Abstract
Aims
Triple-negative breast cancer comprises a clinically aggressive group of invasive carcinomas. We examined a published gene expression screen of a panel of breast cancer cell lines to identify a potential triple-negative breast cancer-specific gene signature, and attempted to verify our findings by performing immunohistochemical analysis on tissue microarrays containing a large cohort of invasive breast carcinomas.
Methods
The microarray dataset for a panel of human breast cancer cell lines was interrogated for triple-negative breast cancer-specific genes. Membranous immunohistochemical expression of the protein product of the AXL gene was assessed semiquantitatively in 569 invasive breast carcinomas grouped according to molecular subgroup by immunohistochemistry.
Results
AXL was significantly upregulated in triple-negative/basal B cell lines compared with luminal or basal A cell lines. No significant difference was observed in the level of immunohistochemical expression of Axl protein between triple-negative breast cancers and other molecular subgroups (p=0.257). Axl expression was significantly associated with lymphovascular invasion (LVI) in all subgroups combined (p=0.033), and within the luminal A (p=0.002) and triple-negative breast cancer subgroups (p=0.026).
Conclusions
Despite preferential upregulation of AXL in triple-negative/basal B cell lines, analysis of Axl protein expression in a large series of patients’ breast tumours revealed no association between Axl expression and triple-negative breast cancer or other subtype. The association of Axl expression with LVI supports previous work that implicates Axl as a promoter of invasiveness in breast cancer cell lines. Further studies are necessary to explore whether Axl expression of individual breast cancer tumours can be clinically useful.
INTRODUCTION
Triple-negative breast cancer comprises a heterogeneous group of tumours that lack expression of oestrogen and progesterone hormone receptors, lack HER-2 overexpression, and overlap with the basal-like intrinsic molecular subtype of breast cancer.1–3 Triple-negative breast cancers are clinically more aggressive and show higher rates of recurrence, earlier recurrence, and lower overall survival compared with other types of breast carcinoma.4–7 Due to the lack of hormone receptor expression and HER-2 overexpression in triple-negative breast cancer, patients with triple-negative breast cancer are not candidates for endocrine therapy or trastu-zumab. Currently, cytotoxic chemotherapy is the mainstay medical treatment option for patients with advanced triple-negative breast cancer. Investigation into triple-negative breast cancer-specific biomarkers is an active area of research.8
The aims of our study were twofold. First, we set out to identify specific genes and potential drivers associated with the triple-negative subtype of breast cancer. Second, based on the finding of AXL as a triple-negative breast cancer cell line-associated gene, we studied a large cohort of patients with invasive breast carcinomas using anti-Axl immunohistochemistry on breast cancer tissue microarrays (TMA) to (1) determine whether Axl is preferentially expressed in triple-negative breast cancer and (2) examine the relationship of Axl expression with clinicopathologic variables of studied patients.
METHODS
Cell lines and reagents
Human breast cancer cell lines SKBR-3, BT474, MDA-MB468, MDA-MB231 and HS578T were cultured and maintained according to laboratory-optimised conditions. The following additional human breast cancer lines were requisitioned from American Type Culture Collection: HCC38 (CRL-2314), MCF 10A (CRL-10317) and MDA-MB-436 (HTB-130) and cultured according to ATCC-recommended procedures.
Protein extraction and immunoblotting
The previously described breast cancer cell lines were harvested and lysed with NP40 buffer supplemented with protease inhibitors. After protein quantification, 50 μg of protein per extract was boiled and denatured in SDS gel loading buffer. Following polyacrylamide gel electrophoresis, protein was transferred to a PVDF membrane, blocked with 2.5% milk and probed with anti-Axl antibody (R&D Systems, Cat #: MAB154) and anti-α tubulin antibody (Proteintech Group, Cat #: 66031). After probing with fluorophore-labelled secondary antibodies, blots were analysed and imaged with Odyssey CLx and Image Studio software (V.3.1).
Microarray analysis
Raw data from Neve et al9 was retrieved from (http://www.ebi.ac.uk/arrayexpress/) at accession number E-TABM-157. Array data was then analysed using Genespring GX (V.11.0.2). In brief, relative probe intensity values were normalised and probe values were filtered on expression (the bottom 20% were discarded), one-way analysis of variance (ANOVA) was performed with asymptotic p value with a cut-off of 0.05 and the Benjamini-Hochberg Method was used to correct for false positives. Interpretations were created to group ‘luminal’, ‘basal A’ and ‘basal B’ cell lines as well as receptor-positive versus triple-negative status within those groups. Significantly upregulated genes were defined as having a twofold increase in relative probe intensity as compared to other groups/ interpretations.
