Abstract
The number of new chemicals that are being synthesized each year has been steadily increasing. While chemicals are of immense benefit to mankind, many of them have a significant negative impact, primarily owing to their inherent chemistry and toxicity, on the environment as well as human health. In addition to chemical exposures, human exposures to numerous non-chemical toxic agents take place in the environment and workplace. Given that human exposure to toxic agents is often unavoidable and many of these agents are found to have detrimental human health effects, it is important to develop strategies to prevent the adverse health effects associated with toxic exposures. Early detection of adverse health effects as well as a clear understanding of the mechanisms, especially at the molecular level, underlying these effects are key elements in preventing the adverse health effects associated with human exposure to toxic agents. Recent developments in genomics, especially transcriptomics, have prompted investigations into this important area of toxicology. Previous studies conducted in our laboratory and elsewhere have demonstrated the potential application of blood gene expression profiling as a sensitive, mechanistically relevant and practical surrogate approach for the early detection of adverse health effects associated with exposure to toxic agents. The advantages of blood gene expression profiling as a surrogate approach to detect early target organ toxicity and the molecular mechanisms underlying the toxicity are illustrated and discussed using recent studies on hepatotoxicity and pulmonary toxicity. Furthermore, the important challenges this emerging field in toxicology faces are presented in this review article.
Keywords: blood, transcriptomics, liver, hepatotoxicity, lungs, pulmonary toxicity
Introduction
Human exposure to toxic agents and the potential adverse effects these agents have on human health are of major concern among healthcare providers, industrial hygienists, and regulatory and public health agencies. While the vast majority of toxic agents commonly encountered in the environment and work-places are chemicals, exposure to toxic agents of a non-chemical origin also contributes significantly to the morbidity and mortality of exposed individuals. Toxic agents, either from the environment or the workplace, fall under various categories including, but not limited to, metals, organic solvents, pesticides, aromatic hydrocarbons, acids, bases and naturally occurring substances, etc. (Thorne, 2007). Even the pharmaceutical industry, whose main objective is to develop, manufacture and sell potentially life-saving drugs, is a source of occupational exposure to toxic agents to tens of thousands of workers.
The potential adverse health effects resulting from human exposure to toxic agents can range from minor discomfort to life-threatening diseases. Ever since the report by Sir Percival Pott in 1775 documenting an association between occupational exposure to soot particles and the incidence of scrotal cancer among chimney sweeps, there have been numerous reports confirming the role played by toxic agents originating in the environment and occupational settings in the diseases and death reported among human beings. Virtually every organ and organ system in the human body, namely skin, nervous system, eye, respiratory system, cardiovascular system, liver, kidney, reproductive system, endocrine system, etc. have been identified as a target for toxicity and diseases resulting from exposure to various toxic agents (Thorne, 2007).
Prevention of morbidity and mortality associated with diseases arising from exposure to toxic agents is an important goal of toxicology research. It has been well recognized that the key elements in the prevention of diseases associated with exposure to toxic agents are a clear understanding of the mechanisms, especially those at the molecular level, responsible for the onset and progression of toxicity as well as the ability to detect the earliest response/effect in the exposed individual. The early pre-clinical effects that occur well before the onset of any clinical symptoms are mostly reversible and, therefore, provide the most appropriate window of opportunity to apply effective intervention strategies based on toxicity mechanisms to prevent the onset of irreversible toxicity and diseases resulting in morbidity and mortality.
The potential risk for exposure to toxic agents and the resulting toxicity can be determined by monitoring for the presence of toxic agents either in the environment or the workplace (external dose) or in the target organs/tissues (internal dose) of the exposed population (Simmons et al., 2005; Van de Sandt et al., 2007). Several biomarkers indicative of the interaction between a toxic agent and its respective target organ have been developed to determine the toxicity resulting from exposure to the toxic agent (Lacour et al., 2005; Rockett & Kim, 2005; Thukral et al., 2005; Wallace et al., 2004). Adducts of chemicals and/or their metabolites with cellular macromolecules such as DNA (Matter et al., 2007; Gyorffy et al., 2008) and protein (Hagmar et al., 2001; Heubi, 2007) have been used as indicators of exposures to toxic agents and the potential toxicity of such exposures. Assaying enzyme activity and quantitation of biochemical constituents in target organs/tissues and biofluids such as blood and urine have also been employed as markers of toxicity (Benyounes et al., 2006; Iseri et al., 2007; Patlolla & Tchounwou, 2005).
Most of the currently available biomarkers are of limited use as effective toxicity markers owing to various limitations. These include, but are not limited to, the lack of specificity and/or lack of sensitivity to predict toxicity well before the onset of clinical symptoms. In addition, most of the currently available toxicity biomarkers do not provide significant information regarding the mechanisms underlying the toxicity. Therefore, there is a need to develop better biomarkers for toxicity that are more sensitive, specific and mechanistically relevant. Ideally, these biomarkers should be measurable, in easily accessible tissues or biofluids which can serve as surrogates for the inaccessible target organs/tissues. The ease and accessibility of using surrogate tissues will facilitate human monitoring of exposure to toxic agents such as those taking place from environmental and occupational sources.
Toxicogenomics, a relatively new branch of toxicology, employs the recent developments in genomics, especially transcriptomics, to study toxicity. Toxicogenomics has the potential to advance the understanding of how multiple genes are involved in cellular responses upon exposure to toxic agents (Afshari et al., 1999; Nuwaysir et al., 1999; Pennie et al., 2000; Olden, 2004). A primary tenet of toxicogenomics is that the effects of toxic agents on cellular functions are mediated through gene expression changes, both as primary and secondary effects. Biological processes are regulated by expression level changes of large numbers of genes organized as specific biological functions, pathways and networks. Toxic agents, upon entering the body, may cause alterations in the expression of one or several genes to result in the functional disruption of the corresponding biological functions, networks and pathways that are vital for normal functioning of the cells/tissues/organs. Therefore, alterations in the expression levels of genes that are involved in specific biological functions, pathways and networks regulating vital functions in the body may be reflective of the toxicity. There is substantial evidence suggesting that gene expression changes in target organs indicative of toxicity appear earlier than the onset of classic toxicity indicators such as biochemical and histological changes (Heinloth et al., 2004). Therefore, determination of gene expression changes in target organs in response to exposure to toxic agents may provide a window of opportunity for the preclinical diagnosis of toxic end points and the application of effective intervention strategies to prevent the resulting adverse health effects.
Gene expression profiling in target organs has been successfully employed to detect, classify and predict toxicity (Buck et al., 2008; Heinloth et al., 2004; Irwin et al., 2004; Konig et al., 2008; Mendrick, 2008). Furthermore, gene expression profiling can provide significant insight regarding the mechanism(s) underlying target organ toxicity (Amin et al., 2004; Beyer et al., 2007; Waring et al., 2001a). In one study, Steiner et al. (2004) determined the gene expression profiles for rats treated with one of 18 chemicals including several well-characterized hepatotoxicants. The aim of their study was to determine whether biological samples from rats treated with a toxic compound could be classified based on their gene expression profiles. The authors were able to differentiate the rats exposed to the hepatotoxic chemicals from those exposed to the non-hepatotoxic chemicals based on liver gene expression profiles. The potential application of gene expression profiling in target organs to classify chemicals based on their toxicity has also been demonstrated by the results obtained from many other investigations (Burczynski et al., 2000; Hamadeh et al., 2002a, 2002b; Thomas et al., 2001).
Gene expression profiling may also provide valuable mechanistic data which can be used in the risk assessment of all chemicals as well as to design and apply effective intervention strategies to prevent toxicity and associated adverse health effects. Waring et al. (2001a, 2001b) treated primary rat hepatocytes with one of 15 independent hepatotoxic chemicals, and the resulting gene expression profiles were determined employing microarrays. The various hepatotoxic chemicals used in the study differed with respect to the underlying mechanisms causing hepatotoxicity such as DNA damage, oxidative stress and induction of cytochrome P450 enzymes. Hierarchical clustering of the hepatotoxicity responsive genes in the treated cells demonstrated distinct gene expression signatures suggesting that the compounds could be classified into various groups based on the underlying mechanism(s) of hepatotoxicity. In another previous study, Hamadeh et al. (2002b) employed gene expression profiling to distinguish chemicals between two types of hepatotoxins based upon their mechanisms of toxicity in rats in vivo. The authors were able to distinguish the toxicity of peroxisome proliferators (clofibrate, Wyeth 14,643 and gemfibrozil) from that of an enzyme inducer (phenobarbital) based on distinct gene expression profiles from the livers of the rats.
A major drawback of the toxicogenomics data reported so far is that most, if not all, were derived from studies which used either cell culture (Waring et al., 2001a, 2001b) or laboratory animals (Amin et al., 2004; Hamadeh et al., 2002b, 2002c) as experimental models. Employing gene expression profiling to monitor toxicant exposure and/or effect in an inaccessible organ/tissue in humans is a difficult prospect. In most cases, direct biopsy of organs/tissues is not feasible.
