Abstract
Genome wide association studies (GWAS) and large scale replication studies have identified common variants in 79 loci associated with breast cancer, explaining ~14% of the familial risk of the disease. To identify new susceptibility loci, we performed a meta-analysis of 11 GWAS comprising of 15,748 breast cancer cases and 18,084 controls, and 46,785 cases and 42,892 controls from 41 studies genotyped on a 200K custom array (iCOGS). Analyses were restricted to women of European ancestry. Genotypes for more than 11M SNPs were generated by imputation using the 1000 Genomes Project reference panel. We identified 15 novel loci associated with breast cancer at P<5×10−8. Combining association analysis with ChIP-Seq data in mammary cell lines and ChIA-PET chromatin interaction data in ENCODE, we identified likely target genes in two regions: SETBP1 on 18q12.3 and RNF115 and PDZK1 on 1q21.1. One association appears to be driven by an amino-acid substitution in EXO1.
Breast cancer is the most common cancer in women worldwide1. The disease aggregates in families, and has an important inherited component. This inherited component is driven by a combination of rare variants, notably in BRCA1, BRCA2, PALB2, ATM and CHEK2 conferring a moderate or high lifetime risk of the disease, together with common variants at more than 70 loci, identified through GWAS and large scale replication studies2–20. Taken together, these loci explain approximately one-third of the excess familial risk of breast cancer.
The majority of susceptibility SNPs has been identified through the Breast Cancer Association Consortium (BCAC), a collaboration involving more than 50 case-control studies. We recently reported the results of a large-scale genotyping experiment within BCAC, which utilised a custom array (iCOGS) designed to study variants of interest for breast, ovarian and prostate cancers. iCOGS comprised more than 200,000 variants, of which 29,807 had been selected from combined analysis of nine breast cancer GWAS involving 10,052 breast cancer cases and 12,575 controls of European ancestry. In total, 45,290 breast cancer cases and 41,880 controls of European ancestry from 41 studies were genotyped with iCOGS, leading to the discovery of 41 novel susceptibility loci16. A parallel analysis identified four loci specific to oestrogen receptor (ER)-negative disease17. However, additional susceptibility loci may have been missed because they were not selected from the original GWAS, or not included on the array.
Genotype imputation is a powerful approach to infer missing genotypes using the genetic correlations defined in a densely genotyped reference panel, thus providing the opportunity to identify novel susceptibility variants even if not directly genotyped21. In this analysis we aimed to identify additional breast cancer susceptibility loci by utilising data from all 200k variants on the iCOGS array, and used imputation to estimate genotypes for more than 11M SNPs. We applied the same approach to data from 11 GWAS. After quality control (QC) exclusions, the dataset comprised 15,748 breast cancer cases and 18,084 controls from GWAS, and 46,785 cases and 42,892 controls from 41 studies genotyped with iCOGS (see Online Methods and Supplementary Tables 1a–1e). All subjects were women of European ancestry.
We imputed genotypes using the 1000 Genomes Project March 2012 release as the reference dataset (see Online Methods) The main analyses were based on ~11.6M SNPs that were imputed with imputation r2 >0.3 and had MAF>0.005 in at least one of the datasets22.
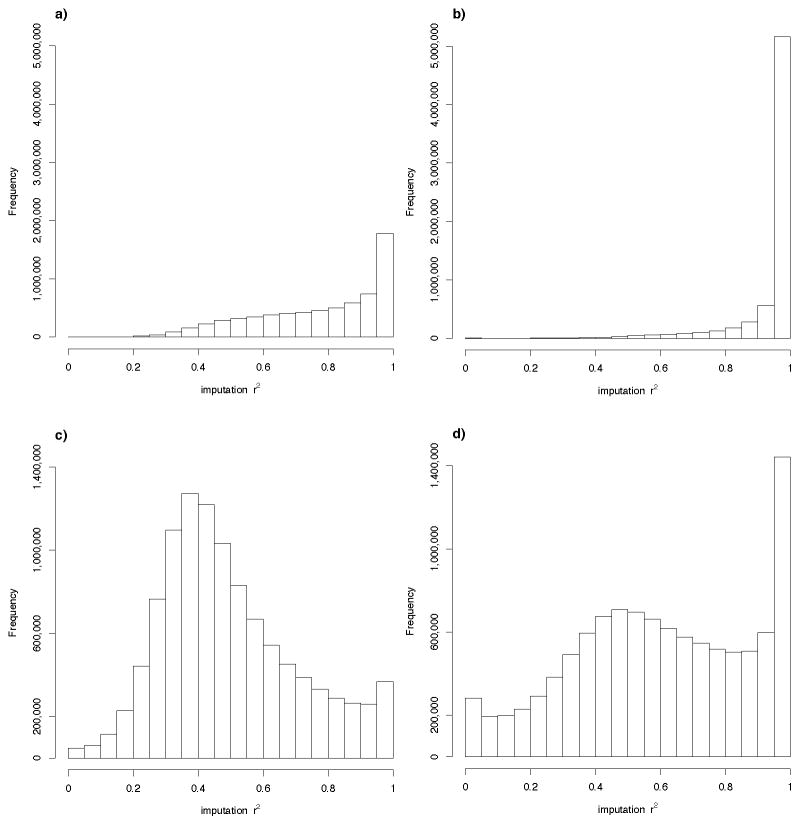
Of common SNPs (MAF>0.05), 88% were imputed from the iCOGS array with r2>0.5; this compared to 99% of variants for the largest GWAS (UK2), which was genotyped using a 670k SNP array (Figure 1a and 1b, Supplementary Table 2). Thirty-seven per cent of common SNPs were imputed on the iCOGS with r2>0.9, compared with 85% for UK2. Thus, despite being designed as a follow-up of GWAS for different diseases rather than a genome-wide array, the majority of common variants could be imputed using the iCOGS, but the overall imputation quality was, poorer that from a standard GWAS array. Imputation quality decreased with decreasing allele frequency (Figure 1c and 1d, Supplementary Table 2).