Patient selection
The study was conducted under an IRB-approved protocol. We retrospectively identified excision specimens from patients with invasive breast carcinoma that had immunohistochemical markers estrogen receptor (ER), progesterone receptor (PR), HER-2 and Ki-67, and fluorescence in situ hybridisation (FISH) for HER-2 gene amplification (when appropriate) performed as part of routine clinical care. Patients who had undergone neoadjuvant chemotherapy were excluded from the study cohort. Formalin-fixed paraffin-embedded tissue blocks were available for all patients and 569 patient samples were retrieved. Based on evaluation of immunohistochemical markers, tumours were categorised as luminal A (ER+/PR±/HER-2-/Ki-67 <14%), luminal B (ER+/PR±/HER-2+ or ER+/PR±/HER-2-/Ki-67 ≥ 14%), HER-2-enriched (ER-/PR-/HER-2+), or triple-negative (ER-/PR-/HER-2−). ER and PR stains were considered positive if ≥ 1% of invasive carcinoma cells showed positive nuclear staining.10 Tumours were considered HER-2-positive if greater than 30% of invasive carcinoma cells showed circumferential 3+ staining or if FISH revealed HER-2 gene amplification.11 Ninety per cent of cases categorised as triple-negative also demonstrated immunoreactivity for at least one basal-like marker (EGFR, CK5, CK14) (previously unpublished data).
Tissue microarray construction
H&E slides were reviewed from each case to confirm the diagnosis and histologic grade. Areas with the most invasive carcinoma cellularity were selected for TMA cores. Each tumour was represented by two 0.6 mm cores which were transferred into the recipient TMA block using the Beecher Manual Tissue Microarrayer Model MTA-1 (Beecher Instruments, Wisconsin, USA). In total, five TMA blocks were constructed (consisting of 569 cases, 1138 cores).
Immunohistochemistry
Immunohistochemical staining of Axl (goat polyclonal, dilution 1:40, R&D Systems) was accomplished using the Bond III Autostainer (Leica Microsystems, Illinois, USA). Formalin-fixed, paraffin-embedded tissue sections were first baked and deparaffinised. Antigen retrieval was accomplished by heating the slides at 99–100°C in Bond Epitope Retrieval Solution 1 for 30 min. Sections were then incubated sequentially with endogenous peroxidase block for 5 min, primary antibody for 30 min, Biotinylated Link Universal (Dako) for 25 min, Streptavidin-HRP for 25 min, diaminobenzidine (DAB) for 10 min, and haematoxylin for 5 min. Finally, the sections were dehydrated in 100% ethanol, and mounted in Cytoseal XYL (Richard-Allan Scientific, Kalamazoo, Michigan, USA). Normal breast tissue, which was used as a positive control, shows membranous Axl staining of luminal ductal cells with variable cytoplasmic staining.
Immunohistochemical interpretation
Membranous Axl immunohistochemical expression was assessed using a semiquantitative approach to generate a H-score for each tumour. The H-score, which produces a value ranging from 0 (no staining) to 300 (diffuse strong staining), represents the sum of percentage of tumour cells staining at each intensity level multiplied by the staining intensity (0=staining, 1=weak staining, 2=moderate staining, and 3=strong staining).12 All cases were scored by two pathologists (TMD, SJS).
Statistical analysis
H-scores obtained were treated as continuous variables. Owing to the non-normal distribution of H-scores, the difference between levels of clinical-pathological features was assessed with usage of non-parametric methods. Wilcoxon rank sum test and Kruskal–Wallis test were used for comparison of two groups and more than two groups, respectively. To evaluate the correlation between two continuous variables, Spearman Correlation Coefficient was used. Multiple testing correction was not carried out due to the exploratory nature of this study. All tests are two-sided, with p≤0.05 considered statistically significant. All analyses were done with statistical software SAS V.9.3 (SAS Institute, Cary, North Carolina, USA).
