Preface
Variation in gene expression is an important mechanism underlying susceptibility to complex disease. The simultaneous genome-wide assay of gene expression and genetic variation allows the mapping of the genetic factors that underpin individual differences in quantitative levels of expression (expression Quantitative Trait Loci, eQTL). The availability of systematically generated eQTL information may provide immediate insight into a biological base for disease associations identified through genome-wide association studies, and can help to identify networks of genes involved in disease pathogenesis. Although there are limitations to current eQTL maps, understanding of disease will be enhanced with novel technologies and international efforts that extend to a wide range of new samples and tissues.
Introduction
Genome wide association (GWA) studies of common complex or multifactorial diseases have been spectacularly successful in the last two years, with many new loci identified with levels of probability that were once thought unattainable. However, the extraordinary levels of significance of the association signals have yet to be translated into a full understanding of the genes or genetic elements that are mediating disease susceptibility at particular loci.
The functional effects of DNA polymorphism on multifactorial disease may be mediated through several mechanisms. Polymorphisms that alter protein function can have very important effects, such as CARD15 (NOD2) mutations in inflammatory bowel disease 1 and FLG mutations in eczema (atopic dermatitis) 2. However, systematic study of complex diseases with known non-synonymous SNPs has not in general yielded highly significant results 3, and variation in gene expression is probably a more important mechanism underlying susceptibility to complex disease. Transcript abundances of genes are directly modified by polymorphism in regulatory elements. Consequently, transcript abundances may be considered as quantitative traits that can be mapped with considerable power. These have been named expression quantitative trait loci (eQTL) 4,5.
The gap between SNP associations from a GWA study and an understanding of how a locus contributes to disease is substantial. Further genotyping and statistical analyses is often necessary to identify causal variants, which are then functionally investigated. This review explores the value of systematic identification of eQTL as one means of characterising the function of loci underlying complex disease traits. The combination of whole-genome genetic association studies and the measurement of global gene expression allows the systematic identification of eQTL. By assaying gene expression and genetic variation simultaneously on a genome-wide basis in a large number of individuals, statistical genetic methods can be applied to map the genetic factors that underpin individual differences in quantitative levels of expression of many thousands of transcripts.
The resulting comprehensive eQTL maps provide an important source of reference for categorising both cis and trans effects of disease-associated SNPs on gene expression. In addition to providing information about the biological control of gene expression, such data aid in interpreting the results of GWA studies. Once the statistical evidence for association of genetic markers to a disease trait has been established, genome-wide eQTL mapping data can be examined to see if the same genetic markers are also associated with quantitative transcript levels of one or more genes. (Such markers are known as “eSNPs”). The availability of systematically generated eQTL information provides immediate insight into a probable biological base for the disease associations, and can help to identify networks of genes involved in disease pathogenesis.
The potential of genome-wide eQTL identification was shown originally in yeast 6 and then in humans, animals and plants 4,7. The history of eQTL mapping has been comprehensively reviewed 7–9, and will not be described in detail here. The present review will instead show how the combination of genetics and global gene expression may be a powerful tool for systematically unravelling the effects of variation in transcription on disease. This review first briefly introduces the principles and current methods of eQTL mapping and describes the basis of eQTL. We then explore the relevance of these results to disease gene identification. The limits of present eQTL mapping data are discussed, as is the expected impact of new technologies, international efforts to extend results to new samples and tissues and how cell lines might be tested with stimuli relevant to disease.
eQTL mapping
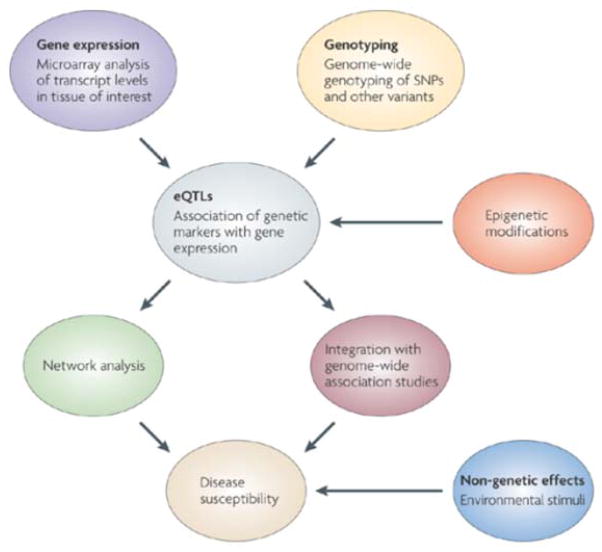
In practical terms, the starting point for eQTL mapping is the measurement of gene expression in a target cell or tissue from multiple individuals (Figure 1). This information is the substrate for investigating the effects of DNA polymorphism (of whatever type) on the expression of individual genes. The use of microarray technology to measure gene expression from many thousand of genes simultaneously has been a principal driving force for systematic mapping of eQTL 7. The field is benefiting from progressively more sophisticated platforms for such studies, which are described in the later sections of this review. Procedures for eQTL mapping rest on the insight that expression levels can be analysed with genetic approaches in the same manner as any other quantitative trait phenotype, such as body weight or blood lipids. In particular, study designs and statistical methods that are used traditionally to map quantitative trait loci can be successfully applied to identifying eQTL 10–12. Interpretation of eQTL data can then be developed further by the incorporation of additional biological information, such as epigenetic modifications, and analysis of regulatory networks, which are discussed later.
Figure 1. eQTL mapping.
Expression QTL mapping begins with the measurement of gene expression in a target cell or tissue from multiple individuals. This information is the substrate for investigating the effects of DNA polymorphism (of whatever type) on the expression of individual genes. Other factors which may alter transcription, such as epigenetic CpG methylation may also be mapped. Network analyses builds upon the strong correlations that are present between transcripts, and allows the identification of modules of genes that mediate complex functions. This information can then be made available to interpret genetic associations and mapping information from the study of complex disease.
EQTLs are influenced not only by genetic polymorphisms, but also a range of other biological factors. These may be dissected systematically, starting with the measurement of heritability (H2).
Heritability
Family studies have demonstrated that many human eQTL are highly heritable 13,14. The linkage approach in which family members are studied has been valuable in demonstrating that genetic factors have widespread and identifiable influences on eQTL in humans, and such studies have provided broad localisation for some of the underlying genetic factors 15,16. GWA mapping of common genetic variants that underlie eQTL has recently become possible due to the wide availability of high-throughput and low-cost SNP genotyping. These results are particularly relevant to disease mapping that is also focused on common SNPs characterised with similar SNP arrays. Moreover, the interpretation of these eQTL data relies strongly on methodologies that have been developed for disease GWA 13. For example a family study of lymphoblastoid cell lines (LCL) identified nearly 15,000 traits (corresponding to individual Affymetrix probes) with an estimated H2 of > 0.3, indicating that genetic influences on gene expression appear to be widespread 13. Other studies have similarly described high heritability of many eQTL in LCL and other tissues 4,17,18.
Genetic factors (with both cis- and trans-acting effects, see below), are often identified for eQTL that have high heritability. For example, in the LCL study mentioned above 13, eQTL for 81% of traits with H2 > 0.8 could be mapped to one or more SNPs at genome-wide significance. However, the SNP map on average accounted for less than 20% of the estimated trait H2, consistent with results obtained by other studies 16. This demonstrates the presence of genetic or other effects affecting familial clustering on transcription that are not detectable in these genetic associations. Factors other than SNPs that might affect H2 are discussed further below. Further understanding of disease phenotypes may also be gained from analysing whether particular types of genes have more heritable variation in expression level (Box 1).
Box 1. Gene ontology analyses.
