Abstract
Objective
To study the main molecular mechanisms responsible for itraconazole resistance in clinical isolates of Candida krusei.
Methods
The 14α-demethylases encoded by ERG11 gene in the 16 C.krusei clinical isolates were amplified by polymerase chain reaction (PCR), and their nucleotide sequences were determined to detect point mutations. Meanwhile, ERG11 and efflux transporters (ABC1 and ABC2) genes were determined by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) for their expression in itraconazole-resistant (R), itraconazole-susceptible dose dependent (SDD) and itraconazole-susceptible (S) C.krusei at the mRNA level.
Results
We found 7-point mutations in ERG11 gene of all the C.krusei clinical isolates, including 6 synonymous mutations and 1 missense mutation (C44T). However, the missense mutation was found in the three groups. The mRNA levels of ERG11 gene in itraconazole-resistant isolates showed higher expression compared with itraconazole-susceptible dose dependent and itraconazole-susceptible ones (P = 0.015 and P = 0.002 respectively). ABC2 gene mRNA levels in itraconazole-resistant group was significantly higher than the other two groups, and the levels of their expression in the isolates appeared to increase with the decrease of susceptibility to itraconazole (P = 0.007 in SDD compared with S, P = 0.016 in SDD with R, and P<0.001 in S with R respectively). While ABC1 gene presented lower expression in itraconazole resistant strains. However, the mRNA levels of ERG11, ABC1 and ABC2 in a C.krusei (CK10) resistant to both itraconazole and voriconazole were expressed highest in all the itraconazole-resistant isolates.
Conclusions
There are ERG11 gene polymorphisms in clinical isolates of C.krusei. ERG11 gene mutations may not be involved in the development of itraconazole resistance in C.krusei. ERG11 and ABC2 overexpression might be responsible for the acquired itraconazole resistance of these clinical isolates.
Introduction
In recent 30 years, the incidence and mortality of fungal infection, especially the opportunistic fungal infection has been on the increase. This is mainly associated with antifungal resistance and the restricted number of available antifungal drugs [1]. Candida species are important opportunistic fungi in the humanbody, mainly colonizing the skin and mucous membrane such as oral cavity, gastrointestinal tract, vagina and so on. It does not cause disease in a healthy body. However, when the body's immune system declines, the extensive use of antibiotics, immunosuppressive agents, hormones, chemotherapy and other drugs, the increasing incidence of leukopenia, tumor incidence, HIV infection, diabetes and other diseases, the widespread use of invasive procedures like indwelling catheters, stapler and artificial valves, the large-scale organ transplants carried out, the increasing number of elderly patients, etc. [1,2], can lead to superficial fungal diseases and invasive candidiasis.
Currently, there are many species known to cause invasive or superficial candidiasis. And more than 90% candidiasis are caused by Candida albicans, Candida glabrata, Candida parapsilosis, Candida tropicalis and Candida krusei [3]. The most common pathogen of Candida spp. is C.albicans. In recent years, the incidence of non-albicans Candida spp. such as C.glabrata, C.tropicalis, C.krusei, C.parapsilosishas increased significantly [4,5]. A recent study conducted by Costa et al. showed that about 35% to 65% of all candidiasis were caused by non-albicans Candida spp. [6]. In North America, Horn et al. reported the incidence of non-albicans Candida spp. was 54.4%, while C.albicans was 45.6% [7]. In Argentina, Cornistein et al. [8] observed that C.albicans was responsible for 43.3% of all clinical specimens, whereas non-albicans Candida spp. were about 56.7%. Thus, C.albicans is the most common pathogenic Candida species, and the incidence of non-albicans Candida species is also on the rise.
Azole is one of the most common anti-Candida drugs, including fluconazole, itraconazole, posaconazole, and voriconazole. The common mechanisms of Candida resistant to azoles include changes in target enzyme and upregulation of multidrug resistance protein (MDR) and so on. The target enzyme of azoles is 14α-lanosterol demethylase (14-DM), encoded by ERG11 gene. ERG11 gene overexpression or mutations have been reported to be involved in the resistance formation of C.albicans, C.glabrata, and C.tropicalis. MDR proteins are kind of efflux pump transporters, including the ATP-binding cassette transporter family (ABC) and the major facilitator super family (MFS). In C.albicans, the efflux pump genes associated with azole resistance include CDR1, CDR2, and MDR1 [9]; in C.glabrata, the resistance genes involved CDR1, CDR2 and SNQ2 [10] while in C.tropicalis, the azole resistance related efflux pump gene is MDR1 [11].
At present, the study of azole resistance mechanisms of Candida has focused on C.albicans, C.glabrata, C.tropicalis. However, few studies talked about resistance mechanisms of C.krusei, especially resistant to itraconazole. C.krusei is naturally resistant to fluconazole. Orozco et al. [12] considered that fluconazole resistance in C.krusei appeared to be mediated predominantly by a reduced susceptibility of 14α-demethylase to this drug. Katiyar et al. [13] reported that C.krusei has found two genes encoding efflux pumps, ABC1, and ABC2. They thought that ABC1 encoded efflux pump played a major role in innate fluconazole resistance of C.krusei, while ABC2 gene expression was insignificant. However, Lamping et al. [14] and Guinea et al. [15] considered that it was mainly the low affinity of 14-DM with fluconazole together with the constitutive but low level of expression of the multidrug efflux pump Abc1p that were responsible for the innate C.krusei resistance to fluconazole.
