Abstract
Association mapping and candidate gene studies within IBD linkage regions, as well as genome-wide association studies in CD have led to the discovery of multiple risk genes, but these only explain a fraction of the genetic susceptibility observed in IBD. We have thus been pursuing a region on chromosome 3p21-22 showing linkage to CD and UC using a gene-centric association mapping approach. We identified twelve functional candidate genes by searching for literature co-citations with relevant keywords and for gene expression patterns consistent with immune/intestinal function. We then performed an association study composed of a screening phase, where tagging SNPs were evaluated in 1020 IBD patients, and an independent replication phase in 745 IBD patients. These analyses identified and replicated significant association to IBD for four SNPs within a 1.2 Mb LD region. We then identified a non-synonymous coding variant (rs3197999, R689C) in the macrophage stimulating 1 (MST1) gene (p-value 3.62×10−6) that accounts for the association signal, and shows association to both CD and UC. MST1 encodes MSP, a protein regulating the innate immune responses to bacterial ligands. R689C is predicted to interfere with MSP binding to its receptor, suggesting a role for this gene in the pathogenesis of IBD.
INTRODUCTION
Crohn’s disease (CD) and ulcerative colitis (UC) are two related but distinct common inflammatory diseases of the gastrointestinal tract, collectively known as the inflammatory bowel diseases (IBD). The epidemiological data on IBD suggests a strong genetic contribution to disease pathogenesis and genetic studies indicate that CD and UC have disease-specific as well as shared susceptibility genes (1–3). Genome-wide linkage mapping studies in IBD have identified and confirmed several genomic intervals conferring risk to either CD, UC or both (4–11). Association mapping, as well as candidate gene association studies, within linkage regions have led to confirmed associations between Crohn’s disease and coding variants in the NOD2 gene on chromosome 16q12 and multiple associated alleles forming a risk haplotype known as IBD5 on chromosome 5q31 (8, 10, 12–14). More recently, genome-wide association (GWA) studies of CD have identified several novel CD associated risk loci, including IL23R, ATG16L1, PTGER4, IRGM, and NKX2-3 (15–19). Because of the unbiased nature of GWA studies, these have identified processes involved in the pathogenesis of CD, which had not been previously implicated. Given that all of the genome-wide association studies to date have been performed specifically on CD, it is not surprising that, with the exception of IL23R, most of the association findings have been CD-specific risk factors. A general conclusion that can be drawn from the genetic studies of IBD performed to date is that the confirmed associated alleles only modestly modify an individual’s risk and that no single study or approach is sufficient to identify all risk factors.
We therefore have been pursuing a gene-centric association mapping approach of a confirmed common IBD locus (ie conferring risk to both CD and UC) on chromosome 3p21-22. This region was first identified in a genome-wide linkage scan of British families (4) and then replicated with the findings from multiple genome-wide linkage scans (8, 11, 20). Subsequent association mapping studies using microsatellite markers provided additional evidence of association and some localization information (20, 21). In an attempt to further narrow down the genomic interval for our search, we have targeted our association study to a 30 Mb overlap region between the reported linked regions. This region contains over 400 annotated loci, several of which may represent interesting candidates for IBD. We selected 12 candidate genes with strong prior probability by searching for literature co-citations with relevant keywords (inflammation, immunity, intestinal disease etc.) and by searching publicly available datasets for gene expression patterns consistent with genes having a role in immune and/or intestinal tissues. We then performed a two-stage association study, composed of (1) a screening phase in a set of 1020 patients with IBD of European descent and (2) a follow-up independent replication phase in 745 patients with IBD from the North American NIDDK IBD Genetics Consortium. We report the identification of four SNPs showing significant evidence of association to IBD and replication in an independent cohort, all located within a single 1.2 Mb genetic interval. We then narrowed the associated interval to 336 Kb region and identified a non-synonymous coding variant (R689C) in the macrophage stimulating 1 gene (MST1) that is a common risk factor for IBD. MSP, the protein encoded by MST1, and its receptor MST1R have been reported to be involved in macrophage chemotaxis and activation (22, 23), as well as regulation of inflammatory responses (such as NO and IL12p40 production) following pro-inflammatory signals (24–27). Taken together these data suggest a role for MST1 in IBD pathogenesis.