Figure 1.
Histograms of the imputation r2 a) Histogram of the imputation r2 for the iCOGS for variants with MAF>0.05 b) Histogram of the imputation r2 for the UK2 GWAS for variants with MAF>0.05 c) Histogram of the imputation r2 for the iCOGS for variants with MAF<=0.05 d) Histogram of the imputation r2 for the UK2 GWAS for variants with MAF<=0.05.
Log odds ratio estimates and standard errors were calculated for each dataset using logistic regression, adjusting for principal components where it was found to reduce substantially the inflation factor. We then combined the results from each dataset for variants with MAF >0.5% using a fixed effects meta-analysis23. More than 7,000 variants with a combined P<5×10−8 for association were identified, the large majority of which was in regions previously shown to be associated with breast cancer susceptibility. Of the 79 previously published breast cancer susceptibility loci identified in women of European ancestry, all but eight show evidence of association at P<5×10−8 for overall, ER-positive or ER-negative disease risk (Supplementary Tables 3a, 3b and 3c). For four of the eight variants, (rs1550623 on 2q31, rs11571833 on 13q13.1, rs12422552 on 12p13.1 and rs11242674 on 6p25.3), slightly weaker evidence of association was observed. One reported variant, rs7726159 did not reach P<5×10−8 in this (P=0.0017) or the previous analysis – it was identified through fine-mapping of the TERT region on 5p15.3318. One other variant in AKAP9, rs6964587 reported previously19 did not reach P<5×10−8 but an alternative correlated with it did (P=3.67×10−8 for chr7:91681597:D; r2 between the two markers = 0.98). The two remaining variants (rs2380205 on 10p15 and rs1045485 at CASP8) were reported in earlier analysis9,24 but did not even reach P<0.0001, suggesting that they may have been false positive reports. An alternative variant at CASP8, rs1830298 (r2=0.06, D’=1 with rs1045485 in 1000G CEU) did reach P<5×10−8 in this dataset25.
To assess evidence for additional susceptibility loci, we removed all SNPs within 500kb of susceptibility variants identified previously in women of European ancestry2–14,16–19, leaving 314 variants from 27 regions associated with breast cancer at P<5×10−8 (Supplementary Figures 1 and 2). The strongest associations were observed in a 610kb (b37 28,314,612- 28,928,858) interval on chromosome 22 (smallest P=8.2×10−22, for rs62237573). This interval lies approximately 100kb centromeric to CHEK2, and further analysis revealed that the associated SNPs were correlated with the CHEK2 founder variant 1100delC (strongest correlation r2=0.39 for SNP rs62235635), CHEK2 1100delC is known to be associated with breast cancer through candidate gene analysis, but has not previously generated an association in GWAS 26,27. We performed an analysis adjusting for CHEK2 1100delC using data on ~40,000 samples that had been genotyped for this variant. The strongest associated variant in this subset was rs140914118; after adjustment for 1100delC the statistical significance diminished markedly (P=3.1×10−9 to P=0.78; Supplementary Figures 3a and 3b), suggesting that this signal is driven by CHEK2 1100delC.
Variants in four regions (DNAJC1, 5p12, PTHLH and MKL1) lay within 2Mb of a previously published susceptibility-associated SNP. In each case, these associations became weaker (no longer P<5×10−8) after adjustment for the previously associated SNP(s) in the region (data not shown). For four other regions, the significant variants were identified in just one GWAS, and failed imputation (r2<0.3) in the remaining datasets, including iCOGS; we did not consider these variants further.