Where humans are concerned, the use of gene expression profiling to determine toxicant exposures or predict possible toxicity outcomes is largely limited to the use of accessible biospecimens. One possible solution is to use easily accessible tissues that reflect similar gene expression changes as a given target organ. These surrogate tissues could offer a non-invasive or minimally invasive and convenient biomonitoring method to provide insight into the effects of toxicants on target organs (Rocket, 2006). The major biospecimens currently being used as surrogate tissues include placenta, hair, nail, milk, urine, blood, breath condensate, buccal cells, saliva, nasal lavage and bronchial lavage (Rockett & Burczynski, 2006).
Blood has several advantages over other surrogate tissues and is often considered the preferred surrogate tissue for inaccessible target organs in the body. The major benefits of using blood as the surrogate tissue for gene expression profiling studies, especially in human, are:
Blood is available from almost everyone and is collected routinely for diagnostic purposes.
Small quantities of blood can yield adequate quantities of high-quality RNA required for gene expression profiling.
Several previous studies conducted in the past have demonstrated the use of blood as the preferred surrogate tissue for gene expression profiling to identify markers of functional impairment in inaccessible organs in the body. Blood gene expression profiling has also been successfully employed as a diagnostic indicator of abnormal organ function under various disease conditions. For example, Burczynski et al. (2005) identified a specific set of predictor genes, capable of distinguishing between the peripheral blood mononuclear cells (PBMCs) of renal cell carcinoma patients and normal volunteers with high accuracy. Similarly, peripheral blood has been successfully employed as a surrogate tissue for gene expression studies to identify disease conditions such as ischemic stroke (Moore et al., 2005), neurologic diseases (Tang et al., 2005), Huntington’s disease (Borovecki et al., 2005), lupus (Baechler et al., 2003) and migraine (Hershey et al., 2004).
There have been a limited number of studies conducted in the past demonstrating the use of gene expression studies using blood as a surrogate tissue to determine chemical exposure and the resulting toxicity. Ember et al. (2000) and Gyongyi et al. (2001) administered 1-nitropyrene and 7,12-dimethylbenz(a) anthrazene, respectively, in rats and determined gene expression profiles in the PBMCs and several internal target organs (lung, liver, lymph nodes, kidneys and spleen). Results of these studies demonstrated that the expression levels of two oncogenes (H-ras and c-myc) and a tumor suppressor (p53) responded to administration of the chemicals, with a good correlation in gene expression between the PBMCs and the target organs. Thus the expression levels of these genes in the PBMCs might be early biomarkers of exposure to the tested chemicals, and PBMCs may be used as an effective surrogate for certain internal target organs. Recently, McHale et al. (2007) employed gene expression profiling in PBMCs as a measure for exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in a population in Seveso, Italy. Higher blood levels of TCDD in the exposed individuals correlated with the expression of specific genes in PBMCs, suggesting that blood gene expression profiling can be used to identify biomarkers for toxicant exposure in humans. Furthermore, the appearance of chloracne in individuals who were accidentally exposed to TCDD was associated with a distinct blood gene expression profile compared with unexposed individuals. Results of an exploratory study conducted by Wang et al. (2005) determined global gene expression profiles in whole blood samples of workers occupationally exposed to metal fume. Several genes involved in biological processes related to inflammatory response, oxidative stress, intracellular signal transduction, cell cycle and programmed cell death were differentially expressed in workers exposed to metal fume compared with non-exposed controls.
Recent studies conducted in our laboratory as well as those by investigators in other laboratories have strengthened the argument that blood gene expression profiling might be a valuable surrogate approach not only to detect early target organ toxicity but also to obtain insight into the molecular mechanisms underlying the target organ toxicity. The remainder of this article is dedicated to discuss the findings obtained from studies in which blood gene expression profiling has been employed as a surrogate approach to detect and study hepatotoxicity and pulmonary toxicity. Efforts are made not only to demonstrate the advantages of employing blood gene expression profiling in detecting and studying target organ toxicity but also to address the challenges this emerging field in toxicology faces.
Blood gene expression profiling and hepatotoxicity
The liver is the primary target organ for a vast majority of toxic agents owing to its central role in chemical metabolism. This is best illustrated in the case of drug-induced liver injury (DILI). DILI is the major reason for both the failure of regulatory approval of new drug candidates and withdrawal or limitation of use of already approved and marketed drugs. Primarily as a result of the inability to obtain human liver samples to determine DILI, there has been immense pressure to develop alternative, sensitive, specific, reliable, mechanistically relevant and non-invasive or minimally invasive biomarkers for DILI. Even though serum levels of transaminases have been routinely employed as surrogate clinical markers of DILI, concerns have been raised especially with respect to their lack of sensitivity. Structural and/or functional impairment of the liver is a prerequisite for observable alterations in the levels of serum transaminases. In addition, elevated serum transaminase levels do not provide any mechanistic insight into DILI. In view of these limitations, investigations have been carried out in recent years to determine the suitability of peripheral blood gene expression profiling as a sensitive, specific and mechanistically relevant surrogate approach to detect and study liver toxicity induced by drugs and other toxic agents.
One of the pioneer studies in this field was carried out by investigators at the National Institute for Environmental Health Sciences (NIEHS) employing acetaminophen (APAP), a drug that has been implicated in numerous cases of human DILI (Bushel et al., 2007). Rats were given toxic or sub-toxic oral doses of APAP; and the resulting hepatotoxicity, or the lack thereof, was determined on the basis of established toxicity markers including serum transaminases, hematological parameters and liver histology. Total RNA was isolated from blood samples obtained from the control and APAP administered rats and global gene expression profiles were determined by microarray analysis to develop blood gene expression signatures capable of predicting APAP exposure and APAP-induced hepatotoxicity. Findings of the study suggested that changes in the whole blood transcriptome, indicative of APAP-induced hepatotoxicity, preceded changes in the traditional hepatotoxicity markers employed in the study. Furthermore, the blood gene expression signatures were able to accurately predict the rats which were given the sub-toxic oral dose of APAP. Subsequently, Zhang et al. (2012) applied the method Extracting Patterns and Identifying co-expressed Genes (EPIG) to the transcriptomics data obtained from the Bushel et al. (2007) study to investigate to what extent changes in blood transcriptome mirror those in the livers of the APAP-administered rats. Similar expression changes were found to be occurring in a sub-set of genes in the blood and liver of the APAP-administered rats. Some of the parallel transcript changes in the blood and liver reflected pathways that are relevant to APAP-induced hepatotoxicity including mitochondrial oxidative phosphorylation and immune function. Although the number of genes that demonstrated coordinate regulation between the liver and blood represented only a small fraction of the total affected genes, the similarities in the pathways and functions linked to these genes support the idea that transcriptomics analysis of blood can provide mechanistic insight into DILI.
A notable finding of the study by Bushel et al. (2007) was the superior sensitivity of blood gene expression markers as predictors of hepatotoxicity compared with the traditional toxicity markers in the rats. This was further confirmed by Lobenhofer et al. (2008) who, using a compendium of eight non-therapeutic hepatotoxic chemicals, investigated whether it was possible to classify histopathological differences, most likely reflecting differences in mechanisms of cell-specific toxicity, using either liver (target organ) or blood (surrogate tissue) transcriptomics. The results of this study demonstrated that it was possible to classify the hepatotoxicants based on the gene expression profiling data derived from either liver or blood. In fact, the blood gene expression data performed slightly better than the liver gene expression data in classifying the hepatotoxicants based on histopathological differences in liver. In a follow-up study, Huang et al. (2010) demonstrated that the transcriptomics markers of blood derived from the Lobenhofer et al. (2008) study were able to accurately predict APAP-induced liver toxicity in rats (accuracy as high as 92.1%).
The potential application of blood gene expression profiling to predict DILI has also been investigated in humans. No hepatotoxicity, as assessed by clinical or biochemical parameters, was seen in healthy volunteers who were treated with a 4-g bolus dose of APAP compared with those receiving placebo (Fannin et al., 2010). In spite of the absence of hepatotoxicity, as evidenced by the lack of any alteration in the established biochemical and clinical markers, significant down-regulation of genes involved in oxidative phosphorylation was noticed in the blood of the APAP-treated individuals. Down regulation of blood genes involved in oxidative phosphorylation noticed in the APAP-treated individuals is consistent with known mechanisms of APAP-induced liver toxicity, as the involvement of mitochondrial injury in APAP-induced hepatotoxicity has been very well established (Heinloth et al., 2004). The potential application of blood gene expression signatures to detect DILI in humans has also been demonstrated by Bushel et al. (2007). The human orthologs of the rat blood discriminatory genes for APAP-induced liver toxicity identified by these authors were able to distinguish APAP-intoxicated patients from control individuals. The cross-species application of blood gene expression markers for toxicity reported by Bushel et al. (2007) is significant and very encouraging in that animal models may be employed, at least, to develop and validate blood gene expression markers that can eventually be tested and applied to monitor human exposure to toxic agents that are commonly found in the environment and workplace.