Materials and Methods
Subject ascertainment and diagnostic classification
The Screening Cohort consists of a combined cohort of Canadian and Italian IBD trios and tetrads (249 father-mother-affected offspring trios), collected from multiple sites in the province of Quebec and at the S. Giovanni Rotondo ‘C.S.S’ (SGRC) Hospital in Italy, and of an Italian IBD case/control cohort (816 cases, 421 controls) also collected at the SGRC hospital. This cohort has previously been used and characterized in several association reports from our group (28–30), and additional cohort information can be found in Supplemental Table 1. The diagnosis of IBD required (a) one or more symptoms of diarrhea, rectal bleeding, abdominal pain, fever, or complicated perianal disease, (b) occurrence of symptoms on two or more occasions separated by at least 8 weeks or ongoing symptoms of at least 6 weeks duration, and (c) objective evidence of inflammation from radiologic, endoscopic, and histologic evaluation. All affected subjects fulfill clinical criteria for IBD (31, 32) and were consented using protocols approved by the institutional review boards of each institution contributing samples.
The Replication Cohort consisted of the combination of an IBD case/control cohort (353 cases and 344 controls) and of an IBD trios cohort (401 trios) collected from the North American NIDDK IBD Genetics Consortium (IBDGC) as previously described (15, 16). Additional information on this cohort can be found in Supplemental table 1 and 2. Cases and geographically matched controls, as well as nuclear families, were ascertained through the Cedars-Sinai Medical Center, Johns Hopkins University, University of Chicago, University of Montreal, University of Pittsburgh, and the University of Toronto Genetics Research Centers (GRCs). In all cases, informed consent was obtained using protocols approved by the local institutional review board in all participating institutions.
Candidate gene selection
The genomic region included in this analysis was selected based on an evaluation of the maximal overlap between previously reported linkage signals for IBD on chromosome 3p (4, 8, 11, 20). Using this approach, a 30 Mb region containing over 400 annotated loci was targeted (position 32185205 to 64772789).
Potential functional candidates were identified by searching publicly available online resources, such as PubMed, OMIM, Unigene and SymAtlas from the Genomic Institute of the Novartis Research Foundation, with a set of predefined search criteria. Specifically, the PubMed and OMIM databases were queried for genes having a link (1) to inflammation (over-/under-expression in inflamed tissues, and pro-/anti-inflammatory role), (2) to immunity (regulation of immune responses or immune cell proliferation), (3) to the development of a bowel disease (malformation, bowel cancer, and intestinal dysfunction), and (4) to experimental models of intestinal inflammation (genetic mouse model or activation/inhibition of experimental models). The Unigene and SymAtlas databases were also queried for genes showing expression patterns compatible with a role in immune and/or intestinal tissues (tissue-specific expression patterns or marked over-expression in tissues of immune or intestinal origin). Genes were given a score of one for each positive attribute and then ranked based on the sum of these scores. The top 12 ranking genes were selected for association analysis (Table 1).
Table 1.
Summary of association results for 12 candidate genes on chromosome 3p.
| IBD | CD | UC | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Locus | SNPs tested* | Associated SNP | Screen P** | Replication P*** | Combined P** | Combined P** | Combined P** |
| MLH1 | 5/8 | - | - | - | - | - | - |
|
| |||||||
| MYD88 | 5/7 | - | - | - | - | - | - |
|
| |||||||
| LTF | 5/7 | - | - | - | - | - | - |
|
| |||||||
| GPX1 | 4/5 | rs9818758 | 0.0098 | 0.0021 | 0.00013 | 0.00065 | 0.0089 |
|
| |||||||
| rs11716445 | 0.0078 | 0.0036 | 0.00016 | 0.0054 | 0.0041 | ||
|
| |||||||
| MST1 | 3/6 | rs9858213 | 0.0046 | 3.59×10−5 | 1.48×10−6 | 6.27×10−5 | 0.0013 |
|
| |||||||
| TRIP | 5/10 | rs9858428 | 0.00092 | 0.0018 | 1.02×10−5 | 0.00055 | 0.0011 |
|
| |||||||
| GNAI2 | 8/9 | rs2298952 | 0.0044 | NR | - | - | - |
|
| |||||||
| CISH | 7/8 | - | - | - | - | - | - |
| TLR9 | 5/5 | - | - | - | - | - | - |
| PRKCD | 12/15 | - | - | - | - | - | - |
| IL17RB | 9/10 | - | - | - | - | - | - |
| IL17RD | 20/20 | - | - | - | - | - | - |
“-” results not significant at p-value threshold 0.01
successful SNP assays/total SNPs selected to tag locus
2-tailed P-values
1-tailed P-value NR, non replicating
SNP selection
Screening phase: SNPs were selected from the HapMap database (www.hapmap.org) to capture the common genetic variation (allele frequency >5%) in and around each of the 12 candidate genes (the coding region plus 10–20 Kb on either side). Specifically, using the Tagger algorithm (33), we selected a set of 110 SNPs that tag all of the common genetic variation in these gene regions with a minimum pair-wise correlation coefficient (r2) of 0.8. Replication phase: The 5 SNPs showing evidence of association (p-value < 0.01 in the screening phase) were selected for replication; of these, 4 were successfully designed and tested on the replication cohort.