To date, the mechanism of acquired drug resistance to azoles of C.krusei is still unclear. The purpose of the present study is to explore the main molecular mechanisms responsible for this species' resistance to itraconazole. Here, ERG11 gene of 16 C.krusei clinical isolates were amplified by polymerase chain reaction (PCR), and their nucleotide sequences were determined to detect point mutations. Meanwhile, ERG11, ABC1, and ABC2 genes were determined by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) for their expression in itraconazole-resistant, itraconazole-susceptible dose dependent and itraconazole-susceptible C.krusei at the mRNA level.
Materials and Methods
2.1. Ethics statement
This study has been approved by the Ethics Committee of Tianjin First Central Hospital. Before collecting clinical isolates from patients, we informed them of our research purposes and signed the consent form with them. Our study is to investigate the molecular mechanisms of Candida krusei clinical isolates resistant to itraconazole, which not only can help fully understand the resistance mechanisms of Candida krusei, but also can provide new ideas and approaches to clinical treatment of fungal infections. We promised to patients that their specimens were for scientific purposes only, and the patients and specimens information were anonymous in order to protect the health, safety and privacy of patients.
2.2. Clinical strains
The isolates of C.krusei included in the present study were from a collection of clinical isolates recovered from Tianjin First Central Hospital and Tianjin Medical University General Hospital. They were identified using Candida chromogenic medium (CHROMagar, France), Vitek2 YBC identification cards (BioMérieux, France) and mass spectrometer (BioMérieux, France). They were tested for their susceptibilities to flucytosine, amphotericin B, fluconazole, itraconazole, and voriconazole by ATB FUNGUS 3 (BioMérieux, France) as recommended by the manufacturer. And all the susceptibility profiles tested by ATB FUNGUS 3 were verified by the Clinical and Laboratory Standard Institute (CLSI) broth microdilution (BMD) method.
These isolates were recovered from various body sites of 16 different patients. Among the isolates studied, there were 5 itraconazole-resistant (R) strains, 8 itraconazole-susceptible dose dependent (SDD), and 3 itraconazole-susceptible (S) isolates (Table 1). In the itraconazole-resistant group, CK10 was recovered from a patient with urinary C.albican infection two weeks ago and administrated fluconazole therapy. All 16 isolates were stored at -80°C on the filter paper.
Table 1. Minimal Inhibitory Concentration (MIC) and susceptibility profile of C.krusei clinical isolates.
| Information of strains | MIC(μg/ml)/ Susceptibility profile | ||||||
|---|---|---|---|---|---|---|---|
| Name | Recovery site | ITR susceptibility category | 5-FC | AMB | FLC | ITR | VRC |
| CK1 | Secretions | SDD | 4/S | 1/S | 64/R | 0.25/SDD | 0.25/S |
| CK2 | Sputum | SDD | 16/SDD | 1/S | 16/SDD | 0.5/SDD | 0.5/S |
| CK4 | Urine | SDD | 16/SDD | 0.5/S | 16/SDD | 0.25/SDD | 0.25/S |
| CK5 | Secretions | R | 16/SDD | 0.5/S | 16/SDD | 1/R | 0.5/S |
| CK6 | Urine | SDD | 16/SDD | 0.5/S | 16/SDD | 0.25/SDD | 0.125/S |
| CK8 | Secretions | SDD | 4/S | 1/S | 64/R | 0.25/SDD | 0.25/S |
| CK9 | Urine | R | 16/SDD | 2/SDD | 64/R | 1/R | 0.5/S |
| CK10 | Urine | R | 16/SDD | 2/SDD | 16/SDD | 4/R | 4/R |
| CK11 | Sputum | R | 16/SDD | 2/SDD | 16/SDD | 2/R | 0.25/S |
| CK12 | Secretions | R | 4/S | 0.5/S | 16/SDD | 1/R | 0.5/S |
| CK13 | Urine | SDD | 16/SDD | 2/SDD | 16/SDD | 0.25/SDD | 0.25/S |
| CK14 | Urine | SDD | 16/SDD | 2/SDD | 32/SDD | 0.5/SDD | 0.5/S |
| CK15 | Sputum | S | 4/S | 0.5/S | 16/SDD | 0.06/S | 0.125/S |
| CK16 | Urine | SDD | 8/SDD | 1/S | 32/SDD | 0.06/S | 0.125/S |
| CK17 | Urine | S | 8/SDD | 2/SDD | 64/R | 0.5/SDD | 0.25/S |
| CK18 | Secretions | S | 16/SDD | 1/S | 16/SDD | 0.125/S | 0.25/S |
CK, Candida krusei; S, susceptible; SDD, susceptible dose dependent; R, resistant; 5-FC, 5-flucytosine, MIC break point: S, ≤4 μg/ml; SDD, 8–16 μg/ml; R, ≥32 μg/ml; AMB, Amphotericin B, MIC break point: S, ≤1 μg/ml; SDD,2 μg/ml; R, >2 μg/ml; FLC, Fluconazole, MIC break point: S, ≤8 μg/ml; SDD, 16–32 μg/ml; R, ≥ 64 μg/ml; ITR, itraconazole, MIC break point: S, ≤0.125 μg/ml; SDD, 0.25–0.5 μg/ml; R, ≥ 1 μg/ml; VRC, voriconazole, MIC break point: S, ≤0.5 μg/ml; SDD,1 μg/ml; R, ≥2 μg/ml
2.3. Antifungal susceptibility testing
2.3.1. ATB FUNGUS 3
Reference antifungal susceptibility testing of the study isolates was performed by ATB FUNGUS 3 according to the manufacturer’s instructions, and visually read the minimal inhibitory concentration (MIC) value [16]. There are five antigungal drugs at different concentrations: 5-flucytosine (4, 16μg/ml), amphotericin B (0.5–16μg/ml), fluconazole (1–128μg/ml), itraconazole (0.125–4μg/ml) and voriconazole (0.06–8μg/ml). Testing on the ATB FUNGUS 3 was performed simultaneously with CLSI BMD method. The susceptibility profiles to itraconazole of the C.krusei isolates were refered to Sanguinetti et al. [10]: susceptible whenever MIC ≤0.125μg/ml, susceptible dosedependent whenever MIC = 0.25 to 0.5 μg/ml and resistant whenever MIC ≥1.0μg/ml. Visual readings were performed after 24h of incubation. Quality control was ensured by testing the NCCLS-recommended strains C.krusei ATCC 6258 and C.parapsilosis ATCC 22019 [17,18].
