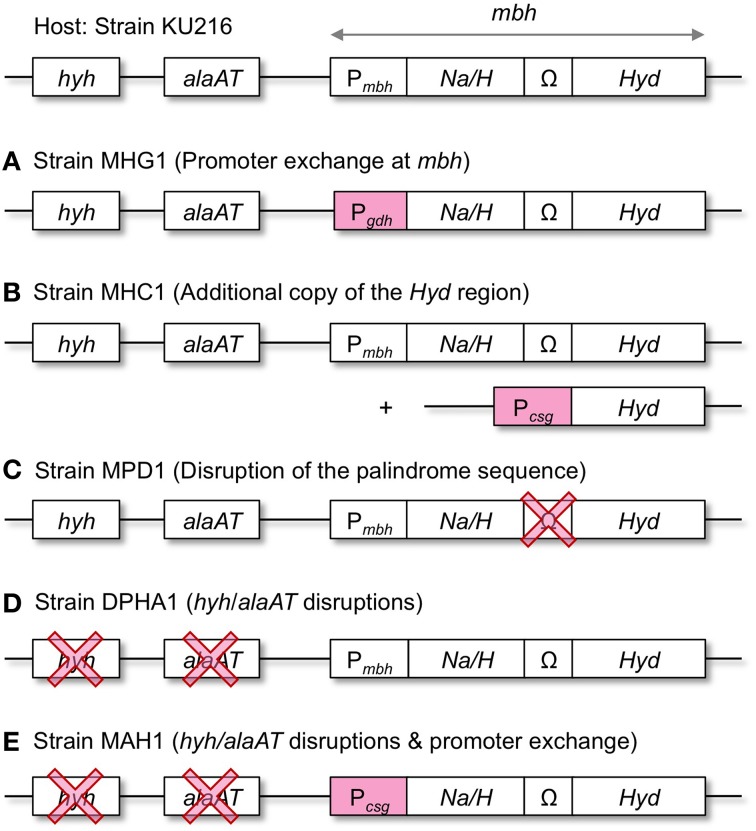
In order to enhance the capacity of H2 production, we took three different genetic approaches aiming to increase the Mbh protein levels in T. kodakarensis (Figure 3). First, the mbh promoter (Pmbh) of the entire operon was exchanged with the strong/constitutive glutamate dehydrogenase gene (TK1431) promoter (Pgdh) (strain MHG1). Second, the Hyd region, which encodes the catalytic subunits, was overexpressed under the control of another strong/constitutive cell-surface glycoprotein gene (TK0895) promoter (Pcsg) (strain MHC1). The construct was inserted into the chiA-locus, which encodes a chitinase (Tanaka et al., 1999), resulting in a strain with a second copy of the Hyd region. Third, the palindrome sequence between mbhI and mbhJ was deleted, as the Hyd gene cluster falls downstream of the palindrome, and removal of the sequence might enhance the expression of the Hyd genes (strain MPD1). All modifications were introduced into the genome of T. kodakarensis strain KU216 by homologous recombination and were confirmed via analytical PCR and sequencing (data not shown).
Figure 3.
Overview of the genetic approaches taken to increase H2 production in T. kodakarensis. (A–C), Mbh overexpression; (D), Disruption of the pyruvate reduction pathway associated with H2 consumption; (E), Combination of both approaches. hyh, cytosolic [NiFe]-hydrogenase gene; alaAT, alanine aminotransferase gene; Na/H, Na+/H+-antiporter region of Mbh (mbhA-I); Ω, palindromic sequence; Hyd, [NiFe]-hydrogenase catalytic region of Mbh (mbhJ-N); Pmbh, mbh promoter; Pgdh, gdh promoter; Pcsg, csg promoter.
Quantification of MbhL protein in the recombinant strains
In order to compare the Mbh production levels of the constructed T. kodakarensis strains, Western blot analysis was performed on the extracts of cells grown in pyruvate medium (MA-YT-Pyr) and compared (Figure 4). Antibodies raised against the large subunit of Mbh (MbhL) were applied to estimate the overexpression of the catalytic Hyd subunits.
Figure 4.
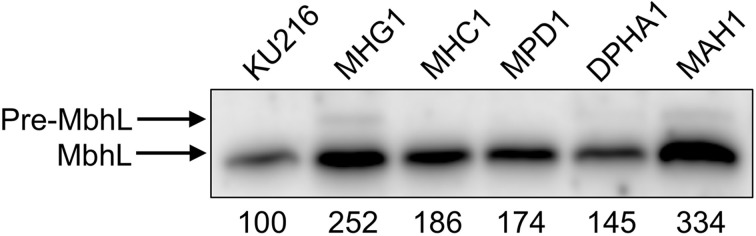
Expression levels of the [NiFe]-hydrogenase large subunit (MbhL) in the constructed T. kodakarensis strains. Numbers below the panel indicate band intensity (%) relative to that of the host strain KU216 (defined as 100%). Pre-MbhL indicates the MbhL precursor (see Discussion).
