Abstract
Background
Study results on the prognostic value of CD11b for acute myeloid leukemia (AML) patients are inconsistent. An up-to-date meta-analysis was conducted to assess the prognostic value of CD11b expression level for AML patients.
Methods
Electronic databases including PubMed, Embase, Cochrane Library, Web of Science and Chinese BioMedical Literature Database (CBM) were searched to identify studies that investigated the association between CD11b expression level and prognosis of AML patients. Pooled hazard ratios (HRs) with 95% confidence intervals (CIs) for overall survival (OS) and disease-free survival (DFS) and pooled odds ratio (OR) with 95% CI for complete remission rate (CRR) were calculated using Revman 5.3 and Stata 11.0.
Results
13 total studies with 2619 patients were included in this meta-analysis. Results of the meta-analysis showed that CD11b positivity was associated with lower CRR (OR = 0.44; 95% CI, 0.25–0.79; p = 0.006) and shorter OS (HR = 0.66; 95% CI, 0.55–0.80; p < 0.0001), but did not affect DFS (HR = 0.67; 95% CI, 0.31–1.48; p = 0.32). Subgroup analysis by ethnicity, cut-off value for CD11b positivity, treatment, subtype and sample preparation method showed no significant interaction between these factors with the prognostic value of CD11b expression level for AML patients. Sensitivity analysis yielded consistent results with the main meta-analysis.
Conclusion
CD11b positivity could predict a poor prognosis for AML patients. Thus, CD11b expression level might be considered a prognostic biomarker for AML patients.
Introduction
Acute myeloid leukemia (AML) is the most common type of leukemia that affects adults, with a prevalence of 3.8 cases per 10,000 adults rising to 17.9 cases per 10,000 adults aged 65 years and older [1]. It is a heterogeneous clonal disorder of hematopoietic stem/progenitor cell which lose the ability to differentiate normally and to respond to normal regulators of proliferation and apoptosis, results in an accumulation of huge amount of immature blasts with variable degrees of myeloid differentiation in the bone marrow and peripheral blood [2,3]. Cell-cell interaction and cell-matrix interaction between AML cells and different tissue/cells is essential for leukemic engraftment, migration and infiltration [4–8]. These biological process are mediated by specific cell surface receptors [9,10].
Cluster of differentiation 11b (CD11b) is a kind of cell surface receptor that are selectively expressed on leukocytes, which is also named as integrin alpha M (ITGAM), complement component 3 receptor alpha chain (CR3a), macrophage-1 antigen alpha subunit or macrophage receptor 1 alpha subunit (MAC1a). In GENE database of national center for biotechnology information (NCBI), this protein is also named as systemic lupus erythematosus type 6 (SLEB6) or MO1A[11, 12,13]. It is one protein subunit that forms the heterodimeric integrin alpha-M beta-2 molecule with cluster of differentiation 18 (CD18), also named as macrophage-1 antigen or macrophage-1 antigen (Mac-1), complement receptor 3 (CR3)or MO1[11, 12,13]. This protein can participate in cell activation, chemotaxis, cytotoxicity, phagocytosis and regulates interaction of leukemic cells with microenvironment through binding to its ligands, such as inactivated complement component 3b (iC3b), intercellular adhesion molecule (ICAM), fibrinogen, beta-glukanes, coagulation factor X etc.[14–19]. Recently, CD11b is also defined as a marker for myeloid-derived suppressor cells, which is reported to be harnessed by malignant cells to restrain antitumor immunity and to promote malignant expansion or refractoriness to treatment [20–22]. So it is presumable that CD11b may participate in the regulation of biology of malignant AML cells and its expression level may affect the prognosis of AML patients.
Actually, CD11b expression level has been considered as an adverse prognostic factor in AML patients since the 90s [23,24]. AML expressing CD11b was even described as a new leukemic syndrome in 1998[25]. Until now, many studies have demonstrated that CD11b positivity is associated with poor prognosis of AML patients[26,27], but still some other studies yielded conflicting results[28], which means that the prognostic value of CD11b for AML patients is controversial. Therefore, we conducted this up-to-date meta-analysis by combining all published literature to assess the prognostic value of CD11b expression level for AML patients.
Materials and Methods
This work was carried out following the Cochrane Handbook of systematic reviews and was reported based on PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) statement [29].
Identification of relevant studies
The following electronic databases were systematically searched for relevant studies from inception to July 2015 without language restrictions: PubMed, Embase, Cochrane Library, Web of Science and Chinese BioMedical Literature Database (CBM). The detailed search strategies for each database are reported in S1 Table.
Study selection
Two authors independently estimated the eligibility of studies by screening the title and abstract of each article identified by above literature search. After excluding obviously irrelevant articles, full-texts were obtained and assessed by the same two authors independently. Disagreements were resolved by consensus.
The inclusion criteria included a) prospective and historical cohort studies; b) studies that evaluated the association between CD11b expression level and the prognosis of AML patients; c) studies that provided sufficient data to estimate hazard ratios (HRs) with 95% confidence intervals (CIs) for overall survival (OS) and disease-free survival (DFS) or odds ratio (OR) with 95% CI for complete remission rate (CRR). When multiple papers reported on the same study, only the most updated one was included.
Data extraction and quality assessment
Data were carefully extracted from all eligible studies independently by two authors including first author, publication year, region, study design, patients’ characteristics, CD11b detection method and predominant treatment regimen for patients.
Methodological quality was assessed by two authors according to the Newcastle-Ottawa Quality Assessment Scale (NOS) which was based on three categories: selection, comparability, and outcome. The full score was 9 points, and a high-quality study in our analysis was defined as a study with ≥7 points [30]. Any disagreement was resolved by consensus.