Genotyping and Quality control analyses
Genotyping for the screening and replication phases of the project were performed in-house (The Laboratory for Genetics and Genomic Medicine of Inflammation, www.inflammgen.org) using primer extension chemistry and mass spectrometric analysis (iPlex assay, Sequenom, San-Diego, USA) on the Sequenom MassArray.
Quality filtering of both samples and SNPs were performed prior to association testing to insure robust data quality for the screening phase. First, all SNPs with less than 75% genotyping call rates were excluded from the analysis. Then, an evaluation of empirical data quality distributions was performed and families and SNPs with an excess of Mendel errors (in the familial cohort) were removed. Finally, only DNAs and SNPs showing a minimum genotyping call rate of greater than 80% were included in the final analyses. The quality filtering brought the overall genotyping rate to 96.7% (ranging from 82.5% to 99.9%). Following these quality control steps, the dataset consisted of 89 SNPs tested on 805 cases and 412 controls and 215 trios (for a total of 1024 IBD cases, see supplemental Table 1 for further cohort information). Five SNPs showing evidence of association in the combined screening cohort (p<0.01) were then tested for replication in the combined NIDDK IBD case/control and trio replication cohort. Quality control thresholds used were the same as for the screening cohort, except for the final DNA and SNP quality threshold which were set at 90%. This quality filtering brought the overall genotyping to 99.6% (ranging from 98.4% to 100%). Following quality control, four of the five putatively associated SNPs were evaluated in 350 cases and 342 controls and on 395 trios (for a total of 745 IBD cases, see supplemental Table 1 for further cohort information). An evaluation of allelic frequencies for the different markers did not identify any significant differences between the different cohorts tested (data not shown).
Association testing
Association testing of single SNPs was performed by the transmission disequilibrium test (TDT), for trios, and by standard Chi-square test carried out on a 2×2 contingency table, for case/control cohorts, as implemented in the Haploview analysis software (34). Combination of the results derived from the case/control and trios/tetrads in the screening and replication cohorts, as well as combination of the results from the screen and replication, was achieved by the calculation of a combined Z-score as described previously (28). Association results (Table 1) are reported as 2-tailed nominal significance p-values for the screening, fine-mapping and combined cohorts, and 1-tailed p-values for the replication. The significance threshold for follow-up replication was set at a p-value < 0.01. Odds ratios and 95% confidence interval are also reported for the combined dataset. Conditional analyses between the different SNPs showing association were performed in the combined cohorts by logistic regression using the WHAP analysis software. The threshold for significant independent association signal was also set at a p-value < 0.01.
RESULTS
Gene-centric association study
Linkage evidence from multiple genome-wide and targeted studies indicates the presence of an IBD susceptibility locus (conferring risk to both CD and UC) in the chromosomal region 3p21-22. We selected a region for our association mapping project, based on the maximal overlap between previously reported linkage intervals. Since this approach defined a 30 Mb region containing over 400 annotated loci, we set out to identify potential functional candidates for inclusion in this project, based on published gene functions and expression patterns (see Materials and Methods). Genes were selected based on their reported roles in inflammation, immunity, bowel dysfunction, or intestinal inflammation in experimental models, as well as on expression patterns compatible with a role in immune or intestinal tissues. With this strategy, 12 candidate genes were selected for the screening phase (Table 1).