Blood gene expression profiling and pulmonary toxicity
The application of blood gene expression profiling in detecting and studying pulmonary toxicity is best illustrated in the case of crystalline silica-induced pulmonary toxicity. Occupational exposure to crystalline silica is a major health hazard affecting millions of workers in the US and elsewhere (Sanderson, 1986). Virtually any activity that involves the movement of earth (e.g. mining, farming, construction, etc.) is considered a potential source for occupational exposure to crystalline silica. However, the major occupations where significant human exposure to crystalline silica takes place are sandblasting, silica milling, surface mining and tunneling. In many cases, occupational exposure to crystalline silica takes place at levels much higher than the National Institute for Occupational Safety and Health (NIOSH) recommended exposure level (REL) of 0.05 mg m−3 (Linch et al., 1998).
The adverse health effects resulting from occupational exposure to crystalline silica have been recognized and identified for a long time. These include, but are not limited to, the development of autoimmune diseases, rheumatoid arthritis, chronic renal diseases, lupus and cancer (IARC, 1997; Parks et al., 1999; Steenland et al., 2001). Silicosis, however, is the adverse health effect that has received the most attention among those exposed occupationally to crystalline silica. Silicosis is an irreversible, but preventable interstitial lung disease, characterized by alveolar proteinosis and diffuse fibrosis resulting in progressively restrictive lung function and death (Castranova & Vallyathan, 2000).
Currently, chest X-ray and pulmonary function tests are routinely employed to detect silicosis. Both chest X-ray and pulmonary function tests rely on structural and/or functional impairment of the lungs associated with the development of silicosis and, therefore, are not capable of predicting or detecting the disease at an early, preventable stage. Therefore, NIOSH has recommended developing highly sensitive and non-invasive or minimally invasive techniques that are capable of predicting or detecting silicosis prior to the onset of clinical symptoms that most likely represent the irreversible stage of the disease (NIOSH, National Insitute for Occupational Safety and Health, 2002).
In compliance with the NIOSH recommendation, a research project was undertaken in our laboratory investigating the potential application of blood gene expression profiling as a highly sensitive surrogate approach to detect and/or to predict crystalline silica exposure and the resulting pulmonary toxicity. The aims of our investigation, which employed a rat silicosis model, were to determine (i) can global gene expression changes in the blood reflect silica-induced pulmonary toxicity, (ii) can blood gene expression changes indicative of silica-induced pulmonary toxicity appear prior to the onset of classic biochemical and histological changes associated with silica-induced pulmonary toxicity, (iii) can bioinformatics analysis of the differentially expressed genes in the blood of the silica exposed rats provide insights into the mechanisms underlying the pulmonary toxicity induced by silica exposure and (iv) can exposure to a sub-toxic concentration of crystalline silica be detected or predicted using a blood gene expression signature. Details regarding the study design of the experiment can be found in our recent publications (Sellamuthu et al., 2011a, 2011b, 2012a, 2012b). Stated briefly, approximately 3-month-old, healthy, male Fischer 344 rats (CDF strain) were exposed to filtered air (control) or an aerosol of respirable crystalline silica (15 mg m−3, 6 h per day for 5 days). After exposure, groups of control and silica exposed rats were sacrificed at post-exposure time intervals of 0, 1, 2, 4, 8, 16, 32 and 44 weeks. Bronchoalveolar lavage fluid (BALF), including the cells within it, and blood and lungs were collected from the rats to determine the effects of pulmonary exposure to crystalline silica as well as to determine lung and blood global gene expression profiles. The major findings of our studies are summarized below:
Inhalation exposure to crystalline silica resulted in pulmonary toxicity in the rats
Lactate dehydrogenase (LDH) activity in the BALF and histological changes in the lungs were evaluated to determine if exposure to crystalline silica resulted in pulmonary toxicity in the rats. In addition, because of the prominent role played by inflammation in silica-induced pulmonary toxicity, the BALF levels of polymorphonuclear leukocytes (PMN) and the pro-inflammatory cytokine, monocyte chemotactic protein 1 (MCP1), were determined in the rats. Our results demonstrated that, with the exception of the 2 weeks post-silica exposure time interval, a significant elevation in BALF LDH activity was detected in the silica-exposed rats compared with the corresponding time-matched control rats. Beyond the 2-week post-exposure time interval, a steady increase in BALF LDH activity in the rats, suggesting a steady progression of silica-induced pulmonary toxicity, was noticed. The BALF levels of PMN and MCP1 were significantly elevated and exhibited the same trend as that of the BALF LDH activity in the rats.
Corraboration of the progression of pulmonary toxicity during the post-silica exposure time intervals was supported by the lung histological changes in the rats. An acute inflammatory response, as evidenced from the accumulation of PMNs around small- and medium-sized airways, was noticed in the silica-exposed rats at the 0-week post-exposure time interval (Sellamuthu et al., 2011a). A further progression in histological changes in the lungs was noticed during the late post-exposure time intervals of 8- and 16-weeks (Sellamuthu et al., 2011a), 32-weeks (Sellamuthu et al., 2012a) and 44-weeks (Sellamuthu et al., 2013). The most prominent histological changes noticed in the lungs of the silica-exposed rats at the late post-exposure time intervals (32- and 44-weeks) included type II pneumocyte hyperplasia and positive staining with trichrome stain, indicative of fibrosis (Sellamuthu et al., 2012a, 2013). These results, therefore, suggested that inhalation exposure to crystalline silica resulted in pulmonary toxicity in the rats, and our rat model was appropriate to investigate whether blood gene expression profiling can be employed as a surrogate approach to detect silica-induced pulmonary toxicity.
Global gene expression changes in the blood correlated with silica-induced pulmonary toxicity in rats
Global gene expression profiles in the lungs and blood samples of the control and silica-exposed rats were determined using the RatRef-12 V1.0 Expression BeadChip Array (Illumina, Inc., San Diego, CA, USA) as described in detail in our original publications (Sellamuthu et al., 2011a, 2012a). The number of significantly differentially expressed genes in the lung and blood samples of the silica-exposed rats, compared with the time-matched control rat samples, was determined at each of the post-silica exposure time intervals. As presented in Table 1, the number of significantly differentially expressed genes in the target organ, lungs and the surrogate tissue, blood, correlated well with markers of pulmonary toxicity (BALF LDH activity) and inflammation (BALF PMN count and MCP1 level). These results, in agreement with several previous publications (Bushel et al., 2007; Huang et al., 2010; Lobenhofer et al., 2008; Umbright et al., 2010), confirmed the potential value of global gene expression changes in the target organ as an indicator of target organ toxicity. However, the better correlation noticed between pulmonary toxicity markers and the number of significantly differentially expressed genes in the blood, compared with that of the lungs, suggested that the gene expression changes taking place in blood, the surrogate tissue, may be better indicators of target organ toxicity.
Table 1.
Correlation coefficients (r2 values) for the relationship between silica-induced pulmonary toxicity (BALF LDH, PMN and MCP-1) and the number of significantly differentially expressed genes (SDEGs) in the lungs and blood of silica-exposed rats
| BALF LDH | BALF PMN | BALF MCP-1 | |
|---|---|---|---|
| Lung SDEGs | 0.776 | 0.879 | 0.927 |
| Blood SDEGs | 0.831 | 0.923 | 0.958 |
The toxicity measurements and the number of differentially expressed genes in the silica-exposed rats at post-exposure time intervals of 0, 1, 2, 4, 8, 16 and 32 weeks after a 1-week exposure were used to determine the correlation coefficients (r2 values) (reproduced with permission from Inhalation Toxicology 2012: 24: 570–579).
