Abstract
Mutations within the tumor suppressor BRCA1 cause the majority of hereditary breast and ovarian cancer cases. The BRCA1 protein is an important regulator of DNA double strand break repair and BRCA1 deficient cells are highly sensitive to ionizing radiation. Furthermore, BRCA1 function may contribute to G2 cell cycle checkpoint enforcement. E3-ubiquitin ligase activity is the only known enzymatic activity of BRCA1, which is mediated by the N-terminal RING finger domain. The C-terminal BRCT repeat domain, which mediates protein-protein interactions, is the only other identified structural domain. By investigating cancer-linked mutations within each domain, we demonstrate that truncation of the BRCT domain greatly impairs the stability and nuclear localization of BRCA1 protein. A missense mutation within the RING domain does not affect these biochemical properties. However, both mutant forms of BRCA1 fail to co-localize in nuclear foci with the known BRCA1-interacting proteins BARD1 and BACH1, which are important for DNA repair. This failure occurs despite the continued ability of the RING mutant protein to interact with BACH1 and the ability of the BRCT mutant to interact with BARD1. Furthermore, neither mutant form of BRCA1 is recruited into DNA-damage-associated foci marked by γH2AX. Therefore, our data suggests that both the RING and BRCT domains of BRCA1 are required for an early step in the function of BRCA1 during DNA repair: recruitment to the sites of DNA damage.
Keywords: BRCA1, mutations, breast cancer
Introduction
BRCA1 is a large, nuclear phospho-protein which regulates the repair of double strand breaks (DSB) in DNA. It contains an N-terminal RING domain, which functions as an E3 ubiquitin ligase when bound to its partner protein BARD1 (1). BRCA1 also contains a C-terminal BRCT repeat domain (2), which functions as phospho-protein binding module (3, 4). BACH1, a helicase involved in DNA repair, is one important interacting partner which binds to the BRCT repeats of BRCA1 (5). Mutations affecting both domains are associated with hereditary cancer (6), suggesting that the function of both domains is necessary for the ability of BRCA1 to suppress tumor formation. However, the functional contribution of each structural domain to the overall ability of BRCA1 to prevent tumorigenesis requires further elucidation.
Multiple factors appear to contribute to nuclear localization of BRCA1. BRCA1 binding to BARD1 appears to increase the nuclear accumulation of BRCA1 (7), presumably by masking the nuclear export sequences of BRCA1 which flank the RING domain (8). BRCA1-BARD1 binding also appears to stabilize BRCA1 protein expression (9). Taken together, these two observations suggest that BARD1 promotion of the nuclear localization of BRCA1 may prevent BRCA1 degradation by cytosolic 26S proteasomes or other proteases. Finally, nuclear localization of BRCA1 appears to be supported by AKT phosphorylation (10).
Nuclear localized BRCA1 organizes into discrete foci both during S phase and following DNA damage (11). Once the DNA damage response is initiated, BRCA1 is recruited to γH2AX foci in a manner dependent on MDC1 (12), where it participates in the regulation of DNA repair. As the repair process concludes, γH2AX is removed from the surrounding region (13).
We were interested in studying whether mutations affecting the RING domain and mutations affecting the BRCT repeat domain have different biochemical consequences for BRCA1 function. We therefore analyzed several characteristics of wild-type, RING mutant (C61G), and BRCT mutant (1853stop) BRCA1 protein expressed in the BRCA1 mutant human breast carcinoma cell line HCC-1937. This cell line carries the 5382insC mutant BRCA1 allele with loss of the wild-type allele and expresses almost no endogenous BRCA1 (14). We found that loss of the BRCT repeat domain decreased the half-life of BRCA1 protein and resulted in mostly cytoplasmic localization. In contrast, the RING domain mutant BRCA1 appeared to have similar protein stability and nuclear localization as wild-type BRCA1. However, mutation of either domain appeared to disrupt co-localization of BRCA1 with its protein binding partners BARD1 and BACH1. Furthermore, both types of mutations prevented the recruitment of BRCA1 protein into γH2AX foci following ionizing radiation.
Materials and Methods
Cell Culture, Chemicals, and Recombinant Human Adenovirus
HCC-1937 cell line is a human breast ductal carcinoma isolated from the primary tumor of a 24 year old caucasian female which carries a 5382insC BRCA1 allele. The wild-type BRCA1 allele is deleted. These cells were maintained at 37°C with 5% CO2 in RPMI medium with 1 mM L-glutamine (Gibco, Carlsbad, CA) supplemented with 10% Fetalclone serum (HyClone, Logan, UT) and 1% insulin-transferrin-selenium-A (Gibco, Carlsbad, CA). Cycloheximide (Calbiochem, San Diego, CA) was dissolved in ethanol and used at a final concentration of 50 μM. Leptomycin B (Calbiochem, San Diego, CA) was dissolved in methanol and used at a final concentration of 10 nM. Radiation treatments were performed with a RS2000 irradiator (Rad Source Technologies, Inc.).
Recombinant human adenoviruses expressing wild type BRCA1 (Ad-BRCA1) 1853stop truncated BRCA1 (Ad-1853), or missense C61G BRCA1 (Ad-C61G) were obtained from Drs. Mel Campbell and Roy Jenson (15, 16). High titer stocks were generated by infection of HEK-293 packaging cells and CsCl banding (17), followed by dialysis into viral storage buffer (10 mM Tris pH 7.4, 10 mM histidine, 75 mM NaCl, 1 mM MgCl2, 0.1 mM EDTA, 0.5% EtOH v/v, 50% glycerol v/v). Viral concentrations were determined by spectrophotometer and the optical titer of virus particles (VP) was translated to plaque forming units (PFU) by the calculation of 100 VP = 1 PFU (18).
Cells were transduced with adenovirus in suspension. Cells were released from their culture vessels with trypsin, recovered in 37°C culture medium (with serum), and pelleted at 1000 rpm. Cells were resuspended in medium (with serum) at a concentration of 2 × 106 cells/mL. Concentrated viral stock was added directly at the multiplicity of infection (MOI) of 100 PFU/cell. Cells were incubated with the virus in suspension at 37°C for 30 minutes and then plated in the appropriate culture dish and medium.
Immunofluorescence Microscopy
Cells were grown on cell culture grade glass cover slips (Fisher Scientific) for experimental treatments. The cells were fixed with 10% neutral buffered formalin for 10 min. and permeabilized for 5 min. in 0.2% Triton X-100/PBS. Cover slips were washed three times in PBS and then blocked in 2% BSA/PBS. Dual BRCA1 staining utilized an N-terminal mouse monoclonal antibody MS13 (1:50; Calbiochem, San Diego, CA) and an exon 11 directed rabbit polyclonal antibody (1:1250; #556443, BD Biosciences/Pharmingen, San Diego, CA) diluted in blocking buffer for 45 min. at room temperature. For co-localization studies, BARD1 (rabbit BL-518, Bethyl Laboratories, Montgomery, TX), BACH1 (rabbit #B 1310, Sigma Aldrich, St. Louis, MO) and γH2AX (rabbit BL-178, Bethyl Laboratories, Montgomery, TX) were all used at 1:800. For γH2AX staining, TBS was substituted for PBS in all buffers and washes.