Given the heritability of many eQTLs, eQTL databases may also be used to identify the types of genes that show most inherited variation in their levels of expression (at least in the cell type studied, usually LCL), by applying Gene Ontology (GO) analyses. The most highly heritable GO biological process for eQTL in LCL in one study was, unexpectedly, “response to unfolded proteins”, a group containing numerous chaperonins and heat shock proteins. The individual variation in response to unfolded proteins may represent an evolutionary response to cellular stress, and these genes could be candidates in the study of neurodegenerative diseases and aging processes. Genes regulating progression through cell cycle, RNA processing, and DNA repair were also exceptionally heritable. The evolutionary advantage of individual variation in these genes is unclear. More expectedly, genes with significant heritability are also enriched in GO categories of immune response 13,19. These highly heritable immune genes may be of particular value for the study of infectious and inflammatory diseases. These most heritable traits can be considered as candidate genes for effects on particular disease traits, but there may also be a place for them to be studied in large population samples, such as those contained in National Biobanks, to investigate their actions on unexpected phenotypes.
Cis and trans effects
Statistical analyses of eQTL need to take into account that the loci identified can influence gene expression either in cis or in trans. The definition of a cis effect is somewhat arbitrary, but cis-acting eQTLs are typically considered to include SNPs within 100Kb up-stream and down-stream of the gene whose expression is affected by that eQTL. This definition becomes more problematic in regions of extended linkage disequilibrium, such as the major histocompatability complex (MHC). Detailed analysis of the position of mapped cis-acting eQTL effects have shown that these are enriched around transcription start sites and within 250 bp upstream of transcription end sites, and rarely reside more than 20kb away from the gene 20. Cis-acting variants also appear to occur more often in exonic SNPs 20. In general, trans effects are weaker than those in cis in humans 4,5 and in rats 21, but are very numerous.
It is not known if trans effects are in general mediated through transcription factor variants or other mechanisms. “Master regulators” are trans-acting factors with multiple effects on gene expression that have been identified in Saccharomyces cerevisiae 22, rat tissues 21, and in the human genome 5. It is of interest that, at least in yeast, master regulators are not enriched for transcription factors, and trans-regulatory variation seems to be broadly dispersed across classes of genes with different molecular functions 22.
Other types of variant
The function of DNA can be altered by many mechanisms in addition to SNPs. Transcription may also be modified by copy number variations (CNV), insertions and deletions, short tandem repeats and single amino acid repeats 23. A systematic investigation of the effects of CNVs in individuals who are part of the International HapMap project showed that SNPs and CNVs captured 84% and 18% of the total detected genetic variation in gene expression, respectively, but the signals from the two types of variation had little overlap. 24. It has been shown that CNVs in regulatory hotspots in the malaria parasite genome dictate transcriptional variation 25. It has also been observed that small-scale copy number variation (on the order of a single or few copies) can lead to multiple orders of magnitude change in gene expression and, in some cases, switches in deterministic control. 26.
Epigenetic factors
In addition to DNA sequence variants, gene transcription is also modulated by epigenetic modifications (see also ‘Limits of mapping studies’, below). For example, non-germline epigenetic methylation of CpG residues that regulate gene expression is common in the human genome 27. In a limited study of three chromosomes 17% of genes may be differentially methylated in their 5′ UTRs and about one-third of the differentially methylated 5′ UTRs are inversely correlated with transcription 27. A further level of complexity comes from post-translational modifications of histones that modulate DNA accessibility and chromatin stability to provide an enormous variety of alternative interaction surfaces for trans-acting factors (reviewed in 28).
EQTL and disease-gene mapping
Combining eQTL and genome-wide association studies
One of the most important consequences of eQTL mapping is the link that it provides between genetic markers of disease identified in GWA studies and the expression of a specific gene or genes. In particular, the power of these studies depends upon the identification of specific genetic markers that are simultaneously associated with disease and eQTL, whereas simply comparing differences in gene expression in cases and controls may not provide sufficient power to detect important differences with the available sample sizes. The value of this is illustrated by several recent investigations in which eQTL analysis was incorporated directly as a component of the GWA study design (included in Table 1). The number of GWA studies continues to rise rapidly. In GWA studies to date, 10–15% of the top hits have also impacted on a known eQTL in a public dataset (Table 1). We will therefore discuss selected instances of these to illustrate the value of the method.
Table 1.
Disease-linked associations with significant expression quantitative loci from the literature and public databases
| Study | Trait | Region | Candidate Gene | Transcript affected by SNP | Transcript Region | LOD |
|---|---|---|---|---|---|---|
| * Gudbjartsson et al 84 | Height | 7p22 | GNA12 | GNA12 | 7p22 | 13 |
| 11q13.2 | Intergenic | CCND1 | 11q13 | 7.4 | ||
| 7q21.3 | LMTK2 |
C17orf37 HSD17B8 NDUFS8 |
17q21 6 11 |
6.0 6.4 6.1 |
||
| 3p14.3 | PXK | RPP14 | 3 | 9.2 | ||
| Göring et al 15 | HDL-C | 6q21 | VNN1 | HDL-C (serum) | 8.0 | |
| Kathiresan et al 40 | Polygenic Dyslipidaemia | 20q13 | PLTP | PLTP | 20q13 | 16 |
| 15q22 | LIPC | LIPC | 15q22 | 17 | ||
| 11q12 | FADS1, FADS2, FADS3 |
FADS1 FADS3 |
11q12 | 35 8.0 |
||
| 9p22 | TTC39B | TTC39B | 9p22 | 7.0 | ||
| 1p13 | CELSR2, PSRC1, SORT1 |
SORT1 PSRC1 CELSR2 |
1p13 | 270 249 80 |
||
| 12q24 | MMAB, MVK | MMAB | 12q24 | 43 | ||
| 1p31 | ANGPLT3 |
DOCK7 ANGPLT3 |
1p31 | 27 11 |
||
| Libioulle et al 37 | Crohn’s Disease | 5p13 | Intergenic | PTGER4 | 5p13 | 3.0 |
| Barrett et al 36 | Crohn’s Disease | 5q31 | OCTN1, SLC22A4, SLC22A5 | SLC22A5 | 5q31 | |
| * Hom et al 85 | SLE | 8p23.1 | C8orf13, BLK |
BLK C8orf13 |
8p23.1 | 20 28 |
| * Harkonason et al 86 | T1D | 12q13 | RAB5B, SUOX, IKZF4 | RPS26 | 12q13 | 33 |
| 1p31.3 | ANGPTL3 | DOCK7 | 1p31.3 | 16 | ||
| * WTCCC 87 | T1D | 12q13.2 | ERBB3 | RPS26 | 12q13.2 | 43.2 |
| * Todd et al 88 | T1D | 12q13.2 | ERBB3 | RPS26 | 12q13.2 | 30.3 |
| * Plenge et al 89 | Rheumatoid arthritis | 9q34 | TRAF1-C5 | LOC253039 | 9q34 | 6.3 |
| Thein et al 90 | HBF production | 6q23.3 | Intergenic | HBS1L | 6q23.3 | 6.0 |
| Moffatt et al 30 | Childhood asthma | 17q21 | Intergenic | ORMDL3 | 17 | 14 |
| * WTCCC 87 | Bipolar disorder | 16p12 | PALB2, NDUFAB1, DCTN5 | DCTN5 | 16p12 | 9.2 |
| 6p21 | NR |
HLA-DQB1 HLA-DRB4 |
6p21 | 8.9 11 |
||
| * Di Bernardo et al 91 | Chronic lymphatic leukaemia | 2q37 | SP140 | SP140 | 2q37 | 8.8 |
Identified through comparison of http://www.genome.gov/gwastudies/ and http://www.sph.umich.edu/csg/liang/asthma/
For example, a recent study generated genome-wide transcriptional profiles of lymphocyte samples from participants in the San Antonio Family Heart Study, and showed that high-density lipoprotein cholesterol concentration was influenced by the cis-regulated vanin 1 (VNN1) gene 15. Similarly, a study of post-mortem brain tissue identified eQTL affecting the MAPT and APOE genes, which play an important role in Alzheimer’s disease 29.