2.3.2. CLSI BMD method
CLSI BMD method was performed in accordance with CLSI M27-A3 and M27-S4 guidelines. The following drugs and concentrations were tested: 5-flucytosine (0.03–64μg/ml), amphotericin B (0.008–16μg/ml), fluconazole (0.03–64 μg/ml), itraconazole (0.03–64μg/ml) and voriconazole (0.015–16μg/ml). Quality control was also ensured by testing the NCCLS-recommended strains C.krusei ATCC 6258 and C.parapsilosis ATCC 22019 [17,18]. The susceptibility profiles conducted by the above two methods were consistent, and shown in Table 1.
2.4. Amplifying and sequencing analysis of ERG11 gene
Prior to experiments, such isolates were subcultured twice on Sabouraud agar (Merck KGaA, Germany) at 37°C for 24 h to revive and ensure the purity of cultures. A single one fresh colonywas transferred to 5ml liquid YPD (yeast extract 1%, peptone 2%, and dextrose 2%) broth at 35°C, 150 rpm, until exponential growth phase. 1ml yeast cells were harvested by centrifugation at room temperature (4000 rpm, 5min), and immediately frozen in liquid nitrogen. Total genomic DNA was extracted using DNA extraction kit (Omega, USA). ERG11 gene product (1873 bp) was amplified by PCR, and primers in Table 2 are refered to Ricardo et al. [19]. The PCR reaction contained: 10μl of 2×Phusion Flash PCR Master Buffer (Thermo, USA), 1μl of 10μM Primer Mix, 1μl of DNA sample, and diethylpyrocarbonate (DEPC) H2O up to 20 μl final volume. All reactions were performed in a GeneAmp PCR system 9700 (Applied Biosystems, USA), and reaction parameters involved an initial 1 min denaturation step at 98°C followed by 35 cycles at 98°C for 10 s, at 65°C for 20s and at 72°C for 30s, with a 1 min final extension step at 72°C. PCR products were used to run 2% agarose gel, and then sent to sequence. All the ERG11 gene sequence results were aligned with a sequence published online (Gene accession number FJ445756) using DNAMAN software.
Table 2. Primers used in this study.
| GenBank Accession no. | Primer Name | Primer Sequences(5’→3’) |
|---|---|---|
| FJ445756 | ERG11-F for sequencing | GGTTGTTTGTTCATTTAATGTGTGT |
| ERG11-R for sequencing | GAAGGGGGAAAGAAAGGGAA | |
| FJ445756 | ERG11-F for RT-PCR | ATTGCGGCCGATGTCCAGAGGTAT |
| ERG11-R for RT-PCR | GCGCAGAGTATAAGAAAGGAATGGA | |
| DQ903907 | ABC1-F | GATAACCATTTCCCACATTTGAGT |
| ABC1-R | CATATGTTGCCATGTACACTTCTG | |
| AF250037 | ABC2-F | CCTTTTGTTCAGTGCCAGATTG |
| ABC2-R | GTAACCAGGGACACCAGCAA | |
| AJ389086 | ACT1-F | TGGGCCAAAAGGATTCTTATG |
| ACT1-R | AGATCTTTTCCATATCATCCCAG |
2.5. ABC1, ABC2 and ERG11 genes expression analysis
2.5.1. Total RNA extraction
Total RNA was extracted as previously described in this paper. After reviving and transferring to 5ml liquid YPD broth overnight, 1ml yeast cells were harvested by centrifugation at room temperature (4000 rpm, 5 min), and immediately frozen in liquid nitrogen. After mechanical disruption of the cells with liquid nitrogen grinding, the fungal powder was collected into a 2ml Eppendorf tube, and 1ml Trizol reagent (Invitrogen, USA) was added and mixed them through a pipette. Incubate the samples for 5 minutes at room temperature. Then add 250μl chloroform to the samples. Shake the tubes vigorously with hand, and incubate the samples for 10 minutes at room temperature. Centrifuge the samples at 12,000 rpm for 15 minutes at 4°C. After centrifugation, the mixture separates into 3 aqueous phases. RNA remains in the upper colorless aqueous phase. Transfer the upper colorless liquid into a new 1.5ml Eppendorf tube, and add same volume isopropyl alcohol into the tube and mix. The the mixture was incubated for at least 4 hours at -20°C to precipitate RNA. Then centrifuge the mixture at 12,000 rpm for 15 minutes at 4°C. Remove the supernatant and wash the RNA with 1ml 75% ethanol (diluted by DEPC water). Then centrifuge the mixture at 12,000 rpm for 1 minute at 4°C. Discard the supernatant and dry the RNA. Finally, RNA was dissolved in DEPC water. RNA integrity was assessed by determination of the OD260/OD280 absorption ratio, and the integrity was considered maintained if the ratio was 1.8 to 2.0. RNA samples were resuspended in DEPC water, with the concentration adjusted to a final value of 200 ng/μl and then stored at -20°C for later use.