Quantification of the bands revealed that protein levels of MbhL were higher in all three recombinant strains compared to that observed in the host strain KU216. MbhL levels in strain MHC1 (addition of Mbh Hyd genes under the control of Pcsg) and strain MPD1 (deletion of the palindrome sequence) increased 1.86-fold and 1.74-fold, respectively. Strain MHG1, whose mbh operon is under the control of Pgdh, displayed even higher levels of MbhL, 2.52-fold higher than that of the host strain. Extracts from this strain revealed an additional band (Pre-MbhL) with a higher molecular weight than that of MbhL (see Discussion).
Hydrogen production under continuous culture conditions
HERs of the Mbh overexpression strains (MHG1, MHC1, and MPD1) were examined and compared with that of the host strain (KU216). If the H2-forming Mbh reaction is the bottleneck of H2 production from pyruvate, increases in MbhL protein might result in increases in SHPR. To investigate this relationship, cell- and culture volume-specific H2 production rates (SHPR, HER) of the T. kodakarensis strains were analyzed under continuous culture conditions using a continuous gas-flow fermenter.
At a dilution rate of 0.27–0.31 h−1, cell densities (OD660) of all strains were between 0.84 and 1.09 (Table 3). In these cultures, HERs ranged from 9.4 to 11.2 mmol L−1 h−1 with the host strain KU216 displaying the lowest H2 production, while the highest production was observed with strain MHC1.
Table 3.
Average cell densities, H2 productivities (HERs and SHPRs), and molecular H2/CO2 ratios of T. kodakarensis strains.
| Strain | D (h−1) | OD660 | HER (mmol L−1 h−1) | SHPR (mmol g-dcw−1 h−1) | H2/CO2 |
|---|---|---|---|---|---|
| KU216 | 0.27 | 0.95 ± 0.01 | 9.4 ± 0.3 | 28.3 ± 0.9 | 0.96 |
| MHG1 | 0.31 | 0.84 ± 0.01 | 10.3 ± 0.1 | 35.3 ± 0.5 | 0.91 |
| MHC1 | 0.27 | 0.98 ± 0.01 | 11.2 ± 0.3 | 32.4 ± 1.2 | 0.93 |
| MPD1 | 0.30 | 1.09 ± 0.01 | 11.1 ± 0.6 | 28.9 ± 1.4 | 0.88 |
| DPHA1 | 0.30 | 1.15 ± 0.01 | 11.7 ± 0.5 | 28.7 ± 1.4 | 0.96 |
| MAH1 | 0.30 | 1.02 ± 0.01 | 13.0 ± 0.3 | 36.2 ± 1.1 | 0.98 |
| MAH1 | 0.59 | 0.91 ± 0.03 | 24.6 ± 0.6 | 76.7 ± 2.4 | 0.99 |
| MAH1 | 0.83 | 0.64 ± 0.03 | 27.1 ± 1.6 | 120 ± 2 | 1.04 |
| MAH1 | 1.07 | 0.41 ± 0.02 | 17.4 ± 0.9 | 120 ± 9 | 1.18 |
D, Dilution rate; Error bars represent standard deviations of at least three measured points at the steady state of each dilution rate.
As the HER depends on the cell densities, differences in SHPR more accurately reflect the impact of genetic modification on H2 production in the cell. The deletion of a palindrome sequence in strain MPD1 caused an increase in MbhL protein (Figure 4), but hardly changed the SHPR (Table 3). For the other strains, on the other hand, there was a general tendency that strains with higher levels of MbhL protein resulted in higher SHPR values; 28.3 (host), 32.4 (strain MHC1) and 35.3 mmol g-dcw−1 h−1 (strain MHG1).
Effect of combining Mbh overexpression with deletion of the pyruvate reduction pathway linked to H2 consumption
Promoter exchange by Pgdh (strain MHG1) exhibited the highest effect among the three Mbh overexpression strains examined. As a next step, we focused on the disruption of the pyruvate reduction pathway to alanine. The pathway is metabolically linked to H2 consumption and its disruption circumvents H2 uptake of T. kodakarensis (Kanai et al., 2011). The double knock out strain (DPHA1) carries hyh and alaAT gene deletions and was previously shown to exhibit a higher SHPR than its host strain KU216 (Kanai et al., 2011). To check whether Mbh overexpression and deletion of hyh and alaAT have an additive effect on H2 production, DPHA1 was further engineered to overexpress the mbh operon via promoter exchange with Pcsg, resulting in strain MAH1.
Levels of MbhL protein in strain DPHA1 and in strain MAH1 were examined via Western blot analysis using anti-MbhL antibodies. As a result, MAH1 exhibited strikingly higher levels of MbhL; 3.34-fold and 2.30-fold higher band intensities were observed when compared to those of the strains KU216 and DPHA1, respectively (Figure 4). The results also indicate that the MbhL protein levels in MAH1 are higher than those in MHG1, and as such, intracellular Pre-MbhL accumulation found in MHG1 was also observed in strain MAH1 (see Discussion).