RESULTS
Receptor tyrosine kinase AXL transcript is highly upregulated in triple-negative breast cancer cell lines
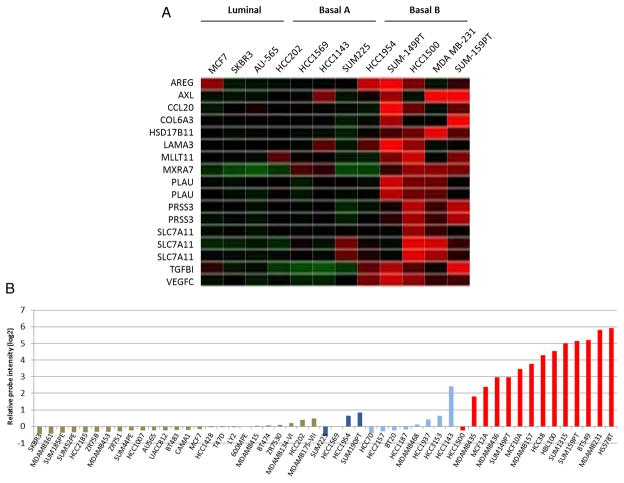
Using gene microarray data from breast cancer cell lines, Neve et al9 described a model to examine the functional contributions of certain genes within each subtype of breast cancer. The transcriptional profiles of cell lines were examined and cell lines were divided into luminal, basal A and basal B. While the luminal subtype had fairly uniform transcriptional profiles, the basal subtypes could be further separated into a basal subtype that fit into the ‘Perou’ categorisation (basal A) and a subtype that exhibited a more stem-like expression pattern (basal B).1 In addition to their distinct expression profile, the basal B-type cells were all classified as triple-negative (cell lines in the basal A group lack hormone receptors but sometimes overexpress HER-2). Therefore, we began our study by searching for genes that were highly upregulated in the basal B group as compared to the other subtypes. In our analysis, we searched for genes encoding cell surface and extracellular proteins as potential markers for optimal triple-negative breast cancer identification and characterisation. Thirteen genes were identified that exhibited twofold or higher relative expression in basal B cells than luminal or basal A cells including: AREG, AXL, CCL20, COL6A3, HSD17B11, LAMA3, MLLT11, MXRA7, PLAU, PRSS3, SLC7A11, TGFBI and VEGFC (figure 1A). As indicated on the heatmap, some of the genes were represented by multiple probes.
Figure 1.
(A) Heat map showing cell membrane and extracellular genes that are upregulated >2-fold higher in basal ‘B; type cells as compared to other subtypes.9 (B) AXL expression in a panel of breast cancer cell lines. Luminal cell lines are in light brown, basal A/HER-2+ cell lines are in dark blue, basal A/triple-negative cell lines are in light blue, and basal-B/triple-negative cell lines are in red.9
Among the genes we identified in our microarray analysis, we chose to focus on AXL expression due to its established role in tumour cell invasion and metastasis, and because of its evident potential as a therapeutic target.13 When we examined the expression profiles of breast cancer cell lines individually, we found a clear trend of AXL upregulation in the basal B (triple-negative breast cancer) cells as compared to the luminal and basal A cell types (figure 1B). On average, AXL expression in the basal B group was over 10-fold higher than the basal A or luminal groups. Despite the triple-negative status of some basal A cell lines, the basal A/triple-negative subgroup demonstrated AXL expression levels that were similar to basal A/HER-2+ cells (figure 1B). We then confirmed that Axl protein is preferentially upregulated in the basal B subtype of breast cancer cells by performing western blot analysis on a panel of breast cancer cell lines (figure 2). According to our analysis, four of the six basal B cell lines we tested exhibited moderate or high levels of Axl protein expression. However, Axl protein was not detected in two of the basal B lines and neither of the luminal lines. (Note: although MCF10A cells were not derived from a breast tumour, they are categorised as basal B according to their transcriptional profile as described in the Neve et al study). Overall, our analysis of in vitro cell lines indicated that increased AXL expression was likely to be associated with the triple-negative subtype of breast cancer in patients.
Figure 2.
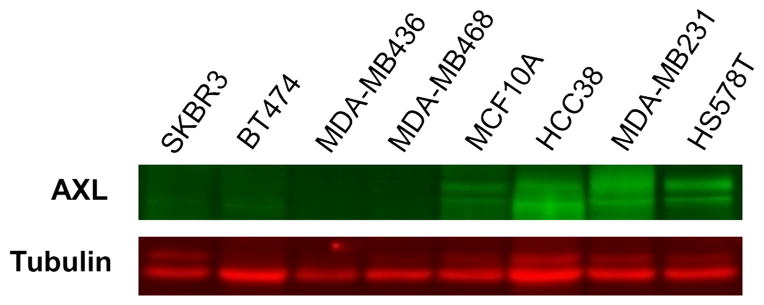
Western blot analysis of Axl protein expression in breast cancer cell lines. Four of the basal-‘B’ triple negative lines we examined (MCF10A, HCC38, MDA-MB231 and HS578T) demonstrated moderate-to-high levels of Axl expression while the MDA-MB436 and MDA-MB468 lines showed no detectable Axl expression. As expected, no Axl protein was detected in the luminal breast cancer cell lines SKBR3 and BT474.