At the same time as the San Antonio study the results of a GWA study of asthma 13,30 identified a series of SNPs in strong linkage disequilibrium and spanning more than 200 kb of chromosome 17q23 and strongly associated with the risk of asthma 30. The region of association contains 19 genes, none of which are obvious candidates to be implicated in disease. Examination of eQTL data derived from Affymetrix HU133A arrays13,30 on the same families showed that the disease associated SNPs had highly significant (P < 10−22) effects in cis on the expression of one the genes called ORMDL3.
This locus illustrates the utility of combining eQTL and disease mapping studies. Despite the highly significant association with both expression and disease, the predicted expression differences in cases and controls, which was averaged over all genotypes, was not expected to be significant given the sample size: this was in agreement with the observed results 30.
In these data, borderline significant effects were also seen on expression of the gene neighbouring ORMDL3, GSDML 30. Subsequent eQTL studies with the Illumina platform and RT-PCR experiments confirmed that the same SNPs determine eQTL with both genes. These results focus attention on one or both of these genes as likely candidates for a role in disease pathology. Many additional studies are now underway to investigate the biological functions of these two genes and their relationship to asthma 31,32–35.
Using eQTL to interpret GWA studies
Such findings have motivated the use of these eQTL data as a general tool for interpreting results from GWA studies. Recent analyses of Crohn’s disease (CD) illustrate this approach 36,37. Initially, markers on Chromosome 5 were shown to be strongly associated with CD in one GWA scan, but their biological effects could not be readily deduced as they reside in a 1.25 Mb gene desert. Examination of the LCL eQTL database showed that one or more of these polymorphisms act as a long-range cis-acting factor influencing expression of PTGER4, a gene that resides approximately 270 kb proximal to the association region 37. The homologue of this gene has been implicated in phenotypes similar to CD in mouse 37,38. Thus, research is now focused on PTGER4 as a primary candidate gene for this disease susceptibility locus.
Subsequently, the eQTL approach has been applied systematically in a meta-analysis of GWA studies of CD, and several other interesting results have been obtained 36. For example, eQTL were used to address an outstanding question in CD genetics related to the identification of the CD susceptibility gene or genes in the cytokine cluster at 5q31, where SNPs have an established association with disease 39. The disease-associated SNPs in the meta-analysis of this region were all shown to be correlated with decreased SLC22A5 mRNA expression levels. Another CD locus identified in the meta-analysis coincided with the asthma risk locus on chromosome 17, in which the disease markers are also correlation with expression of ORMDL3 and GSDML as described above. Thus the same genetic variants contribute to susceptibility to both CD and asthma, possibly by perturbing expression of one or both of these genes. Several additional examples of eQTL within CD susceptibility loci have also been reported 36. These co-localisations greatly exceed the number that would be expected by chance, suggesting that many are reflecting underlying biological processes involved in disease susceptibility 36.
Examination of public GWAS results (http://www.genome.gov/gwastudies/) identifies many other disease associations where eQTL data provides similar insights (Table 1). For example, a recent large study of polygenic dyslipidaemia identified 30 loci with highly significant effects on blood lipid measurements 40. Examination of gene expression in samples of liver from 957 subjects allowed highly significant eQTLs to be identified for 7 of the 30 loci 40 (Table 1). In some cases, the eQTL data gives genetic evidence to support a candidate gene for which a role was previously hypothesised from location and biological hypotheses (such as GNA12 for height on Chromosome 7p22, and BLK and C8orf13 for auto-immune systemic lupus erythematosis on 8p23.1). More often the gene expression data identifies different genes or suggests a particular gene from a number of candidates. Examples of this are the cluster of trans-acting genes from the height locus on 7q21.3, the RPS26 gene from the Type 1 diabetes locus on 12q13.2, and the DCTN5 gene from the bipolar disorder locus on Chromosome 16p12.1.
Not all examples of eQTL findings are straightforward, as exemplified by the association reported between the SH2B1 locus and body mass index (BMI) 41. In this study, a miss-sense SNP in SH2B1 was also associated with significant variation in transcript abundances of EIF3C and TUFM. When mutated, the mouse homologue of SH2B1 leads to extreme obesity in mice, apparently because of a failure for proper regulation of their appetite. The authors speculate that the SH2B1 variant has a causal role but happens to be in LD with a different variant that influences EIF3C and TUFM mRNA levels; alternatively, regulation of EIF3C or TUFM mRNA levels could have a causal role, instead of or in addition to variation in SH2B1 41.
eQTL databases
A database of eQTLs from the asthma studies 13,30 that allows searches by genes, chromosomal regions, and SNPs (http://www.sph.umich.edu/csg/liang/asthma/) illustrates how data from this kind of research can be examined. VarySysDB is another public database based on 190,000 extensively annotated mRNA transcripts from 36,000 loci (http://www.h-invitational.jp/varygene/home.htm). VarySysDB offers information encompassing published human genetic polymorphisms for each of these transcripts separately. In addition to SNP effects on transcription, this database includes deletion-insertion polymorphisms from dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/), copy number variations from Database of Genomic Variants (http://projects.tcag.ca/variation/), short tandem repeats and single amino acid repeats from H-InvDB (http://www.h-invitational.jp/) and linkage disequilibrium regions from D-HaploDB (http://orca.gen.kyushu-u.ac.jp/) 23.
Major Histocompatability Complex
Analysis of eQTL within the MHC is of particular interest for studies of diseases in which infection and autoimmunity is a major component42, Intense study of the MHC over many years has revealed many genes that are duplicated or polymorphic and DNA variants in the MHC have been associated with more diseases than any other region of the human genome 42. Many disease associations have been attributed to selective binding of processed antigen within the antigen-presenting grooves of human leukocyte antigen (HLA) variants.
The results of eQTL studies within the MHC must be interpreted with caution because the high degree of genetic variability and linkage disequilibrium across the MHC locus could introduce some spurious results due to polymorphism in sequences corresponding to probes used for expression measurements (see 43 and below). Nevertheless, global gene expression data has shown very strong effects of particular SNPs on the level of expression of the classical MHC antigens HLA-A, HLA-C, HLA-DP, HLA-DQ and HLA-DR (P<10−20– 10−30) 13. This confirmed the effect of genetic variation on the level of HLA-DQ expression observed previously 44. The strength of these effects suggests that associations of MHC class I and class II polymorphism may be anticipated to depend on the level of gene transcription as much as restriction of response to antigen 13. An example of this may be Type I diabetes, in which the functional effects of the long-recognised association to the class II MHC genes 45 have not been elucidated, despite combined P values < 10−100.
These results suggest that even in this intensively studied region, the investigation of eQTL could add further to understanding of the many known genetic associations.
Additional biological interpretation and validation
A genome exerts its functions not through particular genes or proteins, but instead through highly complex networks that produce a range of responses 46. As perturbations of such networks underlie the pathogenesis of many diseases 47,48, network analysis incorporating eQTL data has recently given important novel insights into mechanisms underlying multifactorial diseases 16,17,49(Box 2).
Box 2. Networks and other analytical tools.
Traditional genetics and cellular biology has rested on the assumption that a single stimulus (or DNA variant) when applied to a cell (or gene) will have a single outcome. The reality is that even a simple stimulus will induce changes in transcription in many genes that interact in complex networks, with an outcome that affects many different transcripts and processes.
The networks may be considered to be made up of multiple pathways that may act at genetic, genomic, cellular, tissue and whole organism levels 46. The technology that is already available to gather global information on gene expression, proteins and metabolites is now making possible the systematic identification of the networks of genes that interact in disease processes 92,93. Analysis of genetic variants that perturb networks through eQTL effects has recently given important novel insights into mechanisms underlying multifactorial diseases 16,17,49. This type of analysis may also lead to systematic identification of transcription modules 94 and the construction of regulatory networks 95. The potential of using genetic mapping approaches to identify networks of genes operating on hematopoietic stem cells 96 and immune responses 97 are amongst the examples that have been discussed.
