Abstract
T cell immunity directed against tumor-encoded amino acid substitutions (AAS) occurs in some melanoma patients. This implicates missense mutations (MM) as a source of patient-specific neoantigens. However, a systematic evaluation of these putative neoantigens as validated targets of anti-tumor immunity is lacking. Moreover, whether vaccination can augment such responses is unknown. Here we show that a dendritic cell vaccine increased naturally occurring and revealed new HLA class I-restricted neoantigens in patients with advanced melanoma. The presentation of neoantigens by HLA-A*02:01 in human melanoma was confirmed by mass spectrometry. Vaccination promoted a diverse neoantigen-specific T cell receptor repertoire in terms of both TCRVβ usage and clonal composition. Our results demonstrate that vaccination directed at tumor AAS broadens the antigenic breadth and clonal diversity of anti-tumor immunity.
Melanoma genomes harbor somatic mutations that are caused by exposure to mutagens such as UV light (1, 2). Tumor missense mutations (MM), translated into amino acid substitutions (AAS), may provide a form of antigens that the immune system perceives as foreign, which elicits tumor-specific T cell immunity (3-6). To examine the immunogenicity of tumor-encoded AAS, three patients (MEL21, MEL38 and MEL218) with stage III resected cutaneous melanoma were consented for genomic analysis of their surgically excised tumors and subsequently enrolled in a phase 1 clinical trial with autologous, functionally mature, interleukin (IL)-12p70-producing dendritic cell (DC) vaccine (Fig. S1) (7). All 3 patients had received prior treatment with ipilimumab (Supplementary Materials, Patient Information). Exome sequencing was performed to identify somatic mutations in tumor samples (Fig. 1A). Multiple metachronous tumors were analyzed from patients MEL21 and MEL38 (Tables S1-S2). Tumor MM, translated as AAS-encoding nonamer peptides, were filtered through in silico analysis to assess HLA-A*02:01 peptide binding affinity (8) and expression of genes encoding predicted HLA-A*02:01 peptide candidates determined by analysis of cDNA capture data (Fig. 1A) (9). Peptide candidates for experimental validation were selected according to the strategy described in Fig. S2 and HLA-A*02:01 binding evaluated using the T2 assay (Fig. S3) (10) and confirmed in the fluorescence polarization-based competitive peptide binding assay (11). Per patient, 7 AAS peptide candidates were selected among validated HLA-A*02:01 binders (Fig. S2, Table S4) for incorporation into a personalized vaccine formulation along with the melanoma gp100-derived peptides G209-2M and G280-9V (as positive controls for vaccination) (7). The expression pattern of mutated genes encoding vaccine candidates is shown in Venn diagrams in Fig. 1A.
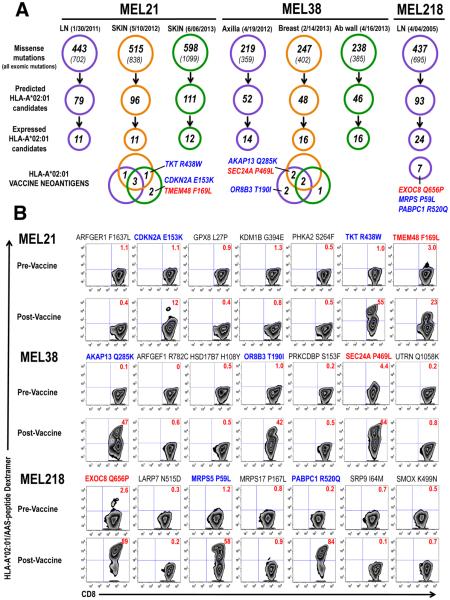
Fig. 1. Vaccine candidate identification and immune monitoring.