Following the primary antibody incubation, slips were washed once in PBS/Tween-20 0.05% for 5 min., once in PBS for 5 min., blocked again for 5 min., and then incubated for 30 min. with anti-mouse 594 (1:1000) and anti-rabbit 488 (1:500) (Molecular Probes/Invitrogen, Carlsbad, CA) secondary antibodies in blocking buffer at room temperature. The washes were repeated, nuclei were stained with DAPI, slips were rinsed in H2O, and mounted with anti-fade fluorescence mounting medium (DAKO). Microscopic images were captured at 600× using a Nikon Eclipse 80i fluorescence microscope and deconvolution was performed with Slidebook software (v4.1, Intelligent Imaging Innovations, Inc., Denver, CO). Quantification was performed manually; for each experimental group at least 200 cells were counted. Cells displaying above-background staining for BRCA1 protein were considered to be “positively stained cells”.
Immunoblot Analysis, Immunoprecipitation, and Densitometry
Whole cell lysates were prepared in modified RIPA buffer (50 mM Tris base, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.5% Na-deoxycholate, 0.1% SDS, 50 mM NaF, 5 mM Na3VO4, plus Roche protease inhibitor cocktail tablets). Lysates were incubated for 30 minutes on ice, clarified by centrifugation at 10,000 rpm at 4°C, and protein concentration determined by Bradford assay (BioRad, Hercules, CA). 100 μg of total protein was loaded per lane for Tris-glycine SDS-PAGE on 8% gels. For analysis of proteins smaller than 50 kD, 12% gels were used. Electrophoresis was performed for approximately 250 volt-hours at 125 V in running buffer (25 mM Tris base, 192 mM glycine, 0.1% SDS). Samples were transferred to PVDF membrane (Millipore, Billerica, MA) for 200 volt-hours at 50 V constant at 16°C in 8% transfer buffer (25 mM Tris base, 192 mM glycine, 8% MeOH v/v). Membranes were blocked in 5% dry non-fat milk (Carnation) dissolved in either PBS or TBS (25mM Tris pH 8.0, 135 mM NaCl, 2.5 mM KCl) to correspond with the diluent use for the primary antibody. Primary antibody incubations were performed overnight at 4°C. The following primary antibodies were diluted in 0.5% dry non-fat milk/PBS-Tween-20 0.1%. BRCA1 (mouse SD118 and MS110, Calbiochem, San Diego, CA) at 1:1,000. BARD1 (rabbit BL-518, Bethyl Laboratories, Montgomery, TX) at 1:5,000. BACH1 (rabbit Brip1, Novus Biologicals, Littleton, CO) at 1:5,000. Cyclin D1 (rabbit monoclonal SP4, Lab Vision Corp., Fremont, CA) at 1:2,500. Tubulin (mouse clone KMX-1, Chemicon, Temecula, CA) at 1:10,000. γH2AX (mouse clone JBW301, Upstate cell signaling solutions, Lake Placid, NY) was diluted in 3% BSA (Santa Cruz Biotechnology, Santa Cruz, CA)/TBS-Tween-20 0.1% at 1:2,500. Anti-mouse and anti-rabbit HRP-conjugated secondary antibodies were obtained from Amersham/GE Healthcare (Piscataway, NJ) and diluted 1:10,000 in the same diluent as the primary antibody.
For immunoprecipitation, cells were harvested by a similar protocol as described above although cells were lysed in IP lysis buffer (20 mM Tris pH 8, 120 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% NP-40, 0.25% Na-deoxycholate, 50 mM NaF, 1 mM Na3VO4, plus Roche protease inhibitor cocktail tablets). 1 mg of total protein was diluted in IP buffer (10 mM Tris pH 7.4, 50 mM NaCl, 5 mM EDTA, 0.5% CHAPS, 50 mM NaF, 1 mM Na3VO4), and 1 μg each of MS110 and SD118 BRCA1 antibodies was added with Exacta-Cruz immunoprecipitation matrix (Santa Cruz Biotechnology) according to manufacturer's protocols. Control immunoprecipitation was performed with 2 μg of anti-tubulin. SDS-PAGE/WB was peformed as described above with the use of Exacta-Cruz secondary antibodies following manufacturer's protocols (Santa Cruz Biotechnology).
Densitometry analysis was performed on 600 dpi TIFF scans of films using Quantity One software (Bio-Rad, Hercules, CA). Global background subtraction was used. Adjusted volume values were normalized by the value for tubulin of that sample as a loading control.
Results
Truncation of the BRCT Domain Decreases BRCA1 Protein Stability and Nuclear Accumulation
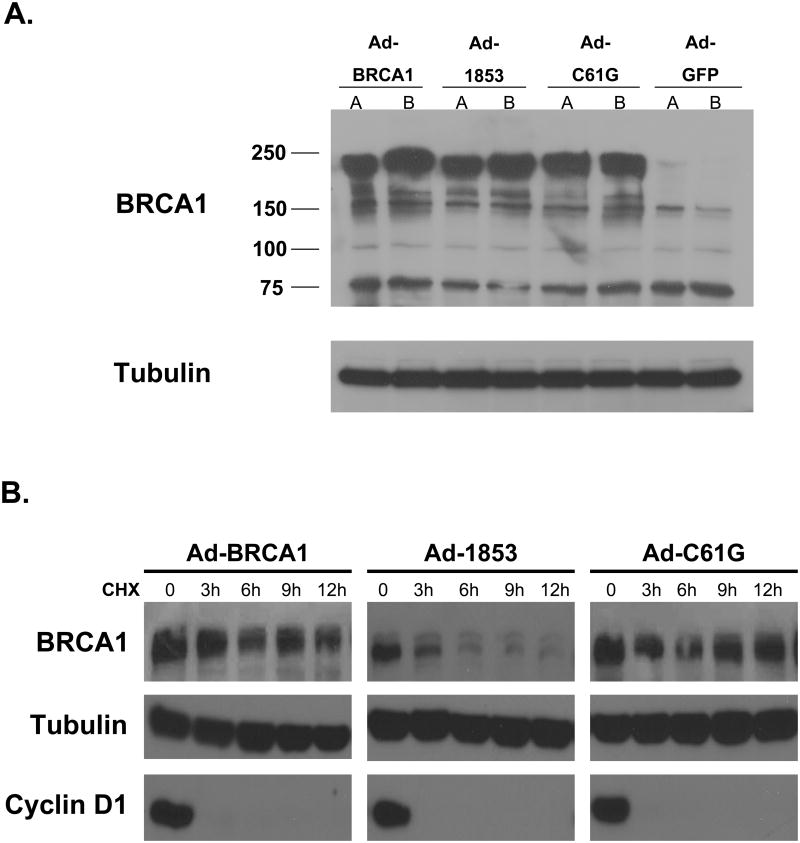
In order to study the biochemical properties of mutant BRCA1 in comparison to wild-type protein we utilized recombinant human adenoviruses expressing BRCT truncated (Ad-1853), RING mutated (Ad-C61G), or wild-type (Ad-BRCA1) protein (16). These adenoviruses contain an R-G-D modification in the fiber knob surface binding protein, which increases the efficiency of infection and downstream expression of the transgene. Transduction of the BRCA1 mutant HCC-1937 human breast carcinoma cell line with these viruses at a multiplicity of infection (MOI) of 100 viral plaque forming units per cell produced robust and approximately equal protein expression of all three forms of BRCA1 48 hours following viral transduction (Figure 1a). Full length BRCA1 is 1863 amino acids in length with a molecular weight of approximately 220 kD. The RING finger mutation C61G does not alter the protein's size. The 1853stop mutation truncates the protein by ten amino acids, but the molecular weight difference is insignificant and cannot be clearly resolved on these gels given the large size of the protein. The full length BRCA1 immunoblot shows several additional bands of lesser molecular weight; some of these may represent alternatively spliced isoforms of BRCA1. Analysis of control cells infected with Ad-GFP showed that almost no endogenous BRCA1 protein can be detected in this cell line (similar to control cells transduced with Ad-LacZ or to mock transduced cells, data not shown). Prior to harvest, quantification of GFP expression in Ad-GFP transduced cell cultures by microscopy indicated that the transduction efficiency is approximately 80% for this cell line under these conditions (data not shown).