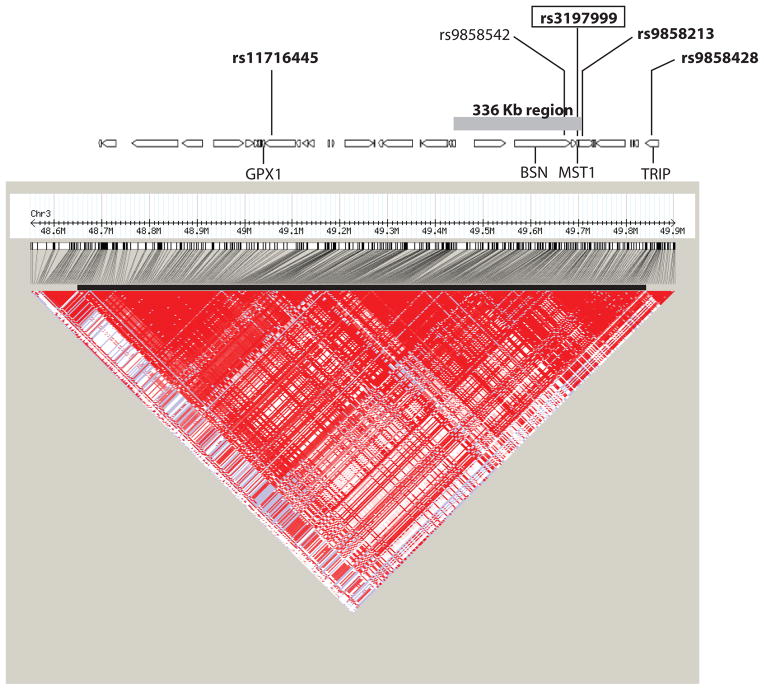
Eighty-nine SNPs tagging the common genetic variation surrounding these twelve candidate genes were typed in 215 father-mother-affected offspring trios and in 805 patients with IBD and 412 healthy controls, all of European ancestry. Allelic tests of association of the individual SNPs identified five SNPs with suggestive evidence of association (P<0.01) in the combined screening cohorts (Table 1). As illustrated in figure 1, four of these 5 SNPs were located within a 1.2 MB region (position 48700711 to 49869034) of high linkage disequilibrium (LD). The fifth SNP is located 392 Kb away from the interval described above.
Figure 1. Association signal and linkage disequilibrium patterns across the 1.2 Mb region of LD.
A candidate-gene approach to association mapping of the linked region on chromosome 3p21-22 suggests a putative association signal located in a 1.2 Mb region of LD containing over 30 genes. The LD plot of pairwise D′ values, derived from HapMap project data, indicates strong LD across the region (lower panel). Loci in the region tagged by the original study design are indicated on the gene track. The associated region of 336 Kb, as defined by the correlation neighborhood at r2>0.8, along with the replicated SNPs in the region are mapped against the gene track. SNPs in bold were identified in this study, the MST1 coding SNP is boxed, and the reported WTCCC SNP (rs9858542, in BSN) is in normal type.
In order to confirm the associations observed in this screening phase, we typed the five SNPs in an independent set of 350 patients with IBD and 342 controls as well as a cohort of 395 IBD families. Genotyping assays were successfully designed for all 5 putatively associated SNPs. All four of the SNPs located within the 1.2 Mb region had evidence of replication (P<0.01) in the combined replication cohorts whereas the fifth SNP outside this region did not replicate (Table 1). Specifically, the strongest association signal observed was to the MST1 tagging SNP rs9858213 (replication P=3.59×10−5; combined P=1.48×10−6; OR 1.33 (95%C.I. 1.18–1.49)). Although this SNP was selected to tag some of the regional variation around MST1, it is located approximately 5.5 Kb centromeric of MST1. The other 3 replicated SNPs had combined p-values ranging from 0.0016 and 1.02×10−5. Limiting the analysis to the CD and UC phenotypes did not improve the evidence for association for any of these SNPs, indicating that the identified association signals are to the general IBD phenotype.
Identification of association to a missense mutation in MST1
Given that there are multiple SNPs that have significant evidence of association in the screen and replication within a region of 1.2 Mb with high LD, it was important to determine whether there was a single or multiple independent association signals in the region. We therefore used the combined screening and replication cohorts, in order to have maximal statistical power, and performed conditional logistic regression analyses of the associated SNPs. Conditioning the data upon rs9858213, the primary association signal tagging MST1, did not identify additional secondary signals in the region (data not shown); indicating that the multiple associated SNPs are likely detecting the same correlated primary signal within this region.