Evaluations of HERs in continuous cultures of DPHA1 and MAH1 were examined at a dilution rate of 0.30 h−1. Unlike the previously reported examination (Kanai et al., 2011), hyh and alaAT deletion (strain DPHA1) only slightly increased SHPR (Table 3). In contrast, strain MAH1 exhibited the highest increases in SHPR with 36.2 mmol g-dcw−1 h−1. The increase of SHPR by 28% is slightly above the increase caused by the promoter exchange with Pgdh in strain MHG1 (25%). This agrees with the higher MbhL protein levels found in strain MAH1 than in MHG1. The higher levels of MbhL in MAH1 compared to those in MHG1 may be due to differences in the strengths of the promoters Pcsg and Pgdh. However, the additional disruption of hyh and alaAT in MAH1 may also have an effect, as the MbhL levels in DPHA1 are higher than those in KU216, even though there are no changes in the promoters governing mbhL expression. In addition to the high SHPR, strain MAH1 also exhibited the highest HER (13.0 mmol L−1 h−1) among the strains examined at a dilution rate of around 0.3 h−1.
Influence of the culture dilution rates on SHPRs
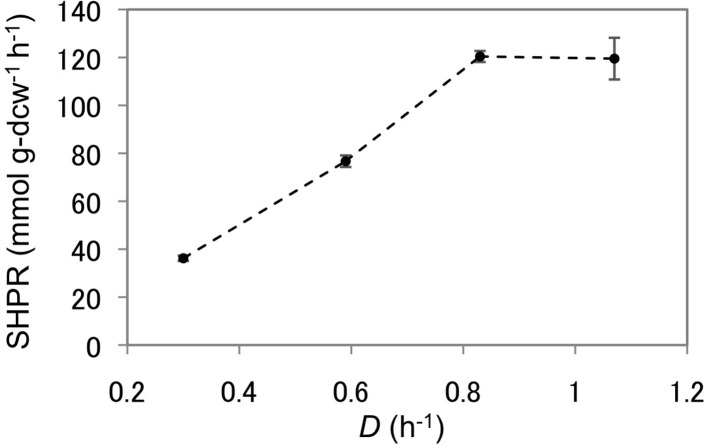
As T. kodakarensis strain MAH1 displayed the highest SHPRs and HERs, this strain was used to analyze the effect of dilution rates on H2 production from pyruvate. The dilution rate was increased stepwise from 0.30 to 0.59, 0.83, and 1.07 h−1. SHPRs as well as HERs increased gradually and both displayed their maxima at a dilution rate of 0.83 (Figure 5, Table 3). The SHPR and HER at this dilution rate were 120 mmol g-dcw−1 h−1 and 27.1 mmol L−1 h−1, respectively. Both values (SHPR and HER) are so far the highest of those reported for T. kodakarensis. At a dilution rate of 1.07 h−1, SHPR maintained a constant value of 120 mmol g-dcw−1, whereas the volume-specific HERs dropped to 17.4 mmol L−1 h−1 as a result of a decrease in cell density.
Figure 5.
SHPRs of strain MAH1 at different dilution rates. Error bars represent standard deviations of at least three measured points during the steady-state of each dilution rate. D, Dilution rate.
Discussion
In this study, different strategies were taken to overproduce the [NiFe]-hydrogenase complex Mbh in T. kodakarensis and to reduce H2-consuming pathways. The H2 production potential of these engineered strains were examined in a continuous culture, where evaluation is possible under steady-state conditions. As a result, we found that the increase in SHPR was highest in strain MAH1, with a 28% increase compared to the host strain at dilution rates of 0.27–0.31 h−1.
In comparison, the SHPR from formate in E. coli increased by 2.8-fold in a batch culture when deleting a negative transcription regulator and overexpressing a transcriptional activator of the formate hydrogenlyase complex (strain SR13 in Table 4) (Yoshida et al., 2005). In T. onnurineus KS0413, also in a batch culture, up to 2.9-fold increased SHPRs were reached by promoter exchange of the carbon monoxide dehydrogenase (CODH) operon including CODH, hydrogenase and an Na+/H+ antiporter with Pcsg (Table 4) (Kim et al., 2013; Lee et al., 2014). In both cases, hydrogenase overexpression yielded much higher increases in SHPR compared to those obtained in this study. This is most likely due to the fact that the substrate to H2 conversion (formate –> H2+ CO2 or CO + H2O –> CO2+ H2) comprises only one enzymatic step which was subjected to overexpression. In contrast, the H2 production from pyruvate in T. kodakarensis involves at least one additional enzyme, POR, and the flux might also be affected by the downstream ACS (Figure 1). As we did observe 25–28% increases in SHPR in strains MHG1 and MAH1, the Mbh reaction seems to be the rate-limiting step for H2 production from pyruvate in the wild type T. kodakarensis. The maximal increase in SHPR upon Mbh overexpression was probably reached, as promoter exchange of Pmbh with Pcsg provided higher protein levels (334%, strain MAH1) than Pgdh (252% strain MHG1), but only slightly increased SHPR values (36 compared to 35 mmol g-dcw−1 h−1). In order to reach higher SHPR values, a simultaneous increase in the levels of Mbh, POR, and ACS may be necessary.
Table 4.