(A) Distribution of somatic (exomic and missense) mutations identified in patients MEL21 and MEL38 metachronous tumors (anatomical location and date of collection indicated) and patient MEL218 tumor are shown. HLA-A*02:01-binding candidate peptides were in silico identified among AAS and expression of gene encoding mutated protein determined from cDNA capture data (Table S1-S3). Venn diagrams show expression, among metachronous tumors, of mutated genes encoding vaccine neoantigens. The identities of the 3 immunogenic neoantigens identified in each patient are depicted in diagrams; color coding identifies naturally occurring (red) and vaccine-induced (blue) neoantigens. (B) Immune-monitoring of neoantigen-specific CD8+ T cell responses. Results are derived from PBMC isolated before DC vaccination (Pre-vaccine) and at peak (Post-Vaccine). PBMCs were cultured in vitro in the presence of peptide and IL-2 for 10 days followed by HLA-A*02:01/AAS-peptide dextramer assay. This immune monitoring strategy allows the reliable detection, as well as, the assessment of replicative potential of vaccine-induced T cell responses (Fig S4A). Color coding according to (A), numbers within dot plots represent percent neoantigen-specific T cells in lymph+/CD8+ gated cells.
To examine the kinetics and magnitude of T cell immunity to AAS peptides upon vaccination, peripheral blood mononuclear cells (PBMC) were collected prior to vaccination and weekly thereafter. The CD8+ T cell response to each peptide was analyzed using a HLA-A*02:01/AAS-peptide dextramer assay after a single round of in vitro stimulation (Fig. S4A) (7). Immune monitoring demonstrated that in each patient, T cell immunity to one AAS peptide could be detected in pre-vaccine PBMC samples after in vitro stimulation (Fig. 1B, MEL21: TMEM48 F169L; MEL38: SEC24A P469L and MEL218: EXOC8 Q656P) although not directly from the blood (Fig. S4B). Pre-existing immunity to these 3 neoantigens was confirmed in ex-vivo expanded pre-vaccine purified CD8+ T cells using dextramer assay (Fig. S4B) and interferon (IFN)-γ production (12) (Fig. S4C).
Vaccination augmented the T cell response to these neoantigens with observed frequencies of 23% TMEM48 F169L+ CD8+ T cells, 64% SEC24A P469L+ CD8+T cells and 89% EXOC8 Q656P+ CD8+ T cells detected, upon culture, at the peak of response (Fig. 1B). Immune monitoring also revealed vaccine-induced T cell immunity to 2 additional neoantigens per patient: TKT R438W and CDKN2A E153K (55% and 12%, respectively) in patient MEL21; AKAP13 Q285K and OR8B3 T190I (47% and 42%, respectively) in patient MEL38, and MRPS5 P59L and PABPC1 R520Q (58% and 84%, respectively) in patient MEL218 (Fig. 1B). Two (MEL21 and MEL218) of the 3 patients had pre-existing immunity to G209-2M and G280-9V peptides, as determined by the presence of gp100-specific T cells in pre-vaccine PBMC samples and their ex-vivo expansion upon antigen stimulation (Fig. S5B). Upon vaccination, these T cell responses were enhanced in patients MEL21 and MEL218 and revealed in patient MEL38 (Fig. S5). No T cell immunity was detected to the remaining 12 AAS peptides. Overall, robust neoantigen T cell immunity was detectable as early as week 2 and peaked at week 8-9 after the initial vaccine dose (Fig. S4A). Neoantigen-specific CD8+ T cells are readily identified by dextramer assay directly in post-vaccine PBMC samples (Fig. S4B) and memory T cells are detected up to 4 months after the final vaccine dose.
Analysis of T cell reactivity among the 3 patients indicated no preferential skewing towards AAS at specific positions in the peptide sequence, that is towards TCR contact residues or primary anchor residues (13). Rather, in each patient, T cell immunity appeared to focus on the 3 AAS candidates exhibiting the highest HLA-A*02:01 binding affinity while the remaining medium-high affinity peptides were non-immunogenic (Table S4) (8, 11). Immunogenic AAS peptides (Fig. 1A) were not preferentially derived from genes with high allelic frequency or expression levels (Tables S1-S3).