To confirm the results for the remaining 18 regions, we performed re-imputation in the iCOGS dataset without phasing (See Online Methods). Fifteen loci remained associated with breast cancer at P<5×10−8 (Table 1 and Supplementary Table 4). For three of the loci, the most significant SNP, or a highly correlated SNP, had been directly genotyped on iCOGS (Supplementary Table 5); one, rs11205277, had been included on the array because it is associated with adult height28, while the other two were selected based on evidence from the combined breast cancer GWAS but failed to reach genome-wide significance in the earlier analyses. We attempted to genotype the 12 remaining variants on a subset of ~4K samples to confirm the quality of the imputation (10 variants could be directly genotyped, for one region an alternative correlated variant was selected (Supplementary Table 5). For the 11 variants that could be assessed, the r2 between the observed and imputed genotypes were close to the r2 estimated in the imputation. Furthermore, the estimated effect sizes in the subset of individuals that we genotyped were similar to those obtained from the imputed genotypes (Supplementary Table 5). These results indicate that the analyses based on imputed genotype data were reliable.
Table 1.
Results for the 15 regions with combined P<5×10−8. Results are shown for the strongest associated variant in the region.
| Best variant | Locus | Position2 | Alleles3 | EAF4 | r25 | GWAS OR (95% CI)6 |
GWAS P7 | iCOGS OR (95% CI) |
iCOGS P | Combined GWAS + iCOGS P |
Genes within +/−2kb |
Enhancers in MCF7/HMEC |
eQTLs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs12405132 | 1q21.1 | 145644984 | C/T | 0.36 | 0.96 | 0.96 (0.92–0.99) | 0.00962 | 0.95 (0.93–0.97) | 2.34×10−7 | 7.92×10−9 | LOC10028814, NBPF10, RNF115 | RNF115, POLR3C,PDZK1, PIAS3 | - |
| rs12048493 | 1q21.2 | 149927034 | A/C | 0.34 | 0.76 | 1.04 (0.99–1.09) | 0.121 | 1.07 (1.05–1.10) | 1.66×10−9 | 1.10×10−9 | - | - | - |
| rs72755295 | 1q43 | 242034263 | A/G | 0.03 | 0.94 | 1.19 (1.03–1.39) | 0.021 | 1.15 (1.09–1.22) | 2.60×10−7 | 1.82×10−8 | EXO1 | - | - |
| rs6796502 | 3p21.3 | 46866866 | G/A | 0.09 | 0.91 | 0.92 (0.87–0.98) | 0.00657 | 0.92 (0.89–0.95) | 8.13×10−7 | 1.84×10−8 | - | - | - |
| rs13162653 | 5p15.1 | 16187528 | G/T | 0.45 | 0.72 | 0.92 (0.88–0.95) | 5.18×10−6 | 0.95 (0.93–0.97) | 1.71×10−6 | 1.08×10−10 | - | - | - |
| rs2012709 | 5p13.3 | 32567732 | C/T | 0.46 | 0.81 | 1.06 (1.02–1.09) | 0.00101 | 1.05 (1.03–1.08) | 1.66×10−6 | 6.38×10−9 | - | - | - |
| rs7707921 | 5q14 | 81538046 | A/T | 0.23 | 0.88 | 0.94 (0.9–0.98) | 0.00302 | 0.93 (0.91–0.95) | 4.09×10−9 | 5.00×10−11 | ATG10 | - | RPS23, ATP6AP1L |
| rs9257408 | 6p22.1 | 28926220 | G/C | 0.38 | 0.92 | 1.05 (1–1.1) | 0.0372 | 1.05 (1.03–1.08) | 4.53×10−7 | 4.84×10−8 | - | - | - |
| rs4593472 | 7q32.3 | 130667121 | C/T | 0.35 | 1.00 | 0.92 (0.88–0.96) | 2.57×10−5 | 0.95 (0.94–0.97) | 3.97×10−6 | 1.83×10−9 | FLJ43663 | - | - |
| rs13365225 | 8p11.23 | 36858483 | A/G | 0.17 | 0.94 | 0.89 (0.85–0.93) | 6.32×10−7 | 0.95 (0.93–0.98) | 0.000159 | 1.06×10−8 | - | - | - |
| rs13267382 | 8q23.3 | 117209548 | G/A | 0.36 | 0.97 | 1.07 (1.03–1.12) | 0.000537 | 1.05 (1.03–1.07) | 4.87×10−6 | 1.72×10−8 | LINC00536 | - | - |
| rs11627032 | 14q32.12 | 93104072 | T/C | 0.26 | 0.73 | 0.94 (0.9–0.98) | 0.00114 | 0.94 (0.92–0.96) | 1.06×10−6 | 4.48×10−9 | RIN3 | - | - |
| chr17:29230520 | 17q11.2 | 29230520 | GGT/G | 0.20 | 0.77 | 0.94 (0.89–0.98) | 0.009 | 0.93 (0.91–0.96) | 1.11×10−6 | 3.34×10−8 | ATAD5 | - | - |
| rs745570 | 17q25.3 | 77781725 | A/G | 0.50 | 0.93 | 0.94 (0.91–0.98) | 0.000754 | 0.95 (0.93–0.97) | 4.52×10−7 | 1.40×10−9 | - | - | - |