In an attempt to delineate the minimal genomic interval defined by our associated signal, and thus potential genes implicated by this association, we evaluated the correlation neighborhood around rs9858213, as defined by all correlated SNPs at a specific correlation coefficient (r2) value of 0.8, in the HapMap dataset. More specifically, inclusion of all highly correlated SNPs (r2 > 0.8) to rs9858213 defined an associated genomic interval of 336Kb (Figure 1). The same 336 Kb region was also identified at a lower r2 threshold of 0.5, further strengthening the likelihood of an IBD susceptibility gene within this interval. This 336 Kb associated interval encompasses 10 genes, two of which (GPX1 and MST1) were selected in our original study design. The association signal for rs9858213 is also highly correlated (r2=0.95) with a signal recently reported by the Wellcome Trust Case Control Consortium (WTCCC) in the same region, providing independent replication evidence of our observation (19, 35). Specifically, the authors report association to a synonymous coding variant (rs9858542, O.R. 1.17 (95%C.I. 1.04–1.31)) of the BSN locus, within a 1 Mb genomic region of LD, but do not provide any additional fine-mapping to narrow down the associated interval. An evaluation of this SNP in our patient cohorts identifies an association signal (combined P=6.66×10−6; OR 1.29 (95%C.I. 1.15–1.49)) that is almost equivalent to the rs9858213 signal described above. In order to have greater statistical power to evaluate the contribution of this region to IBD susceptibility, we were interested in combining our association results with those from the WTCCC for rs9858542. Since there is extensive overlap between our replication cohort and the samples used in the NIDDK GWA scan, these cannot be considered as independent cohorts, and therefore we only included our current results and the WTCCC dataset in the combined analysis. Combining our data with the published WTCCC data unambiguously establishes this region as a risk factor of genome-wide significance for IBD with an OR of 1.24 (95% C.I.1.17–1.32; P= 8.45×10−13).
Having defined a region with unambiguous association to IBD, we then attempted to identify the primary causal allele underlying this association signal. For this, we investigated all highly correlated SNPs (r2 > 0.9) to both our association signal and the WTCCC marker. Using this approach, we identified a single non-synonymous coding variant in MST1 (rs3197999) with correlation coefficients of 0.90 and 0.95 to rs9858213 and rs9858542, respectively. This variant causes an arginine to cysteine amino acid substitution at position 689 (R689C) located in exon 18 of the gene. Typing and association testing of this variant in the combined screening and replication cohorts showed significant evidence of association (combined P=7.25×10−6). This missense coding variant (OR 1.30; 95%C.I. 1.16–1.46) and the rs9858213 SNP (OR 1.33; 95%C.I. 1.18–1.49) had equivalent strength of association. Reciprocal conditional analysis of rs3197999 confirms it as part of the same association signal as rs9858213 (data not shown) and therefore implicates MST1 as a potential risk factor for IBD.
The associated R689C variant and MST1 function
The MST1 locus encodes the macrophage stimulating protein (MSP), a plasminogen-related growth factor containing four Kringle domains and a C-terminal serine protease-like homology domain. This domain of MSP has been suggested to lack enzymatic activity due to amino acid substitutions in the active domain, however, it has been demonstrated to contain the high-affinity binding site for its receptor MST1R (36, 37). The MSP protein was originally identified as a serum protein produced by the liver that could elicit macrophage chemotaxis and activation (22, 23). MSP is secreted as a biologically inactive single chain precursor (pro-MSP) by the liver, and can be activated through proteolytic cleavage by serine proteases in areas of blood coagulation and active inflammation (38). Although the specific activating protease is unknown, a role for macrophage cell-surface proteases has been suggested from in vitro studies with cultured peritoneal macrophages and isolated macrophage membranes (39). MSP is the only known ligand for the MST1R, also known as RON receptor, a disulphide-linked heterodimer composed of an extracellular α chain and a transmembrane β chain with intrinsic kinase activity, expressed predominantly on tissue resident (differentiated) macrophages and epithelial cells (40). Signaling through the MSP/MST1R has been shown to inhibit LPS or cytokine mediated production of inflammatory mediators by macrophages (24–27), through the inhibition of NfkB signaling, while mice deficient in MST1R show increase susceptibility to septic shock following LPS challenge (41, 42). We have also observed, from publicly available microarray datasets, that MST1 gene expression is elevated during intestinal enterocyte differentiation in Caco-2 cells and in the biopsies from non-inflamed ileum of Crohn’s disease patients (data not shown).