Strong microbial H2 producers and their maximal H2 production rates.
| Organism | Substrate | Culture conditions | SHPR | HER | Reference | |
|---|---|---|---|---|---|---|
| Continuous culture | Thermococcus kodakarensis MAH1 | Pyruvate | Gas removal, D: 0.83, T: 85 | 120.4 | 27.1 | This study |
| Pyrococcus furiosus DSM3638 | Maltose | D: 0.6, T: 98 | 102* | – | Schicho et al., 1993 | |
| Thermococcus kodakarensis KOD1 | Pyruvate | Gas removal, D: 0.8, T: 85 | 59.6 | 6.3 | Kanai et al., 2005 | |
| Clostridium sp. No. 2 | Glucose/Xylose | D: 1.2-1.3, T: 36 | 34.0/41.9 | 20.4/15.1 | Taguchi et al., 1995 | |
| Caldicellulosiruptor kristjanssonii DSM12137 | Glucose | D: 0.15, T: 70 | 34.6 | 10.3 | Zeidan et al., 2010 | |
| Klebsiella oxytoca HP1 | Sucrose | T: 38 | 15.2 | 14.4# | Minnan et al., 2005 | |
| Batch culture | Thermococcus onnurineus NA1 | Formate | T: 80 | 351.6 | 85.8 | Bae et al., 2015 |
| Escherichia coli SR13 | Formate | Enriched cells in buffer, substrate feed, T: 37 | 250.0 | 12,351.3# | Yoshida et al., 2005 | |
| Thermococcus onnurineus KS0413 | CO | pH control, CO feed, T: 80 | 207.8 | 88.4 | Lee et al., 2014 | |
| Citrobacter sp. Y19 | Glucose | T: 36 | 32.3 | 4.9# | Oh et al., 2003 | |
| Enterobacter cloacae IIT-BT 08 | Sucrose | pH control, T: 36 | 29.5 | 35.6 | Kumar and Das, 2000 | |
| Ethanoligenens harbinense B49 | Glucose | T: 36 | 27.7 | 7.5* | Xu et al., 2008 | |
| Klebsiella oxytoca HP1 | Glucose | In buffer, T: 35 | 9.6 | 3.6# | Minnan et al., 2005 | |
| Thermoanaerobacterium thermosaccharolyticum W16 | Glucose/Xylose | T: 60 | 9.7/8.8 | 12.9/10.7 | Ren et al., 2008 | |
| Thermotoga elfii DSM9442 | Glucose | T: 65 | 8.9 | 4.5 | van Niel et al., 2002 | |
| Thermotoga neapolitana DSM4359 | Xylose | T: 80 | 0.24 | 1.45 | Eriksen et al., 2011 | |
| Photosynthetic bacteria and algae | Organic acids, sugars | T: 35 | < 6 | < 6 | Hillmer and Gest, 1977 |
SHPR (mmol g-dcw−1 h−1); HER (mmol L−1 h−1); D, Dilution rate (h−1); T, cultivation temperature (°C);
Values estimated from a plot;
Converted from ml/L/h via gas constant at 23°C and 1 atm.
Interestingly, we observed the presence of the precursor of the large Mbh subunit (Pre-MbhL) at high Mbh overexpression levels (strains MHG1 and MAH1 in Figure 4). Posttranslational maturation of the active center of the large Mbh subunit (MbhL) is assisted by the Mbh accessory Hyp proteins (Sasaki et al., 2012, 2013; Watanabe et al., 2012a, 2015; Tominaga et al., 2013), which is completed by the cleavage of the Pre-MbhL protein into the functional MbhL via specific endopeptidases (Forzi and Sawers, 2007; Watanabe et al., 2012b). The increased levels of Pre-MbhL in strains MHG1 and MAH1 may be exceeding the functional capacity of the Hyp proteins, thereby leading to the accumulation of precursor. The Na+/H+ antiporter does not seem to be required for Mbh maturation, as overexpression of the Hyd region without the Na/H region in strain MHC1 resulted in an increase in mature MbhL (Figure 4) and increased H2 production (Table 3).
The increase in SHPR brought about by deletion of hyh and alaAT in this study was lower than those observed elsewhere (Kanai et al., 2011). In batch cultures, three-fold higher cell specific H2 productions from pyruvate were reached when hyh was disrupted and an estimated 9% higher H2 productions when alaAT was disrupted (Santangelo et al., 2011). The continuous removal of H2 from the gas phase in our cultures is probably the reason for the much lower effects of hyh and alaAT disruption on H2 consumption. Large effects of gas removal on H2 production (54% increase) have also been demonstrated in studies with a mixed microbial culture and glucose as a substrate. Gas removal was suggested to prevent H2 (and CO2) consumption by homoacetogenesis (Esquivel-Elizondo et al., 2014). Increased H2 concentrations in the liquid phase caused by higher gas phase pressures were also assumed to influence the equilibrium of the H2 production step in E. cloacae (Mandal et al., 2006).
Among fermentative microorganisms, the T. kodakarensis strain MAH1 exhibits relatively high H2 microbial production rates (Table 4). This demonstrates the high potential of this strain as a host strain for further engineering. Examining the H2 production of this strain grown on cheaper substrates like sugars will be important, as demonstrated with P. furiosus (Schicho et al., 1993), which is also a strong H2 producer. The HER can probably be further enhanced by increasing cell densities, for example by cell immobilization (Zhao et al., 2012). Studies with the E. coli strain SR13 showed that beside genetic modification, the use of concentrated cells results in extremely high H2 yields (Yoshida et al., 2005).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was partially funded by the Core Research for Evolutional Science and Technology program of the Japan Science and Technology Agency to HA within the research area “Creation of Basic Technology for Improved Bioenergy Production through Functional Analysis and Regulation of Algae and Other Aquatic Microorganisms.” This work was also partially funded by JSPS KAKENHI Grant Number 26292038 (to TK).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00847
Strategy for construction of pMHG1. pMHG1 was used to insert Pgdh upstream of the mbhA gene of the mbh operon via homologous recombination using strain KU216 as the host.