Figure 1. Truncation of the BRCT Repeats Decreases the Stability of BRCA1 Protein.
A) Expression of exogenous wild-type and mutant BRCA1 proteins. HCC-1937 cells were transduced with adenoviral vectors expressing wild-type (Ad-BRCA1), BRCT mutant (Ad-1853), or RING mutant (Ad-C61G) BRCA1 protein at an MOI=100 and cultured for 48 hours to achieve steady-state protein expression. Control cells were transduced with Ad-GFP. Immunoblot using SD118 monoclonal anti-BRCA1 antibody is shown. Lanes labeled A and B represent independent viral preparations, and demonstrate consistent protein expression. Full length wild-type BRCA1 and C61G BRCA1 proteins run just below 250 kd (predicted molecular weight approximately 220 kd). 1853stop BRCA1 protein is truncated by only 10 amino acids, and therefore the size difference is not apparent at this resolution. Very little endogenous BRCA1 can be detected in control cells. Tubulin was detected as a control for equal loading. B) HCC-1937 cells were transduced with adenoviral vectors expressing the indicated forms of BRCA1 at MOI=100 and cultured for 48 hours. Cells were then cultured with medium containing 50 μM cycloheximide (CHX) for the indicated times before harvest. Cell lysates were immunoblotted for BRCA1, and densitometry analysis was performed. The t1/2 of wild-type and C61G proteins was similar (9.6 and 9.1 hours respectively), while the t1/2 of 1853stop protein was decreased to 4.7 hours. Cyclin D1 was analyzed to ensure effective inhibition of protein synthesis by CHX treatment. Tubulin serves as a loading control. BRCA1 was not detectable in control Ad-GFP infected cells, similar to Figure 1 (data not shown). The experiment was performed four times with similar results.
We treated similarly transduced cells with the protein synthesis inhibitor cycloheximide (CHX) to determine the protein stability of the mutant BRCA1 proteins in comparison to wild-type. Protein levels of wild-type and C61G BRCA1 appeared stable over the 12 hour time course (Figure 1b). Densitometry calculations indicated the half-life of wild-type BRCA1 is 9.6 hours and the half-life of C61G protein is 9.1 hours. Conversely, levels of 1853stop BRCA1 were noticeably reduced within three hours of CHX treatment, with a calculated half-life of 4.7 hours. These results were consistent among four independent experiments, and indicate that truncation of the C-terminal BRCT repeat domain significantly decreases the stability of the protein.
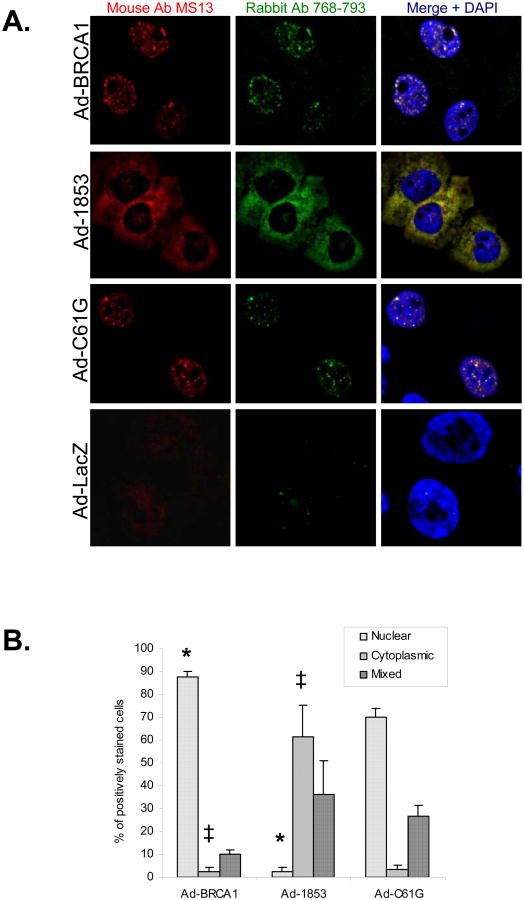
We also determined the subcellular localization of these forms of BRCA1 by immunofluorescence microscopy. A dual anti-BRCA1 antibody protocol was utilized in order to ensure the specificity of the staining protocol (19). C61G BRCA1 was organized into discrete nuclear foci (Figure 2a), similar to the focal nuclear pattern observed with expression of the wild-type protein. However, 1853stop BRCA1 was primarily localized in the cytoplasm (Figure 2a). Similar to immunoblot results (Figure 1a), we found that virtually no endogenous BRCA1 could be detected by this staining protocol (Figure 2a). Quantification was performed on five independent experiments by scoring the subcellular localization of BRCA1 in positively stained cells as either predominantly nuclear, predominantly cytoplasmic, or mixed (Figure 2b). Almost 90% of cells expressing wild-type BRCA1 protein exhibited a predominantly nuclear staining pattern in comparison to only two percent of cells expressing 1853stop protein, a result which was highly significant. Cells expressing C61G BRCA1 demonstrated a staining pattern similar to wild-type, albeit that the proportion of cells with a mixed staining pattern was mildly increased with a concomitant decrease in the percentage of cells with a predominantly nuclear pattern. We attempted to verify these results by sub-cellular fractionation. However we could not obtain consistent and reliable fractionation with the HCC-1937 cell line, and therefore could neither support nor refute our immunofluorescence data by this approach. Overall, these experiments indicate that the BRCT repeat domain is necessary for the efficient nuclear localization of BRCA1 protein, a finding consistent with the previous report of Rodriguez et al. (20).
Figure 2. Truncation of the BRCT Repeats Inhibits Nuclear Accumulation of BRCA1.