Immunohistochemical expression of Axl protein is not restricted to triple-negative breast cancer and is associated with lymphovascular invasion
The immunohistochemical analysis of AXL gene expression was carried out using breast carcinomas from 569 patients who were classified as luminal A (n=142), luminal B (n=203)(ER+/PR ±/HER-2+, 106; ER+/PR±/HER-2-/Ki-67 ≥14%, 97), HER-2-enriched (n=96), and triple-negative (n=128), and the clinicopathologic characteristics of study patients are summarised in tables 1–5. All patients were women ranging in age from 26 to 94 years (mean age, 59, SD±13.8). At the time of diagnosis, 333 (58.5%) patients were stage I, 191 (33.5%) were stage II, and 45 (8%) had stage III disease. One hundred fifty-six (27%) patients had axillary nodal involvement at diagnosis.
Table 1.
AXL immunohistochemical expression compared with clinicopathologic variables for all patients
| Characteristic (n=569) | Number | Axl H-score median (min, max) | p Value |
|---|---|---|---|
| Age (mean years±SD) | 59±13.8 | ||
| Stage | 0.641 | ||
| I | 333 | 15 (0, 300) | |
| II | 191 | 20 (0, 300) | |
| III | 45 | 10 (0, 300) | |
| Tumour size | 0.552 | ||
| T1 | 402 | 15 (0, 300) | |
| T2 | 152 | 17.5 (0, 300) | |
| T3 | 15 | 10 (0, 100) | |
| Node status | 0.229 | ||
| N0 | 413 | 10 (0, 300) | |
| N1 | 118 | 20 (0, 300) | |
| N2 | 25 | 15 (0, 300) | |
| N3 | 13 | 0 (0, 140) | |
| Histologic grade | 0.511 | ||
| 1 | 39 | 20 (0, 300) | |
| 2 | 171 | 15 (0, 300) | |
| 3 | 303 | 15 (0, 300) | |
| ILC | 56 | 5 (0, 300) | |
| Lymphovascular invasion | 0.033 | ||
| Present | 122 | 20 (0, 300) | |
| Absent | 447 | 10 (0, 300) | |
| ER status | 0.294 | ||
| Positive | 345 | 15 (0, 300) | |
| Negative | 224 | 10 (0, 300) |
ILC, invasive lobular carcinoma; ER, oestrogen receptor.
Table 5.
AXL immunohistochemical expression compared with clinicopathologic variables for patients with triple-negative tumours
| Characteristic (n=128) | Number | Axl H-Score median (min, max) | p Value |
|---|---|---|---|
| Age (mean years±SD) | 56±14.3 | ||
| Stage | 0.175 | ||
| I | 65 | 15 (0, 285) | |
| II | 54 | 35 (0, 300) | |
| III | 9 | 60 (0, 300) | |
| Tumour size | 0.997 | ||
| T1 | 84 | 15 (0, 300) | |
| T2 | 41 | 30 (0, 300) | |
| T3 | 3 | 60 (30, 80) | |
| Node status | 0.011 | ||
| N0 | 96 | 20 (0, 285) | |
| N1 | 23 | 80 (0, 300) | |
| N2 | 4 | 137.5 (10, 300) | |
| N3 | 5 | 10 (0, 140) | |
| Histologic grade | 0.784 | ||
| 1 | 2 | 40 (0, 80) | |
| 2 | 17 | 10 (0, 210) | |
| 3 | 107 | 20 (0, 300) | |
| ILC | 2 | 120 (0, 240) | |
| Lymphovascular invasion | 0.026 | ||
| Present | 30 | 40 (0, 270) | |
| Absent | 98 | 17.5 (0, 300) |
ILC, invasive lobular carcinoma.
Among all patients’ tumours, Axl cell membrane expression was observed in 328 (57.6%) cases, while 241 (42.4%) showed no membranous reactivity. The median H-score among all cases was 15 (IQR, 90; range, 0–300). There was no significant difference in the level of Axl expression between the triple-negative subgroup and other subgroups (p=0.257).
Statistical analysis indicated that membranous Axl expression was significantly greater in cases showing lymphovascular invasion (LVI) (figure 3) in all subgroups combined (p=0.033), as well as in the luminal A subgroup (p=0.002) and in the triple-negative subgroup (p=0.026) compared with cases that lacked LVI. Moreover, membranous Axl expression was significantly associated with axillary lymph node positivity (p=0.011).