Statistical analysis
For time-to-event data, OS and DFS, the log HRs and their standard errors were directly extracted from the published articles or indirectly calculated from the reported events and the p value in the log-rank test or from the published Kaplan-Meier curves [31, 32]. We pooled the log HRs and corresponding 95% CIs across studies with the generic inverse-variance method and the weight for each study was calculated by the inverse variances of their effect estimates [33]. For dichotomous data, CRR, we extracted events in each arm and calculated OR and corresponding 95% CI. The Mantel-Haenszel method was used to pool ORs and 95% CIs across studies and the weight for each study was calculated on the size of the study and the number of events [34].
Statistical heterogeneity between studies was assessed by χ2 based Q test with a significant level at p < 0.1 and quantified with I 2 statistic (I 2 = 0–25%: no heterogeneity; I 2 = 25–50%: moderate heterogeneity; I 2 = 50–75%: large heterogeneity; I 2 = 75–100%: extreme heterogeneity) [35]. Fixed-effect model was chosen for summary estimation if heterogeneity was not significant, whereas random-effects model was adopted if heterogeneity was significant. Subgroup analysis and meta-regression were performed to assess the influence of study region, cut-off value for CD11b positivity, treatment, subtype and sample preparation method on the prognostic value of CD11b expression level in patient with AML. Publication bias was assessed using funnel plots [36].
All analyses were conducted in Review Manager Version 5.3 (Revman, the Cochrane Collaboration, Oxford, England) and Stata version 11.0 (STATA Crop, College Station, Texas). A two-sided p-value of ≤ 0.05 was considered significant for all analyses except heterogeneity tests.
Results
Basic characteristics and methodological quality of eligible studies
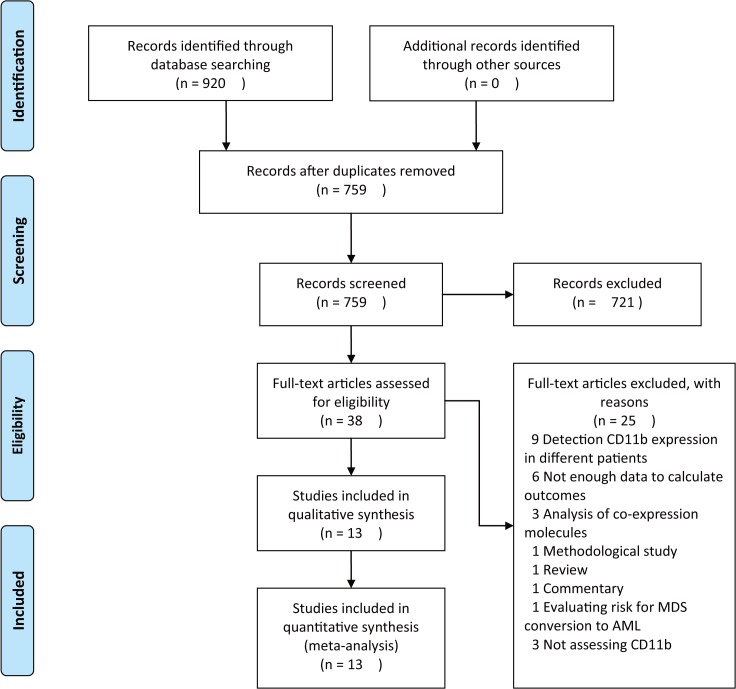
The flow chart of literature search and selection was shown in Fig 1. Totally, 917 reports were retrieved and 13 studies with 2619 patients were eligible for the meta-analysis after screening title and abstract and reviewing the full-text articles [23–28, 37–43]. The main characteristics of the included studies are shown in Table 1. All the included studies were aiming to investigate the prognostic value of CD11b expression level for AML patients. 11 studies suggested that CD11b positivity is associated poor prognosis of AML patents [23–27, 37–41, 43], but two studies yielded conflicting results [28, 42]. Among them, 10 studies [24, 25, 27, 28, 38–43] reported results of CRR, five studies [24, 26–28, 38] reported results of OS and three studies [23, 27, 28] reported results of DFS. Nine studies [23, 24, 26, 28, 37–39, 41, 42] are prospective cohort studies and four studies [25, 27, 40, 43] are retrospective cohort studies. Five studies were conducted in western countries [23–25, 27, 38] and eight studies were conducted in eastern countries [26, 28, 37, 39–43]. Patients in ten studies were treated by standard chemotherapy [23–25, 27, 28, 37–39, 42, 43], patients in two studies were treated by hematopoietic stem cell transplantation (HSCT) [26, 41] and the treatment strategy was not reported in one study [40]. Eight studies [23,27,37–39,41–43] defined positivity of CD11b by a cut-off value of 20%, one study [28] defined positivity of CD11b by a cut-off value of 30%, one study [25] defined positivity of CD11b by a cut-off value of 32%, and the cut-off value for CD11b positivity was not available in the other three studies [24,26,40]. Five studies [23, 28, 37, 39, 42] enrolled all AML patients, one study only enrolled AML-M5 patients [41], two studies excluded AML-M3 patients [38, 43], and the subtype information was not available in the other five studies [24–27, 40]. Four studies [23–25, 28] adopted ficoll-hypaque gradient centrifugation (FHGC) as sample preparation method, three studies [27, 38, 42] adopted red blood cell lysis as sample preparation method, and the other five studies did not reported information about sample preparation method [26, 37, 39–41]. Six studies [25,27,38,41–43] reported the equipment used for detection of CD11b positivity, one study [37] adopted varied equipment because different research centers uses different flow cytometers, and the other six studies [25,26,28,37,39,40] did not reported specific information about equipment used. Seven studies [23, 25, 27, 28, 41] reported the source of antibody used, one study [37] adopted varied antibodies because different research centers uses different antibodies, the other six studies [23, 26, 37, 38, 40] did not reported specific information about antibody used. The score of quality assessment ranges from 5 to 9, and the detailed scoring items of the included 13 studies were shown in Table 2.