The impact of combining eQTL analysis with an investigation of gene networks is illustrated in the recent detection of genetic variants associated with transcript abundance of a macrophage-enriched network and obesity-related traits in human subjects. Parallel studies in mouse and human identified a network module for obesity-related traits that was enriched for genes involved in the inflammatory and immune response. EQTL mapping was then used to identify cis-acting genetic variants associated with this network of genes. The authors characterised these genetic variants in a large cohort of individuals, and showed statistical enrichment for variants that were associated with obesity-related biometrical traits 16. This approach allowed identification of genetic variants that had minor individual effects on the trait, but which can be identified as a group because of the overall perturbation of the network. Three genes in this network, lipoprotein lipase (Lpl), lactamase β (Lactb) and protein phosphatase 1-like (Ppm1l), were validated by gene knockouts, strengthening the association between this network and metabolic disease traits 49.
A bibliography and a range of statistical routines for network analysis can be found at http://www.genetics.ucla.edu/labs/horvath/CoexpressionNetwork/.
Extensive investigations of human populations, animal models and cellular systems are generally required to provide biological validation of the relationship between specific genes and multifactorial disease traits even when identified through eQTL analysis. Given the substantial effort that is required for validation, careful selection of only the strongest candidates is essential. As illustrated in the above examples, the combination of GWA studies and eQTL analysis is a powerful tool for identifying a small number of candidate genes and pathways. With the deployment of new technologies, such as exon arrays and RNA resequencing, and expansion of the tissues covered as described below, we expect future eQTL databases to be even more powerful tools for such identifications.
Potential limitations and future directions
Despite the power of eQTL mapping to help identify the genetic basis of disease, there are many limitations to current methodologies and potential for considerable improvements as technologies develop. The best appreciated technical barriers to optimal eQTL mapping reside in the use of microarrays to measure gene expression (Box 3). Other problems and their potential solutions are given below.
Box 3. Pitfalls with microarrays.
The use of microarrays to measure gene expression has led directly to the development of eQTL analyses. However, the microarray approaches that underlie most eQTL studies to date provide only partial gene coverage and have a limited dynamic range for quantitative detection of expression. Specific problems inherent in the use of these microarrays include the systematic bias that may be introduced during sample preparation, hybridisation and measurement of expression, batch to batch variation in array manufacture, and day to day variation in laboratory conditions 98. These types of effects are probably under-recognised, as exemplified by a report of large scale differences in gene expression between ethnic groups 98,99. In this case the highly significant differences in gene expression that the data had suggested between the groups 98 were found to be due to the separate processing of expression measurements in LCL from subjects of European and Asian ancestry.
Cis-eQTL artefacts may also arise from the overlap of SNPs with transcript probes 100. Alterations in hybridisation efficiency due to the SNP may give an erroneous impression of differences in transcript abundance attributable to the SNP (and other DNA variants with which it may be in linkage disequilibrium) 100. It has been estimated that 15% of microarray probes for any given gene will overlap with SNPs that are polymorphic in the population under study 100. However, most coding SNPs in the human genome are uncommon, and it also appears that measurements of abundances are robust against mismatches between the probe and RNA sequences 101. While evidence for the impact of these artefacts has been presented 43, it is reassuring that in a large study in humans, Emilsson et al. 16 found no evidence of systematic or specific hybridisation artefact from SNPs in their eQTL data. Nevertheless, important findings from microarrays need confirmation by specific assays such as qPCR that avoid polymorphic sequences Statistical methodology to account for batch effects, polymorphism and other sources of artefact is discussed by Alberts et al 102
Most human studies of eQTL have been performed in LCL, primarily because LCL were often created as a source of nucleic acids for genetic studies. However, LCL may exhibit progressive genomic instability with multiple passages of storage and re-growth.
Comparisons between microarray platforms
It has been assumed that different microarray platforms give broadly comparable results 50. However, numerous studies are now showing that the overlap in transcript detection between platforms is only of the order of 30–40%, whether considered as presence or absence of detectable transcripts or the absolute level of transcript abundance 51,50,52,53. The same level of discordance appears whether comparisons are made between Affymetrix arrays and SAGE 52, Affymetrix and Illumina arrays 50, Affymetrix and ABI arrays 53, or across multiple platforms 51.
Some of this discrepancy may be because individual genes are commonly interrogated by different sequences on different platforms. The situation can be improved when matching of genes is sought using genomic sequence rather than sequences inferred from the Unigene database of transcripts (http://www.ncbi.nlm.nih.gov/unigene)54. Concordance between platforms is improved further when probes are compared only when they target overlapping transcript sequence regions on cDNA microarrays or gene-chips 55.
These discrepancies may follow from the complex and unpredictable factors that determine hybridisation of particular nucleic acids to complementary array bound sequences 56,57. In addition, the selection of sequences on microarrays has been strongly biased to the 3′ end of genes, simply because public cDNA databases were first populated with genes identified by 3′ tags.
A consistent conclusion of comparison studies has been that different platforms provide complementary results 51,52, probably because they are all sampling only a selected fraction of the total transcriptome from the cells or tissue under study. The use of multiple platforms to extract all the expression information from a cell or tissue is impractical.
New platforms for measuring gene expression
A more comprehensive measurement of gene expression comes from arrays which interrogate all known human exons. Affymetrix have produced global exon arrays58 which show a high degree of correspondence in terms of fold changes with their pre-existing “classical microarrays”, suggesting that the additional probesets on the exon arrays will provide reliable as well as more detailed coverage of the transcriptome 59. The use of exon arrays allows the identification of tissue-specific alternative splicing events as well as significant expression outside of known exons and well-annotated genes 60. Exon arrays on other platforms are likely to provide similarly robust results.
Many of the problems inherent in the use of microarrays can be solved by massively parallel, ultra-high throughput DNA sequencing systems (reviewed in reference 61). These systems allow direct ultra-high throughput sequencing of RNA, which can then be mapped back to the genome. Sequencing RNA provides a generic tool which can support a family of assays for measuring the global, genome-wide profiles of mRNAs, small RNAs, transcription-factor binding, chromatin structure, DNase hypersensitivity and DNA methylation status 61. RNA splices may also be effectively mapped by sequence-based methods.
Despite the formidable promise, ultra-high throughput sequencing is still not without problems. The machines can produce terabytes of data daily, and make profound demands on bioinformatics for data storage and assembly of reads. Short reads may pose severe problems for interpretation of transcripts arising from gene families with high homology or repetitive regions of the genome. Nevertheless, it can be anticipated that within 2 years many studies will rely on this technology, and that alternative or complementary approaches such as large-scale real-time PCR based expression assays (e.g. Watson et al. 62 http://www.wafergen.com) will continue to evolve.
Limits of mapping studies
As discussed in the section on heritability, presently mapped loci account for only a portion of the estimated heritability of eQTL. A similar degree of unattributed or “dark” heritability has been observed in GWA studies of common complex traits and diseases. A large GWA meta-analysis, for example, recently identified 20 variants significantly associated with adult height. The combined effects of the 20 SNPs explained only 3% of height variation, taking into account such factors as age and population 63. Similarly, a large GWA meta-analysis of Crohn’s disease identified 32 loci significantly impacting on the disease, which together explained only 10% of the overall variance in disease risk and 20% of the genetic risk 36.
A large portion of the unattributed heritability is expected to result from the effects of multiple loci that are too weak to detect using current sample sizes 18. This explanation would be consistent with data in yeast, where only 3% of highly heritable transcript abundances are explained by single-locus (monogenic) inheritance and 50% are consistent with more than five controlling loci of equal effect 64. Although present SNP arrays provide relatively comprehensive coverage of the genome (more than 80%), some of the unattributed heritability will be due to genetic factors that reside in unmapped regions, or variation that is not effectively tagged at present, such as copy number variants (CNVs). Dominance and interaction effects may also account for some of the unattributed heritability, as these may be confounded with additive genetic effects in the heritability estimates with some study designs.
A previously described global eQTL study was based on sib pairs, allowing estimates of heritability for all the transcripts measured 13. The study suggested that dominance had a minimal effect on gene transcription 13. Interestingly it appeared that genetic interactions may have important influences on regulation of expression for some genes, but inclusion of interaction effects had a minimal impact on the overall attributable heritability 13.