Bioinformatics analysis of the differentially expressed genes in blood provided molecular insights into the mechanisms of silica-induced pulmonary toxicity
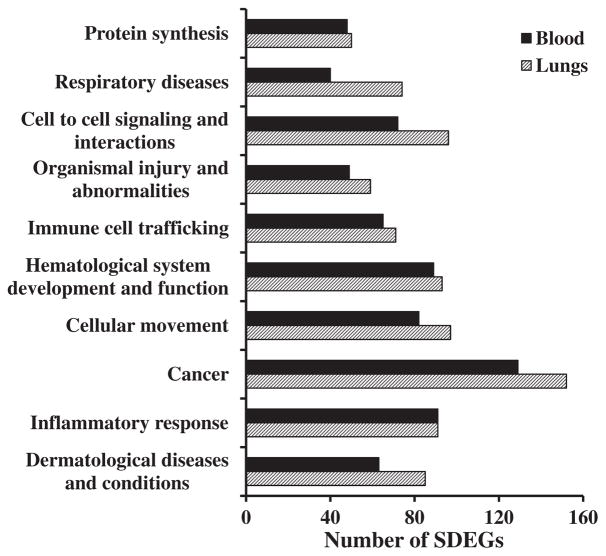
Studies conducted over the past several years have demonstrated the potential of crystalline silica exposure to result in pulmonary toxicity, especially silicosis, autoimmune diseases, renal diseases and cancer (IARC, 1997; Parks et al., 1999; Steenland et al., 2001). These previous studies have also demonstrated that many of the toxic effects associated with crystalline silica exposure are consequences of its ability to result in oxidative stress (Vallyathan et al., 1997), apoptosis (Santarelli et al., 2004), gene expression changes (Hubbard et al., 2002) and inflammation (Porter et al., 2002). Bioinformatics analysis of the differentially expressed genes in the blood and lungs of silica-exposed rats (Sellamuthu et al., 2012a), in addition to supporting the previously recognized toxicity and health effects of silica, provided insights into the molecular mechanisms underlying silica-induced pulmonary toxicity. Bioinformatics analysis of the significantly differentially expressed genes in the lungs and blood of silica-exposed rats showed remarkable similarity in the biological functions, molecular networks and canonical pathways that were significantly enriched in response to silica exposure and the resulting pulmonary toxicity (Sellamuthu et al., 2012a). The top 10 Ingenuity Pathway Analysis (IPA) biological functions that were significantly enriched in the target organ, lungs, were also significantly enriched in the surrogate tissue, blood, of the silica-exposed rats (Fig. 1). Most of the IPA biological function categories that were significantly enriched in the blood of the silica-exposed rats, namely respiratory diseases, cell-to-cell signaling and interaction, immune cell trafficking, cellular movement, cancer, inflammatory response, were functions that are known to be associated with toxicity and health effects of silica exposure. A comparable similarity was noticed with respect to the canonical pathways and molecular networks that were significantly enriched in the lungs and the blood in response to the silica-induced pulmonary toxicity (Sellamuthu et al., 2012a). It has been well established that induction of inflammation plays a central role in the pulmonary effects of crystalline silica exposure in animal models (Castranova, 2004). In fact, the majority of biological functions, molecular networks and canonical pathways that were significantly enriched in the blood of the rats in response to silica-induced pulmonary toxicity were those involved in an inflammatory response (Sellamuthu et al., 2011a, 2011b). Given these results, blood gene expression profiling and bioinformatics analysis of the differentially expressed genes in the blood appears to be a toxicologically relevant surrogate approach to gain insights into the mechanisms of target organ toxicity.
Figure 1.
Enrichment of Ingenuity Pathway Analysis (IPA) biological functions in the lungs and blood of silica-exposed rats. Bioinformatics analysis of the significantly differentially expressed genes identified in the silica-exposed rat lungs and blood was done using IPA software. The top 10 significantly enriched biological functions of the silica-exposed rat lungs compared with the control rat lungs and the same biological functions in the blood samples are presented to demonstrate the similarity in gene expression profile between lungs and blood of the silica exposed rats. Data represents the mean of six rats per group (reproduced with permission from Inhalation Toxicology 2012; 24: 570–579). SDEGs, significantly differentially expressed genes.
Blood gene expression signature predicted exposure to a non-toxic concentration of silica in rats
In a separate set of experiments, a blood gene expression signature was tested for its ability to predict exposure of rats to silica and the resulting pulmonary toxicity. Blood gene expression data obtained from control rats and those exposed to crystalline silica at 15 mg m−3, 6 h per day for 5 days (0-week post-exposure time interval) were used as the training set data to develop a gene expression signature for silica exposure and/or toxicity. A blood gene expression signature consisting of 7 genes was identified and tested in a set of rats which were exposed to lower concentrations of silica (1 or 2 mg m−3, 6 h per day, 5 days). Rats exposed to silica at 2 mg m−3, 6 per day for 5 days resulted in mild pulmonary toxicity as evidenced from the observation of a slight, but statistically significant, elevation in BALF parameters of pulmonary toxicity (LDH activity, albumin and protein content). In contrast, rats exposed to silica at 1 mg m−3, 6 h per day for 5 days did not result in any detectable pulmonary toxicity as evidenced from normal LDH activity and protein and albumin contents in their BALF. The predictive blood gene expression signature developed for silica exposure and toxicity correctly identified 7 out of 8 rats (87.5%) that were exposed to silica at 2 mg m−3, 6 h per day for 5 days and resulted in mild pulmonary toxicity. Six out of eight rats (75%) that were exposed to crystalline silica at 1 mg m−3, 6 h per day for 5 days and did not result in any detectable pulmonary toxicity were correctly identified as silica-exposed rats by the predictive blood gene expression signature. These results, therefore, demonstrated the superior sensitivity of the blood gene expression signature to detect/predict silica exposure and the resulting pulmonary toxicity in the rats. The ability of the predictive blood gene expression signature to detect silica exposure in the absence of pulmonary toxicity detectable by traditional approaches (biochemical and histological toxicity markers) in the rat model may be of significant value with respect to monitoring workers for occupational exposure to crystalline silica and potential health effects. This view is further supported by the earlier report by Bushel et al. (2007) that a blood gene expression signature for hepatotoxicity developed in a rat model was applicable in correctly identifying APAP-induced hepatotoxicity in humans.
Although the vast majority of research with respect to the application of blood gene expression profiling in target organ toxicity has focused on hepatotoxicity and pulmonary toxicity, a few studies have been reported in which blood gene expression profiling was employed to detect toxicity in organs other than liver and lungs.
A blood gene expression signature consisting of eight genes was identified in rats administered a toxic dose of the organophosphorus insecticide, methyl parathion (Umbright et al., 2010). The neurotoxicity signature genes identified in the methyl parathion-administered rats were also significantly differentially expressed in rats administered additional neurotoxic chemicals, ethyl parathion and malathion. None of the blood genes comprising the neurotoxicity signature were significantly differentially expressed in the rats administered any one of the four hepatotoxic chemicals (acetaminophen, carbon tetrachloride, thioacetamide and dimethylnitrobenzene). These findings, therefore, suggest the ability of blood gene expression signatures to distinguish target organ toxicity.
In a study reported by Dadarkar et al. (2010a) the application of blood gene expression markers to detect nephrotoxicity in rats has been described. Rats were administered any one of four predominantly nephrotoxic drugs (cyclophosphamide, amphotericin B, gentamicin and cisplatin). The authors, based on their findings, suggested that changes in expression of the secreted phosphoprtoein 1 (SPP1) gene in whole blood cells could possibly be used as a surrogate marker for drug-induced nephrotoxicity.
Rocket et al. (2002) applied blood gene expression profiling to detect endocrine system perturbation. Overectomized rats were treated with either 17-β-estradiol or vehicle for 3 days, and gene expression profiles were determined by microarray using RNA samples isolated from PBMCs and the uterus of the rats. Of the 1185 genes represented on the microarray, 18 were found differentially expressed in both the PBMCs and the uterus of the rats in response to administration of 17-β-estradiol.
Blood transcriptomics in toxicology: challenges
Like any other new development in toxicology research, the potential application of blood transcriptomics as a relevant surrogate approach to study target organ toxicity faces many challenges. Questions to consider are: Can blood gene expression profiling be employed to monitor human exposure to toxic agents and the resulting toxicity such as those taking place in the environment and workplace? How sensitive are blood gene expression changes as surrogate markers of target organ toxicity compared with the traditional toxicity markers? Can gene expression changes be detected in the blood prior to the onset of clinical symptoms, which most probably, represent irreversible toxicity and disease in the target organ? In other words, can blood gene expression changes representing target organ toxicity be predicted early enough to provide a window of opportunity for intervention to prevent the onset of serious health effects? Typically, human exposure to toxic agents, either in the environment or the workplace, take place at very low concentrations and may not be associated with the onset of immediate target organ toxicity. In addition, co-exposure to multiple toxic agents is likely under conditions of occupational and, especially, environmental exposures. Furthermore, confounding factors such as diseases, cigarette smoking and other life-style factors are likely to affect peripheral blood gene expression profiles in humans. Therefore, can blood gene expression signatures be employed as sensitive and specific biomarkers to detect target organ toxicity in humans?