Fig 1. Flow chart of study selection and identification.
Table 1. Basic characteristics of includes studies.
| First author | Albitar et al | Amirghofran et al | Bradstock et al | Chen et al | Chen et al | Junca et al | Liang et al | Paietta et al | Tucker et al | Xu et al | Xu et al | Yang et al | Zhuang et al |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Publication year | 2011 | 2001 | 1994 | 2013a | 2013b | 2014 | 2001 | 1998 | 1990 | 2006 | 2009 | 2014 | 2011 |
| Region | S.A | Iran | Australia | China | Canada | Spain | China | America | UK | China | China | China | China |
| Study Design | Prospective cohort | Prospective cohort | Prospective cohort | Prospective cohort | Prospective cohort | Retrospective cohort | Prospective cohort | Retrospective cohort | Prospective cohort | Retrospective cohort | Prospective cohort | Prospective cohort | Retrospective cohort |
| No. of Patients | 62 | 70 | 120 | 510 | 233 | 158 | 80 | 382 | 92 | 136 | 113 | 516 | 147 |
| Gender(M/F) | NA | NA | NA | 295/215 | 137/96 | 78/80 | 51/29 | 214/168 | 53/39 | NA | 62/51 | 290/226 | 76/71 |
| Age(median, range; years) | 8 (0.7–14) | 32.7(10–70) | (15–60) | 36(12–83) | 61(18–90) | 56(14–78) | 37(11–67) | 45(15–78) | 42(15–65) | Over 18 | 375(16–68) | 17–88 | 54(15–89) |
| WBC (median, range), 109/L | NA | 61(0.7–650) | NA | NA | 5(0–606) | NA | NA | NA | 33(0–235) | NA | NA | NA | NA |
| Sample type | NA | PB or BM | PB or BM | BM | PB or BM | PB or BM | BM | PB or BM | PB | NA | BM | BM | BM |
| Sample preparation method | NA | FHGC | FHGC | NA | red blood cell lysis | red blood cell lysis | NA | FHGC | FHGC | NA | NA | red blood cell lysis | NA |
| Detection method | FL | FL or IF | FL | FL | FL | FL | APAAP | FL | FL | FL | FL | FL | FL |
| Equipment | NA | NA | Varied* | NA | EPICS XL-MCL | EPICS XL-MCL | NA | NA | Coulter Epics C | NA | FACS Calibur | FACSCanto II | FACS Calibur |
| Source of antibody | NA | Dako | Varied* | NA | NA | Beckman Coulter | BD | BD | NA | NA | ebioscience | BD | BD |
| Cut off value | NA | 30% | NA | 20% | 20% | 20% | 20% | 32% | 20% | NA | 20% | 20% | 20% |
| Dynamic range | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| CD11b+ patients(cases, percent) | 25(40%) | 44(62.9%) | 40(33%) | 23% | 145(70%) | 53(36%) | 24(30%) | 95(25%) | 48(52%) | 71(52.2%) | 83(73.45) | 123(23.8%) | 65(44.2%) |
| FAB type | |||||||||||||
| M0 | NA | 0 | NA | 0 | 23 | NA | 0 | NA | 0 | NA | 0 | 10 | 0 |
| M1 | NA | 12 | NA | 26 | 37 | NA | 11 | NA | 31 | NA | 0 | 34 | 3 |
| M2 | NA | 12 | NA | 147 | 44 | NA | 21 | NA | 19 | NA | 0 | 146 | 107 |
| M3 | NA | 9 | NA | 78 | 0 | NA | 15 | NA | 5 | NA | 0 | 111 | 0 |
| M4 | NA | 30 | NA | 60 | 22 | NA | 13 | NA | 19 | NA | 0 | 49 | 7 |
| M5 | NA | 4 | NA | 109 | 25 | NA | 18 | NA | 14 | NA | 113 | 139 | 20 |
| M6 | NA | 3 | NA | 10 | 4 | NA | 2 | NA | 3 | NA | 0 | 26 | 9 |
| M7 | NA | 0 | NA | 5 | 7 | NA | 0 | NA | 0 | NA | 0 | 1 | 1 |
| Unidentified | NA | NA | NA | 75 | 71 | NA | 0 | NA | 0 | NA | 0 | 0 | 0 |
| Cytogenetics | |||||||||||||
| Favorable | 0 | NA | NA | NA | 0 | 21 | NA | NA | NA | NA | NA | NA | NA |
| Intermediate | 46 | NA | NA | NA | 0 | 97 | NA | NA | NA | NA | NA | NA | NA |
| Unfavorable | 16 | NA | NA | NA | 233 | 33 | NA | NA | NA | NA | NA | NA | NA |
| Treatment (predominant) | HSCT | Standard CT | Standard CT | Standard CT | Standard CT | Standard CT | Standard CT | Standard CT | Standard CT | NA | HSCT | Standard CT | Standard CT |
APAAP = alkaline phosphatase-anti-alkaline phosphatase complex method, HSCT = Hematopoietic stem cell transplantation, S.A = Saudi Arabia, CT = chemotherapy, NA = DATA not available, FHGC = ficoll-hypaque gradient centrifugation, BD = Becton Dickinson.
* means the equipment or antibody varied between different research centers in this study.