Epigenetic modifications and other factors that affect transcript abundance may not be accounted for in SNP-based association studies (see ‘The basis of eQTLs’ above). Genomic imprinting is a particular case of an epigenetic effect with a parent-of-origin dependent pattern. Monoalleleic expression is established at imprinted loci, via epigenetic marks transmitted through the germline. Several common complex diseases exhibit parent of origin effects that might indicate underlying imprinting, including asthma 65, Type I diabetes66,67, rheumatoid arthritis68, psoriasis69, inflammatory bowel disease70 and selective IgA deficiency71, but systematic analysis of parent of origin effects in eQTL data has not yet been reported.
Finally, transcript abundance is a function of transcript stability as well as transcript production. Many factors mediate transcript stability, particularly in trans, either through protein-RNA interaction of through mechanisms mediated through small interacting RNAs (siRNA) 72. It seems clear that future studies of disease susceptibility as well as eQTL will need to take these mechanisms into account.
Gene expression in tissues
While RNA for eQTL analyses would ideally be obtained from a wide variety of tissues, the majority of human studies of eQTL have been performed in LCL, primarily because LCL were often created as a renewable source of nucleic acids for genetic studies. Gene expression in LCL however represents the particular circumstances of EBV infection of B-cells and their subsequent uncontrolled growth. LCL may also exhibit extreme clonality with random patterns of monoallelic expression within single clones 73.
Although only 60% of genes from any particular cell type will also be found in LCL 4,13, it has been established that LCL provide information about gene expression for some genes whose primary function is not in these cells 4,74–76. In addition, a recent comparison of eQTL derived from the analysis of blood and adipose tissue showed little difference in the number of eQTL that could be mapped, and there was about 50% overlap of mapped loci from the two RNA sources 16. Similarly, comparison between four different tissues showed no statistically significant differences in the number of mapped transcripts in experiments involving mapped recombinant inbred strains of mice 18.
Despite the continued utility and convenience of LCL studies of gene expression, it is evident that many of the transcripts expressed in LCL may represent general housekeeping genes, and transcripts that determine specialised cell functions (and modify disease) may be more parsimoniously distributed. In addition LCL are removed from the stimuli that may induce disordered gene transcription in disease, exemplified by the differences that may be observed in gene expression between LCL derived from asthmatics and genes known to be expressed in asthmatic airways 30. These factors all indicate that the direct examination of tissues that may be involved in disease may provide much more information than the LCL alone.
Some eQTL studies of human tissue have already been carried out, notably of liver 17, adipose tissue 49,16 and human brain 29. These show that approximately 60% of the transcriptome is expressed in each tissue and that eQTL from these tissues may be a very valuable source of information for genetic mapping. Data from animal models suggest that tissue samples may allow detection of trans-eQTL that are important in determining the composition of individual tissues 18. Tissue samples also promise the use of network analyses to identify the complex interactions that may underlie disease 17,49,16 (Box 2).
The costs of reagents and limited availability of appropriate tissues have to date restricted studies in humans to at most several hundreds of subjects. While a formal evaluation of optimal study sizes is difficult because of unknown trait heritability we know empirically that studies with a few hundred subjects have consistently identified numerous eQTL with vanishingly small P values 4,5,75. It is also clear that subtle effects, particularly in trans, would be detected more reliably with larger samples.
It is therefore timely that the promise of eQTL as tool for disease genetics has been sufficiently exciting to prompt an NIH proposal for an ambitious genotype-tissue expression database that might include 1000 samples from each of 30 different tissues. The GTEx project is currently running as a 2-year pilot with the primary goal of testing the feasibility of collecting high-quality RNA and DNA from multiple tissues from approximately 160 donors identified through low post-mortem interval autopsy or organ transplant settings. If the pilot phase proves successful, the project will be scaled up to involve approximately 1000 donors, with the eventual creation of a database to house existing and GTEx-generated eQTL data (http://nihroadmap.nih.gov/GTEx/).
The use of tissues poses a number of problems that need to be resolved. Normal and diseased tissue samples may be difficult to access and their use requires careful attention to ethical, legal and social issues. Samples taken at post-mortem from many tissues robustly retain their histological architecture and contain RNA which may be of sufficient quality for measurements of gene expression. However, the changes in gene expression that may accompany death or surgical resection have not yet been documented in any detail. Tissues typically consist of different cell types, and their composition may vary inconsistently in the presence of disease. Finally, tissue-specific DNA methylation profiles may affect 20% of genes 27 and will be expected to be important in understanding tissue eQTL.
Although some of these problems may be expected to degrade the information available from the study of any particular tissue, it should be appreciated that they will not systematically lead to false-positives in eQTL analyses 17, emphasising the robustness of the eQTL approach.
Exercising the genome
Tissue biopsies and other samples extend the “expression space” that may be examined by eQTL studies. They nevertheless still have limitations for functional analyses (particularly in humans as opposed to model organisms) when compared to cells that can be grown freely in culture and manipulated by systematic knockdowns.
Although the transcripts in a particular cell under particular conditions reflect only part of the function of a particular genome, the range of transcripts from a given cell type can be widened by stimulating the cell in a variety of ways. The experimental extension of the genome expression space has been called “exercising the genome” 77, and this strategy can be used to learn much more about gene expression and integrated gene functions. Experimentally, evidence is already emerging that environmental actions on gene expression are profound in humans 78 and model organisms 79,80 (reviewed in 81), and it is reasonable to assume that these components of gene expression may be fruitfully accessed through exposure to relevant stimuli. It is of interest in model organisms that environmentally induced changes in gene expression seem to act through prominent trans effects 79,80 that may not be present in unstressed cells and tissues.
It is therefore desirable that the genome of human LCL or primary cells of particular interest be exercised by stimulating their gene expression in different ways. Model stimuli that could be tested in these systems include pro-inflammatory stresses, metabolic stresses (high or low glucose or hypoxia), the response to radiation, the response to signalling molecules (neurotransmitters, hormones, peptides) and the response to therapeutic and chemotherapeutic agents.
Conclusions
It is now well established that transcript abundances of genes may be considered as quantitative traits that can be mapped with considerable power, and that the assaying gene expression and genetic variation simultaneously on a genome-wide basis in a large number of individuals will provide valuable tools for indentifying the function of previously mapped susceptibility alleles underlying common complex diseases.
Although eQTL have rapidly been shown to be effective in mapping complex traits there are many levels of information that are inherent in the measurement of global gene expression that have yet to be accessed, such as the effects of transcript stability, epigenetic effects, or environmental stimuli. In addition, larger studies involving thousands of subjects may be necessary to identify relatively weak trans effects with the same precision as the more powerful effects often observed in cis. Although trans effects may be relatively weak, the genes they modify (the trans-transcriptome) are likely to contain master regulators with wide effects on key processes that may also appear more strongly in tissues and in cells subjected to particular environmental stimuli. Many genes are only expressed in particular tissues or at particular times during development. Thus, although systematic studies of eQTL are already being planned for a wide variety of tissues, other strategies will need to be formed to study particular cell types and tissues at specific stages of differentiation and development.
The genome of cancer cells and tissues is particularly challenging to understand, because the primary lesions that first drive cellular proliferation are difficult to find when uncontrolled division results in progressive secondary damage to the genome and the transcriptome. EQTL analyses may be of particular value in malignant disease, because they allow a more integrated picture of what is happening in cancer cells (Box 4).
Box 4. eQTL and network analyses of cancer.