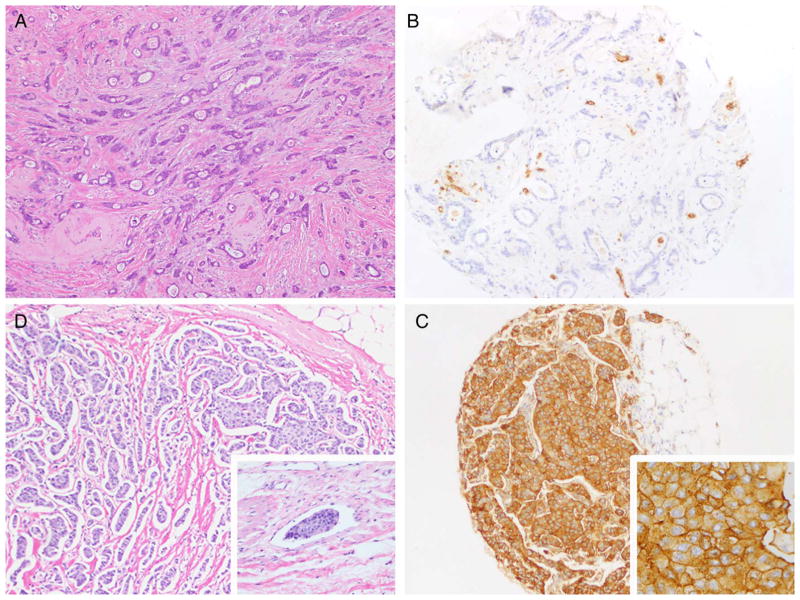
Figure 3.
(A) Well-differentiated invasive ductal carcinoma showing (B) lack of reactivity by Axl immunohistochemical stain. (C) Moderately-differentiated invasive ductal carcinoma showing lymphovascular invasion (inset) and (D) corresponding Axl immunohistochemical stain showing strong membranous as well as cytoplasmic expression.
DISCUSSION
Triple-negative breast cancer and basal-like breast cancers constitute approximately 15% of invasive breast carcinomas, and exhibit distinct clinical, pathologic and molecular features.14,15 In general, triple-negative breast cancers are clinically more aggressive and show higher rates of recurrence and reduced survival compared with other types of breast carcinoma.4–7 Due to the lack of targeted therapies for triple-negative breast cancer, identifying genes that are specific drivers of triple-negative breast cancer is an active area of research. In order to identify potential triple-negative breast cancer-associated genes and drivers, we analysed a published dataset and focused on extra-cellular and membrane-bound gene products (figure 1A).
Among the most upregulated genes in the basal B/triple-negative breast cancer -like subtype was AXL, which belongs to the TAM (Tyro-Axl-Mer) subfamily of receptor tyrosine kinases that was first characterised as a transforming agent isolated from human chronic myelogenous leukemia (CML) cells.16,17 Upregulation of AXL has since been observed in a number of cancers including colon,18 thyroid,19 breast,20,21 lung22 and liver,23 and has been shown to regulate numerous cellular processes relevant to cancer, such as cell survival, proliferation, migration and metastasis.13 Due to AXL’s involvement in cancer and the obvious appeal of receptor tyrosine kinases as drug targets, at least one specific small molecule kinase inhibitor (R428) and an inhibitory monoclonal antibody (YW327.6S2) have been developed to suppress AXL activity.24,25 Studies examining AXL expression in breast cancer have demonstrated preferential upregulation of AXL in triple-negative breast cancer cell lines.9,26 Because of Axl’s role in tumour progression and its upregulation in triple-negative breast cancer, Axl is an attractive candidate therapeutic target for this subset of breast cancers. Despite the preferential upregulation of AXL in triple-negative/basal B cell lines in our study, analysis of Axl protein expression in a large series of patients’ breast tumours revealed no statistically significant association between the level of Axl expression and triple-negative breast cancer or other subtype.
In comparing Axl expression with various clinicopathologic variables, we found a significant difference in Axl expression between patients with and without LVI, where the degree of membranous Axl expression was significantly greater in tumours with LVI, independent of other variables. This association was observed among all groups combined and specifically within the luminal A and triple-negative groups.