Table 2. The assessment of the risk of bias in each cohort study using the Newcastle-ottawa scale.
| Study | Selection | Comparability | Outcome | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| REC | SNEC | AE | DO | SC | AF | AO | FU | AFU | ||
| Albitar 2011 | * | * | - | * | - | - | * | - | * | 5 |
| Amirghofran 2001 | * | * | * | * | - | - | * | * | * | 7 |
| Bradstock 1994 | * | * | - | * | - | - | * | * | * | 6 |
| Chen 2013a | * | * | * | * | - | - | * | * | * | 7 |
| Chen 2013b | * | * | * | * | - | - | * | * | * | 7 |
| Junca 2014 | * | * | * | * | - | - | * | * | * | 7 |
| Liang 2001 | * | * | * | * | * | * | * | * | * | 9 |
| Paietta 1998p | * | * | * | * | - | - | * | * | * | 7 |
| Tucker 1990 | * | * | * | * | - | - | * | * | * | 7 |
| Xu 2006 | * | * | - | * | - | - | * | * | * | 6 |
| Xu 2009 | * | * | * | * | * | - | * | * | * | 8 |
| Yang 2014 | * | * | * | * | - | - | * | * | * | 7 |
| Zhang 2011 | * | * | * | * | - | - | * | * | * | 7 |
REC = representativeness of the exposed cohort, SNEC = selection of the nonexposed cohort, AE = ascertainment of exposure, DO = demonstration that outcome of interest was not present at start of study, SC = study controls for age, subtype, AF = study controls for white blood cell number at diagnosis and treatment, AO = assessment of outcome, FU = follow-up long enough for outcomes to occur (for studies that only assessed CR, ‘long enough’ is defined as 6 month, for studies that assessed survival data, ‘long enough’ is defined as 3 years), AFU = adequacy of follow-up of cohorts (≥80%).
“*” means that the study is satisfied the item and “-” means the opposite situation.
CD11b expression level and CRR of AML patients
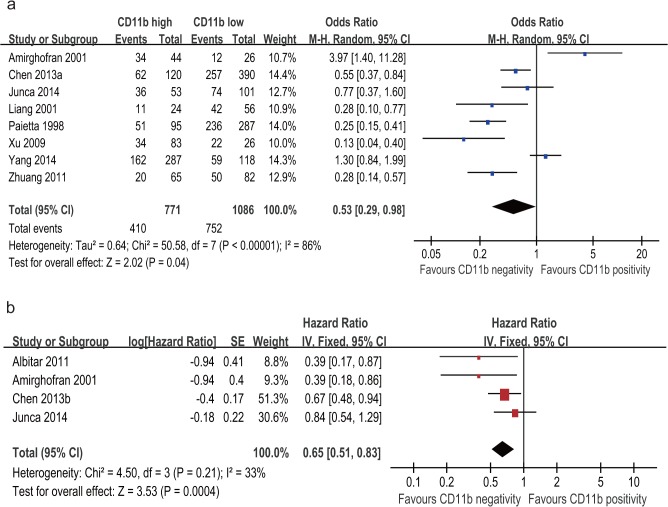
10 studies with 2078 patients assessed the association of CD11b expression level with CRR in AML. The event in each group is defined as acquirement of complete remission for AML patients. The result of meta-analysis for CRR showed that patients with CD11b positivity had a significantly decreased CRR compared with patients with CD11b negativity (OR = 0.44; 95% CI, 0.25–0.79; p = 0.006; Fig 2) although with significant heterogeneity among the studies (I2 = 86%; p < 0.00001).
Fig 2. Forest plot for the association between CD11b expression level and complete remission rate (CRR) of AML patients.
Subgroup analysis showed no significant interaction between the CRR effect of CD11b expression with study country, cut-off value for CD11b positivity, treatment, subtype and sample preparation method (Table 3).
Table 3. Summary of subgroup analysis results for CD11b and prognosis of AML patients.
| Subgroup | Sample size | Effect measures | Heterogeneity | Meta-regression | |||
|---|---|---|---|---|---|---|---|
| HR/OR (95% CI) | p-value | I2(%) | p-value | p-value | |||
| CR | |||||||
| Country | Western | 656 | 0.43(0.21, 0.89) | 0.02 | 70 | 0.04 | 0.98 |
| Eastern | 1422 | 0.44 (0.20, 0.98) | 0.04 | 89 | <0.0001 | ||
| Cut-off value | 20% | 1475 | 0.60 (0.31, 1.15) | 0.12 | 84 | <0.0001 | 0.10 |
| 32% | 382 | 0.25 (0.15, 0.41) | <0.0001 | NA | NA | ||
| NA | 221 | 0.25 (0.04, 1.08) | 0.01 | 86 | <0.0001 | ||
| Treatment | HSCT | 109 | 0.13 (0.04, 0.40) | 0.0004 | NA | NA | 0.04 |
| Standard CT | 1969 | 0.50(0.28,0.90) | 0.02 | 86 | <0.0001 | ||
| Subtype | AML as a whole | 1822 | 0.53 (0.28, 1.02) | 0.06 | 87 | <0.0001 | 0.08 |
| AML without M3 | 147 | 0.28 (0.14, 0.57) | 0.0003 | NA | NA | ||
| AML-M5 | 109 | 0.13 (0.04, 0.40) | 0.0004 | NA | NA | ||
| Sample preparation method | FHGC | 572 | 0.74(0.16,3.35) | 0.70 | 91 | <0.0001 | 0.19 |
| red blood cell lysis | 706 | 0.68(0.27,1.68) | 0.40 | 85 | 0.001 | ||
| NA | 800 | 0.22(0.08, 0.57) | 0.002 | 81 | 0.001 | ||
| OS | |||||||
| Country | Western | 511 | 0.71 (0.58, 0.87) | 0.001 | 0 | 0.71 | 0.05 |
| Eastern | 132 | 0.33 (0.22, 0.68) | 0.001 | 0 | 1.00 | ||
| Cut-off value | 20% | 391 | 0.73 (0.56, 0.95) | 0.02 | 0 | 0.43 | 0.32 |
| 30% | 70 | 0.39 (0.18, 0.86) | 0.02 | NA | NA | ||
| NA | 182 | 0.64 (0.48, 0.86) | 0.003 | 40 | 0.20 | ||
| Treatment | HSCT | 62 | 0.39 (0.17, 0.87) | 0.02 | NA | NA | 0.18 |
| Standard CT | 581 | 0.69 (0.56, 0.83) | 0.0002 | 0 | 0.42 | ||
| Subtype | AML as a whole | 410 | 0.66 (0.53, 0.83) | 0.0005 | 35 | 0.21 | 0.95 |