Mutations that disrupt cell growth control mechanisms are a feature of cancer. In addition, the unchecked cell division that is characteristic of cancer may in time result in many secondary mutations and progressive genomic disorganisation 103. Genetic studies of cancer tissue (“somatic cell genetics”) have been used to try and identify the most common mutations in various tumours. Global gene expression studies have also been used in many cancer types, typically to identify gene signatures that may predict the clinical outcome 104. However, most signature-based outcome predictions have not been replicated by independent studies 104, perhaps due to the innate heterogeneity of cancerous tissue and the problems of deriving statistically stringent results from the measurement of thousands of transcripts in limited numbers of samples. EQTL analyses may be a powerful tool to identify the functional consequences of the numerous CNVs, deletions and epigenetic modifications that are a feature of neoplastic cells. EQTL mapping allows not only the identification of genes underlying malignant processes 105 but also genes modifying disease progression 106 and genes modulating individual responses to chemotherapy 107. Network analyses have not yet been widely applied to the study of cancer, but have already led to interesting findings, such as the identification of the ASPM gene as a molecular target in patients with glioblastoma. The application of network analyses to cancer eQTL may be expected to greatly alleviate problems with multiple comparisons and to lead to easier biological interpretation of results 108,109. Direct comparison of the transcript network architecture of cancerous tissue against normal tissues may also allow much deeper understanding of cancer biology.
Good progress is being made in terms of cataloguing the SNPs and other polymorphisms that regulate transcription, and this may be the basis for a systematic listing of regulatory sequences and regulatory proteins. Greater difficulties seem likely in identifying epigenetic effects, particularly if these are mediated through histone modifications (which are difficult to detect on a large scale) rather than through differential CpG methylation.
The remarkable diversity of human transcriptional regulation raises new questions about the evolutionary value of unexpected variation in genes that mediate basic mechanisms, such as heat shock proteins or genes influencing cell cycle and DNA repair. “Inverse genetics”, could be used to study the SNPs with the strongest effects on expression of such genes to investigate their actions on unexpected phenotypes measured in epidemiological samples.
New analytical techniques, particularly network analyses, promise rapid advances in reducing the complexity of expression data. Modules of co-expressed genes mediating complex functions may also be identified by time-series studies of the response of particular cell types to environmental stimuli 82.
In future, integration of eQTL with data from large-scale approaches for genome resequencing, proteomic and metabolomic analyses, epigenomic studies and functional screening of genes may provide a powerful set of tools to power a systems biology approach to multifactorial disease, and providing a toolbox for identification and biological validation of susceptibility genes 83.
In the future integrated public databases will be needed for the use of complex disease geneticists. Existing databases include http://www.sph.umich.edu/csg/liang/asthma/ and VarySysDB (http://www.h-invitational.jp/varygene/home.htm). A more comprehensive database planned as part of the NIH Genotype-Tissue Expression (GTEx) project (http://nihroadmap.nih.gov/GTEx/), which will house existing as well as GTEx-generated eQTL data. Future databases should include eQTL maps with SNPs, epigenetic marks, trans and cis effects, and effects specific for particular cells, tissues, and environmental stimuli. Ultimately, they will also allow browsing for networks, modules and comparisons with model organisms.
Supplementary Material
ONLINE SUMMARY.
Genome wide association (GWA) studies have identified many new loci, but the association signals have yet to be translated into a proper understanding of which gene or genetic elements are mediating disease susceptibility at particular loci.
The functional effects of DNA polymorphism on multifactorial disease are infrequently mediated through mutations that alter protein function, and variation in gene expression is likely to be a more important mechanism underlying susceptibility to complex disease.
Transcript abundances of genes are directly modified by polymorphism in regulatory elements and transcript abundances may be considered as quantitative traits that can be mapped with considerable power. These have been named expression quantitative trait loci (eQTL).
This review explores the value of systematic identification of eQTL as one means of characterising the function of loci underlying complex disease traits.
The combination of whole-genome genetic association studies and the measurement of global gene expression allows the systematic identification of eQTL.
The resulting comprehensive eQTL maps provide an important source of reference for categorising both cis and trans effects on disease-associated SNPs on gene expression.
In addition to providing information about the biological control of gene expression, such data aid in interpreting the results of GWA studies. The availability of systematically generated eQTL information provides immediate insight into a probable biological base for the disease associations, and can help to identify networks of genes involved in disease pathogenesis.
This review first briefly introduces the principles and current methods of eQTL mapping and describes the basis of eQTL. We then explore the relevance of these results to disease gene identification.
The limits of present eQTL mapping data are discussed, as is the expected impact of new technologies, international efforts to extend results to new samples and tissues and how cell lines might be tested with stimuli relevant to disease.
Acknowledgments
The work was supported by the Wellcome Trust and the EC funded GABRIEL project, the French Ministry of Research and Higher Education and by grants from the National Institutes of Health.
GLOSSARY TERMS
- Genome-wide association study (GWA study)
An examination of common genetic variation across the genome designed to identify associations with traits such as common diseases. Typically, several hundred thousand SNPs are interrogated using microarray or bead chip technologies
- Heritability (H2)
The heritability of an individual trait (H2) is estimated by the ratio of genetic variance to total trait variance, so that 0 indicates no genetic effects on trait variance and 1 indicates that all variance is under genetic control
- Gene ontology
A widely used classification system of gene functions and other gene attributes that uses a standardised vocabulary. The system uses a hierarchical organization of concepts (ontology) with three organizing principles; molecular functions (the tasks done by individual gene products), biological processes (for example, mitosis) and cellular components (examples include the nucleus and the telomere)
- Epigenetic
A mitotically stable change in gene expression that depends not on a change in DNA sequence, but on covalent modifications of DNA or chromatin proteins such as histones
- Major histocompatibility complex (MHC)
A complex locus on chromosome 6p, which comprises numerous genes, including the human leukocyte antigen genes, which are involved in the immune response
- Human leukocyte antigen (HLA)
A glycoprotein, encoded at the MHC locus, found on the surface of antigen-presenting cells that present antigen for recognition by helper T cells
- SAGE (Serial analysis of gene expression)
A method for quantitative and simultaneous analysis of a large number of transcripts
- short sequence tags are isolated
concentrated and cloned; their sequencing reveals a gene-expression pattern that is characteristic of the tissue or cell type from which the tags were isolated
- Additive genetic effects
A mechanism of quantitative inheritance such that the combined effects of genetic alleles at two or more gene loci are equal to the sum of their individual effects
Biographies
Bill Cookson is a Professor of Genomic Medicine and Director of Respiratory Sciences at Imperial College London. He has had a long sending involvement in the genetics of complex disease, particularly asthma.
Miriam Moffatt is a Professor of Genetics at Imperial College, and leads the Respiratory Molecular Genetics group with Bill Cookson. Her interests include complex disease genetics and the application of genomics and gene expression studies to cells and tissues.
Liming Liang is Liming Liang is now a Ph.D. candidate in the Department of Biostatistics at the University of Michigan. His research interests focus on using mathematical modelling, efficient computational tools and statistical inference to tackle a variety of problems posed by gene mapping studies of complex disease. Liming is writing his dissertation with Dr. Gonçalo Abecasis on efficient methods for analysis of genome scale data.
Gonçalo Abecasis is a statistical geneticist. His research group focuses on the development of statistical tools and methods that allow human geneticists to use new high-throughput technologies to understand genetic variation in humans and its contributions to complex disease. Gonçalo completed is doctoral research with Bill Cookson and Lon Cardon at the University of Oxford and is now on the faculty at the University of Michigan.
Mark Lathrop is Director of the Centre National de Genotypage and of the Fondation Jean Dausset-CEPH. His research interests focus on the identification and characterisation of genetic factors implicated in human disease.
Contributor Information
William Cookson, Email: w.cookson@imperial.ac.uk, National Heart and Lung Institute, Imperial College London, SW3 6LY, England.
Liming Liang, Email: lianglim@umich.edu, Center for Statistical Genetics, Dept. of Biostatistics, SPH II, Ann Arbor, MI 48109-2029, USA.
Gonçalo Abecasis, Email: goncalo@umich.edu, Center for Statistical Genetics, Dept. of Biostatistics, SPH II, Ann Arbor, MI 48109-2029, USA.
Miriam Moffatt, Email: m.moffatt@imperial.ac.uk, National Heart and Lung Institute, Imperial College London, SW3 6LY, England.
