Abstract
We characterized the complete genome of a putative novel Usutu virus (USUV) strain (Usutu-BONN) detected in a dead blackbird from Germany. Genomic analysis revealed several unique amino acid substitutions among the polyprotein gene. Phylogenetic analyses demonstrated that Usutu-BONN constitutes a putative novel African USUV lineage, which was probably recently introduced to central Europe.
Keywords: African Usutu virus, complete genome, novel lineage, phylogeny, viruses, Central Europe
Originally isolated from a Culex neavei mosquito in South Africa in 1959 (1,2), Usutu virus (USUV) was subsequently detected in different mosquito and bird species throughout Sub-Saharan countries (3). USUV has recently been introduced to Europe, where it caused widespread deaths among resident bird populations, established a local transmission cycle, and became a resident pathogen (4–6). USUV is maintained in an enzootic cycle involving mosquitoes as vectors and birds as the main amplifying hosts; humans are considered incidental or dead-end hosts. We have demonstrated that bats could also be infected with USUV and might act as amplifying hosts (7), and there is increasing evidence that USUV is pathogenic for humans, thus becoming a potential public health problem (8,9). On the basis of genetic differences, in comparison with the USUV strains from Africa, the USUV strains from Europe, except those from Spain, form a distinct clade within USUV phylogeny (7). The detection and isolation of USUV from different bird species and mammalophilic mosquitoes during the 2011 epizootic in Germany raised questions regarding the USUV host range. Thus, as a part of the German Arbovirus Surveillance Program (10), we continued the monitoring of the mosquitoes, birds, and bats for the presence of USUV.
The Study
From May to October 2014, ≈23,300 female mosquitoes from different parts of Germany were trapped, morphologically identified, and pooled (up to 250 mosquitoes/pool). During January–November 2014, a total of 8 dead Pipistrellus bats and 32 dead birds (mainly blackbirds) from different regions of the country were subjected to complete necropsy, and samples were collected for virus detection. Total RNA and DNA from homogenized mosquito pools and tissue samples (brain, liver, lung, and heart) from bats and birds were extracted by using an RTP DNA/RNA Virus Mini Kit (STRATEC Biomedical, Birkenfeld, Germany) according the manufacturer’s instructions. Extracted samples were analyzed for the presence of flavivirus RNA by using a modified pan-flavivirus reverse transcription PCR (5). Positive results were found for 5 mosquito pools (C. pipiens biotype pipiens), 5 blackbirds (Turdus merula), and 1 bat (Pipistrellus pipistrellus). Direct sequencing of the pan-flavivirus PCR amplicons showed USUV nucleic acid sequences in each sample. The positive samples were further subjected to PCRs for the amplification of a partial segment of USUV envelope and nonstructural (NS) 5 gene. Sequencing results showed that all samples, except 1 blackbird-derived USUV sequence, were identical (data not shown) and originated from southwest Germany, corresponding to the previously described USUV-endemic area (Figure 1) (5,10,11). The USUV-positive blackbird sample, which exhibited numerous nucleotide and amino acid changes compared with other sequences, had been found outside of the USUV-endemic area, at the beginning of August in Sankt Augustin (50°46′12′′N, 7°11′12′′E), a city located near Bonn.
Figure 1.
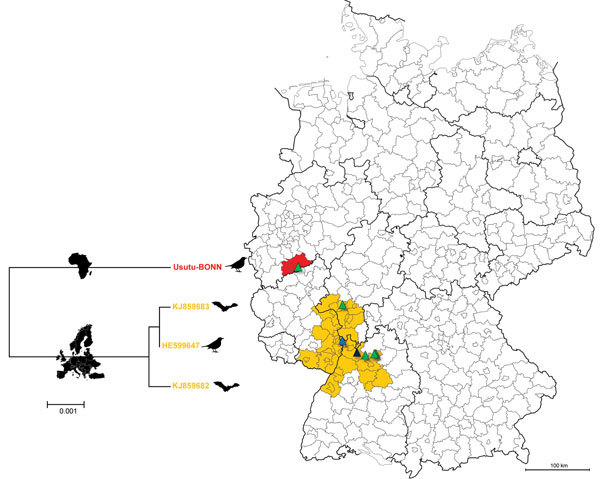
Region of Germany where Usutu virus (USUV) is endemic (orange) and location where the putative novel USUV strain Usutu-BONN was detected (red). Phylogenetic tree illustrates the genetic relationship between the strains circulating in the USUV-endemic region of Germany (belonging to the European USUV clade) and Usutu-BONN (belonging to the African USUV clade) (7), based on complete amino acid sequences of the polyprotein-encoding gene. Triangles indicate locations of the USUV-positive samples according to hosts; blue, mosquitoes; green, blackbirds; black, bat. Scale bar on tree indicates amino acid substitutions per site.