A) HCC-1937 cells were transduced at MOI=100 with adenoviral vectors expressing the indicated forms of BRCA1 or with a control virus expressing β-gal (Ad-LacZ). Cells were cultured for 48 hours and then fixed for immunostaining of BRCA1 protein with both a BRCA1 N-terminal directed mouse monoclonal (red, clone MS13) and a BRCA1 exon 11 directed rabbit polyclonal (green) antibody. Overlap of both signals produces a yellow signal in the merged images, which demonstrates specific staining for BRCA1. Nuclei are stained with DAPI. Both wild-type and C61G BRCA1 proteins stain predominantly in nuclear foci, while 1853stop BRCA1 appears to be primarily localized in the cytoplasm. B) Quantification of subcellular localization of wild-type and mutant BRCA1 proteins. 200 positively stained nuclei were counted for each experiment and scored as predominantly nuclear, predominantly cytoplasmic, or mixed. N=5 independent experiments, error bars represent SEM. (*) represents p<0.001 and (‡) represents p=0.005 by student's t-test.
Inhibition of Nuclear Export Stabilizes BRCT-truncated BRCA1 Protein
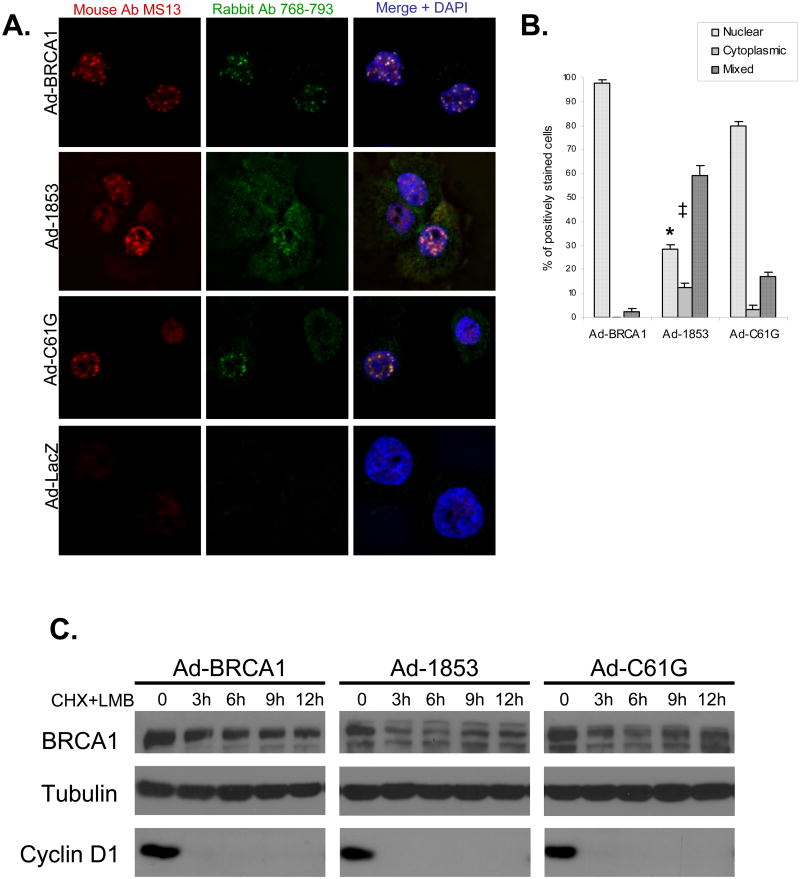
We hypothesized that the lack of nuclear accumulation of 1853stop BRCA1 may be related to its decreased protein stability in comparison to wild-type and C61G BRCA1. In order to test this hypothesis, we treated cells with the nuclear export inhibitor Leptomycin B (LMB). LMB treatment of cells expressing 1853stop increased the number of cells exhibiting a predominantly nuclear pattern on immunofluorescence staining (Figure 3a). This experiment was performed three times in parallel with staining of untreated cells (representing three of the five experiments represented in Figure 2). Statistical comparison of these matched groups indicated that LMB treatment increased the percentage of cells expressing 1853stop protein in a predominantly nuclear pattern from 2.4% (Figure 2b) to 28.4% (Figure 3b, p<0.001). Correspondingly, the fraction of 1853stop expressing cells with a predominantly cytoplasmic staining pattern dropped from 61.3% (Figure 2b) to 12.3% (Figure 3b, p=0.02) following LMB treatment. As well, nuclear foci were observed in a proportion of cells expressing 1853stop BRCA1, although these appeared to be larger and less defined than wild-type foci (Figure 3a), suggesting that the BRCT repeat domain may also be required for the organization of these structures once the protein is localized to the nucleus. LMB treatment of cells expressing wild-type and C61G BRCA1 (Figure 3a) did not appear to alter the focal nuclear staining pattern compared to untreated cells (Figure 2a), and mildly increased the proportion of cells exhibiting a predominantly nuclear staining pattern in both transduction groups. These results suggest that 1853stop BRCA1 can be imported into the nucleus, and that an accelerated rate of nuclear export is in part responsible for its cytoplasmic mislocalization in comparison to wild-type and C61G BRCA1.
Figure 3. Inhibition of Nuclear Export Promotes the Nuclear Accumulation and Improves the Protein Stability of BRCT-truncated BRCA1.
A) HCC-1937 cells were transduced at MOI=100 with adenoviral vectors expressing the indicated forms of BRCA1 or β-gal (Ad-LacZ) as a control. Cells were cultured for 48 hours to allow for protein expression prior to treatment with 10 nM Leptomycin B (LMB) for 10 hours prior to fixation and immunostaining with two anti-BRCA1 antibodies. Overlap of red and green channels in the merge produces a yellow signal which indicates specific staining of BRCA1. Nuclei are stained with DAPI. Focal nuclear staining of truncated 1853stop BRCA1 protein is increased following LMB treatment in comparison to untreated cells (Figure 2). The experiment was performed independently three times concurrently with three of the five experiments represented in Figure 2. B) Quantification of subcellular localization of wild-type and mutant BRCA1 proteins following LMB treatment. 200 positively stained nuclei were counted for each experiment, N=3, error bars represent SEM. In comparison to untreated cells analyzed at the same time, LMB treatment significantly increased (*, p<0.001) the amount of nuclear and decreased the amount of cytoplasmic (‡, p=0.02) 1853stop BRCA1 protein. Concurrent untreated samples are represented within the composite of Figure 2b. Comparison performed by student's t-test. C) HCC-1937 cells were transduced with adenoviral vectors and cultured for 48 hours. Cells were then pretreated with LMB at 10 nM for 1 hour, and next treated with a combination of 10 nM LMB and 50 μM cycloheximide (CHX) for the indicated time course. Cell lysates were immunoblotted for BRCA1, and densitometry analysis was performed. The t1/2 of 1853stop BRCA1 protein increased to 6.7 hours following LMB treatment in comparison to untreated cells (4.7 hours, Figure 1B). The t1/2 of wild-type and C61G BRCA1 proteins (9.4 and 8.9 hours respectively) following LMB treatment showed minimal variation in comparison to untreated cells (Figure 1B). Immunoblot for tubulin indicates equal loading and cyclin D1 was analyzed to ensure the effectiveness of CHX treatment. BRCA1 protein was not detectable in Ad-GFP transduced control cells (data not shown). The experiment was repeated independently three times with similar results.