Mark Lathrop, Email: mark@cng.fr, CEA/Centre National de Genotypage, 91057 Evry, France.
References
- 1.Hugot JP, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 2.Palmer CN, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–6. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 3.Burton PR, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–37. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. This paper dramatically showed the power of eQTL analysis in humans. [DOI] [PubMed] [Google Scholar]
- 5.Morley M, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–7. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brem RB, Yvert G, Clinton R, Kruglyak L. Genetic dissection of transcriptional regulation in budding yeast. Science. 2002;296:752–5. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 7.Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–72. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 8.Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–15. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia Z, Xu S. Mapping quantitative trait loci for expression abundance. Genetics. 2007;176:611–23. doi: 10.1534/genetics.106.065599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlborg O, et al. Methodological aspects of the genetic dissection of gene expression. Bioinformatics. 2005;21:2383–93. doi: 10.1093/bioinformatics/bti241. [DOI] [PubMed] [Google Scholar]
- 11.Kendziorski CM, Chen M, Yuan M, Lan H, Attie AD. Statistical methods for expression quantitative trait loci (eQTL) mapping. Biometrics. 2006;62:19–27. doi: 10.1111/j.1541-0420.2005.00437.x. [DOI] [PubMed] [Google Scholar]
- 12.Schliekelman P. Statistical power of expression quantitative trait loci for mapping of complex trait loci in natural populations. Genetics. 2008;178:2201–16. doi: 10.1534/genetics.107.076687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007 doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 14.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era--concepts and misconceptions. Nat Rev Genet. 2008;9:255–66. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 15.Goring HH, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–16. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 16.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–8. doi: 10.1038/nature06758. This paper illustrates the power of eQTL and network analysis in unravelling complex trait genetics. [DOI] [PubMed] [Google Scholar]
- 17.Schadt EE, et al. Mapping the Genetic Architecture of Gene Expression in Human Liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petretto E, et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monks SA, et al. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75:1094–105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veyrieras JB, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubner N, et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet. 2005;37:243–53. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 22.Yvert G, et al. Trans-acting regulatory variation in Saccharomyces cerevisiae and the role of transcription factors. Nat Genet. 2003;35:57–64. doi: 10.1038/ng1222. [DOI] [PubMed] [Google Scholar]
- 23.Shimada MK, et al. VarySysDB: a human genetic polymorphism database based on all H-InvDB transcripts. Nucleic Acids Res. 2008 doi: 10.1093/nar/gkn798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzales JM, et al. Regulatory hotspots in the malaria parasite genome dictate transcriptional variation. PLoS Biol. 2008;6:e238. doi: 10.1371/journal.pbio.0060238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mileyko Y, Joh RI, Weitz JS. Small-scale copy number variation and large-scale changes in gene expression. Proc Natl Acad Sci U S A. 2008;105:16659–64. doi: 10.1073/pnas.0806239105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. This paper shows the extent and distribution of methylation in the human genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krebs JE. Moving marks: dynamic histone modifications in yeast. Mol Biosyst. 2007;3:590–7. doi: 10.1039/b703923a. [DOI] [PubMed] [Google Scholar]
- 29.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–9. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 30.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 31.Bouzigon E, et al. Effect of 17q21 Variants and Smoking Exposure in Early-Onset Asthma. N Engl J Med. 2008 doi: 10.1056/NEJMoa0806604. [DOI] [PubMed] [Google Scholar]
- 32.Duan S, et al. Genetic architecture of transcript-level variation in humans. Am J Hum Genet. 2008;82:1101–13. doi: 10.1016/j.ajhg.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galanter J, et al. ORMDL3 gene is associated with asthma in three ethnically diverse populations. Am J Respir Crit Care Med. 2008;177:1194–200. doi: 10.1164/rccm.200711-1644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sleiman PM, et al. ORMDL3 variants associated with asthma susceptibility in North Americans of European ancestry. J Allergy Clin Immunol. 2008 doi: 10.1016/j.jaci.2008.06.041. [DOI] [PubMed] [Google Scholar]
- 35.Tavendale R, Macgregor DF, Mukhopadhyay S, Palmer CN. A polymorphism controlling ORMDL3 expression is associated with asthma that is poorly controlled by current medications. J Allergy Clin Immunol. 2008;121:860–3. doi: 10.1016/j.jaci.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 36.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. A substantial meta-analysis of susceptibility loci underlying Crohn’s disease that illustrates the problem of unattributed heritability and the utility of eQTL data in understanding the function of disease-associated SNPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Libioulle C, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabashima K, et al. The prostaglandin receptor EP4 suppresses colitis, mucosal damage and CD4 cell activation in the gut. J Clin Invest. 2002;109:883–93. doi: 10.1172/JCI14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rioux JD, et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet. 2001;29:223–8. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- 40.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horton R, et al. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–99. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 43.Alberts R, et al. Sequence polymorphisms cause many false cis eQTLs. PLoS ONE. 2007;2:e622. doi: 10.1371/journal.pone.0000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beaty JS, West KA, Nepom GT. Functional effects of a natural polymorphism in the transcriptional regulatory sequence of HLA-DQB1. Mol Cell Biol. 1995;15:4771–82. doi: 10.1128/mcb.15.9.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nejentsev S, et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–92. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sieberts SK, Schadt EE. Moving toward a system genetics view of disease. Mamm Genome. 2007;18:389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 48.Goh KI, et al. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–90. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–35. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barnes M, Freudenberg J, Thompson S, Aronow B, Pavlidis P. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 2005;33:5914–23. doi: 10.1093/nar/gki890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedotti P, et al. Can subtle changes in gene expression be consistently detected with different microarray platforms? BMC Genomics. 2008;9:124. doi: 10.1186/1471-2164-9-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Ruissen F, et al. Evaluation of the similarity of gene expression data estimated with SAGE and Affymetrix GeneChips. BMC Genomics. 2005;6:91. doi: 10.1186/1471-2164-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bosotti R, et al. Cross platform microarray analysis for robust identification of differentially expressed genes. BMC Bioinformatics. 2007;8 (Suppl 1):S5. doi: 10.1186/1471-2105-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji Y, et al. RefSeq refinements of UniGene-based gene matching improve the correlation of expression measurements between two microarray platforms. Appl Bioinformatics. 2006;5:89–98. doi: 10.2165/00822942-200605020-00003. [DOI] [PubMed] [Google Scholar]
- 55.Carter SL, Eklund AC, Mecham BH, Kohane IS, Szallasi Z. Redefinition of Affymetrix probe sets by sequence overlap with cDNA microarray probes reduces cross-platform inconsistencies in cancer-associated gene expression measurements. BMC Bioinformatics. 2005;6:107. doi: 10.1186/1471-2105-6-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sohail M, Akhtar S, Southern EM. The folding of large RNAs studied by hybridization to arrays of complementary oligonucleotides. Rna. 1999;5:646–55. doi: 10.1017/s1355838299982195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Southern E, Mir K, Shchepinov M. Molecular interactions on microarrays. Nat Genet. 1999;21:5–9. doi: 10.1038/4429. This review, by the inventor of DNA microarrays, highlights the complexity and unpredictability of the interactions between nucleic acids in solution and target sequences on solid supports. [DOI] [PubMed] [Google Scholar]