2.5.2. Reverse transcriptase PCR (RT-PCR)
One-step RT-PCR reaction was performed with a 20μl volume containing the following reagents: 6μl of Reverse Transcriptase Premixture (Thermo,USA), 7μl of 200 ng/μl RNA sample, and DEPC water up to the final volume. RT-PCR reaction was carried out in a GeneAmp PCR system 9700 (Applied Biosystems, USA), and reaction parameters were as follows: an initial 15min step at 25°C followed by 40min step at 55°C, with a final step at 85°C for 5min. The cDNAs were kept at -20°C for later use.
2.5.3. Quantitative Real-Time PCR (qRT-PCR)
Quantitative expression of the ERG11, ABC1, and ABC2 genes was conducted by real-time RT-PCR with a LightCycler 96 system (Roche, Switzerland). For the target genes and ACT1 reference gene, they were amplified using the following primers in Table 2 refered to Ricardo et al. [19]. The qRT-PCR reaction mixture contained: 7.5μl of 2×KAPA SYBR FAST Mix (Kapa, USA), 1μl of 10μM Primer Mix, 1μl of 10ng cDNA sample, and DEPC water up to the final 15μl volume. Each reaction was run in triplicate. The qRT-PCR reaction condition was subjected to an initial predegeneration step at 95°C for 30s, followed by 40 cycles of 95°C for 3s, 60°C for 30s. Fluorescence data were collected and analyzed with the LightCycler 96 software.
2.6. Statistical analysis
Result analysis was performed with the GraphPad Prism program (GraphPad Software, Inc. San Diego, CA). The two-tailed Student's t-test was used to analyze significant differences between gene expression displayed by the distinct C.krusei strains; P<0.05 was considered statistically significant.
Results
3.1. Amplifying and sequencing analysis of ERG11 gene
3.1.1. Amplifying analysis of ERG11 gene
Total genomic DNA of all the 16 C.krusei clinical isolates were extracted to amplify ERG11 gene, and the PCR products were used to run 2% agarose gel electrophoresis. We observed that the entire purpose band (1873bp) clearly at the same level near 2000bp.
3.1.2. Sequencing analysis of ERG11 gene
Several ERG11 gene mutations have been previously described to be associated with azole resistance in C.albicans [20]; therefore, C.krusei ERG11 gene was sequenced in our strains.
In the present study, we found 7-point mutations aligned with the online published sequence (Gene Accession Number FJ445756). And the results are presented in Table 3. There are 6 synonymous mutations and 1 missense mutation. The 6 synonymous mutations can appear in both non-resistant and resistant strains, and all the C.krusei strains presented C642T, T1389C, and G1536C mutations. The only one missense mutation occurred at 44bp (C→T), resulting inan amino acid change from alanine to valine. However, such a missense mutation can be found in non-resistant and resistant C.krusei, suggesting that the C44T missense mutation might not be associated with azoles drug resistance of C.krusei.
Table 3. ERG11 gene point mutations in C.krusei clinical isolates.
| Information of strains | ERG11 gene mutation sites | |||||||
|---|---|---|---|---|---|---|---|---|
| Name | ITR susceptibility category | 44 | 51 | 642 | 756 | 939 | 1389 | 1536 |
| FJ445756 | C | C | C | A | T | T | G | |
| CK1 | SDD | — | — | T | — | — | C | C |
| CK2 | SDD | T | — | T | — | — | C | C |
| CK4 | SDD | — | T | T | — | — | C | C |
| CK5 | R | T | T | T | — | C | C | C |
| CK6 | SDD | T | — | T | — | C | C | C |
| CK8 | SDD | — | — | T | — | — | C | C |
| CK9 | R | — | — | T | — | — | C | C |
| CK10 | R | T | T | T | — | C | C | C |
| CK11 | R | — | — | T | T | — | C | C |
| CK12 | R | T | — | T | — | C | C | C |
| CK13 | SDD | T | — | T | T | — | C | C |
| CK14 | SDD | — | — | T | — | — | C | C |
| CK15 | S | — | T | T | — | — | C | C |
| CK16 | SDD | T | — | T | — | C | C | C |
| CK17 | S | — | — | T | — | — | C | C |
| CK18 | S | T | — | T | — | — | C | C |
3.2. ABC1, ABC2 and ERG11 genes expression analysis
3.2.1. ABC1 gene expression analysis (N = 16)
16 C.krusei clinical isolates ABC1 gene mRNA relative expression levels are shown in Table 4 and Fig 1A. Individual data of ABC1 gene mRNA relative expression in all the C.krusei is listed in S1 Table. However, in our study, the expression level of ABC1 gene mRNA in itraconazole-resistant isolates is significantly lower than itraconazole-susceptible dose dependent and itraconazole-susceptible strains (P = 0.007 and P = 0.034 respectively, Fig 1B), and there is no significance between itraconazole-susceptible dose dependent and itraconazole-susceptible strains (P = 0.876, Fig 1B). In the itraconazole-resistant strains, a Candida krusei named CK10, both resistant to itraconazole and voriconazole, has the highest ABC1 mRNA expression level in this group.