Therefore, we tested whether the increased nuclear localization of 1853stop BRCA1 in cells treated with LMB affected the apparent stability of the protein. Cells transduced with the BRCA1 expression vectors were pretreated with LMB for one hour, and then subjected to a CHX time course with samples taken at 3, 6, 9, and 12 hours. LMB was maintained in the medium during the entire CHX treatment course. Although protein levels of 1853stop did initially decline between the zero time point and 3 hours of CHX treatment, the level of protein remained stable for the rest of the treatment course (Figure 3c). Densitometry analysis indicated that the half-life of 1853stop protein across the time course was 6.7 hours, increased from 4.7 hours in untreated cells (Figure 1b). Levels of both wild-type and C61G BRCA1 proteins appeared stable during the LMB/CHX treatment course (Figure 3c), and had half-lives similar to those determined in parallel transduction groups treated with CHX only (Figure 1b). In total, these results demonstrate that wild-type BRCA1 is efficiently translocated into the nucleus where the protein is relatively stable. Mutation of the RING domain (C61G) does not appear to effect the nuclear accumulation of stable BRCA1 protein, but loss of the BRCT domain by truncation severely compromises the nuclear retention of the protein and results in rapid degradation.
BRCT Domain and RING Domain Mutant BRCA1 Fail to Co-localize with BARD1 and BACH1
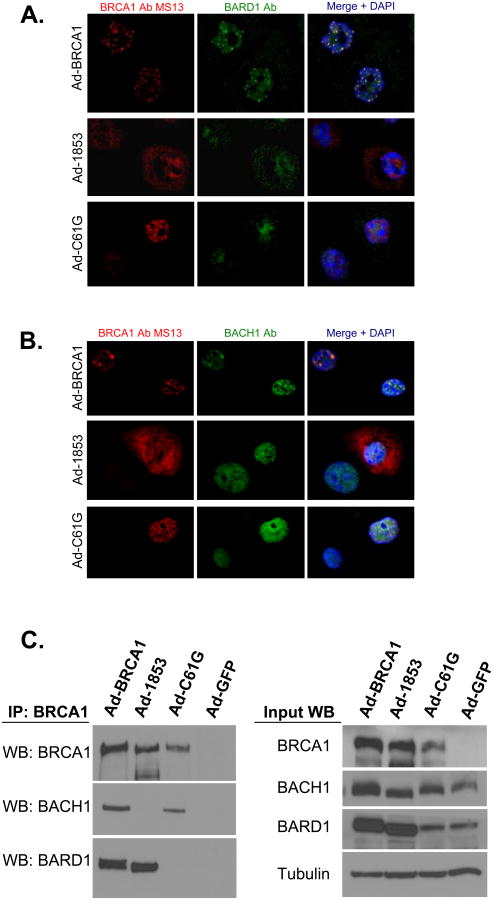
We next investigated whether either mutant form of BRCA1 demonstrated nuclear co-localization with the BRCA1-interacting protein BARD1 with dual immunofluorescence microscopy. Staining of cells expressing wild-type protein showed clear co-localization of BRCA1 and BARD1 in nuclear foci (Figure 4a), a finding expected based on previously published work (7, 21). Staining for 1853stop BRCA1 did not appear to overlap with BARD1 staining, even in cells with higher amounts of nuclear localized 1853stop protein. Furthermore, staining for BRCA1 and BARD1 in cells transduced with Ad-C61G demonstrated that these two proteins do not co-localize in vivo despite the fact that C61G BRCA1 was organized into discrete nuclear foci. BARD1 staining in cells expressing either mutant form of BRCA1 was diffusely nuclear in comparison to the focal staining observed in cells expressing wild-type BRCA1, suggesting that recruitment of BARD1 into these nuclear structures may be dependent on BRCA1.
Figure 4. BRCT Domain and RING Domain Mutant BRCA1 Proteins Fail to Co-localize With Both BARD1 and BACH1.
HCC-1937 cells were transduced with adenoviral vectors expressing the specified forms of BRCA1 at MOI=100 and cultured for 48 hours prior to fixation. A) Cells were stained with monoclonal antibody MS13 against BRCA1 (red) and a polyclonal antibody against BARD1 (green). Yellow signal in the merged image indicates co-localization of the two proteins. B) Cells were stained with monoclonal antibody MS13 against BRCA1 (red) and a polyclonal antibody against BACH1 (green). Yellow signal in the merged image indicates co-localization of the two proteins. Nuclei are stained with DAPI in both A and B. Neither C61G (RING mutant) or 1853stop (BRCT mutant) BRCA1 proteins appear to co-localize with either BARD1 or BACH1. Both BARD1 and BACH1 co-localize with wild-type BRCA1. Experiments in A and C were performed three times independently, with similar results. C) Co-immunoprecipitation of BRCA1, BARD1, and BACH1. Lysates were prepared from cells expressing the indicated forms of BRCA1 or control GFP protein and immunoprecipitation of BRCA1 was performed with a combination of monoclonal antibodies (clones MS110 and SD118). Wild-type BRCA1 co-precipitates both BARD1 and BACH1. 1853stop BRCA1 is only capable of interacting with BARD1, while C61G BRCA1 is only capable of interacting with BACH1.
We also investigated whether the mutant forms of BRCA1 were capable of co-localizing with BACH1. The interaction between BRCA1 and BACH1 is dependent on BACH1 phosphorylation and is mediated by the BRCT repeats of BRCA1 (5). Therefore 1853stop BRCA1 should not co-localize with BACH1, an observation which we confirmed in our system (Figure 4b). BACH1 and BRCA1 proteins did form distinct co-localized nuclear foci in cells expressing wild-type BRCA1. However, we observed that C61G BRCA1 and BACH1 did not appear to co-localize within the nucleus. Despite the focal nuclear staining of C61G BRCA1, the staining for BACH1 had no clear overlap with the BRCA1 signal. We noted that BACH1 staining was diffusely nuclear in cells expressing either mutant BRCA1 protein, although some faint focal staining was observed, suggesting that organization of BACH1 into distinct nuclear structures may be only partially dependent on BRCA1.
We next sought to determine if a failure of direct interaction between mutant BRCA1 proteins and either BARD1 or BACH1 might explain the failure to co-localize by immunofluorescence. The 1853stop mutation has previously been shown to disrupt the direct interaction between BRCA1 and BACH1 (22). In regard to BARD1-BRCA1 binding, initial work demonstrated that C61G BRCA1 and BARD1 did not interact via co-immunoprecipitation and yeast 2 hybrid approaches (1). However, a subsequent publication showed that the C61G mutation does not disrupt binding to BARD1 using RING domain fragments to study the detailed structure of the interaction (23). We utilized our system to determine whether wild-type or mutant BRCA1 proteins expressed in vivo were capable of co-immunoprecipitating both BARD1 and BACH1 from whole cell lysates (Figure 4c). Wild-type BRCA1 effectively co-immunoprecipitated both BARD1 and BACH1 proteins. 1853stop BRCA1 was capable of pulling down BARD1, but failed to interact with BACH1 as expected. C61G BRCA1 co-immunoprecipitated BACH1 protein efficiently, but was incapable of pulling down BARD1. Therefore, our results clearly demonstrate that the complete 1853stop mutant BRCA1 protein does not interact with BACH1 and the complete C61G mutant protein does not interact with BARD1 when expressed in vivo. Conversely, interactions are maintained between 1853stop BRCA1 and BARD1 and between C61G BRCA1 and BACH1.