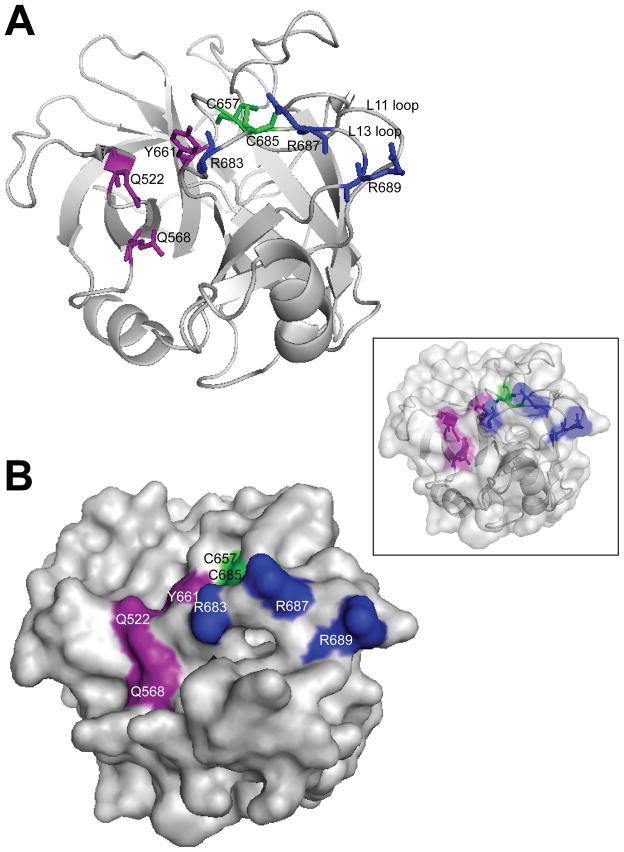
An alignment of MSP protein sequences from different species shows that this non-conservative amino acid substitution affects a residue that is conserved in mammals (Figure 2). By examining the MSP crystal structure data (PDB ID: 2ASU) (43), we note that R689 is located on the surface of the MSP β-chain and within a positively charged patch (L13 loop) that consists of a cluster of arginine residues including R683, which has been shown to be essential for MST1R binding (44) (Figure 3). The L13 loop borders the S1 specificity pocket (involving Y661, Q522 and Q568) and together, form the putative high-affinity binding surface for MST1R, resembling the structurally determined interaction between hepatocyte growth factor (scatter factor), which is structurally related to MSP, and its receptor, MET (45, 46). Furthermore, the proximity of R689C to a conserved cysteine residue (C685) that establishes a critical disulfide bond with C657 may introduce aberrant bond formations and disrupt the structural integrity of the adjacent receptor binding surface. Hence, the strategic location of the R689C variant will likely have a substantial impact on receptor binding or activation; the functional implication of which is an active focus of our on-going studies.
Figure 2. Sequence conservation of the arginine residue at position 689 of MSP across different species.
Amino acid sequence is shown as a single letter code (uppercase). The amino acid modified by the associated coding variant at position 689 (R689C) is in bold and demonstrates conservation of the arginine across mammalian species.
Figure 3. Three-dimensional representation of MSP.
(A) Ribbon diagram and (B) molecular surface representations of the MSP β chain. A merged superimposed image of the two representations is shown in the inset. The amino acid residues discussed in the text are highlighted in strong colors and labeled. The residues (Y661, Q522 and Q568) corresponding to the ‘catalytic triad’ of bona fide serine proteinases are colored red. The cluster of arginine residues (R689, R687 and R683) located within a positively charged patch on the L13 loop that forms a continuous region with the S1 specificity pocket, generating the putative primary (high-affinity) binding surface for its receptor, are colored blue. The two cysteine residues (C657 on loop L11 and C685 on loop L13) connected by a disulfide bond that lies in close proximity to the cluster of arginine residues, are colored green. The side-chains of the highlighted residues are displayed in the ribbon diagram as sticks. The structural representations were constructed with PyMOL (DeLano 2002) and the data was obtained from the Protein Data Bank (pdb: 2ASU) containing 3-D structure coordinates from X-ray crystallographic analysis of 225 residues of the MSP β chain, 4 residues of the α chain and 154 water molecules. Reference DeLano WL (2002). The PyMOL molecular graphics system. DeLano Scientific, Palo Alto, CA, USA.
DISCUSSION
Over the last 20 years, genome-wide linkage scans of both CD and UC have identified several chromosomal regions conferring susceptibility to these diseases (4–10). Two main strategies have been used to identify susceptibility loci within linkage intervals: (1) association fine mapping of the entire linked interval, or (2) positional candidate gene (gene-centric) association studies. Both of these approaches were independently used to successfully identify NOD2 variants within the IBD1 linkage peak in the 16q12 region (8, 10, 12–14). Recent genome-wide association (GWA) studies have identified and replicated multiple novel susceptibility genes for CD, and promise to identify many more genes in the near future (15–19). Interestingly, to date few of these novel susceptibility loci can explain the linkage peaks previously defined from the genome-wide linkage analyses, suggesting that complementary approaches will be necessary to identify the full complement of susceptibility genes for CD and UC.