Strategy for construction of pMHC1. pMHC1 was used to introduce an additional Hyd gene region under the control of Pcsg into the chitinase region of strain KU216 via homologous recombination.
Strategy for construction of pMPD1. pMPD1 was used to replace the palindrome sequence between the Na/H- and Hyd regions of strain KU216 with a non-coding sequence that does not form a stem loop structure via homologous recombination.
Strategy for construction of pMAH1. pMAH1 was used to introduce Pcsg upstream of the mbhA gene of the mbh operon via homologous recombination. Strain DPHA1 was used as the host in order to combine Mbh overexpression with alaAT and hyh deletion.
References
- Akkerman I., Janssen M., Rocha J., Wijffels R. H. (2002). Photobiological hydrogen production: photochemical efficiency and bioreactor design. Int. J. Hydrogen Energy 27, 1195–1208. 10.1016/S0360-3199(02)00071-X [DOI] [Google Scholar]
- Atomi H., Fukui T., Kanai T., Morikawa M., Imanaka T. (2004). Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1, 263–267. 10.1155/2004/204953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S. S., Lee H. S., Jeon J. H., Lee J. H., Kang S. G., Kim T. W. (2015). Enhancing bio-hydrogen production from sodium formate by hyperthermophilic archaeon, Thermococcus onnurineus NA1. Bioprocess Biosyst. Eng. 38, 989–993. 10.1007/s00449-014-1336-9 [DOI] [PubMed] [Google Scholar]
- Blamey J. M., Adams M. W. (1993). Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim. Biophys. Acta 1161, 19–27. 10.1016/0167-4838(93)90190-3 [DOI] [PubMed] [Google Scholar]
- Bräsen C., Esser D., Rauch B., Siebers B. (2014). Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 78, 89–175. 10.1128/MMBR.00041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant F. O., Adams M. W. (1989). Characterization of hydrogenase from the hyperthermophilic archaebacterium, Pyrococcus furiosus. J. Biol. Chem. 264, 5070–5079. [PubMed] [Google Scholar]
- Chittibabu G., Nath K., Das D. (2006). Feasibility studies on the fermentative hydrogen production by recombinant Escherichia coli BL-21. Process Biochem. 41, 682–688. 10.1016/j.procbio.2005.08.020 [DOI] [Google Scholar]
- Consalvi V., Chiaraluce R., Politi L., Vaccaro R., De Rosa M., Scandurra R. (1991). Extremely thermostable glutamate dehydrogenase from the hyperthermophilic archaebacterium Pyrococcus furiosus. Eur. J. Biochem. 202, 1189–1196. 10.1111/j.1432-1033.1991.tb16489.x [DOI] [PubMed] [Google Scholar]
- Endoh T., Kanai T., Sato Y. T., Liu D. V., Yoshikawa K., Atomi H., et al. (2006). Cell-free protein synthesis at high temperatures using the lysate of a hyperthermophile. J. Biotechnol. 126, 186–195. 10.1016/j.jbiotec.2006.04.010 [DOI] [PubMed] [Google Scholar]
- Eriksen N. T., Riis M. L., Holm N. K., Iversen N. (2011). H2 synthesis from pentoses and biomass in Thermotoga spp. Biotechnol. Lett. 33, 293–300. 10.1007/s10529-010-0439-x [DOI] [PubMed] [Google Scholar]
- Esquivel-Elizondo S., Chairez I., Salgado E., Aranda J. S., Baquerizo G., Garcia-Pena E. I. (2014). Controlled continuous bio-hydrogen production using different biogas release strategies. Appl. Biochem. Biotechnol. 173, 1737–1751. 10.1007/s12010-014-0961-8 [DOI] [PubMed] [Google Scholar]
- Forzi L., Sawers R. G. (2007). Maturation of [NiFe]-hydrogenases in Escherichia coli. Biometals 20, 565–578. 10.1007/s10534-006-9048-5 [DOI] [PubMed] [Google Scholar]
- Fukui T., Atomi H., Kanai T., Matsumi R., Fujiwara S., Imanaka T. (2005). Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15, 352–363. 10.1101/gr.3003105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasemacher J., Bock A. K., Schmid R., Schönheit P. (1997). Purification and properties of acetyl-CoA synthetase (ADP-forming), an archaeal enzyme of acetate formation and ATP synthesis, from the hyperthermophile Pyrococcus furiosus. Eur. J. Biochem. 244, 561–567. 10.1111/j.1432-1033.1997.00561.x [DOI] [PubMed] [Google Scholar]
- Hallenbeck P. C. (2009). Fermentative hydrogen production: principles, progress, and prognosis. Int. J. Hydrogen Energy 34, 7379–7389. 10.1016/j.ijhydene.2008.12.080 [DOI] [Google Scholar]
- Hillmer P., Gest H. (1977). H2 metabolism in the photosynthetic bacterium Rhodopseudomonas capsulata: H2 production by growing cultures. J. Bacteriol. 129, 724–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata A., Kanai T., Santangelo T. J., Tajiri M., Manabe K., Reeve J. N., et al. (2008). Archaeal RNA polymerase subunits E and F are not required for transcription in vitro, but a Thermococcus kodakarensis mutant lacking subunit F is temperature-sensitive. Mol. Microbiol. 70, 623–633. 10.1111/j.1365-2958.2008.06430.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai T., Imanaka H., Nakajima A., Uwamori K., Omori Y., Fukui T., et al. (2005). Continuous hydrogen production by the hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1. J. Biotechnol. 116, 271–282. 10.1016/j.jbiotec.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Kanai T., Imanaka T., Atomi H. (2013). Hydrogen production by the hyperthermophilic archaeon Thermococcus kodakarensis. J. Jpn. Petrol Inst. 56, 267–279. 10.1627/jpi.56.26721515783 [DOI] [Google Scholar]