| rs6507583 | 18q12.3 | 42399590 | A/G | 0.07 | 0.96 | 0.91 (0.85–0.98) | 0.00803 | 0.91 (0.88–0.95) | 1.21×10−6 | 3.20×10−8 | SETBP1 | SETBP1 | - |
Chromosome
Build 37 position
Reference/effect allele, based on the forward strand
Mean effect allele frequency over all controls
Imputation r2 in the iCOGS samples (calculated by the average info score from IMPUTEv2)
Per allele odds ratio for the minor allele relative to the major allele
P value for the 1df trend test
There was little or no evidence of heterogeneity in the per-allele odds ratios (ORs) among studies genotyped using iCOGS (Supplementary Table 6 and Supplementary Figure 4). There was little evidence for departure from a log-additive model for any locus, except for a borderline departure for rs6796502 (P=0.049) for which the ORs for heterozygotes and homozygotes for the risk associated allele were similar (Supplementary Table 6).
The estimated ORs for invasive versus in-situ disease were similar for all the loci (P>0.05) (Supplementary Table 7). For four of the loci, rs12405132, rs12048493, rs4593472 and rs6507583 the association was stronger for ER positive disease (case only P<0.05) (Supplementary Table 8). Seven of the loci were associated with ER-negative disease (P<0.05) but none had a stronger association for ER-negative than ER-positive disease. Two of the loci showed significant trends in the OR by age at diagnosis: for rs13162653, the OR was higher at younger ages (P=0.007), while for rs6507583, the OR was higher at older ages (P=0.006) (Supplementary Table 9). One of the variants, chr17:29230520:D in ATAD5 is correlated with a variant that has also been shown to be associated with serous ovarian cancer in a meta-analysis29 (r2=0.93 between chr17:29230520:D and chr17:29181220:I).
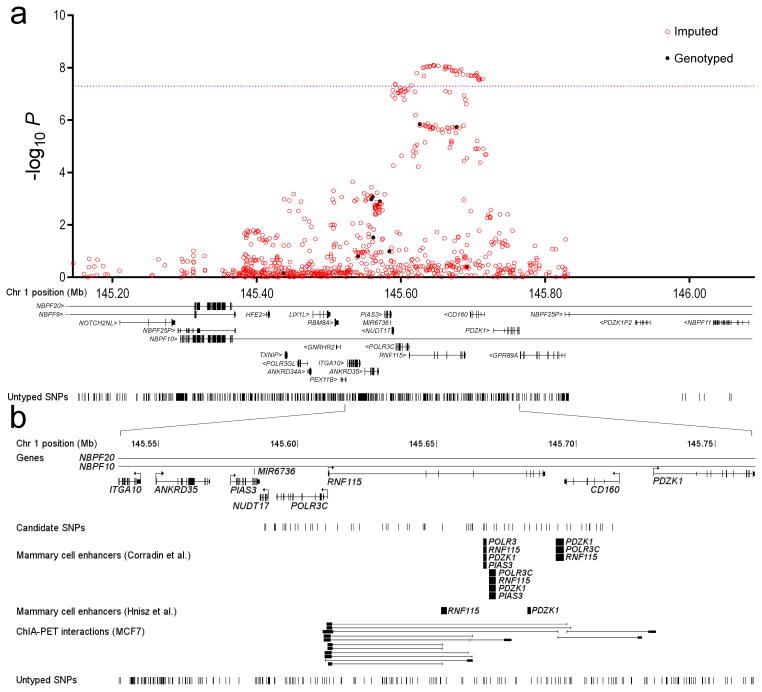
To approach the task of identifying the likely causal variants and genes underlying these associations, we first defined the set of all SNPs correlated with each of the 15 lead SNPs and that could not be ruled out as potentially causal (based on a likelihood ratio 100:130), resulting in a subset of 522 variants (Supplementary Table 10). One of the variants, rs72755295, lies in an intron of EXO1, encoding a protein involved in mismatch repair. It is strongly correlated with only one other variant, rs4149909, coding for an amino-acid substitution in EXO1 (p.Asn279Ser; CADD score 3331), suggesting that this variant is likely to be functionally related to breast cancer risk. None of the remaining SNPs lay within gene coding sequences, consistent with previous observations that most common cancer susceptibility variants are regulatory. For each of the remaining 520 variants, we then looked for enhancer elements in mammary cell lines, based on ENCODE ChIP-Seq data32,33. To identify potential gene targets, we combined this information with ENCODE ChIA-PET chromatin interaction data. We identified two regions in which the associated variants overlapped with putative enhancer sequences and for which consistent promoter interactions were predicted (Table 1). For rs12405132 at 1q21.1, we identified four potential interacting genes, RNF115, POLR3C, PDZK1 and PIAS3 (Figure 2). Of these, the strongest evidence was for RNF115 and PDZK1; three of the 64 potentially causal variants lay in interacting enhancer regions. RNF115 (also known as BCA2) is an E3 ubiquitin ligase RING finger protein that is overexpressed in ER-positive breast cancers34. PDZK1 is a scaffold protein that connects plasma membrane proteins and regulatory components, regulating their surface expression in epithelial cells apical domains, and has been proposed to act as an oncogene in breast cancer35.