Although data from breast cancer tumour samples did not support the proposition that AXL is preferentially upregulated in triple-negative breast cancers, as seen in breast cancer cell lines, the over-representation of Axl expression in tumours showing LVI has potential clinical implications. In determining the need for adjuvant chemotherapy and/or radiation in breast cancer patients, clinicians rely on clinicopathologic factors, such as axillary lymph node status, tumour size, grade, stage and hormone receptor status. LVI, a histologic parameter that is routinely assessed in breast cancer, also has important prognostic and therapeutic implications, and may be a deciding factor as to whether or not a patient receives additional therapies. LVI has been shown to be an adverse prognostic factor, independent of axillary lymph node status or other variables, in numerous studies. Rates of locoregional recurrence (LRR), in particular, have been shown to be increased in patients with LVI. In a cohort of 763 women with stage pT1-2, pN0 breast cancer treated with modified radical mastectomy and adjuvant chemotherapy, the presence of LVI conferred a higher risk of LRR, distant recurrence, and lower overall survival.27 Pinder et al28 reported LVI to be a significant independent predictor of LRR, but not overall survival, in a group of 776 patients treated by mastectomy or wide excision without adjuvant chemotherapy or radiation. Other studies have highlighted LVI as an independently poor prognostic indicator.29–33 The association of Axl protein expression with the presence of LVI is consistent with AXL’s role in tumour cell migration, invasion and specifically, metastasis. In a study examining AXL expression in breast cancer, Gjerdrum et al34 showed that AXL is upregulated by EMT-inducing transcription factors and is necessary for tumour cell invasion. Knockdown of AXL in orthotopically injected mammary carcinoma in mice inhibited the development of metastasis, but not the formation of large primary tumours, implicating AXL’s role in metastasis.
In conclusion, we examined the expression of Axl in over 500 breast tumours with the hypothesis that Axl is preferentially expressed in the group of triple-negative breast cancers, based on previously published in vitro cell line transcriptional profile data. We found that Axl expression was fairly uniform in its distribution in this heterogeneous group of tumours. However, we found Axl to be significantly associated with aggressive histopathologic features. Our work indicates the limitations of cell lines as predictors of tumour phenomena as they occur in patients as well as their usefulness in supplying preliminary data which can be expanded upon. Studies of Axl expression in breast cancer patients with long-term clinical outcome would be informative in further understanding its role in the progression of breast cancer.
Table 2.
AXL immunohistochemical expression compared with clinicopathologic variables for patients with luminal A tumours
| Characteristic (n=142) | Number | Axl H-Score median (min, max) | p Value |
|---|---|---|---|
| Age (mean years±SD) | 64±13.4 | ||
| Stage | 0.346 | ||
| I | 109 | 10 (0, 300) | |
| II | 31 | 20 (0, 300) | |
| III | 2 | 130 (0, 300) | |
| Tumour size | 0.691 | ||
| T1 | 118 | 20 (0, 300) | |
| T2 | 22 | 50 (0, 300) | |
| T3 | 2 | 10 (0, 20) | |
| Node status | 0.528 | ||
| N0 | 116 | 15 (0, 300) | |
| N1 | 24 | 20 (0, 300) | |
| N2 | 2 | 130 (60, 200) | |
| Histologic grade | 0.381 | ||
| 1 | 30 | 22.5 (0, 300) | |
| 2 | 72 | 20 (0, 300) | |
| 3 | 11 | 22.5 (0, 180) | |
| ILC | 29 | 30 (0, 300) | |
| Lymphovascular invasion | 0.002 | ||
| Present | 17 | 150 (0, 300) | |
| Absent | 125 | 10 (0, 300) |
ILC, invasive lobular carcinoma.
Table 3.
AXL immunohistochemical expression compared with clinicopathologic variables for patients with luminal B tumours
| Characteristic (n=203) | Number | Axl H-Score median (min, max) | p Value |
|---|---|---|---|
| Age (mean years±SD) | 58±13 | ||
| Stage | 0.427 | ||
| I | 117 | 15 (0, 300) | |
| II | 68 | 15 (0, 300) | |
| III | 18 | 10 (0, 80) | |
| Tumour size | 0.745 | ||
| T1 | 142 | 17.5 (0, 300) | |
| T2 | 55 | 10 (0, 300) | |
| T3 | 6 | 10 (0, 20) | |
| Node status | 0.652 | ||
| N0 | 143 | 15 (0, 300) | |
| N1 | 45 | 30 (0, 300) | |
| N2 | 12 | 10 (0, 80) | |
| N3 | 3 | 40 (0, 80) | |
| Histologic grade | 0.523 | ||
| 1 | 7 | 10 (0, 200) | |
| 2 | 69 | 15 (0, 300) | |
| 3 | 107 | 15 (0, 300) | |
| ILC | 20 | 10 (0, 300) | |
| Lymphovascular invasion | 0.981 | ||
| Present | 45 | 20 (0, 270) | |
| Absent | 158 | 10 (0, 300) |
ILC, invasive lobular carcinoma.
Table 4.