- Kanai T., Ito S., Imanaka T. (2003). Characterization of a cytosolic NiFe-hydrogenase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185, 1705–1711. 10.1128/JB.185.5.1705-1711.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai T., Matsuoka R., Beppu H., Nakajima A., Okada Y., Atomi H., et al. (2011). Distinct physiological roles of the three [NiFe]-hydrogenase orthologs in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 193, 3109–3116. 10.1128/JB.01072-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M. S., Bae S. S., Kim Y. J., Kim T. W., Lim J. K., Lee S. H., et al. (2013). CO-dependent H2 production by genetically engineered Thermococcus onnurineus NA1. Appl. Environ. Microbiol. 79, 2048–2053. 10.1128/AEM.03298-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Seol E., Oh Y. K., Wang G. Y., Park S. (2009). Hydrogen production and metabolic flux analysis of metabolically engineered Escherichia coli strains. Int. J. Hydrogen Energy 34, 7417–7427. 10.1016/j.ijhydene.2009.05.053 [DOI] [Google Scholar]
- Klein M., Ansorge-Schumacher M. B., Fritsch M., Hartmeier W. (2010). Influence of hydrogenase overexpression on hydrogen production of Clostridium acetobutylicum DSM 792. Enzyme Microb. Technol. 46, 384–390. 10.1016/j.enzmictec.2009.12.015 [DOI] [Google Scholar]
- Kumar N., Das D. (2000). Enhancement of hydrogen production by Enterobacter cloacae IIT-BT 08. Process Biochem. 35, 589–593. 10.1016/S0032-9592(99)00109-0 [DOI] [Google Scholar]
- Kumar N., Das D. (2001). Continuous hydrogen production by immobilized Enterobacter cloacae IIT-BT 08 using lignocellulosic materials as solid matrices. Enzyme Microb. Technol. 29, 280–287. 10.1016/S0141-0229(01)00394-5 [DOI] [Google Scholar]
- Lee S. H., Kim M. S., Bae S. S., Choi A. R., Lee J. W., Kim T. W., et al. (2014). Comparison of CO-dependent H2 production with strong promoters in Thermococcus onnurineus NA1. Appl. Microbiol. Biotechnol. 98, 979–986. 10.1007/s00253-013-5448-y [DOI] [PubMed] [Google Scholar]
- Lipscomb G. L., Schut G. J., Thorgersen M. P., Nixon W. J., Kelly R. M., Adams M. W. W. (2014). Engineering hydrogen gas production from formate in a hyperthermophile by heterologous production of an 18-subunit membrane-bound complex. J. Biol. Chem. 289, 2873–2879. 10.1074/jbc.M113.530725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y. C., Chen C. Y., Lee C. M., Chang J. S. (2010). Sequential dark-photo fermentation and autotrophic microalgal growth for high-yield and CO2-free biohydrogen production. Int. J. Hydrogen Energy 35, 10944–10953. 10.1016/j.ijhydene.2010.07.090 [DOI] [Google Scholar]
- Ma K., Weiss R., Adams M. W. W. (2000). Characterization of hydrogenase II from the hyperthermophilic archaeon Pyrococcus furiosus and assessment of its role in sulfur reduction. J. Bacteriol. 182, 1864–1871. 10.1128/JB.182.7.1864-1871.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai X., Adams M. W. W. (1996). Purification and characterization of two reversible and ADP-dependent acetyl coenzyme A synthetases from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 178, 5897–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal B., Nath K., Das D. (2006). Improvement of biohydrogen production under decreased partial pressure of H2 by Enterobacter cloacae. Biotechnol. Lett. 28, 831–835. 10.1007/s10529-006-9008-8 [DOI] [PubMed] [Google Scholar]
- Mars A. E., Veuskens T., Budde M. A. W., van Doeveren P. F. N. M., Lips S. J., Bakker R. R., et al. (2010). Biohydrogen production from untreated and hydrolyzed potato steam peels by the extreme thermophiles Caldicellulosiruptor saccharolyticus and Thermotoga neapolitana. Int. J. Hydrogen Energy 35, 7730–7737. 10.1016/j.ijhydene.2010.05.063 [DOI] [Google Scholar]
- Melis A., Zhang L. P., Forestier M., Ghirardi M. L., Seibert M. (2000). Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol. 122, 127–135. 10.1104/pp.122.1.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnan L., Jinli H., Xiaobin W., Huijuan X., Jinzao C., Chuannan L., et al. (2005). Isolation and characterization of a high H2-producing strain Klebsiella oxytoca HP1 from a hot spring. Res. Microbiol. 156, 76–81. 10.1016/j.resmic.2004.08.004 [DOI] [PubMed] [Google Scholar]