To characterize the function of vaccine-induced neoantigen-specific T cells, short-term expanded CD8+ T cell lines were established and antigen specificity confirmed by dextramer assay (Fig. S4B) (7, 12). Neoantigen-specific T cells displayed significant levels of cytotoxic activity at AAS peptide concentrations of 1 to 10nM, a finding that is consistent with high avidity T cell recognition of antigen (Fig. 2A). OR8B3 T190I-specific T cells could not discriminate between AAS and wild-type (WT) peptide when presented on T2 cells, while all of the remaining T cell lines showed clear specificity for AAS peptide sequences (Fig. 2A). Next, we sought to characterize the cytokine production profile of these T cells as a previous report suggests that IL-12p70-producing DC promote type 1 CD8+ T immunity, which in turn, correlates with increased clinical benefit (7, 14). Upon antigen stimulation, most vaccine-induced neoantigen-specific T cells produced high amounts of IFN-γ relative to IL-4, IL-5 and IL-13, a pattern that is indicative of a type 1 phenotype (Fig. S6). However, SEC24A P469L-specific T cells exhibited a type 2-skewed phenotype (high IL-4, IL-5 and IL-13 levels relative to IFN-γ), and TMEM48 F169L specific T cells showed a mixed phenotype with only higher IL-13 (but not IL-4 or IL-5) levels relative to IFN-γ (Fig. S6).
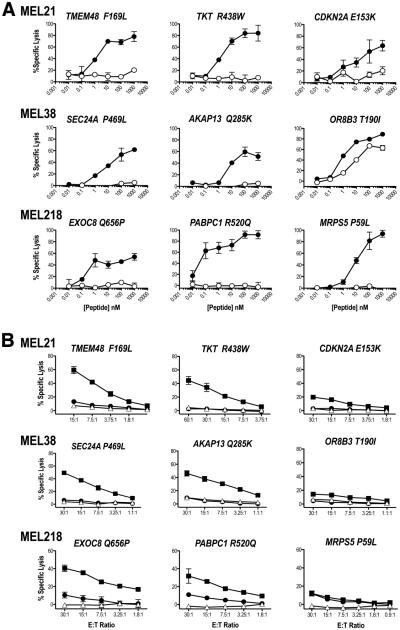
Fig. 2. Antigenic determinants recognized by vaccine-induced T cells.
(A) Neoantigen-specific T cells recognition of AAS (closed circles) and WT (open circles) peptides was determined in a standard 4h 51Cr-release assay using peptide titrations on T2 (HLA-A*02:01) cells. Percent specific lysis of triplicates (mean ± standard deviation) is shown for each peptide concentration; spontaneous lysis was <5%. Results are shown at 10:1 E: T ratios for all T cell lines except TMEM48 F169L and CDKN2A E153K T cells which are shown at 60:1 E:T ratio. A representative experiment of two independent evaluations is shown. (B) Neoantigen processing and presentation. Neoantigen-specific T cells were co-cultured with DM6 expressing AAS- (closed rectangles) or WT- (closed circles) TMC in a 4h 51Cr-release assay. Open triangles represent lysis obtained with parental DM6 cells. Percent specific lysis of triplicates (mean ± standard deviation) is shown for each E:T ratio; spontaneous lysis was <5%. A representative experiment of two independent evaluations is shown.
We next transfected DM6, a HLA-A*02:01+ melanoma cell line (15), with tandem minigene constructs (TMC) to evaluate neoantigen processing and presentation. Each minigene encoded an AAS, or the corresponding WT AA, embedded in 19-21 amino acids derived from the normal gene product (Fig. S7A, Table S5). TMC also encoded the West Nile Virus (WNV) SVG9 (16) and melanoma G280 (17) antigenic determinants as controls (Fig. S5 and S7B). Seven (TMEM48 F169L, TKT R438W, CDKN2A E153K, SEC24A P469L, AKAP13 Q285K, EXOC8 Q656P and PABPC1 R520Q) of the 9 immunogenic neoantigens are processed and presented as evidenced by cytotoxic activity (Fig. 2B) and IFN-γ production (Fig. S7C) by corresponding neoantigen-specific T cells upon co-culture with DM6 expressing AAS-encoding TMC. In contrast, neither cytotoxic activity (Fig. 2B) nor IFN-γ production (Fig. S7C) was observed upon co-culture of OR8B3 T190I- and MRPS5 P59L-specific T cells with DM6 expressing AAS-encoding TMC suggesting that these neoantigens are not processed and presented from endogenously expressed protein. None of the neoantigen-specific T cells recognized WT-encoding TMC (Fig. 2B and S7C). Based on these findings and the immune monitoring results (Fig. 1B), the 9 neoantigens identified in this study fall into 3 distinct antigenic determinant categories (18, 19). TMEM48 F169L, SEC24A P469L, and EXOC8 Q656P represent dominant antigens as T cell immunity was detected prior to vaccination (naturally occurring) (Fig. 1B) and these neoantigens are processed and presented from endogenously expressed protein (Fig. 2B). TKT R438W, CDKN2A E153K, AKAP13 Q285K and PABPC1 R520Q are characterized as subdominant antigens as T cell immunity required peptide vaccination (Fig. 1B) and these neoantigens are processed and presented from endogenously expressed protein (Fig. 2B). And finally, OR8B3 T190I and MRPS5 P59L constitute cryptic antigens since peptide vaccination elicited T cell immunity but these neoantigens are not processed from endogenously expressed protein.