Table 4. ABC1, ABC2 and ERG11 genes relative level of mRNA expression in C.krusei clinical isolates.
| Information of strains | Relative level of mRNA a (M±SD) | Log10+3 fold increase of expression b | |||||
|---|---|---|---|---|---|---|---|
| Name | ITR susceptibility category | ABC1 | ABC2 | ERG11 | ABC1 | ABC2 | ERG11 |
| CK1 | SDD | 0.0458 ± 0.0097 | 0.0733 ± 0.0056 | 0.0483 ± 0.0066 | 1.66 | 1.87 | 1.68 |
| CK2 | SDD | 0.0918 ± 0.0056 | 0.1141 ± 0.0006 | 0.0506 ± 0.0035 | 1.96 | 2.06 | 1.70 |
| CK4 | SDD | 0.1811 ± 0.0304 | 0.0729 ± 0.0161 | 0.0469 ± 0.0160 | 2.26 | 1.86 | 1.67 |
| CK5 | R | 0.0304 ± 0.0041 | 0.0902 ± 0.0047 | 0.0525 ± 0.0093 | 1.48 | 1.96 | 1.72 |
| CK6 | SDD | 0.0491 ± 0.0027 | 0.0506 ± 0.0058 | 0.0802 ± 0.0113 | 1.69 | 1.70 | 1.90 |
| CK8 | SDD | 0.0336 ± 0.0041 | 0.1191 ± 0.0197 | 0.0955 ± 0.0089 | 1.53 | 2.08 | 1.98 |
| CK9 | R | 0.0237 ± 0.0001 | 0.1419 ± 0.0600 | 0.1036 ± 0.0068 | 1.37 | 2.15 | 2.02 |
| CK10 | R | 0.0319 ± 0.0067 | 0.2338 ± 0.0337 | 0.1654 ± 0.0063 | 1.50 | 2.37 | 2.22 |
| CK11 | R | 0.0175 ± 0.0021 | 0.1336 ± 0.0149 | 0.1525 ± 0.0125 | 1.24 | 2.13 | 2.18 |
| CK12 | R | 0.0151 ± 0.0027 | 0.1298 ± 0.0134 | 0.0875 ± 0.0118 | 1.18 | 2.11 | 1.94 |
| CK13 | SDD | 0.0532 ± 0.0079 | 0.0818 ± 0.0249 | 0.0421 ± 0.0036 | 1.73 | 1.91 | 1.62 |
| CK14 | SDD | 0.0801 ± 0.0071 | 0.0279 ± 0.0015 | 0.0122 ± 0.0003 | 1.90 | 1.45 | 1.09 |
| CK15 | S | 0.0446 ± 0.0091 | 0.0408 ± 0.0049 | 0.0228 ± 0.0017 | 1.65 | 1.61 | 1.36 |
| CK16 | S | 0.1819 ± 0.0097 | 0.0136 ± 0.0178 | 0.0207 ± 0.0015 | 2.26 | 1.13 | 1.32 |
| CK17 | SDD | 0.0586 ± 0.0125 | 0.0532 ± 0.0042 | 0.0325 ± 0.0083 | 1.77 | 1.73 | 1.51 |
| CK18 | S | 0.0280 ± 0.0078 | 0.0296 ± 0.0037 | 0.0305 ± 0.0049 | 1.45 | 1.47 | 1.48 |
Note:
a Quantification was performed by real-time RT-PCR. The values are averages of triplicate wells. M: mean; SD: standard deviation.
b The data was transformed from the values of relative level of mRNA, and obeyed the normal distribution and variance homogeneity.
Fig 1. ABC1 relative gene expression levels in three groups of C.krusei clinical isolates.
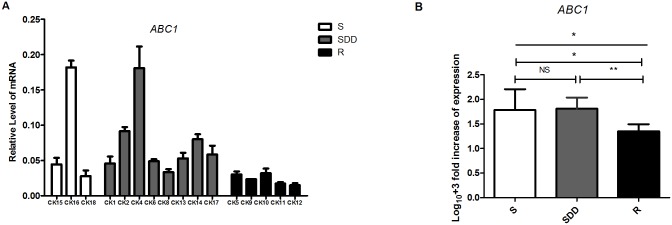
(A) Relative levels of ABC1 mRNA in all the C.krusei clinical isolates. ABC1 gene expression levels was quantified and normalized relative to the housekeeping gene, ACT1; S, itraconazole-susceptible; SDD, itraconazole-susceptible dose dependent; R, itraconazole-resistant. (B) Log10 +3 fold increase of gene expression levels in three groups. (NS, no significance in SDD compared with S; *P<0.05 in R with S; **P<0.01 in R with SDD).
3.2.2. ABC2 gene expression analysis (N = 16)
16 clinical isolates of C.krusei ABC2 gene mRNA relative expression levels are presented in Table 4 and Fig 2A. Individual data of ABC2 gene mRNA relative expression in all the C.krusei is listed in S2 Table. Our data shows that the expression level of ABC2 gene mRNA in itraconazole-resistant isolates is significantly higher than itraconazole-susceptible dose dependent (P = 0.016, Fig 2B) and itraconazole-susceptible strains (P<0.001, Fig 2B). The mRNA level in itraconazole-susceptible dose dependent group is also significantly higher compared to itraconazole-susceptible one (P = 0.007, Fig 2B). And in the itraconazole-resistant strains, the ABC2 mRNA expression level of CK10 is highest in this group.
Fig 2. ABC2 relative gene expression levels in three groups of C.krusei clinical isolates.
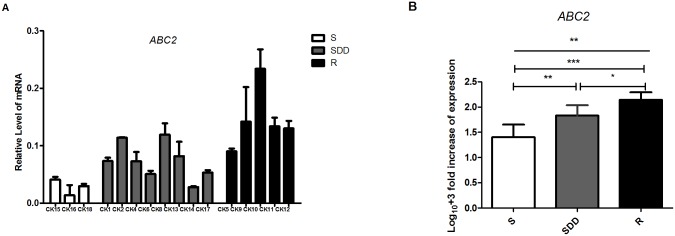
(A) Relative levels of ABC2 mRNA in all the C.krusei clinical isolates. ABC2 gene expression levels was quantified and normalized relative to the housekeeping gene, ACT1; S, itraconazole-susceptible; SDD, itraconazole-susceptible dose dependent; R, itraconazole-resistant. (B) Log10 +3 fold increase of gene expression levels in three groups. (*P<0.05 in R compared with SDD; **P<0.01 in SDD with S; ***P<0.001 in R with S).