AXL immunohistochemical expression compared with clinicopathologic variables for patients with tumours in the HER-2 subgroup
| Characteristic (n=96) | Number | Axl H-Score median (min, max) | p Value |
|---|---|---|---|
| Age (mean years±SD) | 57±14 | ||
| Stage | 0.719 | ||
| I | 42 | 0 (0, 300) | |
| II | 40 | 0 (0, 300) | |
| III | 14 | 0 (0, 200) | |
| Tumour size | 0.405 | ||
| T1 | 58 | 0 (0, 300) | |
| T2 | 34 | 5 (0, 300) | |
| T3 | 4 | 20 (0, 100) | |
| Node status | 0.696 | ||
| N0 | 58 | 0 (0, 300) | |
| N1 | 26 | 5 (0, 200) | |
| N2 | 7 | 0 (0, 200) | |
| N3 | 5 | 0 (0, 10) | |
| Histologic grade | 0.481 | ||
| 1 | 0 | NA | |
| 2 | 13 | 10 (0, 300) | |
| 3 | 78 | 0 (0, 240) | |
| ILC | 5 | 0 (0, 00) | |
| Lymphovascular invasion | 0.379 | ||
| Present | 30 | 0 (0, 240) | |
| Absent | 66 | 0 (0, 300) |
ILC, invasive lobular carcinoma.
Take home messages.
AXL encodes a receptor tyrosine kinase and has been found to be upregulated in a variety of malignancies from various sites, including the breast.
Axl protein is expressed in most invasive breast carcinomas, regardless of molecular subgroup, despite cell line studies showing preferential upregulation of AXL in triple-negative breast cancer.
Axl expression was significantly associated with lymphovascular invasion in our study, an independent histologic prognostic marker in breast cancer. Studies examining the significance of this finding in relationship to tumour behaviour and recurrence would be informative.
Acknowledgments
We thank Fanming Kong and Lisa Prevedel for their help in collection of breast cancer paraffin blocks for the construction of tissue microarrays.
Footnotes
Provenance and peer review Not commissioned; externally peer reviewed.
Contributors All authors are justifiably credited with authorship, according to the authorship criteria. In detail — TMD: conception, design, analysis and interpretation of data, drafting of the manuscript, final approval given; JH: acquisition of data, analysis and interpretation of data, drafting of the manuscript, final approval given; ZC: statistical support, final approval given; YL: immunohistochemical workup and staining, drafting of the manuscript, final approval given; PZ: conception, design, analysis and interpretation of data, final approval given; SJS: conception, design, analysis and interpretation of data, final approval given.
Competing interests This study was supported by the National Institute of Health grant 1R01 CA159925 to PZ and SJS, and 5R01 CA098210 to PZ. At the onset of the study, JH was supported by a Ruth L. Kirschstein National Service Award (NRSA) Institutional Research Training Grant (T32 GM008539).
References
- 1.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gazinska P, Grigoriadis A, Brown JP, et al. Comparison of basal-like triple-negative breast cancer defined by morphology, immunohistochemistry and transcriptional profiles. Mod Pathol. 2013;26:955–66. doi: 10.1038/modpathol.2012.244. [DOI] [PubMed] [Google Scholar]
- 4.Haffty BG, Yang Q, Reiss M, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24:5652–7. doi: 10.1200/JCO.2006.06.5664. [DOI] [PubMed] [Google Scholar]
- 5.Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 6.Bauer KR, Brown M, Cress RD, et al. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer. 2007;109:1721–8. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 7.Rakha EA, El-Sayed ME, Green AR, et al. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- 8.O’Toole SA, Beith JM, Millar EK, et al. Therapeutic targets in triple negative breast cancer. J Clin Pathol. 2013;66:530–42. doi: 10.1136/jclinpath-2012-201361. [DOI] [PubMed] [Google Scholar]
- 9.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/ College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–95. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolff AC, Hammond ME, Schwartz JN, et al. American Society of Clinical Oncology/ College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–45. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 12.Cohen DA, Dabbs DJ, Cooper KL, et al. Interobserver agreement among pathologists for semiquantitative hormone receptor scoring in breast carcinoma. Am J Clin Pathol. 2012;138:796–802. doi: 10.1309/AJCP6DKRND5CKVDD. [DOI] [PubMed] [Google Scholar]
- 13.Paccez JD, Vogelsang M, Parker MI, et al. The receptor tyrosine kinase Axl in cancer: Biological functions and therapeutic implications. Int J Cancer. 2014;134:1024–33. doi: 10.1002/ijc.28246. [DOI] [PubMed] [Google Scholar]