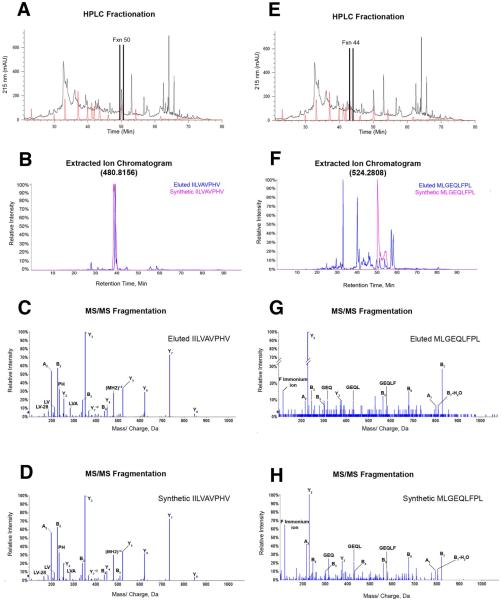
To validate neoantigen processing and presentation, proteomic analysis was performed on peptides eluted from soluble HLA-A*02:01 molecules isolated from melanoma cells expressing a TMC encoding AAS candidates from patient MEL218 tumor (18, 19). Reverse phase HPLC was used to reduce the complexity and determine the elution profile of the pool of soluble HLA-A*02:01 restricted peptides presented by melanoma cells, as well as, the synthetic AAS peptide mixture (Fig 3A,E). The fractions corresponding to each synthetic peptide were subjected to LC/MS. Extracted ion chromatograms revealed the presence of an eluted peptide with a retention time within 2 minutes of synthetic EXOC8 Q656P peptide in fraction 50 (Fig. 3B). MS/MS fragmentation pattern comparison of the eluted and the synthetic peptides ensured EXOC8 Q656P sequence identity and confirmed HLA-A*02:01 presentation of this dominant neoantigen (Fig. 3C,D). A similar analysis of fraction 44 demonstrated the HLA-A*02:01 presentation of subdominant neoantigen PABPC1 R520Q (Fig. 3E-H). Altogether, these results show that two of the 7 neoantigens included in patient MEL218 vaccine, along with antigen controls WNV SVG9 and G280 (Fig. S8), are processed and presented in the context of HLA-A*02:01 molecules.
Fig. 3. Processing and presentation of tumor neoantigens.
(A) RP-HPLC fractionation of HLA-A*02:01 peptides eluted from the AAS-TMC expressing melanoma cell line (black trace) and the synthetic peptide mixture containing MEL218 neoantigen candidates (red trace), with fraction 50 indicated. (B) Extracted ion chromatogram of the parent ion with the theoretical m/z of 480.8156 (+2) in HPLC fraction 50 from the HLA-A*02:01 eluted peptides (blue) overlaid with the EXOC8 Q656P synthetic peptide (pink). MS/MS fragmentation pattern of (C) the EXOC8 Q656P ion eluted from HLA-A*02:01 identified as IILVAVPHV, and (D) the corresponding synthetic peptide. (E) Same as in (A), with fraction 44 indicated. (F) Extracted ion chromatogram of the parent ion with the theoretical m/z 524.2808 (+2) in HPLC fraction 44 from the HLA-A*02:01 eluted peptides (blue) overlaid with the PABPC1 R520Q synthetic peptide (pink). MS/MS fragmentation pattern of (G) the PABPC1 R520Q ion eluted from HLA-A*02:01 identified as MLGEQLFPL and (H) the corresponding synthetic peptide.