| AML without M3 | 233 | 0.67 (0.48, 0.94) | 0.02 | NA | NA | ||
| Sample preparation method | FHGC | 190 | 0.64 (0.48, 0.35) | 0.003 | 43 | 0.19 | 0.33 |
| red blood cell lysis | 391 | 0.73 (0.56, 0.95) | 0.02 | 0 | 0.43 | ||
| NA | 62 | 0.39 (0.17, 0.87) | 0.01 | NA | NA | ||
95% CI = 95% confidence interval, DFS = disease-free survival, HR = hazard ratio, NA = data not available, OR = odds ratio, OS = overall survival.
CD11b expression level and OS of AML patients
Five studies with 643 patients assessed the association of CD11b expression level with OS in AML. The result of meta-analysis for OS showed that patients with CD11b positivity had a significantly shorter OS compared with patients with CD11b negativity (HR = 0.66; 95% CI, 0.55–0.80; p < 0.00001; Fig 3) with no significant heterogeneity among the studies (I2 = 13%; p = 0.33).
Fig 3. Forest plot for the association between CD11b expression level and overall survival (OS) of AML patients.

Subgroup analysis showed no significant interaction between the OS effect of CD11b expression with study country, cut-off value for CD11b positivity, treatment, subtype and sample preparation method (Table 3).
CD11b expression level and DFS of AML patients
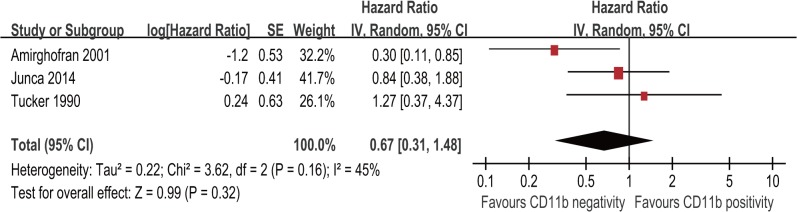
Three studies with 320 patients assessed the association of CD11b expression level with DFS in AML. The result of meta-analysis for DFS showed that patients with CD11b positivity had a similar DFS compared with patients with CD11b negativity (HR = 0.67; 95% CI, 0.31–1.48; p = 0.32, Fig 4) with no significant heterogeneity among the studies (I2 = 45%; p = 0.16). Since only three studies were included in this meta-analysis, subgroup analysis was not conducted.
Fig 4. Forest plot for the association between CD11b expression level and disease-free survival (DFS) of AML patients.
Sensitivity analysis and publication bias
A sensitivity analysis for CRR and OS was conducted by only including high NOS score studies to assess the effect of study quality on the stability of this meta-analysis, the results of sensitivity analysis is consistent with the main meta-analysis, suggesting that the results of this meta-analysis is reliable (Fig 5). Since all studies included in the meta-analysis for DFS are with high quality, so we didn’t perform this sensitivity analysis for this outcome.
Fig 5. Forest plot for sensitivity analysis by only including high quality score studies for the association between CD11b expression level and CRR of AML patients (a) and for the association between CD11b expression level and OS of AML patients (b).
Another sensitivity analysis, in which one study was removed at a time, was also conducted. The pooled HRs or ORs were not significantly changed, further indicating the stability of our analyses (Table 4).
Table 4. Sensitivity analysis by omitting each of the included studies in different outcomes.
| Outcomes | Omitted Study | HR or OR | 95% CI | P | I2(%) | Ph |
|---|---|---|---|---|---|---|
| CR | Amirghofran 2001 | 0.36 | 0.21–0.63 | 0.0003 | 84% | <0.00001 |
| Bradstock 1994 | 0.44 | 0.23–0.83 | 0.01 | 88% | <0.00001 | |
| Chen 2013a | 0.43 | 0.21–0.86 | 0.02 | 88% | <0.00001 | |
| Junca 2014 | 0.42 | 0.22–0.79 | 0.007 | 87% | <0.00001 | |
| Liang 2001 | 0.46 | 0.25–0.86 | 0.02 | 87% | <0.00001 | |
| Paietta 1998 | 0.48 | 0.26–0.89 | 0.02 | 86% | <0.00001 | |
| Xu 2006 | 0.52 | 0.30–0.92 | 0.02 | 84% | <0.00001 | |
| Xu 2009 | 0.47 | 0.25–0.88 | 0.02 | 87% | <0.00001 | |
| Yang 2014 | 0.39 | 0.22–0.68 | 0.001 | 81% | <0.00001 | |
| Zhang 2011 | 0.50 | 0.28–0.90 | 0.02 | 86% | <0.00001 | |
| OS | Albitar 2011 | 0.69 | 0.56–0.83 | 0.0002 | 0% | 0.42 |
| Amirghofran 2001 | 0.69 | 0.57–0.84 | 0.0002 | 0% | 0.44 | |
| Bradstock 1994 | 0.65 | 0.51–0.83 | 0.004 | 33% | 0.21 | |
| Chen 2013b | 0.66 | 0.53–0.83 | 0.0005 | 35% | 0.21 | |
| Junca 2014 | 0.63 | 0.51–0.78 | <0.0001 | 8% | 0.35 | |
| DFS | Amirghofran 2001 | 0.95 | 0.49–1.87 | 0.89 | 0% | 0.59 |
| Junca 2014 | 0.59 | 0.15–2.43 | 0.47 | 67% | 0.08 | |
| Tucker 1990 | 0.53 | 0.20–1.45 | 0.22 | 58% | 0.12 |
CRR = complete remission rate, DFS = disease free survival, HR = hazard ratio, Ph = p for heterogeneity, OR = odds ratio, OS = overall survival.