The immunofluorescence studies presented in Figures 4a and 4b were performed three times independently. At least 200 cells were analyzed for each condition in each experiment; no instances of multiple, distinct co-localized foci were observed for either mutant form of BRCA1 with either BARD1 or BACH1. Despite this observation, each mutation only disrupts the biochemical interaction of BRCA1 with the protein binding partner of the corresponding domain (RING mutant with BARD1; BRCT mutant with BACH1). Therefore our results suggest that additional factors are required for the efficient recruitment and maintenance of BRCA1 into nuclear foci with BARD1 and BACH1 beyond direct interaction of these proteins. Mutations affecting either the RING or BRCT domains appear to disrupt BRCA1 co-localization with both of these important protein-interaction partners. Furthermore, this failure occurs despite the similarity of nuclear staining patterns observed between C61G mutant and wild-type BRCA1.
BRCT Domain and RING Domain BRCA1 Proteins are not Effectively Recruited to Sites of DNA Damage
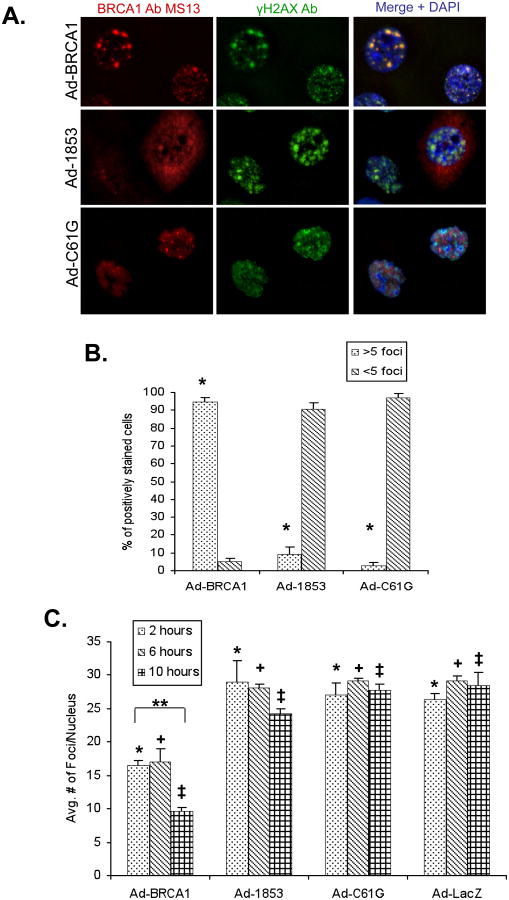
Early in the DNA damage response, BRCA1 is recruited to sites of damage which are marked by the phosphorylation of the variant Histone 2AX (γH2AX) (12). In these damage foci, BRCA1 appears to regulate DNA repair in a manner that is dependent upon the interaction of BRCA1 with BARD1 (24) and BACH1 (5). Due to the abnormal subcellular localization of C61G and 1853stop BRCA1 mutants to interact with either BARD1 or BACH1, we speculated that the recruitment of these mutant BRCA1 proteins to sites of DNA damage might also be impaired. To test this idea, we treated cells expressing either wild-type or mutant BRCA1 with 2 Gy of ionizing radiation (IR), cultured these cells for an additional two hours, and then immunostained the cells for BRCA1 and γH2AX. Numerous foci positive for both proteins were observed in cells expressing wild-type BRCA1 (Figure 5a). Quantification of three independent experiments demonstrated that 95% of these cells had greater than five nuclear foci positive for both BRCA1 and γH2AX (Figure 5b). On the other hand, less than 10% of cells expressing either 1853stop or C61G BRCA1 protein had greater than five nuclear foci staining positively for both the BRCA1 protein and γH2AX. Furthermore, we continued to observe focal nuclear BRCA1 staining in cells expressing C61G protein after ionizing radiation, but these foci only rarely showed overlap with γH2AX staining (Figure 5a). These results indicate that both the RING domain and BRCT domain mutant forms of BRCA1 fail to localize at sites of DNA damage.
Figure 5. BRCT Domain and RING Domain Mutant BRCA1 Proteins Are Not Effectively Recruited to γH2AX Foci Following Ionizing Radiation.
HCC-1937 cells were transduced at MOI=100 with adenoviral vectors expressing the indicated proteins and cultured for 48 hours prior to treatment. A) Cells expressing the indicated forms of BRCA1 protein were exposed to 2 Gy of ionizing radiation (IR) and cultured for an additional 2 hours at 37°C prior to fixation. Cells were stained with a monoclonal antibody against BRCA1 (red) and a polyclonal antibody against γH2AX (green), which is localized to sites of DNA damage. Wild-type BRCA1 is recruited to γH2AX foci, but both mutant forms of BRCA1 fail to co-localize with γH2AX. The experiment was performed three times. B) Quantification of co-localized BRCA1 and γH2AX foci at 2 hours following 2 Gy IR. 200 cells stained positively for BRCA1 were scored for the number of nuclear foci demonstrating co-localization of BRCA1 and γH2AX proteins. N=3, error bars represent SEM. Wild-type BRCA1 expressing cells consistently showed greater than 5 foci per nucleus, while cells expressing either 1853stop or C61G BRCA1 demonstrated less than 10% of nuclei with greater than 5 foci (*, p<0.001 for both WT vs. 1853stop and WT vs. C61G, student's t-test). C) Quantification of the average number of γH2AX foci per nucleus at 2, 6, and 10 hours post 2 Gy IR. The number of foci per nuclei was counted in 10 representative high powered fields for each group within each experiment (approximately 50 nuclei per group). N=3, error bars represent SEM. The number of foci/nucleus was significantly lower in BRCA1 groups versus 1853stop, C61G, and LacZ at 2 (*), 6 (+), and 10 (‡) hours (p≤0.02 for all comparisons, student's t-test). The decreased number of foci/nucleus in BRCA1 groups between 2 and 10 hours (**) was also significant (p=0.001, student's t-test). The 2 h vs. 10 h comparison in 1853stop groups was not significant.
We also observed that the number of γH2AX foci per cell two hours following 2 Gy IR was less in cells expressing wild-type BRCA1 versus cells expressing either mutant protein or control LacZ. To further characterize this observation, we analyzed additional time points at six and ten hours following 2 Gy IR. We performed the experiment three times, and the number of γH2AX foci per nucleus was counted in at least 50 nuclei for each treatment group (in ten representative high power fields). The average number of γH2AX foci was significantly lower at all time points analyzed in cells expressing wild-type BRCA1 in comparison to cells expressing either mutant form of the protein or control LacZ (Figure 5c). Importantly, cells expressing wild type BRCA1 also demonstrated a significant decrease in the average number of γH2AX foci per nucleus between the two hour and ten hour time points, suggesting that resolution of DNA damage foci was occurring. Neither 1853stop nor C61G BRCA1 expressing cells demonstrated this result.