- Morikawa M., Izawa Y., Rashid N., Hoaki T., Imanaka T. (1994). Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60, 4559–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y. K., Raj S. M., Jung G. Y., Park S. (2011). Current status of the metabolic engineering of microorganisms for biohydrogen production. Bioresour. Technol. 102, 8357–8367. 10.1016/j.biortech.2011.04.054 [DOI] [PubMed] [Google Scholar]
- Oh Y. K., Seol E. H., Kim J. R., Park S. (2003). Fermentative biohydrogen production by a new chemoheterotrophic bacterium Citrobacter sp Y19. Int. J. Hydrogen Energy 28, 1353–1359. 10.1016/S0360-3199(03)00024-7 [DOI] [Google Scholar]
- Pisa K. Y., Huber H., Thomm M., Müller V. (2007). A sodium ion-dependent A1A0 ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus. FEBS J. 274, 3928–3938. 10.1111/j.1742-4658.2007.05925.x [DOI] [PubMed] [Google Scholar]
- Ren N. Q., Cao G. L., Wang A. J., Lee D. J., Guo W. Q., Zhu Y. H. (2008). Dark fermentation of xylose and glucose mix using isolated Thermoanaerobacterium thermosaccharolyticum W16. Int. J. Hydrogen Energy 33, 6124–6132. 10.1016/j.ijhydene.2008.07.107 [DOI] [Google Scholar]
- Rittmann S., Herwig C. (2012). A comprehensive and quantitative review of dark fermentative biohydrogen production. Microb. Cell Fact. 11:115. 10.1186/1475-2859-11-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittmann S. K., Lee H. S., Lim J. K., Kim T. W., Lee J. H., Kang S. G. (2015). One-carbon substrate-based biohydrogen production: microbes, mechanism, and productivity. Biotechnol. Adv. 33, 165–177. 10.1016/j.biotechadv.2014.11.004 [DOI] [PubMed] [Google Scholar]
- Robb F. T., Park J. B., Adams M. W. W. (1992). Characterization of an extremely thermostable glutamate dehydrogenase: a key enzyme in the primary metabolism of the hyperthermophilic archaebacterium, Pyrococcus furiosus. Biochim. Biophys. Acta 1120, 267–272. 10.1016/0167-4838(92)90247-B [DOI] [PubMed] [Google Scholar]
- Santangelo T. J., Cubonová L., Reeve J. N. (2011). Deletion of alternative pathways for reductant recycling in Thermococcus kodakarensis increases hydrogen production. Mol. Microbiol. 81, 897–911. 10.1111/j.1365-2958.2011.07734.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra R., Bagramyan K., Adams M. W. W. (2003). A simple energy-conserving system: proton reduction coupled to proton translocation. Proc. Natl. Acad. Sci. U.S.A. 100, 7545–7550. 10.1073/pnas.1331436100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra R., Verhagen M. F., Adams M. W. W. (2000). Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 182, 3423–3428. 10.1128/JB.182.12.3423-3428.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki D., Watanabe S., Kanai T., Atomi H., Imanaka T., Miki K. (2012). Characterization and in vitro interaction study of a [NiFe] hydrogenase large subunit from the hyperthermophilic archaeon Thermococcus kodakarensis KOD1. Biochem. Biophys. Res. Commun. 417, 192–196. 10.1016/j.bbrc.2011.11.083 [DOI] [PubMed] [Google Scholar]
- Sasaki D., Watanabe S., Matsumi R., Shoji T., Yasukochi A., Tagashira K., et al. (2013). Identification and structure of a novel archaeal HypB for [NiFe] hydrogenase maturation. J. Mol. Biol. 425, 1627–1640. 10.1016/j.jmb.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Sato T., Fukui T., Atomi H., Imanaka T. (2003). Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185, 210–220. 10.1128/JB.185.1.210-220.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Fukui T., Atomi H., Imanaka T. (2005). Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71, 3889–3899. 10.1128/AEM.71.7.3889-3899.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schicho R. N., Ma K., Adams M. W. W., Kelly R. M. (1993). Bioenergetics of sulfur reduction in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 175, 1823–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schut G. J., Nixon W. J., Lipscomb G. L., Scott R. A., Adams M. W. W. (2012). Mutational analyses of the enzymes involved in the metabolism of hydrogen by the hyperthermophilic archaeon Pyrococcus furiosus. Front. Microbiol. 3:163. 10.3389/fmicb.2012.00163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva P. J., van den Ban E. C., Wassink H., Haaker H., de Castro B., Robb F. T., et al. (2000). Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur. J. Biochem. 267, 6541–6551. 10.1046/j.1432-1327.2000.01745.x [DOI] [PubMed] [Google Scholar]
- Taguchi F., Mizukami N., Taki T. S., Hasegawa K. (1995). Hydrogen-production from continuous fermentation of xylose during growth of Clostridium sp strain No-2. Can. J. Microbiol. 41, 536–540. 10.1139/m95-071 [DOI] [Google Scholar]