3.2.3. ERG11 gene expression analysis (N = 16)
16 C.krusei clinical isolates ERG11 gene mRNA relative expression levels are shown in Table 4 and Fig 3A. Individual data of ERG11 gene mRNA relative expression in all the C.krusei is listed in S3 Table. Our data shows that the expression level of ERG11 gene mRNA in itraconazole-resistant isolates is also significantly higher than itraconazole-susceptible dose dependent (P = 0.015, Fig 3B) and itraconazole-susceptible strains (P = 0.002, Fig 3B). But there is insignificant between itraconazole-susceptible dose dependent isolates and itraconazole-susceptible ones (P = 0.12, Fig 3B). However, in the itraconazole-resistant strains, the ERG11 mRNA expression level of CK10 is still the highest in this group.
Fig 3. ERG11 relative gene expression levels in three groups of C.krusei clinical isolates.
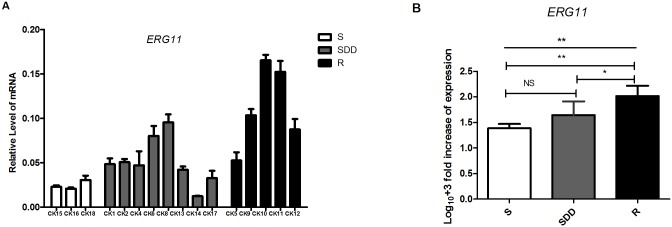
(A) Relative levels of ERG11 mRNA in all the C.krusei clinical isolates. ERG11 gene expression levels was quantified and normalized relative to the housekeeping gene, ACT1; S, itraconazole-susceptible; SDD, itraconazole-susceptible dose dependent; R, itraconazole-resistant. (B) Log10 +3 fold increase of gene expression levels in three groups. (NS, no significance in SDD compared with S; *P<0.05 in R with SDD; **P<0.01 in R with S).
Discussion
Azole antifungal agents are ergosterol biosynthesis inhibitors. On one hand, azole combines with target enzyme 14-DM to inhibit ergosterol biosynthesis. On the other, it can lead to accumulation of toxic steroid through bypass. These two ways synergize to exert antifungal effect on Candida. However, due to its inhibition effect on Candida, it can cause the emergence of drug resistance after a long course of treatment or repeated dose administration. The common mechanisms of Candida resistant to azoles include changes in target enzyme and upregulation of multidrug resistance protein (MDR). These could occur through (1) increased ERG11 gene overexpression, which can cause target enzyme to be produced more, leading to a higher concentration of azoles to block the biosynthesis of 14-DM; (2) ERG11 gene existed mutations, which can cause structural change of 14-DM, resulting in affinity decline between azoles and 14-DM; (3) efflux pumps of MDR upregulation, which can decrease the azole concentration within the cell, making it unable to inhibit fungi cell effectively, thus leading to the emergence of drug resistance. To date, there are two kinds of efflux transporters associated with C.krusei resistance: Abc1p and Abc2p, encoded by the gene ABC1 and ABC2 respectively.
Currently, studies on azole resistance mechanisms of Candida mainly focused on C.albicans, C.glabrata, and C.tropicalis. However, there are few researches discussing the resistance mechanisms of C.krusei. In addition, the mechanisms of Candida resistant to different azoles also showed distinctly. Research results suggested that ERG11, ABC1 and ABC2 genes were associated with azole resistance of C.krusei [13, 21]. However, the strains studied by researchers were mostly from the same patient at a different stage, with strictly and genetically matched fluconazole-resistant and fluconazole-sensitive ones. In this case, the number of patients or the number of strains is always limited. The strains used in other studies were from lab-induced resistant ones. However, in clinical practice, it is always difficult to obtain strictly and genetically matched isolates. In addition, whether the resistance mechanisms of lab-induced strains are similar to clinical isolates from different patients is still unclear. As we all know, the resistance mechanisms of strictly and genetically paired strains cannot completely explain the mechanisms of azole resistance in unmatched isolates. Therefore, it is very important to study the resistance mechanisms of unmatched isolates of C.krusei from different patients, which can help fully understand the mechanisms of resistance formation.
In the present study, we recovered 16 C.krusei clinical isolates from different patients and divided them into three groups according to their susceptibility to itraconazole. There were 5 strains in the itraconazole-resistant group, 8 in the itraconazole-susceptible dose dependent group, and 3 in the itraconazole-susceptible one. Here, to explore the relation between ERG11, ABC1 and ABC2 genes and resistance formation of itraconazole in clinical isolates of C.krusei, we used PCR to amplify ERG11 gene and sequenced it in order to find new mutations. Then, we performed qRT-PCR to detect the differences of the resistant gene expression levels in the three groups.
In order to fully understand ERG11 gene mutation of C.krusei, we amplified the PCR product containing the whole open reading frame of ERG11 gene and sent it to sequence in our study. The PCR product is about 1873bp, including 1587bp of ERG11 gene, which encodes 529 amino acids. Our data showed that there were 7-point mutations existing in ERG11 gene of C.krusei, including 6 synonymous mutations (C51T, C642T, A756T, T939C, T1389C, and G1536C) and 1 missense mutation (C44T). The only missense mutation can result in the 15th amino acid changing from alanine to valine. However, such a missense mutation can be found in the three groups, indicating that C44T may not be involved in itraconazole resistance of C.krusei. Moreover, in our study, all the non-resistant and resistant C.krusei strains presented C642T, T1389C, G1536C mutations. Ricardo et al. [19] also reported that T1389C mutation occurred in all the experimental strains. Besides, T939C mutation have been reported by Tavakoli et al. [21]. In addition, all the point mutations found in our experiment are consistent with ERG11 gene single nucleotide polymorphism (SNP) sites reported by Lamping [14] except for C51T, a new synonymous mutation which has not been reported before.