Figure 2.
The chromosome 1 locus tagged by rs12405132 a) The Manhattan Plot displays the strength of genetic association (−log10 P) versus chromosomal position (Mb), where each dot presents a genotyped (solid black dot) or imputed (red circle) SNP (in the iCOGS stage). The purple horizontal line represents the threshold for genome-wide significance (P=5×10−8). Gene structures are depicted as well as the location of SNPs with MAF>0.01 which were neither imputed reliably nor genotyped. b) Mammary cell enhancer locations as defined in Corradin et al.32, and Hnisz et al.33, are shown where elements overlapping the best associated SNPs are labelled with their predicted target genes. A subset of ChiA-PET interactions in MCF7 cells (mediated by either RNApolII or ERa) between enhancers and their target gene promoters are also shown.
SNPs correlated with rs6507583 at 18q12.3 lay in regions interacting with the promoter of SETBP1 (Supplementary Figure 5). The encoded protein has been shown to bind the SET nuclear oncogene which is involved in DNA replication.
We utilised data from TCGA to assess associations between the 15 novel susceptibility variants and expression of neighbouring genes in breast tumors and normal breast tissue. One SNP, rs7707921, was strongly associated with RPS23 expression in all tissues (Supplementary Table 11, Supplementary Figure 6). However, stronger associations with expression were observed with more telomeric SNPs that were less strongly associated with disease risk (top eQTL SNP rs3739: P=10−23, P-risk=5.28×10−7), suggesting that this association may be coincidental. SNP, rs7707921 was also more weakly associated with expression of ATP6AP1L (P=5.6×10−5 in tumours, P=0.066 in normal tissue).
Based on the estimated ORs in the iCOGS stage (all but one of which were in the range 1.05–1.10), the 15 novel loci identified here would explain a further ~2% of the 2-fold familial risk of breast cancer. Taken together with previously identified loci, more than 90 independent common susceptibility loci for breast cancer have been identified, explaining ~16% of the familial risk. We estimate assuming a log-additive model that, based on genotypes for variants at these loci, approximately 5% of women in the general population have a >2 fold increased risk of breast cancer and 0.7% of women have a >3 fold increased risk. In the current analyses, more than 50% of variants with MAF>0.005 in subjects of European ancestry were well imputable (r2>0.5) These results suggest that, while there may be further susceptibility variants with comparable associated effects that were not well imputed, the identification of many additional loci will require larger association studies. In the meantime, inclusion of these additional loci in polygenic risk scores will improve our ability to discriminate between high and low risk individuals, potentially improving breast cancer screening and prevention.
Online Methods
Details of the subjects, genotyping and QC measures for the GWAS and iCOGS data are described elsewhere12,14,16,36,37. All participating studies were approved by their appropriate ethics review board and all subjects provided informed consent. Analyses were restricted to women of European ancestry. All imputations were performed using the 1000 Genomes Project March 2012 release as the reference panel. Of the 11 GWAS, 8 (C-BCAC) plus a subset of the BPC3 GWAS (CGEMS) were used in the combined GWAS analysis that nominated 29,807 SNPs for the array. The BPC3 and TNBCC GWAS nominated additional SNPs with evidence for association with ER-negative or triple-negative (ER-, PR- and HER2- negative) breast cancer. The EBCG GWAS was not used to nominate SNPs for the iCOGS array.