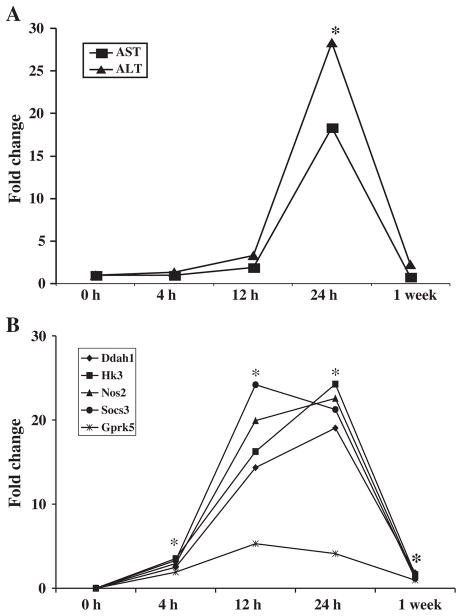
An unequivocal observation throughout the studies conducted so far in the area of blood transcriptomics in toxicology is the finding of superior sensitivity of blood gene expression changes compared with the histological, biochemical and hematological changes as indicators of target organ toxicity. In a study conducted in our laboratory (Umbright et al., 2010), rats were administered a hepatotoxic dose of APAP. Transaminase activity and expression levels of hepatotoxicity marker genes were determined in the blood at post-exposure time intervals ranging from 4 h to 1 week. As presented in Fig. 2, significant differential expression of the hepatotoxicity marker genes in the blood was detectable as early as 4 h following administration of APAP. On the other hand, the earliest post-exposure time interval when significant elevations in transaminase activities appeared in the blood, suggesting the onset of hepatotoxicity, was 24 h. As described above, superior sensitivity of blood gene expression signatures, compared with histological, biochemical and hematological changes, as biomarkers for APAP-induced hepatotoxicity have been demonstrated by several investigators in rats (Bushel et al., 2007; Huang et al., 2008, 2010; Lobenhofer et al., 2008; Zhang et al., 2012). The superior sensitivity of blood gene expression changes indicative of hepatotoxicity induced by APAP, compared with clinical and biochemical markers of hepatotoxicity, have also been demonstrated in humans (Fannin et al., 2010). In a recent study published by Kim et al. (2011), blood gene expression signatures for hepatotoxicity induced by volatile organic compounds (VOCs) were developed in rats, and their sensitivity was compared with the traditional toxicity markers. In agreement with previous studies (Bushel et al., 2007; Huang et al., 2008; Lobenhofer et al., 2008; Umbright et al., 2010; Zhang et al., 2012), the blood gene expression markers identified by Kim et al. (2011) outperformed the traditional toxicity markers with respect to their sensitivity to detect hepatotoxicity in rats. Similar to hepatotoxicity, superior sensitivity of blood gene expression signatures, compared with well-established traditional toxicity markers, has also been demonstrated in the case of pulmonary toxicity (Sellamuthu et al., 2011a) and neurotoxicity (Umbright et al., 2010).
Figure 2.
Gene expression profiling is more sensitive than the traditional toxicity endpoints. A single acute toxic dose of acetaminophen was administered to rats. At various time intervals ranging from 4 h to 1 week after administration of the chemical, blood was analyzed for toxicity based on the activities of aspartate transaminase (AST) and alanine transaminase (ALT) (A) and expression of marker genes for hepatotoxicity (B). Significant alterations in the expression of the selected hepatotoxicity marker genes were observed in the blood before any significant change in the activities of transaminases suggesting the superior sensitivity of gene expression changes as indicators of target organ toxicity (reproduced with permission from Molecular and Cellular Biochemistry 2010; 335: 223–234. *p<0.05 compared with time matched controls.
Similar to sensitivity, the issue of specificity is very important in the case of blood gene expression signatures as surrogate markers of target organ toxicity. Is it possible to distinguish the toxic agent and the target organ involved in the toxicity based on blood gene expression marker(s)? Unfortunately, not many studies have been conducted to date directly investigating whether the blood gene expression markers are specific to either the toxic agent or the target organ involved. In our previous study (Umbright et al., 2010), rats were administered either APAP or methyl parathion to induce hepatotoxicity and neurotoxicity, respectively. Microarray analysis of total RNA isolated from blood samples of the rats resulted in the identification of a panel of hepatotoxicity and neurotoxicity marker genes. The capability of the marker genes to detect and distinguish hepatotoxicity and neurotoxicity was tested in rats administered additional hepatotoxic (thioacetamide, dimethylnitrobenzene, and carbon tetrachloride) or neurotoxic (ethyl parathion and malathion) chemicals. The hepatotoxicity marker genes, identified using APAP as the model compound, were found differentially expressed in the blood samples derived from rats that were administered the additional hepatotoxic chemicals and not in the blood samples of any of the rats administered the neurotoxic chemicals. Therefore, in spite of the limited number of chemicals tested in the study, our results suggested that the blood gene expression markers may be specific to the type of target organ toxicity involved. Dadarkar et al. (2010b) performed gene expression analysis and hierarchical clustering in RNA samples obtained from human PBMCs treated with either hepatotoxic (acetaminophen, rosiglitazone, fluconazole and isoniazid) or nephrotoxic (cyclophosphamide, amphotericin B, gentamicin and cisplatin) drugs and identified a set of 365 genes that could discriminate the two classes of drugs. Further support to the specificity of blood gene expression markers to target organ toxicity is provided by Kim et al. (2011) who investigated blood gene expression profiles in rats administered each of three different VOCs (dichloromethane, ethylbenzene and trichloroethylene). Supervised analysis identified 1217 outlier genes as a distinct molecular signature distinguishing VOC exposure from controls while unsupervised gene expression analysis resulted in a characteristic molecular signature for each VOC.
Although experimental evidence is lacking, a critical review of the literature suggests that the blood gene expression signature identified in our laboratory may be specific to silica-induced pulmonary toxicity (Sellamuthu et al., 2011a). The blood gene expression signature identified for the silica-induced pulmonary toxicity is quite different from that reported by Bushel et al. (2007) for hepatotoxicity in spite of the predominance of inflammatory response genes in both the signatures. In spite of the central role played by inflammation in silica-induced pulmonary toxicity (Castranova, 2004) and the pulmonary effects associated with cigarette smoking (Bhalla et al., 2009), the effects of these two pulmonary toxic agents on blood transcriptome appear to be different. It is worth mentioning that none of the genes we identified as part of the blood gene expression signature for silica-induced pulmonary toxicity were present among the 342 genes that were reported to be differentially expressed in the blood of current cigarette smokers (Charlesworth et al., 2010). Similarly, none of the genes identified as part of the blood gene expression signature for silica-induced pulmonary toxicity were found differentially expressed in the blood under conditions of inflammation induced by exposure to endotoxin (Calvano et al., 2005) or diesel exhaust particles (Peretz et al., 2007), further suggesting that the blood gene expression signature identified in our laboratory may be specific to silica exposure and the resulting pulmonary toxicity. However, additional experimental evidence is required to make a definite conclusion regarding the very important issue concerning specificity of blood gene expression markers either for the toxic agent or the target organ toxicity.
Blood gene expression profiling to monitor human exposure to toxic agents and potential adverse health effects
The ultimate objective of any research focused on blood gene expression profiling in toxicology is to prevent adverse health effects resulting from human exposure to toxic agents present in the environment and workplace. This will require, as described above, early detection of exposure to toxic agents and a clear understanding of the molecular mechanism(s) underlying the toxic effects. The results of studies conducted so far, mostly by employing animal models, have been encouraging and have demonstrated blood gene expression changes as sensitive indicators of early toxicity (Bushel et al., 2007; Sellamuthu et al., 2011a; Umbright et al., 2010).
The very same superior sensitivity of the blood transcriptome to respond to exposure to toxic agents may limit its potential application in monitoring human exposure to toxic agents. It has been fairly well recognized that the blood transcriptome is highly dynamic in nature and may be influenced by a plethora of endogenous and exogenous factors. Some factors cannot be controlled. For example, circadian rhythm is known to affect gene expression profiles in tissues (Almon et al., 2008; Sukumaran et al., 2010) and probably in the blood. In addition, various life style factors, for example, cigarette smoking (Charlesworth et al., 2010) and various disease conditions (Baechler et al., 2003; Borovecki et al., 2005; Hershey et al., 2004; Moore et al., 2005; Tang et al., 2005) may affect the human blood transcriptome. Obviously, the question arises whether such a dynamic system is suitable to monitor human exposure to toxic agents that may be present in very small quantities in the environment and the workplace.
It is, therefore, apparent that more research is needed in order to determine the suitability of blood gene expression profiling as a surrogate approach to monitor human exposure to toxic agents and the resulting toxicity. However, it is encouraging to notice that mechanistically relevant gene expression changes were detectable in the peripheral blood of individuals who had received a bolus of APAP that did not result in biochemical and clinical changes indicative of liver toxicity (Fannin et al., 2010). Similarly, the cross-species application of the rat blood gene expression signature for APAP-induced hepatotoxicity in human (Bushel et al., 2007) is also encouraging. Blood gene expression signatures can be developed and characterized for sensitivity and specificity using animal models and may be subsequently tested for their suitability to monitor human exposure to toxic agents and the resulting adverse health effects. Blood gene expression profiling may, therefore, turn out to be a valuable, practical first step to prevent morbidity and mortality associated with exposure to toxic agents present in the environment and the workplace.