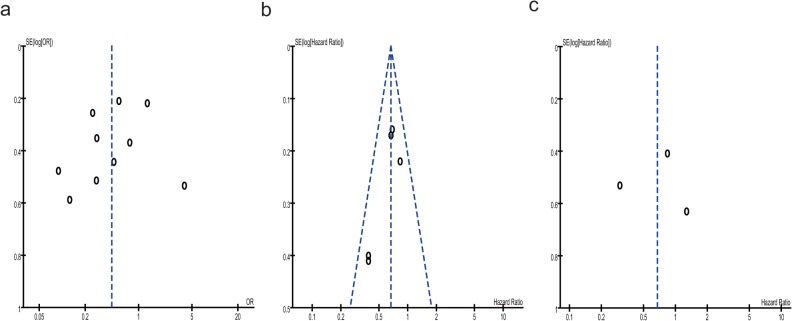
The funnel plots were largely symmetric suggesting that there were no publication biases in this meta-analysis of CD11b expression level and prognosis of AML patients (Fig 6).
Fig 6. The funnel plots were largely symmetric suggesting there were no publication biases in the meta-analysis of CD11b expression level and prognosis of AML patients.
The funnel plot from ten studies assessed the association between CD11b expression level and CRR of AML patients (a). The funnel plot from five studies assessed the association between CD11b expression level and OS of AML patients (b). The funnel plot from three studies assessing the association between CD11b expression level and DFS of AML patients (c).
Discussion
Although CD11b expression level has long been recognized with prognostic value for AML patients, the results are controversial between different studies. This may be attributed to the statistical limitation (e.g., small sample size) of individual study, different ethnicity of included participants, different antibody or equipment used or varied cut-off value for CD11b positivity. Thus, we performed this meta-analysis with subgroup analysis and sensitivity analysis to pool these relevant studies together to resolve this controversial issue and provide up-to-date clinical evidence for adopting CD11b expression level as a prognostic biomarker for AML patients.
To the best to our knowledge, this is the first meta-analysis that evaluates the role of CD11b expression level for predicting the prognosis of AML patients. Results of our meta-analysis showed that compared with AML patients with CD11b negativity, AML patients with CD11b positivity are associated with lower CRR, shorter OS, but has no significant effect on DFS.
Previous studies evaluating the prognostic role of CD11b expression level in AML patients have enrolled participants with different ethnicity and different subtypes, adopted varied cut-off value for CD11b positivity ranging from 20% to 32% and conducted different treatment for recruited participants. Thus, we undertook subgroup analyses according to these factors to investigate the interaction between these factors with the results of this meta-analysis. We also conducted sensitivity analyses by only including high quality score studies and by omitting each study. Results of different subgroup or sensitivity analyses are consistent with the main meta-analyses, indicating the results of this meta-analysis are reliable. Taken together, these results clearly demonstrated that CD11b expression level might be regarded as a prognostic biomarker for AML patients.
CD11b is a protein subunit of integrin alpha-M beta-2 molecule which is essential for cell-cell interaction between leukemic cells with its microenvironment [8, 10], and then participates in regulation of biological activities of leukemic cells [13–19]. Currently, CD11b is also defined as a marker for myeloid-derived suppressor cells, which is reported to be involved in restraining antitumor immunity of the host and promoting expansion and drug-resistance of hematological malignant cells [20, 22, 44]. So it is mechanistically reasonable that CD11b expression level should be regarded as a prognostic biomarker for AML patients.
Meta-analysis of large amount of patients can provide direct and definite evidence for assessing the prognostic biomarkers for AML patients. This meta-analysis integrated the data from different clinical studies evaluating the prognostic value of CD11b expression level for AML patients in different countries for the first time, hence the statistical power is increased and the applicability is widened. What is more, most of the included cohort studies are with high quality and no statistically significant publication bias for each outcome was noted which also ensure reliability of this meta-analysis. Last but not the least, although sample preparation method, equipment and antibody used for detection of CD11b varied between studies, results of subgroup analysis according to sample preparation method showed no significant interaction between these factors with results, which suggests that the prognostic value of CD11b expression level is valid.
However, there are some limitations of this meta-analysis. Firstly, this meta-analysis is based on summary data rather than individual patients’ data, although we have undertaken subgroup analysis trying to evaluate the prognostic value of CD11b expression level in different subgroup of patients, but we could not explore more detailed or even patient-level prognostic value of CD11b expression level. Secondly, different length of follow-up among included studies might affect the evaluation of this meta-analysis. Thirdly, heterogeneity cannot be avoided in certain analysis which forced us to use the relatively conservative random effect model in these conditions. Last, the meta-analysis for DFS only included three studies, so this result should be interpreted with caution.