- 58.Kapur K, Xing Y, Ouyang Z, Wong WH. Exon arrays provide accurate assessments of gene expression. Genome Biol. 2007;8:R82. doi: 10.1186/gb-2007-8-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okoniewski MJ, Hey Y, Pepper SD, Miller CJ. High correspondence between Affymetrix exon and standard expression arrays. Biotechniques. 2007;42:181–5. doi: 10.2144/000112315. [DOI] [PubMed] [Google Scholar]
- 60.Clark TA, et al. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wold B, Myers RM. Sequence census methods for functional genomics. Nat Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- 62.Watson RM, Griaznova OI, Long CM, Holland MJ. Increased sample capacity for genotyping and expression profiling by kinetic polymerase chain reaction. Anal Biochem. 2004;329:58–67. doi: 10.1016/j.ab.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 63.Weedon MN, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–83. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brem RB, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. Proc Natl Acad Sci U S A. 2005;102:1572–7. doi: 10.1073/pnas.0408709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moffatt M, Cookson W. The genetics of asthma. Maternal effects in atopic disease. Clin Exp Allergy. 1998;28 (Suppl 1):56–61. doi: 10.1046/j.1365-2222.1998.0280s1056.x. [DOI] [PubMed] [Google Scholar]
- 66.Bennett S, Todd J. Human type 1 diabetes and the insulin gene: principles of mapping polygenes. Annu Rev Genet. 1996;30:343–70. doi: 10.1146/annurev.genet.30.1.343. [DOI] [PubMed] [Google Scholar]
- 67.Warram JH, Krolewski AS, Gottlieb MS, Kahn CR. Differences in risk of insulin-dependent diabetes in offspring of diabetic mothers and diabetic fathers. N Engl J Med. 1984;311:149–52. doi: 10.1056/NEJM198407193110304. [DOI] [PubMed] [Google Scholar]
- 68.Koumantaki Y, et al. Family history as a risk factor for rheumatoid arthritis: a case-control study. J Rheumatol. 1997;24:1522–6. [PubMed] [Google Scholar]
- 69.Burden A, et al. Genetics of psoriasis: paternal inheritance and a locus on chromosome 6p [see comments] J Invest Dermatol. 1998;110:958–60. doi: 10.1046/j.1523-1747.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 70.Akolkar PN, et al. Differences in risk of Crohn’s disease in offspring of mothers and fathers with inflammatory bowel disease. Am J Gastroenterol. 1997;92:2241–4. [PubMed] [Google Scholar]
- 71.Vorechovsky I, Webster AD, Plebani A, Hammarstrom L. Genetic linkage of IgA deficiency to the major histocompatibility complex: evidence for allele segregation distortion, parent-of-origin penetrance differences, and the role of anti-IgA antibodies in disease predisposition. Am J Hum Genet. 1999;64:1096–109. doi: 10.1086/302326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grosshans H, Filipowicz W. Molecular biology: the expanding world of small RNAs. Nature. 2008;451:414–6. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- 73.Plagnol V, et al. Extreme clonality in lymphoblastoid cell lines with implications for allele specific expression analyses. PLoS ONE. 2008;3:e2966. doi: 10.1371/journal.pone.0002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 75.Cheung VG, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–5. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 76.Gretarsdottir S, et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat Genet. 2003;35:131–8. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 77.Kohane IS, Kho AT, Butte AJ. Microarrays for an integrative genomics. MIT Press; Cambridge, Massachusetts: 2002. [Google Scholar]
- 78.Idaghdour Y, Storey JD, Jadallah SJ, Gibson G. A genome-wide gene expression signature of environmental geography in leukocytes of Moroccan Amazighs. PLoS Genet. 2008;4:e1000052. doi: 10.1371/journal.pgen.1000052. Although the paper describes a relatively small study, it shows the profound effects of different environments on gene expression in peripheral blood lymphocytes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y, et al. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2006;2:e222. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith EN, Kruglyak L. Gene-environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. doi: 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gibson G. The environmental contribution to gene expression profiles. Nat Rev Genet. 2008;9:575–81. doi: 10.1038/nrg2383. [DOI] [PubMed] [Google Scholar]
- 82.Reis BY, Butte AS, Kohane IS. Extracting knowledge from dynamics in gene expression. J Biomed Inform. 2001;34:15–27. doi: 10.1006/jbin.2001.1005. This paper shows the utility of using time-series measurements of gene expression to identify co-regulated modules of genes. [DOI] [PubMed] [Google Scholar]
- 83.Schadt EE, Lum PY. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Reverse engineering gene networks to identify key drivers of complex disease phenotypes. J Lipid Res. 2006;47:2601–13. doi: 10.1194/jlr.R600026-JLR200. [DOI] [PubMed] [Google Scholar]
- 84.Gudbjartsson DF, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–15. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 85.Hom G, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–9. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 86.Hakonarson H, et al. A novel susceptibility locus for type 1 diabetes on Chr12q13 identified by a genome-wide association study. Diabetes. 2008;57:1143–6. doi: 10.2337/db07-1305. [DOI] [PubMed] [Google Scholar]
- 87.Genome-wide association study of 14, 000 cases of seven common diseases 3, 000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Todd JA, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Plenge RM, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357:1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thein SL, et al. Intergenic variants of HBS1L-MYB are responsible for a major QTL on chromosome 6q23 influencing HbF levels in adults. PNAS. 2007 doi: 10.1073/pnas.0611393104. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Di Bernardo MC, et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat Genet. 2008;40:1204–10. doi: 10.1038/ng.219. [DOI] [PubMed] [Google Scholar]
- 92.Gardner TS, di Bernardo D, Lorenz D, Collins JJ. Inferring genetic networks and identifying compound mode of action via expression profiling. Science. 2003;301:102–5. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- 93.Sontag E, Kiyatkin A, Kholodenko BN. Inferring dynamic architecture of cellular networks using time series of gene expression, protein and metabolite data. Bioinformatics. 2004;20:1877–86. doi: 10.1093/bioinformatics/bth173. [DOI] [PubMed] [Google Scholar]
- 94.Li H, et al. Integrative genetic analysis of transcription modules: towards filling the gap between genetic loci and inherited traits. Hum Mol Genet. 2006;15:481–92. doi: 10.1093/hmg/ddi462. [DOI] [PubMed] [Google Scholar]
- 95.Keurentjes JJ, et al. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Natl Acad Sci U S A. 2007;104:1708–13. doi: 10.1073/pnas.0610429104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gerrits A, Dykstra B, Otten M, Bystrykh L, de Haan G. Combining transcriptional profiling and genetic linkage analysis to uncover gene networks operating in hematopoietic stem cells and their progeny. Immunogenetics. 2008;60:411–22. doi: 10.1007/s00251-008-0305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.de Koning DJ, Carlborg O, Haley CS. The genetic dissection of immune response using gene-expression studies and genome mapping. Vet Immunol Immunopathol. 2005;105:343–52. doi: 10.1016/j.vetimm.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 98.Akey JM, Biswas S, Leek JT, Storey JD. On the design and analysis of gene expression studies in human populations. Nat Genet. 2007;39:807–8. doi: 10.1038/ng0707-807. author reply 808–9. [DOI] [PubMed] [Google Scholar]
- 99.Spielman RS, et al. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226–31. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–91. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hughes TR, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–7. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 102.Alberts R, Terpstra P, Bystrykh LV, de Haan G, Jansen RC. A statistical multiprobe model for analyzing cis and trans genes in genetical genomics experiments with short-oligonucleotide arrays. Genetics. 2005;171:1437–9. doi: 10.1534/genetics.105.045930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 104.Sun Z, Wigle DA, Yang P. Non-overlapping and non-cell-type-specific gene expression signatures predict lung cancer survival. J Clin Oncol. 2008;26:877–83. doi: 10.1200/JCO.2007.13.1516. [DOI] [PubMed] [Google Scholar]
- 105.Walker BA, et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood. 2006;108:1733–43. doi: 10.1182/blood-2006-02-005496. [DOI] [PubMed] [Google Scholar]
- 106.Lastowska M, et al. Identification of candidate genes involved in neuroblastoma progression by combining genomic and expression microarrays with survival data. Oncogene. 2007;26:7432–44. doi: 10.1038/sj.onc.1210552. [DOI] [PubMed] [Google Scholar]
- 107.Huang RS, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci U S A. 2007;104:9758–63. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article 17. doi: 10.2202/1544-6115.1128. This paper lays out a statistical approach to network analyses and provides a set of software tools for their implementation. [DOI] [PubMed] [Google Scholar]
- 109.Horvath S, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103:17402–7. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.