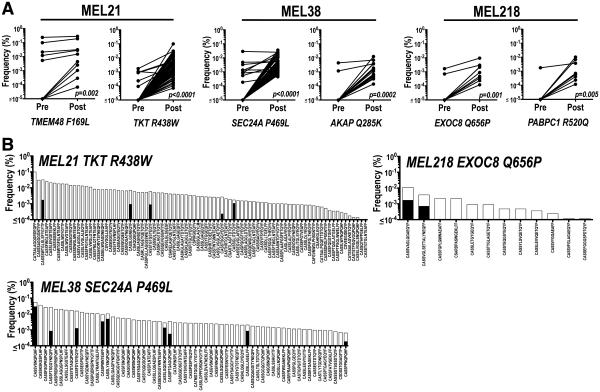
Little is known about the composition and diversity of neoantigen-specific T cells (20, 21) and the effect vaccination may have on these repertoires. To address this question, reference T cell receptor-β (TCRβ) complementarity-determining region 3 (CDR3) sequence libraries (Fig. S9, Tables S6-10) were generated from short-term expanded sorted neoantigen-specific T cells (97-99% dextramer-positive, Fig. S10) and used to characterize neoantigen TCRβ clonotypes in purified CD8+ T cells isolated from pre- and post-vaccine PBMC samples (22-24). In pre-vaccination CD8+ T cell populations, as few as one and as many as 10 unique TCRβ clonotypes per neoantigen were identified (Fig. 4A). Vaccination increased the frequency of most existing pre-vaccine TCRβ clonotypes and revealed new clonotypes for all 6 neoantigens (Fig. 4A). For both dominant and subdominant neoantigens, the TCRβ repertoire was increased significantly after vaccination (Fig. 4). For example, 84 clonotypes representing 19 TCRβ families are detected for TKT R438W, 61 clonotypes representing 12 TCRβ families are detected for SEC24A P469L and 12 clonotypes representing 8 TCRβ families are detected for EXOC8 Q656P (Fig. 4B). Thus, peptide vaccination with functionally mature DC may promote the expansion of a highly diverse neoantigen TCR repertoire.
Fig. 4. Vaccination promotes a diverse neoantigen-specific T cell repertoire.
(A) Summary of TCRβ clonotypes identified, using neoantigen-specific TCRβ CDR3 reference libraries (see Tables S6-S10), in CD8+ T cell populations isolated from PBMC obtained before and after vaccination. Each symbol represents a unique TCRβ sequence and its frequency (%) in pre- and post-vaccine samples. Wilcoxon-signed rank test was performed and p values indicated in figure. (B) TCRβ CDR3 sequence of clonotypes (Tables S6-S10) identified in pre- (black bars) and post- (white bars) vaccine CD8+ T cell populations for neoantigens TKT R438W (pre=5, post=84 clonotypes); SEC24A P469L (pre=9, post=61) and EXOC8 Q656P (pre=2, post =12). Frequency of each unique clonotype is reported as percentage of total read counts.