However, T418C missense mutation reported by Ricardo [19] has not been found in our study, further illustrating the mechanism differences between in vivo or in vitro induced resistance and drug resistance in clinical isolates of C.krusei from different persons. Previous researches [22,23] reported that, ERG11 gene missen mutation was associated with C.albicans and C.tropicalis azole resistance, which can cause chages in amino acid sequence resulting in structural change of target enzyme 14-DM. This can lead to decreased affinity with fluconazole and the emergence of resistance. In our view, C44T missense mutation may occur outside the active site of 14-DM, which doesn’t affect the interaction of azole and 14-DM or a single missense mutation is not enough to change the affinity of the 14-DM to azole. On the other hand, the target enzyme structure of C.krusei may be a lot different from other Candida, and its ERG11 gene polymorphism is more complicated.
Our study also found that, ERG11 and ABC2 genes mRNA expression levels of itraconazole-resistant group in C.krusei clinical isolates were significantly higher than itraconazole non-resistant strains. And the mRNA levels of ABC2 gene expression in the isolates appeared to increase with the decrease of susceptibility to itraconazole. While ABC1 gene mRNA expression level in resistant group was significantly lower than itraconazole non-resistant groups. In addition, an isolate in the resistant group named CK10, recovered from a patient pre-exposed to fluconazole, which was resistant to itraconazole and voriconazole, has its ERG11, ABC1 and ABC2 genes mRNA expression levels as the highest in this group. We speculate that the resistance mechanisms of itraconazole and voriconazole in C.krusei clinical isolates may be different. However, Lamping et al. [14] considered that Abc1p played a major role in innate fluconazole resistance of C.krusei. Similar findings were previously described by Ricardo et al. [19] for C.krusei. They considered that ABC2 can be more rapidly activated than ABC2 when exposure to voriconazole [19]. When involving long term voriconazole tolerance, Abc1p efflux pump seemed to be more efficient in antifungal expelling and played a late role in the development of resistance. And their data also showed that, “ERG11 gene overexpression seems to be relevant to the development of voriconazole resistance only at an early stage, as an initial adaptation mechanism. Later on, other distinct mechanisms such as the acquisition of point mutations may take the lead [19].” Venkateswarlu et al. [24] reported that the accumulation of itraconazole in resistant C.krusei was less than that in sensitive strains, but the two strains showing different sensitivity to itraconazole were both highly resistant to fluconazole, suggesting that the resistance mechanism between fluconazole and itraconazole in C.krusei may vary from each other. They thought that C.krusei resistant to itraconazole was due to reduced drug accumulation in fungal cells and guessed that there may exist one or more efflux pumps contributing to C.krusei itraconazole resistance, which can be well explained in our study. Our data presented that Abc2p may play a more important role in C.krusei itraconazole resistance, instead of Abc1p.
In conclusion, ERG11, ABC1 and ABC2 genes of C.krusei clinical isolates play different roles in the development of itraconazole resistance. However, in this study, we conclude that ERG11 and ABC2 genes upregulation contribute mostly to itraconazole resistance of C.krusei clinical isolates while ERG11 gene mutations and ABC1 gene may not be associated with the development of itraconazole resistance in our study. Due to our limited number of experimental strains, it is unreasonable to rule out the participation of ERG11 mutations in the emergence of resistance. Whether ERG11 mutations affect ERG11 gene expression is still unknown, and this needs to be further studied.
Supporting Information
Ratio: the target gene ABC1 mRNA levels relative to the reference gene ACT1.
(PDF)
Ratio: the target gene ABC2 mRNA levels relative to the reference gene ACT1.
(PDF)
Ratio: the target gene ERG11 mRNA levels relative to the reference gene ACT1.
(PDF)
Acknowledgments
This study was supported by the National Natural Sciences Foundation of China (81041043, 81400092), the National Natural Science Foundation of Tianjin (13JCYBJC23400), and Science and Technique Foundation of Tianjin (13KG106), as well as Tianjin Medical University and Tianjin First Central Hospital.
We thank Jinyan Chen and Jianlei Zhang (Department of Clinical Laboratory, Tianjin First Central Hospital) for providing strains and helpful suggestion.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the National Natural Sciences Foundation of China (81041043, 81400092), the National Natural Science Foundation of Tianjin (13JCYBJC23400), and Science and Technique Foundation of Tianjin (13KG106), as well as Tianjin Medical University and Tianjin First Central Hospital.