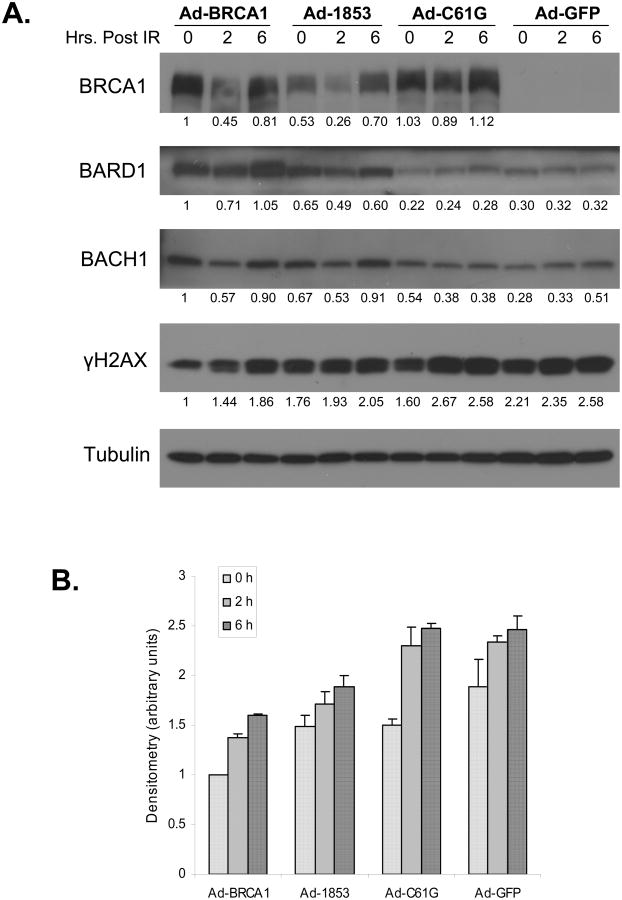
We further investigated this observation by immunoblot analysis of similarly transduced cells at two and six hours post IR in comparison to cells which had not been irradiated. Untreated cells (represented by the zero time point) reconstituted with wild-type BRCA1 had less total γH2AX protein than cells expressing either mutant form of BRCA1 or control GFP (Figure 6a, lanes 1, 4, 7, 10, and Figure 6b). The data presented in Figures 5 and 6 is consistent with the DNA repair defect of the HCC-1937 cell line (14); as these cells traverse S phase of the cell cycle they accumulate numerous double strand breaks which are not adequately repaired, a phenotype which is rescued by wild-type BRCA1. Two hours following 2 Gy IR, all transduction groups demonstrate an increase from baseline in the total level of γH2AX protein (Figure 6a, lanes 2, 5, 8, 11). Similar to our immunofluorescence data (Figure 5a,c), cells expressing wild-type BRCA1 had the lowest level of γH2AX protein. 1853stop expressing cells had a modestly higher amount of γH2AX protein, and both C61G and control GFP groups showed even greater amounts. Interestingly, total BRCA1 protein levels showed a biphasic response following IR, decreasing at two hours post treatment and then increasing back toward or even greater than baseline levels by six hours. BACH1 levels were slightly higher in wild-type and 1853stop expressing groups but this was not a consistent finding over three independent experiments. On the other hand, BARD1 protein levels were consistently increased by expression of either wild-type or 1853stop BRCA1, and BARD1 levels varied similarly following IR in parallel with BRCA1. The protein-protein interaction between either wild-type or 1853stop BRCA1 and BARD1 (Figure 4c) may explain this observation, as previous work has shown that the BRCA1-BARD1 interaction stabilizes expression of both proteins (9).
Figure 6. Total γH2AX Protein Levels Before and After DNA Damage are Lowest in Cells Expressing Wild-type BRCA1.
A) HCC-1937 cells were transduced with adenoviral vectors expressing the specified forms of BRCA1 or GFP control at MOI=100. Cells were cultured for 48 hours prior to treatment with 2 Gy of ionizing radiation (IR). Cells were returned to culture for the indicated times prior to harvest for immunoblot analysis. The zero time point represents untreated cells which were harvested at the time of IR treatment. Cell lysates were immunoblotted with antibodies against the specified proteins. Tubulin was analyzed as a control for loading. Densitometry was performed for all proteins, normalized to tubulin, and expressed as a ratio of the value for lane 1. Levels of γH2AX protein were lowest at all time points in cells expressing wild-type BRCA1 in comparison to cells expressing 1853stop, C61G, or GFP control. B) Normalized densitometry values for γH2AX protein expression are presented. N=3 biologically independent experiments. Error bars represent SEM.
Discussion
The ability of BRCA1 to regulate DNA repair is likely a critical aspect of its tumor suppressor function. Mutations within both structural domains of BRCA1 predispose individuals to hereditary cancer syndromes (6), which indicates that the function of both domains is important for the suppression of tumorigenesis. In this work, we show that mutations which affect either the RING domain or the BRCT repeats both disrupt a critical step in the response to DNA damage: recruitment of BRCA1 to γH2AX positive foci at sites of damage. We also demonstrate that mutation in either the RING domain or the BRCT repeats decreases the co-localization of BRCA1 with two important protein-binding partners, BARD1 and BACH1.
Interestingly, the phenotype of RING mutant (C61G) BRCA1 protein is quite similar to wild-type protein in regard to nuclear localization and protein stability despite the lack of clear co-localization between BRCA1 and BARD1. This observation appears to conflict with a previous report that suggests BARD1 is responsible for the nuclear retention and stabilization of BRCA1 (7). In our model, C61G mutant BRCA1 cannot interact directly with BARD1, nor does it demonstrate nuclear co-localization with BARD1 on immunofluorescence staining. Nevertheless, the C61G protein itself does translocate to the nucleus, aggregate into nuclear foci, and it appears to have a similar half-life as the wild-type protein. Therefore, our results suggest that the interaction of BRCA1 with BARD1 is not absolutely necessary for nuclear retention or regulation of protein stability. However, the failure of C61G BRCA1 to interact with BARD1 may underlie the lack of C61G recruitment to sites of DNA damage following ionizing radiation and the higher levels of γH2AX observed in this transduction group.
The failure of C61G BRCA1 to co-localize with BACH1 also has interesting implications for the in vivo regulation of BRCA1 sub-nuclear targeting. The phospho-protein binding pocket created by the C-terminal BRCT repeats of BRCA1 mediates the direct protein-protein contacts with BACH1, and the RING mutation does not disrupt the direct interaction (Figure 4c). The fact that neither mutant BRCA1 protein localizes with either BACH1 or BARD1 suggests that additional regulatory mechanisms, which require both BRCA1 structural domains, likely target BRCA1 into nuclear structures with its functional binding partners BARD1 and BACH1.