In summary, vaccination with high affinity patient-specific tumor-derived mutant peptides augments T cell immunity directed at naturally occurring (dominant) neoantigens and expanded the breadth of the anti-tumor immune response by revealing subdominant neoantigens (25). Vaccination against tumor neoantigens appears safe as all 3 patients are alive and well with no autoimmune adverse events. Inclusion of subdominant neoantigens into any therapeutic strategy would be expected to exert pressure to reduce the selection of antigen loss variants, especially in the setting of clonal tumor evolution (26) which occurs with targeted agents such as BRAF inhibition in melanoma (27, 28). The revelation of a highly diverse TCRβ repertoire specific for dominant and subdominant neoantigens was surprising and points to a potentially rich pool of naïve tumor-specific T cells that remain ignorant unless activated by vaccination. The effect of prior ipilimumab exposure on the pre-vaccine T cell repertoire in the 3 patients reported is unknown. However, recent data (29) indicates that anti-CTLA-4 monoclonal antibody administration can influence TCR repertoire diversity in patients and suggests a new therapeutic strategy testing checkpoint inhibitors, including ipilimumab, together with neoantigen vaccine formulations in order to further improve clinical outcomes. The paradigm described in this report could be applied to other malignancies presenting high mutational burdens such as lung, bladder and colorectal cancers (1). Other categories of genomic alterations such as deletions, nonsense and frame shift mutations may also generate potential neoantigens; mining these neoantigens may be particularly relevant in low mutational burden malignancies such as leukemias (30). Personalized immunotherapies targeting private somatic tumor alterations may become feasible in the near future.
Supplementary Material
Acknowledgments
We thank T. Hansen, D. Link, and J. DiPersio for critical review; T. Ley, M. Walter and J. Welch for advice and helpful discussions; R. Austin, R. Demeter, W. Eades, S. McGrath, W. Swaney, T. Vickery, J. Walker, and T. Wylie for technical assistance; L. Cornelius, R. Fields, R. Aft, J. Moley, A. Schaffer, and J. Steel for clinical collaboration. We gratefully acknowledge A. Alyasiry, S. Kalathiveetil, I. King, M. Schellhardt, and C. Rush for clinical research support and B. Fritz from Adaptive Biotechnologies for technical guidance. This work was supported by Barnes-Jewish Hospital Foundation, Siteman Cancer Frontier Fund, Our Mark on Melanoma (MOM) Foundation, Come Out Swinging (COS) Foundation, Blackout Melanoma Foundation and NCI (P30 CA91842 and R21 CA179695, B. Carreno, PI). We acknowledge The Genome Institute at Washington University School of Medicine for the sequencing production pipeline that generated data, established by NIH/NHGRI 5U54HG00307 (R. Wilson, PI). The Siteman Cancer Center provided the use of the Flow Cytometry Core, Clinical Trials Core, Tissue Procurement Core and the Biologic Therapy Core. The data presented in this manuscript is tabulated in the main paper and the supplementary materials. The raw exome and transcriptome sequence data are available on the Sequence Read Archive database: Bioproject PRJNA278450. The raw proteomic data are available at MassIVE database ID MSV000079125. Washington University and the authors (B.M.C., V.M., E.R.M. and G.P.L.) have filed a patent application (US62/050,195) related to the use of tumor somatic mutations for cancer immunotherapy entitled “Cancer Immunotherapy, Methods, Compositions and Uses Thereof. We thank the patients and families for their dedicated participation in this study.