References
- 1. Silva NC, Nery JM, Dias AL. Aspartic proteinases of Candida spp.: role in pathogenicity and antifungal resistance.Mycoses.2014; 57:1–11. [DOI] [PubMed] [Google Scholar]
- 2. Bondaryk M, Kurzątkowski W, Staniszewska M. Antifungal agents commonly used in the superficial and mucosal candidiasis treatment: mode of action and resistance development.Postepy Dermatol Alergol.2013; 30:293–301. 10.5114/pdia.2013.38358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pfaller MA, Diekema DJ, Procop GW, Rinaldi MG. Multicenter comparison of the VITEK 2 antifungal susceptibility test with the CLSI broth microdilution reference method for testing amphotericin B, flucytosine, and voriconazole against Candida spp.J Clin Microbiol.2007; 45:3522–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sardi JC,Scorzoni L, Bernardi T, Fusco-Almeida AM, Mendes Giannini MJ. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options.J Med Microbiol.2013; 62:10–24. 10.1099/jmm.0.045054-0 [DOI] [PubMed] [Google Scholar]
- 5. Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance.FEMS Microbiol Rev.2012; 36:288–305. 10.1111/j.1574-6976.2011.00278.x [DOI] [PubMed] [Google Scholar]
- 6. Costa E, Inês A, Mendes-Faia A, Saavedra MJ, Mendes-Ferreira A. Potential virulence factors of Candida spp. isolated from clinical and food sources.J Hosp Infect.2010; 75:240–1. 10.1016/j.jhin.2010.03.001 [DOI] [PubMed] [Google Scholar]
- 7. Horn DL,Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, et al. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry.Clin Infect Dis.2009; 48:1695–703. 10.1086/599039 [DOI] [PubMed] [Google Scholar]
- 8. Cornistein W, Mora A, Orellana N, Capparelli FJ, del Castillo M. Candida: epidemiology and risk factors for non-albicans species. Enferm Infecc Microbiol Clin.2013; 31:380–4. 10.1016/j.eimc.2012.09.011 [DOI] [PubMed] [Google Scholar]
- 9. Chen LM, Xu YH, Zhou CL, Zhao J, Li CY, Wang R. Overexpression of CDR1 and CDR2 genes plays an important role in fluconazole resistance in Candida albicans with G487T and T916C mutations.J Int Med Res.2010; 38:536–45. [DOI] [PubMed] [Google Scholar]
- 10. Sanguinetti M, Posteraro B, Fiori B, Ranno S, Torelli R, Fadda G. Mechanisms of azole resistance in clinical isolates of Candida glabrata collected during a hospital survey of antifungal resistance.Antimicrob Agents Chemother.2005;49:668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang C, Dong D, Yu B, Cai G, Wang X, Ji Y, et al. Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China.J Antimicrob Chemother.2013; 68:778–85. 10.1093/jac/dks481 [DOI] [PubMed] [Google Scholar]
- 12. Orozco AS, Higginbotham LM, Hitchcock CA, Parkinson T, Falconer D, Ibrahim AS, et al. Mechanism of fluconazole resistance in Candida krusei .Antimicrob Agents Chemother.1998; 42:2645–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katiyar SK, Edlind TD. Identification and expression of multidrug resistance-related ABC transporter genes in Candida krusei .Med Mycol.2001; 39:109–16. [DOI] [PubMed] [Google Scholar]
- 14. Lamping E, Ranchod A, Nakamura K, Tyndall JD, Niimi K, Holmes AR, et al. Abc1p is a multidrug efflux transporter that tips the balance in favor of innate azole resistance in Candida krusei .Antimicrob Agents Chemother.2009; 53:354–69. 10.1128/AAC.01095-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guinea J, Sánchez-Somolinos M, Cuevas O, Peláez T, Bouza E. Fluconazole resistance mechanisms in Candida krusei: the contribution of efflux-pumps.Med Mycol.2006; 44:575–8. [DOI] [PubMed] [Google Scholar]
- 16. Zhang L, Wang H, Xiao M, Kudinha T, Mao LL, Zhao HR, et al. The widely used ATB FUNGUS 3 automated readings in China and its misleading high MICs of Candida spp. to azoles: challenges for developing countries' clinical microbiology labs.PLoS One.2014; 9:e114004 10.1371/journal.pone.0114004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barry AL, Pfaller MA, Brown SD, Espinel-Ingroff A, Ghannoum MA, Knapp C, et al. Quality control limits for broth microdilution susceptibility tests of ten antifungal agents.J Clin Microbiol.2000; 38:3457–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. CLSI, Wayne PA.Reference Method for Broth DilutionAntifungal Susceptibility Testing of Yeasts. Approved standard—third editionCLSI document M27-A3. Clinical and Laboratory Standard Institute; 2008. [Google Scholar]
- 19. Ricardo E, Miranda IM, Faria-Ramos I, Silva RM, Rodrigues AG, Pina-Vaz C. In vivo and in vitro acquisition of resistance to voriconazole by Candida krusei .Antimicrob Agents Chemother.2014; 58:4604–11. 10.1128/AAC.02603-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morio F, Loge C, Besse B, Hennequin C, Le Pape P. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature.Diagn Microbiol Infect Dis.2010; April; 66:373–84. 10.1016/j.diagmicrobio.2009.11.006 [DOI] [PubMed] [Google Scholar]
- 21. Tavakoli M, Zaini F, Kordbacheh M, Safara M, Raoofian R, Heidari M. Upregulation of the ERG11 gene in Candida krusei by azoles.Daru.2010; 18:276–80. [PMC free article] [PubMed] [Google Scholar]
- 22. Vandeputte P, Larcher G, Bergès T, Renier G, Chabasse D, Bouchara JP. Mechanisms of azole resistance in a clinical isolate of Candida tropicalis .Antimicrob Agents Chemother.2005; 49:4608–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strzelczyk JK, Slemp-Migiel A, Rother M, Gołąbek K, Wiczkowski A. Nucleotide substitutions in the Candida albicansERG11 gene of azole-susceptible and azole-resistant clinical isolates.Acta Biochim Pol.2013; 60:547–52. [PubMed] [Google Scholar]
- 24. Venkateswarlu K, Denning DW, Manning NJ, Kelly SL. Reduced accumulation of drug in Candida krusei accounts for itraconazole resistance.Antimicrob Agents Chemother.1996; 40:2443–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ratio: the target gene ABC1 mRNA levels relative to the reference gene ACT1.
(PDF)
Ratio: the target gene ABC2 mRNA levels relative to the reference gene ACT1.
(PDF)
Ratio: the target gene ERG11 mRNA levels relative to the reference gene ACT1.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