Footnotes
Disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
References
- Afshari CA, Nuwaysir EF, Barrett JC. Application of complementary DNA microarray technology to carcinogen identification, toxicology, and drug safety evaluation. Cancer Res. 1999;59:4759–4760. [PubMed] [Google Scholar]
- Almon RR, Yang E, Lai W, Androulakis IP, DuBois DC, Jusko WJ. Circadian variations in rat liver gene expression: relationships to drug actions. J Pharmacol Exp Ther. 2008;326:700–716. doi: 10.1124/jpet.108.140186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin RP, Vickers AE, Sistare F, Thompson KL, Roman RJ, Lawton M, Kramer J, Hamadeh HK, Collins J, Grissom S, Bennett L, Tucker CJ, Wild S, Kind C, Oreffo V, Davis JW, 2nd, Curtiss S, Naciff JM, Cunningham M, Tennant R, Stevens J, Car B, Bertram TA, Afshari CA. Identification of putative gene based markers of renal toxicity. Environ Health Perspect. 2004;112:465–479. doi: 10.1289/ehp.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyounes M, Sempoux C, Daumerie C, Rahier J, Geubel AP. Propylthiouracyl-induced severe liver toxicity: an indication for alanine aminotransferase monitoring? World J Gastroenterol. 2006;12:6232–6234. doi: 10.3748/wjg.v12.i38.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer RP, Fry RC, Lasarev MR, McConnachie LA, Meira LB, Palmer VS, Powell CL, Ross PK, Bammler TK, Bradford BU, Cranson AB, Cunningham ML, Fannin RD, Higgins GM, Hurban P, Kayton RJ, Kerr KF, Kosyk O, Lobenhofer EK, Sieber SO, Vliet PA, Weis BK, Wolfinger R, Woods CG, Freedman JH, Linney E, Kaufmann WK, Kavanagh TJ, Paules RS, Rusyn I, Samson LD, Spencer PS, Suk W, Tennant RJ, Zarbl H Members of the Toxicogenomics Research. Multicenter study of acetaminophen hepatotoxicity reveals the importance of biological endpoints in genomic analyses. Toxicol Sci. 2007;99:326–337. doi: 10.1093/toxsci/kfm150. [DOI] [PubMed] [Google Scholar]
- Bhalla DK, Hirata F, Rishi AK, Gairola CG. Cigarette smoke, inflammation, and lung injury: a mechanistic perspective. J Toxicol Environ Health B Crit Rev. 2009;12:45–64. doi: 10.1080/10937400802545094. [DOI] [PubMed] [Google Scholar]
- Borovecki F, Lovrecic L, Zhou J, Jeong H, Then F, Rosas HD, Hersch SM, Hogarth P, Bouzou B, Jensen RV, Krainc D. Genome-wide expression profiling of human blood reveals biomarkers for Huntington’s disease. Proc Natl Acad Sci USA. 2005;102:11023–11028. doi: 10.1073/pnas.0504921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck WR, Waring JF, Blomme EA. Use of traditional end points and gene dysregulation to understand mechanisms of toxicity: toxicogenomics in mechanistic toxicology. Methods Mol Biol. 2008;460:23–44. doi: 10.1007/978-1-60327-048-9_2. [DOI] [PubMed] [Google Scholar]
- Burczynski ME, McMillian M, Ciervo J, Li L, Parker JB, Dunn RT, 2nd, Hicken S, Farr S, Johnson MD. Toxicogenomics-based discrimination of toxic mechanism in HepG2 human hepatoma cells. Toxicol Sci. 2000;58:399–415. doi: 10.1093/toxsci/58.2.399. [DOI] [PubMed] [Google Scholar]
- Burczynski ME, Twine NC, Dukart G, Marshall B, Hidalgo M, Stadler WM, Logan T, Dutcher J, Hudes G, Trepicchio WL, Strahs A, Immermann F, Slonim DK, Dorner AJ. Transcriptional profiles in peripheral blood mononuclear cells prognostic of clinical outcomes in patients with advanced renal cell carcinoma. Clin Cancer Res. 2005;11:1181–1189. [PubMed] [Google Scholar]
- Bushel PR, Heinloth AN, Li J, Huang L, Chou JW, Boorman GA, Malarkey DE, Houle CD, Ward SM, Wilson RE, Fannin RD, Russo MW, Watkins PB, Tennant RW, Paules RS. Blood gene expression signatures predict exposure levels. Proc Natl Acad Sci USA. 2007;104:18211–18216. doi: 10.1073/pnas.0706987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Chen RO, Brownstein BH, Cobb JP, Tschoeke SK, Miller-Graziano C, Moldawer LL, Mindrinos MN, Davis RW, Tompkins RG, Lowry SF Inflammation and Host Response to Injury Large Scale Collabortion Research Program. A network-based analysis of systemic inflammation in humans. Nature. 2005;437:1032–1037. doi: 10.1038/nature03985. [DOI] [PubMed] [Google Scholar]
- Castranova V. Signaling pathways controlling the production of inflammatory mediators in response to crystalline silica exposure: role of reactive oxygen/nitrogen species. Free Radic Biol Med. 2004;37:916–925. doi: 10.1016/j.freeradbiomed.2004.05.032. [DOI] [PubMed] [Google Scholar]
- Castranova V, Vallyathan V. Silicosis and coal workers’ pneumoconiosis. Environ Health Perspect. 2000;108(Suppl 4):675–684. doi: 10.1289/ehp.00108s4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth JC, Curran JE, Johnson MP, Goring HH, Dyer TD, Diego VP, Kent JW, Jr, Mahaney MC, Almasy L, MacCluer JW, Moses EK, Blangero J. Transcriptomic epidemiology of smoking: the effect of smoking on gene expression in lymphocytes. BMC Med Genomics. 2010;3:29. doi: 10.1186/1755-8794-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadarkar SS, Fonseca LC, Mishra PB, Lobo AS, Doshi LS, Dagia NM, Rangasamy AK, Padigaru M. Phenotypic and genotypic assessment of concomitant drug-induced toxic effects in liver, kidney and blood. J Appl Toxicol. 2010a;31:117–130. doi: 10.1002/jat.1562. [DOI] [PubMed] [Google Scholar]
- Dadarkar SS, Fonseca LC, Thakkar AD, Mishra PB, Rangasamy AK, Padigaru M. Effect of nephrotoxicants and hepatotoxicants on gene expression profile in human peripheral blood mononuclear cells. Biochem Biophys Res Commun. 2010b;401:245–250. doi: 10.1016/j.bbrc.2010.09.039. [DOI] [PubMed] [Google Scholar]
- Ember I, Pusztai Z, Gyongyi Z, Kiss I. 1-Nitropyrene induces elevated expression of oncogenes and tumor suppressor genes 24 hours after treatment in CBA/Ca mice. Anticancer Res. 2000;20:1563–1566. [PubMed] [Google Scholar]
- Fannin RD, Russo M, O’Connell TM, Gerrish K, Winnike JH, Macdonald J, Newton J, Malik S, Sieber SO, Parker J, Shah R, Zhou T, Watkins PB, Paules RS. Acetaminophen dosing of humans results in blood transcriptome and metabolome changes consistent with impaired oxidative phosphorylation. Hepatology. 2010;51:227–236. doi: 10.1002/hep.23330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyongyi Z, Ember I, Kiss I, Varga C. Changes in expression of onco-and suppressor genes in peripheral leukocytes--as potential biomarkers of chemical carcinogenesis. Anticancer Res. 2001;21:3377–3380. [PubMed] [Google Scholar]
- Gyorffy E, Anna L, Kovacs K, Rudnai P, Schoket B. Correlation between biomarkers of human exposure to genotoxins with focus on carcinogen-DNA adducts. Mutagenesis. 2008;23:1–18. doi: 10.1093/mutage/gem043. [DOI] [PubMed] [Google Scholar]
- Hagmar L, Tornqvist M, Nordander C, Rosen I, Bruze M, Kautiainen A, Magnusson AL, Malmberg B, Aprea P, Granath F, Axmon A. Health effects of occupational exposure to acrylamide using hemoglobin adducts as biomarkers of internal dose. Scand J Work Environ Health. 2001;27:219–226. doi: 10.5271/sjweh.608. [DOI] [PubMed] [Google Scholar]
- Hamadeh HK, Bushel PR, Jayadev S, DiSorbo O, Bennett L, Li L, Tennant R, Stoll R, Barrett JC, Paules RS, Blanchard K, Afshari CA. Prediction of compound signature using high density gene expression profiling. Toxicol Sci. 2002a;67:232–240. doi: 10.1093/toxsci/67.2.232. [DOI] [PubMed] [Google Scholar]
- Hamadeh HK, Bushel PR, Jayadev S, Martin K, DiSorbo O, Sieber S, Bennett L, Tennant R, Stoll R, Barrett JC, Blanchard K, Paules RS, Afshari CA. Gene expression analysis reveals chemical-specific profiles. Toxicol Sci. 2002b;67:219–231. doi: 10.1093/toxsci/67.2.219. [DOI] [PubMed] [Google Scholar]