The current study has adopted a positional candidate gene association strategy similar to the one used in the identification of NOD2, for examining the confirmed chromosome 3p21-22 IBD-specific linkage peak, although testing 12 different genes within the linked region. This approach takes advantage of the power of association testing while limiting the extent of statistical testing by focusing on a genomic region that has been implicated by multiple linkage studies and by targeting a limited set of genes with higher prior probability of being true IBD loci.
The results of the current study identify and replicate significant evidence of association to 4 SNPs located in a 1.2 Mb region of high linkage disequilibrium. Conditional logistic regression analysis of these associated SNPs defines a single association signal in the region. An analysis of the minimal associated interval for this signal, based on based on the evaluation of the correlation neighborhood at an r2>0.8 around the top associated signal (rs9858213), defines a narrower region of 336Kb containing only 10 genes. In a recently reported GWA study from the WTCCC, a single synonymous SNP (rs9858542) located within the BSN gene was significantly associated to CD. Testing of this SNP in our own screening and replication cohorts identifies an association signal that is equivalent in strength and that is highly correlated to rs9858213, with combined results providing overwhelming evidence that this region contains a gene for IBD (p value < 10−12). A second recent GWA association study (47), also reports a significant association signal within the same region for marker rs11718165, an intronic SNP also located in BSN. The authors report several associated SNPs across a 1.2 Mb region but do not narrow down the association further. Our current study, however, further identifies a single non-synonymous arginine to cysteine coding variant at base pair position 689 within exon 18 of the MST1 gene that can account for the association to this region. While the association results and the identification of a coding variant suggest a potential role for MST1 in susceptibility to both CD and UC, we cannot however formally exclude other variants in this region having an impact on disease susceptibility.
The R689C variant is located within an important region that forms part of the putative high-affinity receptor-binding surface with the S1 specificity pocket located in the serine proteinase homology domain of MSP. Several aspects of MSP activation and signaling make it an attractive functional candidate for IBD. First, proteolytic activation of MSP have been proposed to occur in areas of active inflammation through cell-surface proteases located on macrophages (38), (39). Second, while MSP was originally identified as a protein that could elicit macrophage chemotaxis and activation, signaling though the MSP/MST1R has recently been shown to inhibit LPS or cytokine mediated production of inflammatory mediators (such as NO, COX2, PGE2 and IL-12p40) by macrophages, through the inhibition of NfkB signaling (24–27). Interestingly, increased NfkB signaling activity, as well as increased levels on NO and expression of IL-12p40, have all been observed in the inflamed mucosa of patients with IBD (48–50). Finally, mice deficient in MST1R show increase susceptibility to septic shock following LPS challenge, highlighting the importance of the MSP/MST1R signaling pathway in controlling inflammation triggered from innate immune responses (41, 42). This dampening effect of the MSP/MST1R pathway on macrophages activation in response to microbial ligands, may be of particular interest in the intestinal epithelium where the innate immune system is under constant challenge, due to close contact with a variety of microorganisms. The loss of macrophage hyporesponsiveness or anergy has been proposed as a potential mechanism for triggering local inflammation and ultimately aspects of adaptive immunity implicated in IBD (51–53). The capacity of MSP to control IL-12p40 gene expression, a subunit of the heterodimeric IL-23, is also interesting in the context of IBD given the recent observations showing that IL23 regulation plays a central role in the control of intestinal inflammation (54, 55) and that neutralizing anti-IL-12p40 antibodies prevent the development of disease in murine models of IBD (56, 57). Finally, a recent report from the NIDDK IBD Genetic Consortium has identified a variant in a subunit of the IL23 receptor (IL-23R) that is associated with reduced risk of IBD, highlighting the importance of the IL23 signaling pathway in this disease.
Our identification of a missense coding variant in MST1 within the narrow interval defined by our association signal and the reported role of MSP in the regulation of innate immune responses implicate the MST1 gene, and its product MSP, in the pathogenesis of IBD. Interestingly, MST1 is one of a growing number of loci involved in the regulation of innate and adaptive immune responses, including NOD2, IL23R, IRGM and ATG16L1, which have been identified to impact on IBD. Further studies evaluating the functional significance of this coding variant on innate immune responses, on the MSP/RON signaling pathway in IBD and on its interaction with IL23R signaling will be essential for better understanding the mechanisms leading to IBD. It is interesting that both the MST1 and IL23R variant are among the few susceptibility genes that have been shown to impact on both CD and UC susceptibility. Their functional proximity in the regulation of IL23 signaling may point to common pathological mechanisms of intestinal inflammation postulated from epidemiological studies of IBD.