For eight GWAS (C-BCAC), genotypes were imputed in a two-stage procedure, using SHAPEIT to derive phased genotypes and IMPUTEv2 to perform the imputation on the phased data 22. We performed the imputation using 5Mb non-overlapping intervals for the whole genome. OR estimates and standard errors where obtained using logistic regression with SNPTEST 21. For two of the studies we adjusted for the 3 leading principal components as it was found to reduce materially the inflation factor; for the rest of the studies no such adjustment was necessary. For the remaining three GWAS (BPC3, TNBCC and EBCG), imputation was performed using MACH and Minimac23. Genomic control adjustment was applied to each GWAS as previously described16. The iCOGS data were also imputed in a two-stage procedure using SHAPEIT and IMPUTEv2, again using 5Mb non-overlapping intervals. We split the ~90K samples into 10 subsets, where possible keeping subjects from the same study in the same subset. We obtained OR estimates and standard errors using logistic regression adjusting for study and 9 principal components.
For the regions showing evidence of association we repeated the imputation in iCOGS, using IMPUTEv2 but without pre-phasing in SHAPEIT to improve imputation accuracy. We also increased the number of MCMC iterations from 30 to 90, and increased the buffer region from 250kb to 500kb.
Meta-analysis
OR estimates and standard errors were combined in a fixed effects inverse variance meta-analysis using METAL23. For the GWAS, results were included in the analysis for all SNPs with MAF>0.01 and imputation r2>0.3, except for the TN GWAS where the criteria were r2>0.9 and MAF>0.05. For iCOGS, we included all SNPs with r2>=0.3 and MAF>0.005.
Confirmatory genotyping
The best variant in each region after the re-imputation and meta-analysis was genotyped in 4123 samples from SEARCH, using Taqman according to the manufacturer’s instructions. The squared correlations between the observed genotypes and the genotypes estimated by imputation are shown in Supplementary Table 5. For all the imputed SNPs the squared correlations was greater than 0.7, the call-rates were >=0.98 and there was no evidence of departure of genotype frequencies from those expected under HWE (p>0.1).
eQTL analyses
Germline genotype, mRNA expression, and somatic copy number data for samples taken from breast tumours and tumour-adjacent normal tissue were obtained from The Cancer Genome Atlas38. The copy number and genotype data were measured using the Affymetrix Genome-Wide Human SNP 6.0 platform. For the mRNA expression data, we used the expression profiles obtained using the Agilent G4502A-07-3 microarray. The genotype data were subjected to the following quality control filters. SNPs were excluded in case of low frequency (MAF < 1%), low call rate (< 95%,) or departure from Hardy-Weinberg equilibrium at P < 1 × 1013. Individuals were excluded based on low call rate (< 95%), or high heterozygosity (false discovery rate < 1%). Furthermore, individuals were also excluded in case of non-European ancestry, or male gender. Quality control and intersection with the other genomic data types resulted in 380 tumour samples and 56 normal samples.
The genotype data were imputed as described above. eQTL analysis was performed using linear regression with SNPTEST, regressing the mRNA expression of selected candidate genes on the imputed genotype. For each gene, we performed the eQTL analysis against every microarray probe that uniquely maps to that gene. We adjusted the analyses for somatic copy number of the gene, and for SNPs that intersect the probe sequence, provided that their MAF exceeds 1% in individuals of European ancestry in the 1,000 Genomes data.
Enhancer analyses
Maps of enhancer regions with predicted target genes were obtained from Hnisz et al.33, and Corradin et al.32. Enhancers active in the mammary cell types MCF7, HMEC and HCC1954 were intersected with candidate causal variants using Galaxy. ENCODE ChIA-PET chromatin interaction data from MCF7 cells (mediated by RNApolII and ERα) were downloaded using the UCSC Table browser. Galaxy was used to identify ChIA-PET interactions between an implicated mammary cell enhancer (containing a strongly associated variant) and a predicted gene promoter (defined as regions 3 kb upstream and 1 kb downstream of the transcription start site).
Supplementary Material
Acknowledgments
The authors wish to thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. BCAC is funded by Cancer Research UK [C1287/A10118, C1287/A12014] and by the European Community’s Seventh Framework Programme under grant agreement n° 223175 (HEALTH-F2-2009-223175) (COGS). Meetings of the BCAC have been funded by the European Union COST programme [BM0606]. Genotyping of the iCOGS array was funded by the European Union (HEALTH-F2-2009-223175), Cancer Research UK (C1287/A10710, C8197/A16565), the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” program, and the Ministry of Economic Development, Innovation and Export Trade of Quebec – grant # PSR-SIIRI-701. Combining the GWAS data was supported in part by The National Institute of Health (NIH) Cancer Post-Cancer GWAS initiative grant: No. 1 U19 CA 148065-01 (DRIVE, part of the GAME-ON initiative). For a full description of funding and acknowledgments, see Supplementary Note.