Full-length genome sequence of this putative novel USUV strain, designated Usutu-BONN (GenBank accession no. KM659876), was successfully obtained by using a previously described protocol (7). The genome contained 11,065 nt with a 96-nt 5′ untranslated region and a 664-nt 3′ untranslated region. The single open reading frame encoded a polyprotein of 3,434 aa. Depending on the USUV strain, the nucleotide sequence similarity ranged from 81% to 98%, whereas amino acid sequence conservation ranged from 94.7% to 99.2% of pairwise identity (Table 1). Comparison of the Usutu-BONN complete polyprotein sequence with the other USUV strains showed several identical synonymous and nonsynonymous mutations characteristic for African USUV strains and some unique substitutions (Table 2). Usutu-BONN contained 15 aa substitutions, 7 of which were unique mutations (Table 2). Most were located in the envelope (E) and NS2a proteins (Table 2). We also found 1 putative change in the C/anchC cleavage site of the polyprotein (Table 2); this mutation was observed also in the highly divergent African USUV strain (ArB1803; GenBank accession no. KC754958). We detected 2 more unique amino acid substitutions in the domain II of the E protein. Strikingly, no potential N-glycosylation sites or substitutions were found in well-known features of the protein E fusion peptide or antibody binding sites. Unique mutations (V142 and V189) in the NS2a protein were also observed (Table 2). Alignment of the deduced amino acid sequences of the complete polyprotein (including all previously described strains for which complete polyprotein-encoding sequences are available) by using the MAFFT plugin in Geneious 7.1.5 (Biomatters, Ltd, Auckland, New Zealand) and subsequent phylogenetic reconstruction by using a maximum-likelihood tree (JTT+G model) with 1,000 bootstrap replicates in PhyML (12) and parallel Bayesian Markov Chain Monte Carlo method implemented in MrBayes 3.0 software (13) (data not shown) demonstrated that Usutu-BONN forms a separate lineage (basal position of the African clade) within the USUV phylogeny (Figure 2). Furthermore, the phylogenetic tree showed moderate genetic relatedness of the Usutu-BONN strain with USUV strains circulating in central Europe.
Table 1. Comparison of Usutu-BONN virus with Usutu virus strains from other countries.
| Strain/GenBank accession no. | Country of origin | Host | Year of detection/isolation | Usutu-BONN* |
|
|---|---|---|---|---|---|
| % Identity of nt sequence | % Identity of aa sequence | ||||
| ArD192495/KC754957 | Senegal | Mosquito | 2007 | 97.3 | 99.2 |
| HB81P08/KC754955 | Central African Republic | Human | 1981 | 98.0 | 99.2 |
| MB119/06/KF573410 | Spain | Mosquito | 2006 | 95.7 | 98.5 |
| ArD19848/KC754954 | Senegal | Mosquito | 1974 | 96.5 | 99.0 |
| SAAR-1776/AY453412 | South Africa | Mosquito | 1959 | 96.5 | 98.9 |
| ArD101291/KC754956 | Senegal | Mosquito | 1993 | 97.5 | 99.2 |
| BH65/11–02–03/HE599647 | Germany | Avian | 2011 | 97.2 | 99.2 |
| BAT2USUTU-BNI/KJ859683 | Germany | Bat | 2013 | 97.2 | 99.1 |
| BAT1USUTU-BNI/KJ859682 | Germany | Bat | 2013 | 97.2 | 99.1 |
| Bologna 2009/HM569263 | Italy | Human | 2009 | 97.3 | 99.1 |
| Italia 2009/JF266698 | Italy | Avian | 2009 | 97.3 | 99.1 |
| Vienna 2001/AY453411 | Austria | Avian | 2001 | 97.5 | 99.1 |
| Budapest/EF206350 | Hungary | Avian | 2005 | 97.4 | 99.1 |
| Meise H/JQ219843 | Austria | Avian | 2002 | 97.4 | 99.1 |
| ArB1803/KC754958 | Central African Republic | Mosquito | 1969 | 81.0 | 94.7 |
*Usutu-BONN strain was isolated from a blackbird in Germany in 2014.