- Hamadeh HK, Knight BL, Haugen AC, Sieber S, Amin RP, Bushel PR, Stoll R, Blanchard K, Jayadev S, Tennant RW, Cunningham ML, Afshari CA, Paules RS. Methapyrilene toxicity: anchorage of pathologic observations to gene expression alterations. Toxicol Pathol. 2002c;30:470–482. doi: 10.1080/01926230290105712. [DOI] [PubMed] [Google Scholar]
- Heinloth AN, Irwin RD, Boorman GA, Nettesheim P, Fannin RD, Sieber SO, Snell ML, Tucker CJ, Li L, Travlos GS, Vansant G, Blackshear PE, Tennant RW, Cunningham ML, Paules RS. Gene expression profiling of rat livers reveals indicators of potential adverse effects. Toxicol Sci. 2004;80:193–202. doi: 10.1093/toxsci/kfh145. [DOI] [PubMed] [Google Scholar]
- Hershey AD, Tang Y, Powers SW, Kabbouche MA, Gilbert DL, Glauser TA, Sharp FR. Genomic abnormalities in patients with migraine and chronic migraine: preliminary blood gene expression suggests platelet abnormalities. Headache. 2004;44:994–1004. doi: 10.1111/j.1526-4610.2004.04193.x. [DOI] [PubMed] [Google Scholar]
- Heubi JE. Measurement of serum acetaminophen-protein adducts in patients with acute liver failure. J Pediatrp Gastoenterol Nutr. 2007;44:513–515. doi: 10.1097/MPG.0b013e3180399464. [DOI] [PubMed] [Google Scholar]
- Huang L, Heinloth AN, Zeng ZB, Paules RS, Bushel PR. Genes related to apoptosis predict necrosis of the liver as a phenotype observed in rats exposed to a compendium of hepatotoxicants. BMC Genomics. 2008;9:288. doi: 10.1186/1471-2164-9-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Shi W, Zhang J, Chou JW, Paules RS, Gerrish K, Li J, Luo J, Wolfinger RD, Bao W, Chu TM, Nikolsky Y, Nikolskaya T, Dosymbekov D, Tsyganova MO, Shi L, Fan X, Corton JC, Chen M, Cheng Y, Tong W, Fang H, Bushel PR. Genomic indicators in the blood predict drug-induced liver injury. Pharmacogenomics J. 2010;10:267–277. doi: 10.1038/tpj.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard AK, Timblin CR, Shukla A, Rincon M, Mossman BT. Activation of NF-kappaB-dependent gene expression by silica in lungs of luciferase reporter mice. Am J Physiol Lung Cell Mol Physiol. 2002;282:L968–975. doi: 10.1152/ajplung.00327.2001. [DOI] [PubMed] [Google Scholar]
- IARC. Monograph on the Evaluation of Carcinogenic Risk to Human. Vol. 68. International Agency for Research on Cancer; Lyon, France: 1997. pp. 1–475. [Google Scholar]
- Irwin RD, Boorman GA, Cunningham ML, Heinloth AN, Malarkey DE, Paules RS. Application of toxicogenomics to toxicology: basic concepts in the analysis of microarray data. Toxicol Pathol. 2004;32 (Suppl 1):72–83. doi: 10.1080/01926230490424752. [DOI] [PubMed] [Google Scholar]
- Iseri S, Ercan F, Gedik N, Yuksel M, Alican I. Simvastatin attenuates cisplatin-induced kidney and liver damage in rats. Toxicology. 2007;230:256–264. doi: 10.1016/j.tox.2006.11.073. [DOI] [PubMed] [Google Scholar]
- Kim JK, Jung KH, Noh JH, Eun JW, Bae HJ, Xie HJ, Jang JJ, Ryu JC, Park WS, Lee JY, Nam SW. Identification of characteristic molecular signature for volatile organic compounds in peripheral blood of rat. Toxicol Appl Pharmacol. 2011;250:162–169. doi: 10.1016/j.taap.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Konig R, Ca P, Guo X, Ansari GA. Transcriptomic analysis reveals early signs of liver toxicity in female MRL +/+ mice exposed to the acylating chemicals dichloroacetyl chloride and dichloroacetic anhydride. Chem Res Toxicol. 2008;21:572–582. doi: 10.1021/tx7002728. [DOI] [PubMed] [Google Scholar]
- Lacour S, Gautier JC, Pallardy M, Roberts R. Cytokines as potential biomarkers of liver toxicity. Cancer Biomark. 2005;1:29–39. doi: 10.3233/cbm-2005-1105. [DOI] [PubMed] [Google Scholar]
- Linch KD, Miller WE, Althouse RB, Groce DW, Hale JM. Surveillance of respirable crystalline silica dust using OSHA compliance data (1979–1995) Am J Ind Med. 1998;34:547–558. doi: 10.1002/(sici)1097-0274(199812)34:6<547::aid-ajim2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Lobenhofer EK, Auman JT, Blackshear PE, Boorman GA, Bushel PR, Cunningham ML, Fostel JM, Gerrish K, Heinloth AN, Irwin RD, Malarkey DE, Merrick BA, Sieber SO, Tucker CJ, Ward SM, Wilson RE, Hurban P, Tennant RW, Paules RS. Gene expression response in target organ and whole blood varies as a function of target organ injury phenotype. Genome Biol. 2008;9:R100. doi: 10.1186/gb-2008-9-6-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter B, Guza R, Zhao J, Li ZZ, Jones R, Tretyakova N. Sequence distribution of acetaldehyde-derived N2-ethyl-dG adducts along duplex DNA. Chem Res Toxicol. 2007;20:1379–1387. doi: 10.1021/tx7001146. [DOI] [PubMed] [Google Scholar]
- McHale CM, Zhang L, Hubbard AE, Zhao X, Baccarelli A, Pesatori AC, Smith MT, Landi MT. Microarray analysis of gene expression in peripheral blood mononuclear cells from dioxin-exposed human subjects. Toxicology. 2007;229:101–113. doi: 10.1016/j.tox.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Mendrick DL. Genomic and genetic biomarkers of toxicity. Toxicology. 2008;245:175–181. doi: 10.1016/j.tox.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Moore DF, Li H, Jeffries N, Wright V, Cooper RA, Jr, Elkahloun A, Gelderman MP, Zudaire E, Blevins G, Yu H, Goldin E, Baird AE. Using peripheral blood mononuclear cells to determine a gene expression profile of acute ischemic stroke: a pilot investigation. Circulation. 2005;111:212–221. doi: 10.1161/01.CIR.0000152105.79665.C6. [DOI] [PubMed] [Google Scholar]
- NIOSH (National Insitute for Occupational Safety and Health) Hazard Review: Health Effects of Occupational Exposures to Crystalline Silica. US Department of Health and Human Services (NIOSH); 2002. [accessed on 15 October 2012]. Publication No. 2002-129. URL http://www/cdc/gov/niosh/docs/2002-129/02-129a.html. [Google Scholar]
- Nuwaysir EF, Bittner M, Trent J, Barrett JC, Afshari CA. Microarrays and toxicology: the advent of toxicogenomics. Mol Carcinog. 1999;24:153–159. doi: 10.1002/(sici)1098-2744(199903)24:3<153::aid-mc1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Olden K. Genomics in environmental health research--opportunities and challenges. Toxicology. 2004;198:19–24. doi: 10.1016/j.tox.2004.01.015. [DOI] [PubMed] [Google Scholar]
- Parks CG, Conrad K, Cooper GS. Occupational exposure to crystalline silica and autoimmune disease. Environ Health Perspect. 1999;107(Suppl 5):793–802. doi: 10.1289/ehp.99107s5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patlolla AK, Tchounwou PB. Serum acetyl cholinesterase as a biomarker of arsenic induced neurotoxicity in sprague-dawley rats. Int J Environ Res Public Health. 2005;2:80–83. doi: 10.3390/ijerph2005010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennie WD, Tugwood JD, Oliver GJ, Kimber I. The principles and practice of toxigenomics: applications and opportunities. Toxicol Sci. 2000;54:277–283. doi: 10.1093/toxsci/54.2.277. [DOI] [PubMed] [Google Scholar]
- Peretz A, Peck EC, Bammler TK, Beyer RP, Sullivan JH, Trenga CA, Srinouanprachnah S, Farin FM, Kaufman JD. Diesel exhaust inhalation and assessment of peripheral blood mononuclear cell gene transcription effects: an exploratory study of healthy human volunteers. Inhal Toxicol. 2007;19:1107–1119. doi: 10.1080/08958370701665384. [DOI] [PubMed] [Google Scholar]
- Porter DW, Ye J, Ma J, Barger M, Robinson VA, Ramsey D, McLaurin J, Khan A, Landsittel D, Teass A, Castranova V. Time course of pulmonary response of rats to inhalation of crystalline silica: NF-kappa B activation, inflammation, cytokine production, and damage. Inhal Toxicol. 2002;14:349–367. doi: 10.1080/08958370252870998. [DOI] [PubMed] [Google Scholar]