Footnotes
Competing Financial Interests
The authors confirm that they have no competing financial interests
URLs
BCAC http://ccge.medschl.cam.ac.uk/consortia/bcac/index.html
COGS http://http://www.cogseu.org/
ENCODE http://www.genome.gov/encode/, genome.ucsc.edu/ENCODE/
iCOGS http://www.nature.com/icogs/, http://ccge.medschl.cam.ac.uk/research/consortia/icogs/
IMPUTE http://mathgen.stats.ox.ac.uk/impute/impute.html
MACH http://www.sph.umich.edu/csg/abecasis/MACH/
SHAPEIT https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html
TCGA http://cancergenome.nih.gov/1000 genomes: http://www.1000genomes.org/
Author Contributions
K. Michailidou and D.F.E. performed the statistical analysis and drafted the manuscript. D.F.E. conceived and coordinated the synthesis of the iCOGS array and led the BCAC. P.H. coordinated the Collaborative Oncological Gene-Environment Study (COGS). J.Benitez led the iCOGS genotyping working group. A.G.-N., G.P., M.R.A., J. Benitez, D.V., F.B., D.C.T., J. Simard, A.M.D., C.L., C. Baynes, S.A, C.S.H and M.J.M. co-ordinated genotyping of the iCOGS array. M.G-C., P.D.P.P. and M.K.S. led the BCAC pathology and survival working group. J.C-C. led the BCAC risk factor working group. A.M.D. and G.C.-T. led the iCOGS quality control working group. J. Beesley, J.D and M.J.L. provided bioinformatics support. M.K.B. and Q. Wang provided data management support for BCAC. S. Canisius provided analysis of the TCGA expression data. J.L.H, M.C.S, H.T. and C.A co-ordinated ABCFS. M.K.S, A.B., S.V and S. Cornelissen co-ordinated ABCS. K. Muir, A. Lophatananon, S.S.-B and P.S. co-ordinated ACP. P.A.F., A.H., M.W.B. and L.H. co-ordinated BBCC. J.P., I.d.S.S., O.F. and L.G. co-ordinated BBCS. E.J.S., I.T., M.J.K. and N.M. co-ordinated BIGGS. P.K, D.J.H., S.L., S.M.G., M.M.G., W.R.D., C.A.H., F.S., B.E.H., L.L.M., C.D.B., S.C, J.F. and R.N.H co-ordinated BPC3. B.B., F.M., H.S. and C. Sohn co-ordinated BSUCH. P.G, T.T, C. Mulot and M. Sanchez co-ordinated CECILE. S.E.B, B.G.N, H.F. and S.F.N. coordinated CGPS. A.G.-N., J. Benitez, M.P.Z. and J.I.A.P co-ordinated CNIO-BCS. H.A-C. and S.L.N. coordinated CTS. H.Brenner, A.K.D., V.A and C. Stegmaier co-ordinated ESTHER. A. Meindl, R.K.S, C. Sutter and R.Y co-ordinated GC-HBOC. H. Brauch, U.H. and T.B. co-ordinated GENICA. H.N., T.A.M, K.A., C.Blomqvist, K.A. and S.K. co-ordinated HEBCS. K. Matsuo, H. Ito, H. Iwata and K.T. co-ordinated HERPACC. T.D. and N.V.B. co-ordinated HMBCS. A. Lindblom and S. Margolin co-ordinated KARBAC. A. Mannermaa, V. Kataja, V-M.K. and J.M.H. co-ordinated KBCP. G.C.-T. and J. Beesley co-ordinated kConFab/AOCS. A.H.W., C-C.T., D.V.D.B and D.O.S co-ordinated LAABC. D.L., P.N., H.W. and E.V.L. coordinated LMBC. J.C-C. D.F-J., U.E., S.B. and A.R. co-ordinated MARIE. P.R., P.P., S. Manoukian and L. Bernard co-ordinated MBCSG. F.J.C., J.E.O., E.H. and C.V. co-ordinated MCBCS. G.G.G., R.L.M. and C. McLean co-ordinated MCCS. C.A.H., B.E.H., F.S. and L.L.M. co-ordinated MEC. J. Simard, M.S.G., F.L. and M.D. co-ordinated MTLGEBCS. S.H.T., C.H.Y., Y.-C.T and N.A.M.T. co-ordinated MYBRCA. V. Kristensen, G.I.G.A., S.N. and A-L.B-D. co-ordinated NBCS. W.Z., S.L.H., M. Shrubsole and J. Long coordinated NBHS. R.W., K.P., A.J-V. and M.G co-ordinated OBCS. I.L.A., J.A.K., G.G. and A.M.M. coordinated OFBCR. P.D., R.A.E.M.T, C. Seynaeve and C.J.V.A. co-ordinated ORIGO. M.G-C., J.F., S.J.C. and L. Brinton co-ordinated PBCS. K.C., H.D., M.E. and J. Brand co-ordinated pKARMA. J.W.M.M. and J.M.C. co-ordinated RBCS. P. Hall, J. Li, J. Liu and K.H. co-ordinated SASBAC. X.-O.S, W.L., Y.-T.G. and H.C. co-ordinated SBCGS. A.C., S.S.C. and M.W.R. Reed co-ordinated SBCS. W.B., L.B.S. and Q.C. coordinated SCCS. M. Shah and B.J.B. co-ordinated SEARCH. D.K., J-Y.C., S.K.P. and K-Y.Y. co-ordinated SEBCS. M.H., H.M., K.S.C. and C.W.C. co-ordinated SGBCC. U.H., M. Kabisch and D. Torres coordinated SKKDKFZS. A.J., J. Lubinski, K.J. and T.H., co-ordinated SZBCS. S. Sangrajrang, V.G., P.B. and J.M. co-ordinated TBCS. F.J.C, S. Slager, A.E.T, C.B.A. and D.Y. co-ordinated the TNBCC. C.-Y.S, C.-N.H., P.-E.W. and M.-F.H. co-ordinated TWBCS. A.S., A.A., N.O. and M.J.S. co-ordinated UKBGS. H.A., M.G.K., A.S.W., E.M.J., K.E.M., M.D.G., R.M.S., G.U., E.M., D.F.S and G.C. co-ordinated EBCG GWAS. Q.W, H.M-H., M.A.A. and R.B.v.d.L co-ordinated DFBBCS GWAS. D.F.E., N.H. and C.T. co-ordinated UK2 GWAS. F.C., D.Trichopoulos, P.P., E.L., M.Sund, K-T.K., M.J.G, D.P., L.D., J-M.H and L.M.M coordinated EPIC. All authors provided critical review of the manuscript.