Table 2. Comparison of amino acid substitutions of Usutu-BONN strain with those of all available complete Usutu virus polyprotein sequences*.
| Protein | Amino acid substitution Usutu-BONN | Unique substitutions | Total substitutions | Changed putative cleavage sites |
|---|---|---|---|---|
| C | No | No | 0/0 | No |
| anchC | S105†→G; A120→V | No | 0/2 | TKKKR/S†NNGP |
| PrM | N120→Y | N120 | 1/1 | No |
| M | N28→Y | N28 | 1/1 | No |
| E | L231→S; T238→ I/L | L231; T238 | 2/2 | No |
| NS1 | V146→ A/G | No | 0/1 | No |
| NS2A | A91→ V/T; L123→F; V142→A; V189→ A/S | V142; V189 | 2/4 | No |
| NS2B | No | No | 0/0 | No |
| NS3 | F46→ L; V338→A/T | No | 0/2 | No |
| NS4A | No | No | 0/0 | No |
| 2K | No | No | 0/0 | No |
| NS4B | F189→ L | F189 | 1/1 | No |
| NS5 | S274→T/A | No | 0/1 | No |
*USUTU-Bonn strain was isolated from a blackbird in Germany in 2014. †Substitution in cleavage sites. GenBank accession numbers of Usutu virus polyproteins used for sequence alignments are shown in Table 1.
Figure 2.
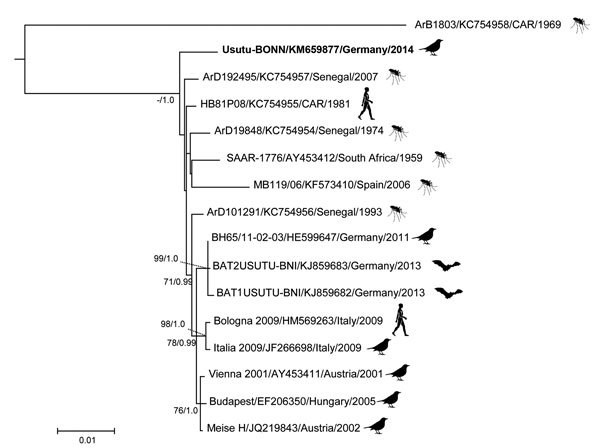
Phylogenetic relationship of the Usutu virus Usutu-BONN strain (from Germany, 2014) and other Usutu viruses, based on complete amino acid sequences of the polyprotein-encoding gene. The phylogenetic tree was constructed by using the maximum-likelihood method PhyML 3.0 (12) with 1,000 pseudoreplicates and, in parallel, Bayesian Markov chain Monte Carlo tree-sampling methods by using MrBayes 3.1.2 (13). The Akaike information criterion was chosen as the model selection framework, and the Johnes-Taylor-Thorton model of sequence evolution with gamma-distributed rate variation among sites (JJT + Γ) was chosen as the best model. Maximum-likelihood bootstrap replicate scores (>70%) and posterior probabilities of the Bayesian analysis (>90%) are shown next to the nodes. Taxon information includes strain designation, GenBank accession number, country of origin, and year of detection/isolation. Boldface indicates the Usutu-BONN strain. Scale bar indicates amino acid substitutions per site.
Conclusions
We detected and genetically characterized a putative novel USUV strain (Usutu-BONN) by determining its complete genome sequence and comparing it with USUV strains for which complete polyprotein-encoding sequences are available. We demonstrated that the Usutu-BONN strain from Germany constitutes a putative novel USUV lineage. The unique synonymous mutations detected in the E and NS2a genes of Usutu-BONN strains may suggest an adaptive evolution. In this strain, 1 putative cleavage site of the viral polyprotein responsible for processing of structural proteins was changed. Given that Usutu-BONN has not led to massive deaths among birds and has not yet been found in other hosts or mosquito vectors, it seems evident that this strain was recently introduced into Germany and evolved in another geographic region, probably Africa (Figure 1, 2). The possibility of an African origin of this virus strain is strengthened by the fact that phylogenetic analysis of complete polyprotein sequence established a separate basal lineage for the Usutu-BONN strain in a sister relationship with the African USUV strains, suggesting that Usutu-BONN has evolved in parallel with strains from Africa sharing a recent common ancestor. This putative novel USUV strain was introduced into Europe probably as other strains, via viremic migratory birds returning from winter migration from Africa to Europe or through ship- or aircraft-borne transportation of USUV-infected mosquitoes from Africa. However, identification of the possible sources (e.g., infected mosquitoes, resident or short-ranging migratory birds) of this new USUV strain will require sequence information from neighboring countries where USUV has been detected. The detection of USUV in a Pipistrellus bat 1 year after the first detection of USUV in bats from the same area in 2013 further strengthens our previous hypothesis that bats may contribute to the epizootic by serving as amplifying/reservoir hosts (7). The unique mutations (V142 and V189) in the NS2a protein of the Usutu-BONN strain are located very close to the identical mutation observed in the previously described bat-derived USUV strains. Although the biological consequences of these mutations are not known, similar mutations in West Nile virus were responsible for inhibition of interferon signaling (14).