Footnotes
Supplementary Materials
Materials and Methods
Supplementary Text
Figs. S1 to S10
Tables S4 to S10
References (31-36)
Data Files (Tables S1 to S3)
References and Notes
- 1.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfel T, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269:1281. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 4.Coulie PG, et al. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc. Natl. Acad. Sci. U. S. A. 1995;92:7976. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Rooij N, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J. Clin. Oncol. 2013;31:e439. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robbins PF, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med. 2013;19:747. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carreno BM, et al. IL-12p70-producing patient DC vaccine elicits Tc1-polarized immunity. J. Clin. Invest. 2013;123:3383. doi: 10.1172/JCI68395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen M, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabanski CR, et al. cDNA hybrid capture improves transcriptome analysis on low-input and archived samples. J Mol Diagn. 2014;16:440. doi: 10.1016/j.jmoldx.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elvin J, Potter C, Elliott T, Cerundolo V, Townsend A. A method to quantify binding of unlabeled peptides to class I MHC molecules and detect their allele specificity. J. Immunol. Methods. 1993;158:161. doi: 10.1016/0022-1759(93)90210-x. [DOI] [PubMed] [Google Scholar]
- 11.Buchli R, et al. Development and validation of a fluorescence polarization-based competitive peptide-binding assay for HLA-A*0201--a new tool for epitope discovery. Biochemistry. 2005;44:12491. doi: 10.1021/bi050255v. [DOI] [PubMed] [Google Scholar]
- 12.Carreno BM, et al. Amino-terminal extended peptide single-chain trimers are potent synthetic agonists for memory human CD8+ T cells. J. Immunol. 2012;188:5839. doi: 10.4049/jimmunol.1103647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y, Sette A, Peters B. Applications for T-cell epitope queries and tools in the Immune Epitope Database and Analysis Resource. J. Immunol. Methods. 2011;374:62. doi: 10.1016/j.jim.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer. 2012;12:298. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 15.Darrow TL, Slingluff CL, Jr., Seigler HF. The role of HLA class I antigens in recognition of melanoma cells by tumor-specific cytotoxic T lymphocytes. Evidence for shared tumor antigens. J. Immunol. 1989;142:3329. [PubMed] [Google Scholar]
- 16.McMurtrey CP, et al. Epitope discovery in West Nile virus infection: Identification and immune recognition of viral epitopes. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2981. doi: 10.1073/pnas.0711874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox AL, et al. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1994;264:716. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 18.Sercarz EE, et al. Dominance and crypticity of T cell antigenic determinants. Annu. Rev. Immunol. 1993;11:729. doi: 10.1146/annurev.iy.11.040193.003501. [DOI] [PubMed] [Google Scholar]
- 19.Assarsson E, et al. A quantitative analysis of the variables affecting the repertoire of T cell specificities recognized after vaccinia virus infection. J. Immunol. 2007;178:7890. doi: 10.4049/jimmunol.178.12.7890. [DOI] [PubMed] [Google Scholar]
- 20.Gros A, et al. PD-1 identifies the patient-specific CD8(+) tumor-reactive repertoire infiltrating human tumors. J. Clin. Invest. 2014;124:2246. doi: 10.1172/JCI73639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robins H, et al. Ultra-sensitive detection of rare T cell clones. J. Immunol. Methods. 2012;375:14. doi: 10.1016/j.jim.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lossius A, et al. High-throughput sequencing of TCR repertoires in multiple sclerosis reveals intrathecal enrichment of EBV-reactive CD8+ T cells. Eur. J. Immunol. 2014;44:3439. doi: 10.1002/eji.201444662. [DOI] [PubMed] [Google Scholar]
- 24.Robins HS, et al. Digital genomic quantification of tumor-infiltrating lymphocytes. Sci. Transl. Med. 2013;5:214ra169. doi: 10.1126/scitranslmed.3007247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat. Rev. Cancer. 2014;14:135. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 26.Gerlinger M, McGranahan N, Dewhurst SM, Burrell RA, Tomlinson I, Swanton C. Cancer: Evolution within a lifetime. Annual Reviews in Genetics. 2014;48 doi: 10.1146/annurev-genet-120213-092314. [DOI] [PubMed] [Google Scholar]
- 27.Shi H, et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Allen EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cha E, et al. Improved survival with T cell clonotype stability after anti-CTLA-4 treatment in cancer patients. Sci. Transl. Med. 2014;6:238ra70. doi: 10.1126/scitranslmed.3008211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rajasagi M, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124:453. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Service SK, et al. Re-sequencing expands our understanding of the phenotypic impact of variants at GWAS loci. PLoS Genet. 2014;10:e1004147. doi: 10.1371/journal.pgen.1004147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundegaard C, et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res. 2008;36:W509. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flicek P, Ahmed I, Barrel D, et al. Ensembl 2013. Nucleic Acid Research. 2013;D48 doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S, et al. Single-chain HLA-A2 MHC trimers that incorporate an immundominant peptide elicit protective T cell immunity against lethal West Nile virus infection. J. Immunol. 2010;184:4423. doi: 10.4049/jimmunol.0903955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaabinejadian S, et al. Identification of class I HLA T cell control epitopes for West Nile virus. PLoS One. 2013;8:e66298. doi: 10.1371/journal.pone.0066298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlson CS, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.