- 14.Badve S, Dabbs DJ, Schnitt SJ, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24:157–67. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- 15.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 16.O’Bryan JP, Frye RA, Cogswell PC, et al. axl, a transforming gene isolated from primary human myeloid leukemia cells, encodes a novel receptor tyrosine kinase. Mol Cell Biol. 1991;11:5016–31. doi: 10.1128/mcb.11.10.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janssen JW, Schulz AS, Steenvoorden AC, et al. A novel putative tyrosine kinase receptor with oncogenic potential. Oncogene. 1991;6:2113–20. [PubMed] [Google Scholar]
- 18.Craven RJ, Xu LH, Weiner TM, et al. Receptor tyrosine kinases expressed in metastatic colon cancer. Int J Cancer. 1995;60:791–7. doi: 10.1002/ijc.2910600611. [DOI] [PubMed] [Google Scholar]
- 19.Ito T, Ito M, Naito S, et al. Expression of the Axl receptor tyrosine kinase in human thyroid carcinoma. Thyroid. 1999;9:563–7. doi: 10.1089/thy.1999.9.563. [DOI] [PubMed] [Google Scholar]
- 20.Berclaz G, Altermatt HJ, Rohrbach V, et al. Estrogen dependent expression of the receptor tyrosine kinase axl in normal and malignant human breast. Ann Oncol. 2001;12:819–24. doi: 10.1023/a:1011126330233. [DOI] [PubMed] [Google Scholar]
- 21.Meric F, Lee WP, Sahin A, et al. Expression profile of tyrosine kinases in breast cancer. Clin Cancer Res. 2002;8:361–7. [PubMed] [Google Scholar]
- 22.Shieh YS, Lai CY, Kao YR, et al. Expression of axl in lung adenocarcinoma and correlation with tumor progression. Neoplasia. 2005;7:1058–64. doi: 10.1593/neo.05640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsou AP, Wu KM, Tsen TY, et al. Parallel hybridization analysis of multiple protein kinase genes: identification of gene expression patterns characteristic of human hepatocellular carcinoma. Genomics. 1998;50:331–40. doi: 10.1006/geno.1998.5338. [DOI] [PubMed] [Google Scholar]
- 24.Holland SJ, Pan A, Franci C, et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res. 2010;70:1544–54. doi: 10.1158/0008-5472.CAN-09-2997. [DOI] [PubMed] [Google Scholar]
- 25.Ye X, Li Y, Stawicki S, et al. An anti-Axl monoclonal antibody attenuates xenograft tumor growth and enhances the effect of multiple anticancer therapies. Oncogene. 2010;29:5254–64. doi: 10.1038/onc.2010.268. [DOI] [PubMed] [Google Scholar]
- 26.Mackiewicz M, Huppi K, Pitt JJ, et al. Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA. Breast Cancer Res Treat. 2011;130:663–79. doi: 10.1007/s10549-011-1690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong PT, Yong CM, Abnousi F, et al. Lymphovascular invasion is associated with reduced locoregional control and survival in women with node-negative breast cancer treated with mastectomy and systemic therapy. J Am Coll Surg. 2005;200:912–21. doi: 10.1016/j.jamcollsurg.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Pinder SE, Ellis IO, Galea M, et al. Pathological prognostic factors in breast cancer. III. Vascular invasion: relationship with recurrence and survival in a large study with long-term follow-up. Histopathology. 1994;24:41–7. doi: 10.1111/j.1365-2559.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 29.Lauria R, Perrone F, Carlomagno C, et al. The prognostic value of lymphatic and blood vessel invasion in operable breast cancer. Cancer. 1995;76:1772–8. doi: 10.1002/1097-0142(19951115)76:10<1772::aid-cncr2820761014>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 30.Nime FA, Rosen PP, Thaler HT, et al. Prognostic significance of tumor emboli in intramammary lymphatics in patients with mammary carcinoma. Am J Surg Pathol. 1977;1:25–30. doi: 10.1097/00000478-197701010-00003. [DOI] [PubMed] [Google Scholar]
- 31.Bettelheim R, Penman HG, Thornton-Jones H, et al. Prognostic significance of peritumoral vascular invasion in breast cancer. Br J Cancer. 1984;50:771–7. doi: 10.1038/bjc.1984.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rakha EA, Martin S, Lee AH, et al. The prognostic significance of lymphovascular invasion in invasive breast carcinoma. Cancer. 2012;118:3670–80. doi: 10.1002/cncr.26711. [DOI] [PubMed] [Google Scholar]
- 33.Sundquist M, Thorstenson S, Klintenberg C, et al. Indicators of loco-regional recurrence in breast cancer. The South East Swedish Breast Cancer Group. Eur J Surg Oncol. 2000;26:357–62. doi: 10.1053/ejso.1999.0898. [DOI] [PubMed] [Google Scholar]
- 34.Gjerdrum C, Tiron C, Hoiby T, et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc Natl Acad Sci USA. 2010;107:1124–9. doi: 10.1073/pnas.0909333107. [DOI] [PMC free article] [PubMed] [Google Scholar]