In conclusion, besides the limitations mentioned above, our meta-analysis indicates that CD11b expression level is closely related to the prognosis of AML patients and should be considered as a prognostic biomarker for stratifying AML patients. It might be also promising to develop drugs that target CD11b for improving the prognosis of AML patients.
Supporting Information
(DOCX)
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by National Science Foundation of China (81270605, 81470324), Youth Cultivation Project of Medical Science and Technology of Chinese PLA (14QNP089), Postgraduate Education Reform Project of Chongqing (yjg123114), and Third Military Medical University Clinic and Science Great Fund Project (2012XLC03). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ferrara F, Schiffer CA (2013) Acute myeloid leukaemia in adults. Lancet 381: 484–495. 10.1016/S0140-6736(12)61727-9 [DOI] [PubMed] [Google Scholar]
- 2. Hildreth CJ, Lynm C, Glass RM (2010) JAMA patient page. Acute myeloid leukemia. JAMA 304: 2759 10.1001/jama.302.22.2759 [DOI] [PubMed] [Google Scholar]
- 3. Kohnke T, Sauter D, Ringel K, Hoster E, Laubender RP, Laubender RP, et al. (2015) Early assessment of minimal residual disease in AML by flow cytometry during aplasia identifies patients at increased risk of relapse. Leukemia 29: 377–386. 10.1038/leu.2014.186 [DOI] [PubMed] [Google Scholar]
- 4. Konopleva MY, Jordan CT (2011) Leukemia stem cells and microenvironment: biology and therapeutic targeting. J Clin Oncol 29: 591–599. 10.1200/JCO.2010.31.0904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sison EA, Brown P (2011) The bone marrow microenvironment and leukemia: biology and therapeutic targeting. Expert Rev Hematol 4: 271–283. 10.1586/ehm.11.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Foss B, Tronstad KJ, Bruserud O (2010) Connexin-based signaling in acute myelogenous leukemia (AML). Biochim Biophys Acta 1798: 1–8. 10.1016/j.bbamem.2009.10.014 [DOI] [PubMed] [Google Scholar]
- 7. Johrer K, Hofbauer SW, Zelle-Rieser C, Greil R, Hartmann TN (2012) Chemokine-dependent B cell-T cell interactions in chronic lymphocytic leukemia and multiple myeloma—targets for therapeutic intervention? Expert Opin Biol Ther 12: 425–441. 10.1517/14712598.2012.664128 [DOI] [PubMed] [Google Scholar]
- 8. Tohda S (2014) NOTCH signaling roles in acute myeloid leukemia cell growth and interaction with other stemness-related signals. Anticancer Res 34: 6259–6264. [PubMed] [Google Scholar]
- 9. Buzzai M, Licht JD (2008) New molecular concepts and targets in acute myeloid leukemia. Curr Opin Hematol 15: 82–87. 10.1097/MOH.0b013e3282f3ded0 [DOI] [PubMed] [Google Scholar]
- 10. Kawamoto H, Minato N (2004) Myeloid cells. Int J Biochem Cell Biol 36: 1374–1379. [DOI] [PubMed] [Google Scholar]
- 11. Hickstein DD, Ozols J, Williams SA, Baenziger JU, Locksley RM, Roth GJ. (1987) Isolation and characterization of the receptor on human neutrophils that mediates cellular adherence. J Biol Chem 262: 5576–5580. [PubMed] [Google Scholar]
- 12. Arnaout MA, Gupta SK, Pierce MW, Tenen DG (1988). Amino acid sequence of the alpha subunit of human leukocyte adhesion receptor Mo1 (complement receptor type 3). J Cell Biol 106:2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arnaout MA, Lanier LL, Faller DV (1988). Relative contribution of the leukocyte molecules Mo1, LFA-1, and p150,95 (LeuM5) in adhesion of granulocytes and monocytes to vascular endothelium is tissue- and stimulus-specific. J Cell Physiol 137:305–309. [DOI] [PubMed] [Google Scholar]
- 14. Fan ST, Edgington TS (1991) Coupling of the adhesive receptor CD11b/CD18 to functional enhancement of effector macrophage tissue factor response. J Clin Invest 87: 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coombe DR, Watt SM, Parish CR (1994) Mac-1 (CD11b/CD18) and CD45 mediate the adhesion of hematopoietic progenitor cells to stromal cell elements via recognition of stromal heparan sulfate. Blood 84: 739–752. [PubMed] [Google Scholar]
- 16. Kusunoki T, Tsuruta S, Higashi H, Hosoi S, Hata D, Sugie K, et al. (1994) Involvement of CD11b/CD18 in enhanced neutrophil adhesion by Fc gamma receptor stimulation. J Leukoc Biol 55: 735–742. [DOI] [PubMed] [Google Scholar]
- 17. Simon DI, Ezratty AM, Francis SA, Rennke H, Loscalzo J (1993) Fibrin(ogen) is internalized and degraded by activated human monocytoid cells via Mac-1 (CD11b/CD18): a nonplasmin fibrinolytic pathway. Blood 82: 2414–2422. [PubMed] [Google Scholar]
- 18. Ueda T, Rieu P, Brayer J, Arnaout MA (1994) Identification of the complement iC3b binding site in the beta 2 integrin CR3 (CD11b/CD18). Proc Natl Acad Sci U S A 91: 10680–10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee JO, Rieu P, Arnaout MA, Liddington R (1995) Crystal structure of the A domain from the alpha subunit of integrin CR3 (CD11b/CD18). Cell 80: 631–638. [DOI] [PubMed] [Google Scholar]