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–50. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Easton DF, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stacey SN, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2008;40:703–6. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 5.Stacey SN, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39:865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 6.Ahmed S, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–90. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng W, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–8. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas G, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat Genet. 2009;41:579–84. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbull C, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat Genet. 2010;42:504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antoniou AC, et al. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor-negative breast cancer in the general population. Nat Genet. 2010;42:885–92. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher O, et al. Novel breast cancer susceptibility locus at 9q31.2: results of a genome-wide association study. J Natl Cancer Inst. 2011;103:425–35. doi: 10.1093/jnci/djq563. [DOI] [PubMed] [Google Scholar]
- 12.Haiman CA, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nat Genet. 2011;43:1210–4. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghoussaini M, et al. Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat Genet. 2012;44:312–8. doi: 10.1038/ng.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siddiq A, et al. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet. 2012;21:5373–84. doi: 10.1093/hmg/dds381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long J, et al. Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet. 2012;8:e1002532. doi: 10.1371/journal.pgen.1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michailidou K, et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45:353–61. doi: 10.1038/ng.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Closas M, et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392–8. doi: 10.1038/ng.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bojesen SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013;45:371–84. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milne RL, et al. Common non-synonymous SNPs associated with breast cancer susceptibility: findings from the Breast Cancer Association Consortium. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Q, et al. Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nat Genet. 2014;46:886–90. doi: 10.1038/ng.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 22.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–9. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox A, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–8. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 25.Lin WY, et al. Identification and characterisation of novel associations in the CASP8/ALS2CR12 region on chromosome 2 with breast cancer risk. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meijers-Heijboer H, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31:55–9. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 27.CHEK2 Breast Cancer Case-Control Consortium. CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10 860 breast cancer cases 9 065 controls from 10 studies. Am J Hum Genet. 2004;74:1175–82. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudbjartsson DF, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–15. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 29.Kuchenbaecker KB, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet. 2015 doi: 10.1038/ng.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udler MS, Tyrer J, Easton DF. Evaluating the power to discriminate between highly correlated SNPs in genetic association studies. Genet Epidemiol. 2010;34:463–8. doi: 10.1002/gepi.20504. [DOI] [PubMed] [Google Scholar]
- 31.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corradin O, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, et al. RNF115/BCA2 E3 ubiquitin ligase promotes breast cancer cell proliferation through targeting p21Waf1/Cip1 for ubiquitin-mediated degradation. Neoplasia. 2013;15:1028–35. doi: 10.1593/neo.13678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim H, et al. PDZK1 is a novel factor in breast cancer that is indirectly regulated by estrogen through IGF-1R and promotes estrogen-mediated growth. Mol Med. 2013;19:253–62. doi: 10.2119/molmed.2011.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahsan H, et al. A genome-wide association study of early-onset breast cancer identifies PFKM as a novel breast cancer gene and supports a common genetic spectrum for breast cancer at any age. Cancer Epidemiol Biomarkers Prev. 2014;23:658–69. doi: 10.1158/1055-9965.EPI-13-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens KN, et al. 19p13.1 is a triple-negative-specific breast cancer susceptibility locus. Cancer Res. 2012;72:1795–803. doi: 10.1158/0008-5472.CAN-11-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.