Further monitoring studies are necessary to evaluate the pathogenic potential of this newly introduced USUV strain in central Europe for susceptible and receptive avian/mammalian hosts and for humans. This information could be used to predict future epidemics and to implement adequate preventive and control measures.
Acknowledgments
We thank Alexandra Bialonski, Martin Rudolf, Claudia Poggensee, Mathis Petersen, Birgit Hüsing, Corinna Thomè, and Sabine Köhler for excellent technical assistance.
Biography
Dr. Cadar is a postdoctoral researcher at the Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany. His research interests include the arbovirus ecology, evolutionary genetics of viral emergence, and arbovirus discovery.
Footnotes
Suggested citation for this article: Cadar D, Bosch S, Jöst H, Börstler J, Garigliany MM, Becker N, et al. Putative lineage of novel African Usutu virus, central Europe. Emerg Infect Dis. 2015 Sep [date cited]. http://dx.doi.org/10.3201/eid2109.142026
References
- 1.Williams MC, Simpson DI, Haddow AJ, Knight EM. The isolation of West Nile virus from man and of Usutu virus from the bird-biting mosquito Mansonia aurites (Theobald) in the Entebbe area of Uganda. Ann Trop Med Parasitol. 1964;58:367–74 . [DOI] [PubMed] [Google Scholar]
- 2.McIntosh BM. Usutu (SA Ar 1776), nouvel arbovirus du groupe B. International Catalogue of Arboviruses. 1985;3:1059–60. [Google Scholar]
- 3.Nikolay B, Diallo M, Boye CS, Sall AA. Usutu virus in Africa. Vector Borne Zoonotic Dis. 2011;11:1417–23 . 10.1089/vbz.2011.0631 [DOI] [PubMed] [Google Scholar]
- 4.Weissenböck H, Kolodziejek J, Url A, Lussy H, Rebel-Bauder B, Nowotny N. Emergence of Usutu virus, an African mosquitoborne flavivirus of the Japanese encephalitis virus group, central Europe. Emerg Infect Dis. 2002;8:652–6. 10.3201/eid0807.020094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker N, Jöst H, Ziegler U, Eiden M, Höper D, Emmerich P, et al. Epizootic emergence of Usutu virus in wild and captive birds in Germany. PLoS ONE. 2012;7:e32604. http://dx.doi.org/10.1371/ journal.pone.0032604. [DOI] [PMC free article] [PubMed]
- 6.Savini G, Monaco F, Terregino C, Di Gennaro A, Bano L, Pinoni C, et al. Usutu virus in Italy: an emergence or a silent infection? Vet Microbiol. 2011;151:264–74. 10.1016/j.vetmic.2011.03.036 [DOI] [PubMed] [Google Scholar]
- 7.Cadar D, Becker N, Campos Rde M, Börstler J, Jöst H, Schmidt-Chanasit J. Usutu virus in bats, Germany, 2013. Emerg Infect Dis. 2014;20:1771–3. 10.3201/eid2010.140909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pecorari M, Longo G, Gennari W, Grottola A, Sabbatini A, Tagliazucchi S, et al. First human case of Usutu virus neuroinvasive infection, Italy, August–September 2009. Euro Surveill. 2009;14:19446 . [PubMed] [Google Scholar]
- 9.Vilibic-Cavlek T, Kaic B, Barbic L, Pem-Novosel I, Slavic-Vrzic V, Lesnikar V, et al. First evidence of simultaneous occurrence of West Nile virus and Usutu virus neuroinvasive disease in humans in Croatia during the 2013 outbreak. Infection. 2014;42:689–95. 10.1007/s15010-014-0625-1 [DOI] [PubMed] [Google Scholar]
- 10.Becker N, Krüger A, Kuhn C, Plenge-Bönig A, Thomas SM, Schmidt-Chanasit J, et al. Mosquitoes as vectors for exotic pathogens in Germany. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2014;57:531–40. 10.1007/s00103-013-1918-8 [DOI] [PubMed] [Google Scholar]
- 11.Jöst H, Bialonski A, Maus D, Sambri V, Eiden M, Groschup MH, et al. Isolation of Usutu virus in Germany. Am J Trop Med Hyg. 2011;85:551–3. 10.4269/ajtmh.2011.11-0248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21 . 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 13.Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- 14.Liu WJ, Wang XJ, Mokhonov VV, Shi PY, Randall R, Khromykh AA. Inhibition of interferon signaling by the New York 99 strain and Kunjin subtype of West Nile virus involves blockage of STAT1 and STAT2 activation by nonstructural proteins. J Virol. 2005;79:1934–42 . 10.1128/JVI.79.3.1934-1942.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]