- 20. Shojaei F, Wu X, Malik AK, Zhong C, Baldwin ME, Schanz S, et al. (2007) Tumor refractoriness to anti-VEGF treatment is mediated by CD11b+Gr1+ myeloid cells. Nat Biotechnol 25: 911–920. [DOI] [PubMed] [Google Scholar]
- 21. De Veirman K, Van Valckenborgh E, Lahmar Q, Geeraerts X, De Bruyne E, Menu E, et al. (2014) Myeloid-derived suppressor cells as therapeutic target in hematological malignancies. Front Oncol 4: 349 10.3389/fonc.2014.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Younos IH, Abe F, Talmadge JE (2015) Myeloid-derived suppressor cells: their role in the pathophysiology of hematologic malignancies and potential as therapeutic targets. Leuk Lymphoma: 1–13. [DOI] [PubMed] [Google Scholar]
- 23. Tucker J, Dorey E, Gregory WM, Simpson AP, Amess JA, Lister TA, et al. (1990) Immunophenotype of blast cells in acute myeloid leukemia may be a useful predictive factor for outcome. Hematol Oncol 8: 47–58. [DOI] [PubMed] [Google Scholar]
- 24. Bradstock K, Matthews J, Benson E, Page F, Bishop J (1994) Prognostic value of immunophenotyping in acute myeloid leukemia. Australian Leukaemia Study Group. Blood 84: 1220–1225. [PubMed] [Google Scholar]
- 25. Paietta E, Andersen J, Yunis J, Rowe JM, Cassileth PA, Tallman MS, et al. (1998) Acute myeloid leukaemia expressing the leucocyte integrin CD11b—a new leukaemic syndrome with poor prognosis: result of an ECOG database analysis. British Journal of Haematology 100: 265–272. [DOI] [PubMed] [Google Scholar]
- 26. Albitar M, Alseraihy A, Ayas M, Al-Jefri A, Al-Ahmari A, Asim F Belgaumi, et al. (2011) CD11b Expression Is An Independent Adverse Prognostic Factor in Pediatric Acute Myeloid Leukemia Treated with Allogeneic Stem Cell Transplantation. Blood 118: 1747–1747. [Google Scholar]
- 27. Junca J, Garcia-Caro M, Granada I, Rodriguez-Hernandez I, Torrent A, Morgades M, et al. (2014) Correlation of CD11b and CD56 expression in adult acute myeloid leukemia with cytogenetic risk groups and prognosis. Annals of Hematology 93: 1483–1489. 10.1007/s00277-014-2082-4 [DOI] [PubMed] [Google Scholar]
- 28. Amirghofran Z, Zakerinia M, Shamseddin A (2001) Significant association between expression of the CD11b surface molecule and favorable outcome for patients with acute myeloblastic leukemia. International Journal of Hematology 73: 502–506. [DOI] [PubMed] [Google Scholar]
- 29. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement.BMJ. 2009;339:b2535 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25: 603–605. 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 31. Parmar MK, Torri V, Stewart L (1998) Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 17: 2815–2834. [DOI] [PubMed] [Google Scholar]
- 32. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR (2007) Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 8: 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 34. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 35. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen F, Huang F, Yi Z, Zheng Z, Wei X, Wei Y, et al. (2013) Immunophenotype and prognosis in 510 patients with acute myeloid leukemia: a single-center study. J Clin Hematol (China) 26: 375–378. [Google Scholar]
- 38. Chen M-H, Atenafu E, Craddock KJ, Brandwein J, Chang H (2013) CD11b expression correlates with monosomal karyotype and predicts an extremely poor prognosis in cytogenetically unfavorable acute myeloid leukemia. Leukemia Research 37: 122–128. 10.1016/j.leukres.2012.09.019 [DOI] [PubMed] [Google Scholar]
- 39. Liang J, Zhang H, Chang Y, Yu F, Zou Z (2001) Clinical significance of CD11b expression in acute myeloid leukemia. Jiangsu Med J 27: 259–260. [Google Scholar]
- 40. Xu J, Sun XJ, Li YH, Zhaung GY, He JJ (2006) CD11b expression on acute promyelocytic leukemia cells is associated with poor clinical outcome. Cytometry Part B-Clinical Cytometry 70B: 377–377. [Google Scholar]
- 41. Xu N, Liu XL, Du QF, Liu Z, Zhong M, Lin R, et al. (2009) [CD56 and CD11b antigen expressions in patients with acute monocytic leukemia and the clinical implications]. Nan Fang Yi Ke Da Xue Xue Bao 29: 1605–1608. [PubMed] [Google Scholar]
- 42. Yang LL, Liu X, Li Q, Zhu XY, Wang XB, Zhu WB. (2014) [Immunophenotyping characteristics of AML and their correlation with the curative effects]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 22: 1–5. 10.7534/j.issn.1009-2137.2014.01.001 [DOI] [PubMed] [Google Scholar]
- 43. Zhuang G, Xu J, Sun X, Liu C, Wan S, Hui W, et al. (2011) Expression of leukemia associated phenotype on non-M3 acute myeloid leukemia cells and its relationship with prognosis. Beijing Med J 33: 929–931. [Google Scholar]
- 44. Ahn GO, Tseng D, Liao CH, Dorie MJ, Czechowicz A, Brown JM. (2010) Inhibition of Mac-1 (CD11b/CD18) enhances tumor response to radiation by reducing myeloid cell recruitment. Proc Natl Acad Sci U S A 107: 8363–8368. 10.1073/pnas.0911378107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.