- Rockett JC. Blood derived transcritpomic profiles as a means to monitor levels of toxicant exposure and the effects of toxicants on inaccessible target tissues. In: Burczynski ME, Rockett JC, editors. Surrogate Tissue Analysis. Taylor & Francis; Bocca Raton: 2006. pp. 65–76. [Google Scholar]
- Rockett JC, Kavlock RJ, Lambright CR, Parks LG, Schmid JE, Wilson VS, Wood C, Dix DJ. DNA arrays to monitor gene expression in rat blood and uterus following 17beta-estradiol exposure: biomonitoring environmental effects using surrogate tissues. Toxicol Sci. 2002;69:49–59. doi: 10.1093/toxsci/69.1.49. [DOI] [PubMed] [Google Scholar]
- Rockett JC, Kim SJ. Biomarkers of reproductive toxicity. Cancer Biomark. 2005;1:93–108. doi: 10.3233/cbm-2005-1110. [DOI] [PubMed] [Google Scholar]
- Rockett JC, Burczynski ME. Introduction to surrogate tissue analysis. In: Burczynski ME, Rockett JC, editors. Surrogate Tissue Analysis. Taylor & Francis; Bocca Raton: 2006. pp. 3–11. [Google Scholar]
- Sanderson W. The U.S. population-at-risk to occupational respiratory diseases. In: Merchan JA, editor. Occupational Respiratory Diseases. Department of Health and Human Services (NIOSH); Washington: 1986. Publication 86–102. [Google Scholar]
- Santarelli L, Recchioni R, Moroni F, Marcheselli F, Governa M. Crystalline silica induces apoptosis in human endothelial cells in vitro. Cell Biol Toxicol. 2004;20:97–108. doi: 10.1023/b:cbto.0000027935.45070.75. [DOI] [PubMed] [Google Scholar]
- Sellamuthu R, Umbright C, Roberts JR, Chapman R, Young SH, Richardson D, Leonard H, McKinney W, Chen B, Frazer D, Li S, Kashon M, Joseph P. Blood gene expression profiling detects silica exposure and toxicity. Toxicol Sci. 2011a;122:253–264. doi: 10.1093/toxsci/kfr125. [DOI] [PubMed] [Google Scholar]
- Sellamuthu R, Umbright C, Li S, Kashon M, Joseph P. Mechanisms of crystalline silica-induced pulmonary toxicity revealed by global gene expression profiling. Inhal Toxicol. 2011b;23:927–937. doi: 10.3109/08958378.2011.625995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellamuthu R, Umbright C, Roberts JR, Chapman R, Young SH, Richardson D, Cumpston J, McKinney W, Chen BT, Frazer D, Li S, Kashon M, Joseph P. Transcriptomics analysis of lungs and peripheral blood of crystalline silica-exposed rats. Inhal Toxicol. 2012a;24:570–579. doi: 10.3109/08958378.2012.697926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellamuthu R, Umbright C, Roberts JR, Cumpston A, McKinney W, Chen BT, Frazer D, Li S, Kashon M, Joseph P. Molecular insights into the progression of crystalline silica-induced pulmonary toxicity in rats. J App Toxicol. 2012b doi: 10.1002/jat.2733. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellamuthu R, Umbright C, Roberts JR, Young SH, Richardson D, McKinney W, Chen B, Frazer D, Li S, Kashon M, Joseph P. Pulmonary Toxicity and Global Gene Expression Profile in Response to Crystalline Silica Exposure in Rats. Society of Toxicology Annual Meeting; San Antonio, TX. March 10–14, 2013.2013. [Google Scholar]
- Simmons JE, Evans MV, Boyes WK. Moving from external exposure concentration to internal dose: duration extrapolation based on physiologically based pharmacokinetic derived estimates of internal dose. J Toxciol Environ Health A. 2005;68:927–950. doi: 10.1080/15287390590912586. [DOI] [PubMed] [Google Scholar]
- Steenland K, Mannetje A, Boffetta P, Stayner L, Attfield M, Chen J, Dosemeci M, DeKlerk N, Hnizdo E, Koskela R, Checkoway H International Agency for Research on Cancer. Pooled exposure-response analyses and risk assessment for lung cancer in 10 cohorts of silica-exposed workers: an IARC multicentre study. Cancer Causes Control. 2001;12:773–784. doi: 10.1023/a:1012214102061. [DOI] [PubMed] [Google Scholar]
- Steiner G, Suter L, Boess F, Gasser R, de Vera MC, Albertini S, Ruepp S. Discriminating different classes of toxicants by transcript profiling. Environ Health Perspect. 2004;112:1236–1248. doi: 10.1289/txg.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran S, Xue B, Jusko WJ, Dubois DC, Almon RR. Circadian variations in gene expression in rat abdominal adipose tissue and relationship to physiology. Physiol Genomics. 2010;42A:141–152. doi: 10.1152/physiolgenomics.00106.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Gilbert DL, Glauser TA, Hershey AD, Sharp FR. Blood gene expression profiling of neurologic diseases: a pilot microarray study. Arch Neurol. 2005;62:210–215. doi: 10.1001/archneur.62.2.210. [DOI] [PubMed] [Google Scholar]
- Thomas RS, Rank DR, Penn SG, Zastrow GM, Hayes KR, Pande K, Glover E, Silander T, Craven MW, Reddy JK, Jovanovich SB, Bradfield CA. Identification of toxicologically predictive gene sets using cDNA microarrays. Mol Pharmacol. 2001;60:1189–1194. doi: 10.1124/mol.60.6.1189. [DOI] [PubMed] [Google Scholar]
- Thorne PS. Occupational Toxicology. In: Klaassen CD, editor. Casarett and Doull’s Toxicology, The Basic Science of Toxicology. 7. McGraw Hill; New York: 2007. pp. 1273–1292. [Google Scholar]
- Thukral SK, Nordone PJ, Hu R, Sullivan L, Galambos E, Fitzpatrick VD, Healy L, Bass MB, Cosenza ME, Afshari CA. Prediction of nephrotoxicant action and identification of candidate toxicity-related biomarkers. Toxicol Pathol. 2005;33:343–355. doi: 10.1080/01926230590927230. [DOI] [PubMed] [Google Scholar]
- Umbright C, Sellamuthu R, Li S, Kashon M, Luster M, Joseph P. Blood gene expression markers to detect and distinguish target organ toxicity. Mol Cell Biochem. 2010;335:223–234. doi: 10.1007/s11010-009-0272-5. [DOI] [PubMed] [Google Scholar]
- Vallyathan V, Leonard S, Kuppusamy P, Pack D, Chzhan M, Sanders SP, Zweir JL. Oxidative stress in silicosis: evidence for the enhanced clearance of free radicals from whole lungs. Mol Cell Biochem. 1997;168:125–132. doi: 10.1023/a:1006850920080. [DOI] [PubMed] [Google Scholar]
- Van de Sandt JJ, Dellarco M, Van Hemmen JJ. From dermal exposure to internal dose. J Expo Sci Environ Epidemiol. 2007;17(Suppl 1):S38–47. doi: 10.1038/sj.jes.7500579. [DOI] [PubMed] [Google Scholar]
- Wallace KB, Hausner E, Herman E, Holt GD, MacGregor JT, Metz AL, Murphy E, Rosenblum IY, Sistare FD, York MJ. Serum troponins as biomarkers of drug-induced cardiac toxicity. Toxicol Pathol. 2004;32:106–121. doi: 10.1080/01926230490261302. [DOI] [PubMed] [Google Scholar]
- Wang Z, Neuburg D, Li C, Su L, Kim JY, Chen JC, Christiani DC. Global gene expression profiling in whole-blood samples from individuals exposed to metal fumes. Environ Health Perspect. 2005;113:233–241. doi: 10.1289/txg.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring JF, Ciurlionis R, Jolly RA, Heindel M, Ulrich RG. Microarray analysis of hepatotoxins in vitro reveals a correlation between gene expression profiles and mechanisms of toxicity. Toxicol Lett. 2001a;120:359–368. doi: 10.1016/s0378-4274(01)00267-3. [DOI] [PubMed] [Google Scholar]
- Waring JF, Jolly RA, Ciurlionis R, Lum PY, Praestgaard JT, Morfitt DC, Buratto B, Roberts C, Schadt E, Ulrich RG. Clustering of hepatotoxins based on mechanism of toxicity using gene expression profiles. Toxicol Appl Pharmacol. 2001b;175:28–42. doi: 10.1006/taap.2001.9243. [DOI] [PubMed] [Google Scholar]
- Zhang L, Bushel PR, Chou J, Zhou T, Watkins PB. Identification of Identical Transcript Changes in Liver and Whole Blood during Acetaminophen Toxicity. Front Genet. 2012;3:162. doi: 10.3389/fgene.2012.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]