Finally, our results indicate that both the RING domain and the BRCT repeats are required for the targeting of BRCA1 to DNA damage sites following ionizing radiation. This finding reinforces the concept of additional regulatory mechanisms of BRCA1 sub-nuclear localization, consistent with a previously published report (25). Total levels of γH2AX protein are increased in the HCC-1937 cell line secondary to the BRCA1-null DNA repair deficiency (14). Wild-type BRCA1 expression decreases total γH2AX levels at baseline to a much greater degree than either mutant protein in comparison to controls. γH2AX levels are also lower in wild-type expressing cells at both time points following ionizing radiation in comparison to either mutant group or control, suggesting that DNA repair efficiency is improved in the wild-type group.
Our observation of a focal pattern of BRCA1 staining in the absence of ionizing radiation is consistent with the results shown by Au (25) but expands on this study by showing differences between the effects of BRCA1 mutants and wildtype BRCA1 on γH2AX. This study differs from Rodriguez (20) by showing that both the RING domain and the BRCT domain appear to be required for the targeting of BRCA1 to nuclear damage sites and by indicating that neither mutant protein localizes with BARD1 or BACH1 suggesting that additional regulatory mechanisms are responsible for proper BRCA1 function.
Importantly, this work demonstrates that despite significant differences in the biological characteristics of 1853stop BRCA1 compared to C61G BRCA1, both mutant forms are not recruited to sites of DNA damage and both fail to co-localize with important functional binding partners. Further work addressing the mechanisms of how mutations in either the RING or the BRCT domain severely disrupt the nuclear trafficking of BRCA1 will increase our understanding of tumorigenesis associated with BRCA1 mutation.
Acknowledgments
Grant Support: This work was funded by a grant from the Department of Defense Breast Cancer Research Program W81XWH-06-1-0335 to ACN.
Human recombinant adenoviruses encoding wild-type, 1853stop, and C61G BRCA1 were provided by Dr. Mel Campbell and Dr. Roy Jensen. Ad-GFP and Ad-LacZ were provided by Dr. Jerome Schaack. The authors wish to thank Drs. David Orlicky and James McManaman for their assistance with immunofluorescence microscopy.
References
- 1.Wu LC, Wang ZW, Tsan JT, Spillman MA, Phung A, Xu XL, Yang MC, Hwang LY, Bowcock AM, Baer R. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV, Altschul SF, Bork P. BRCA1 protein products … Functional motifs. Nat Genet. 1996;13:266–268. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 3.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT Repeats as Phosphopeptide-Binding Modules Involved in Protein Targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 4.Yu X, Chini CCS, He M, Mer G, Chen J. The BRCT Domain is a Phospho-Protein Binding Domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 5.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 6.Friedman LS, Ostermeyer EA, Szabo C, Dowd P, Lynch ED, Rowell SE, King MC. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nature Genetics. 1994;8:399–404. doi: 10.1038/ng1294-399. [DOI] [PubMed] [Google Scholar]
- 7.Fabbro M, Rodriguez JA, Baer R, Henderson BR. BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J Biol Chem. 2002;277:21315–21324. doi: 10.1074/jbc.M200769200. [DOI] [PubMed] [Google Scholar]
- 8.Thompson ME, Robinson-Benion CL, Holt JT. An amino-terminal motif functions as a second nuclear export sequence in BRCA1. J Biol Chem. 2005;280:21854–21857. doi: 10.1074/jbc.M502676200. [DOI] [PubMed] [Google Scholar]
- 9.Choudhury AD, Xu H, Baer R. Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression. J Biol Chem. 2004;279:33909–33918. doi: 10.1074/jbc.M403646200. [DOI] [PubMed] [Google Scholar]
- 10.Hinton CV, Fitzgerald LD, Thompson ME. Phosphatidylinositol 3-kinase/Akt signaling enhances nuclear localization and transcriptional activity of BRCA1. Exp Cell Res. 2007;313:1735–1744. doi: 10.1016/j.yexcr.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scully R, Chen J, Ochs R, Keegan K, Hoekstra M, Feunteun J, Livingston DM. Dynamic Changes of BRCA1 Subnuclear Location and Phosphorylation State Are Inititated by DNA Damage. Cell. 1997;90:425–435. doi: 10.1016/s0092-8674(00)80503-6. [DOI] [PubMed] [Google Scholar]
- 12.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 13.Foster ER, Downs JA. Histone H2A phosphorylation in DNA double-strand break repair. Febs J. 2005;272:3231–3240. doi: 10.1111/j.1742-4658.2005.04741.x. [DOI] [PubMed] [Google Scholar]
- 14.Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 15.Campbell M, Aprelikova ON, Meet Rvd, Woltjer RL, Yee CJ, Liu ET, Jensen RA. Construction and characterization of recombinant adenoviruses expressing human BRCA1 or murine BRCA1 genes. Cancer Gene Therapy. 2001;8:231–239. doi: 10.1038/sj.cgt.7700291. [DOI] [PubMed] [Google Scholar]
- 16.Campbell M, Qu S, Wells S, Sugandha H, Jensen RA. An adenoviral vector containing an arg-gly-asp (RGD) motif in the fiber knob enhances protein product levels from transgenes refractory to expression. Cancer Gene Therapy. 2003;10:559–570. doi: 10.1038/sj.cgt.7700599. [DOI] [PubMed] [Google Scholar]
- 17.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein BA. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orlicky DJ, Schaack J. Adenovirus transduction of 3T3-L1 cells. J Lipid Res. 2001;42:460–466. [PubMed] [Google Scholar]
- 19.Scully R, Ganesan S, Brown M, De Caprio JA, Cannistra SA, Feunteun J, Schnitt S, Livingston DM. Location of BRCA1 in human breast and ovarian cancer cells. Science. 1996;272:123–126. doi: 10.1126/science.272.5258.123. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez JA, Au WW, Henderson BR. Cytoplasmic mislocalization of BRCA1 caused by cancer-associated mutations in the BRCT domain. Exp Cell Res. 2004;293:14–21. doi: 10.1016/j.yexcr.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 21.Jin Y, Xu XL, Yang MC, Wei F, Ayi TC, Bowcock AM, Baer R. Cell cycle-dependent colocalization of BARD1 and BRCA1 proteins in discrete nuclear domains. Proc Natl Acad Sci U S A. 1997;94:12075–12080. doi: 10.1073/pnas.94.22.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol. 2004;11:512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 23.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 24.Westermark UK, Reyngold M, Olshen AB, Baer R, Jasin M, Moynahan ME. BARD1 participates with BRCA1 in homology-directed repair of chromosome breaks. Mol Cell Biol. 2003;23:7926–7936. doi: 10.1128/MCB.23.21.7926-7936.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Au WW, Henderson BR. The BRCA1 RING and BRCT domains cooperate in targeting BRCA1 to ionizing radiation-induced nuclear foci. J Biol Chem. 2005;280:6993–7001. doi: 10.1074/jbc.M408879200. [DOI] [PubMed] [Google Scholar]