- Tanaka T., Fujiwara S., Nishikori S., Fukui T., Takagi M., Imanaka T. (1999). A unique chitinase with dual active sites and triple substrate binding sites from the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1. Appl. Environ. Microbiol. 65, 5338–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T., Watanabe S., Matsumi R., Atomi H., Imanaka T., Miki K. (2013). Crystal structures of the carbamoylated and cyanated forms of HypE for [NiFe] hydrogenase maturation. Proc. Natl. Acad. Sci. U.S.A. 110, 20485–20490. 10.1073/pnas.1313620110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel E. W. J., Budde M. A. W., de Haas G. G., van der Wal F. J., Claasen P. A. M., Stams A. J. M. (2002). Distinctive properties of high hydrogen producing extreme thermophiles, Caldicellulosiruptor saccharolyticus and Thermotoga elfii. Int. J. Hydrogen Energy 27, 1391–1398. 10.1016/S0360-3199(02)00115-5 [DOI] [Google Scholar]
- Verhees C. H., Kengen S. W., Tuininga J. E., Schut G. J., Adams M. W. W., de Vos W. M., et al. (2003). The unique features of glycolytic pathways in Archaea. Biochem. J. 375, 231–246. 10.1042/BJ20021472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D. E., Kengen S. W., van der Oost J., de Vos W. M. (2000). Purification and characterization of the alanine aminotransferase from the hyperthermophilic Archaeon Pyrococcus furiosus and its role in alanine production. J. Bacteriol. 182, 2559–2566. 10.1128/JB.182.9.2559-2566.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Kawashima T., Nishitani Y., Kanai T., Wada T., Inaba K., et al. (2015). Structural basis of a Ni acquisition cycle for [NiFe] hydrogenase by Ni-metallochaperone HypA and its enhancer. Proc. Natl. Acad. Sci. U.S.A. 112, 7701–7706. 10.1073/pnas.1503102112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S., Matsumi R., Atomi H., Imanaka T., Miki K. (2012a). Crystal structures of the HypCD complex and the HypCDE ternary complex: transient intermediate complexes during [NiFe] hydrogenase maturation. Structure 20, 2124–2137. 10.1016/j.str.2012.09.018 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Sasaki D., Tominaga T., Miki K. (2012b). Structural basis of [NiFe] hydrogenase maturation by Hyp proteins. Biol. Chem. 393, 1089–1100. 10.1515/hsz-2012-0197 [DOI] [PubMed] [Google Scholar]
- Xu L. Y., Ren N. Q., Wang X. Z., Jia Y. F. (2008). Biohydrogen production by Ethanoligenens harbinense B49: nutrient optimization. Int. J. Hydrogen Energy 33, 6962–6967. 10.1016/j.ijhydene.2008.09.005 [DOI] [Google Scholar]
- Yokooji Y., Sato T., Fujiwara S., Imanaka T., Atomi H. (2013). Genetic examination of initial amino acid oxidation and glutamate catabolism in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 195, 1940–1948. 10.1128/JB.01979-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Nishimura T., Kawaguchi H., Inui M., Yukawa H. (2005). Enhanced hydrogen production from formic acid by formate hydrogen lyase-overexpressing Escherichia coli strains. Appl. Environ. Microbiol. 71, 6762–6768. 10.1128/AEM.71.11.6762-6768.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Nishimura T., Kawaguchi H., Inui M., Yukawa H. (2007). Efficient induction of formate hydrogen lyase of aerobically grown Escherichia coli in a three-step biohydrogen production process. Appl. Microbiol. Biotechnol. 74, 754–760. 10.1007/s00253-006-0721-y [DOI] [PubMed] [Google Scholar]
- Zeidan A. A., Rådström P., van Niel E. W. (2010). Stable coexistence of two Caldicellulosiruptor species in a de novo constructed hydrogen-producing co-culture. Microb. Cell Fact. 9:102. 10.1186/1475-2859-9-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Cao G. L., Wang A. J., Guo W. Q., Liu B. F., Ren H. Y., et al. (2012). Enhanced bio-hydrogen production by immobilized Clostridium sp T2 on a new biological carrier. Int. J. Hydrogen Energy 37, 162–166. 10.1016/j.ijhydene.2011.09.103 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strategy for construction of pMHG1. pMHG1 was used to insert Pgdh upstream of the mbhA gene of the mbh operon via homologous recombination using strain KU216 as the host.
Strategy for construction of pMHC1. pMHC1 was used to introduce an additional Hyd gene region under the control of Pcsg into the chitinase region of strain KU216 via homologous recombination.
Strategy for construction of pMPD1. pMPD1 was used to replace the palindrome sequence between the Na/H- and Hyd regions of strain KU216 with a non-coding sequence that does not form a stem loop structure via homologous recombination.
Strategy for construction of pMAH1. pMAH1 was used to introduce Pcsg upstream of the mbhA gene of the mbh operon via homologous recombination. Strain DPHA1 was used as the host in order to combine Mbh overexpression with alaAT and hyh deletion.