Supplementary Material
Acknowledgments
The North American IBD Genetics Consortium is funded by multiple grants from the National Institutes of Diabetes, Digestive and Kidney diseases (NIDDK): DK62431 (S.R.B.), DK62422/DK62429 (J.H.C.), DK62420 (R.H.D.), DK62432 (J.D.R.), DK62423 (M.S.S.), and DK62413 (K.D.T.). The current work was funded by an NIDDK grant (DK064869) to J.D.R and an NIAID grant (AI062773) to R.J.X. A.N. is the recipient of a fellowship from the Crohn’s and Colitis Foundation of America. The authors would like to thank Catherine Labbe for her contribution to the genotying performed as part of this project.
References
- 1.Binder V. Genetic epidemiology in inflammatory bowel disease. Dig Dis. 1998 Nov-Dec;16(6):351–5. doi: 10.1159/000016891. [DOI] [PubMed] [Google Scholar]
- 2.Orholm M, Munkholm P, Langholz E, Nielsen OH, Sorensen TI, Binder V. Familial occurrence of inflammatory bowel disease. N Engl J Med. 1991 Jan 10;324(2):84–8. doi: 10.1056/NEJM199101103240203. [DOI] [PubMed] [Google Scholar]
- 3.Probert CS, Jayanthi V, Hughes AO, Thompson JR, Wicks AC, Mayberry JF. Prevalence and family risk of ulcerative colitis and Crohn’s disease: an epidemiological study among Europeans and south Asians in Leicestershire. Gut. 1993 Nov;34(11):1547–51. doi: 10.1136/gut.34.11.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Satsangi J, Parkes M, Louis E, Hashimoto L, Kato N, Welsh K, et al. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet. 1996 Oct;14(2):199–202. doi: 10.1038/ng1096-199. [DOI] [PubMed] [Google Scholar]
- 5.Hampe J, Schreiber S, Shaw SH, Lau KF, Bridger S, Macpherson AJ, et al. A genomewide analysis provides evidence for novel linkages in inflammatory bowel disease in a large European cohort. Am J Hum Genet. 1999 Mar;64(3):808–16. doi: 10.1086/302294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho JH, Nicolae DL, Gold LH, Fields CT, LaBuda MC, Rohal PM, et al. Identification of novel susceptibility loci for inflammatory bowel disease on chromosomes 1p, 3q, and 4q: evidence for epistasis between 1p and IBD1. Proc Natl Acad Sci U S A. 1998 Jun 23;95(13):7502–7. doi: 10.1073/pnas.95.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y, Ohmen JD, Li Z, Bentley LG, McElree C, Pressman S, et al. A genome-wide search identifies potential new susceptibility loci for Crohn’s disease. Inflamm Bowel Dis. 1999 Nov;5(4):271–8. doi: 10.1097/00054725-199911000-00005. [DOI] [PubMed] [Google Scholar]
- 8.Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, McLeod RS, Griffiths AM, et al. Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet. 2000 Jun;66(6):1863–70. doi: 10.1086/302913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duerr RH, Barmada MM, Zhang L, Pfutzer R, Weeks DE. High-density genome scan in Crohn disease shows confirmed linkage to chromosome 14q11-12. Am J Hum Genet. 2000 Jun;66(6):1857–62. doi: 10.1086/302947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, et al. Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature. 1996 Feb 29;379(6568):821–3. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 11.Paavola P, Helio T, Kiuru M, Halme L, Turunen U, Terwilliger J, et al. Genetic analysis in Finnish families with inflammatory bowel disease supports linkage to chromosome 3p21. Eur J Hum Genet. 2001 May;9(5):328–34. doi: 10.1038/sj.ejhg.5200626. [DOI] [PubMed] [Google Scholar]
- 12.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001 May 31;411(6837):599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 13.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001 May 31;411(6837):603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 14.Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, et al. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet. 2001 Oct;29(2):223–8. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- 15.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006 Dec 1;314(5804):1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007 May;39(5):596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007 Apr 20;3(4):e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007 Feb;39(2):207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 19.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007 Jul;39(7):830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hampe J, Lynch NJ, Daniels S, Bridger S, Macpherson AJ, Stokkers P, et al. Fine mapping of the chromosome 3p susceptibility locus in inflammatory bowel disease. Gut. 2001 Feb;48(2):191–7. doi: 10.1136/gut.48.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duerr RH, Barmada MM, Zhang L, Achkar JP, Cho JH, Hanauer SB, et al. Evidence for an inflammatory bowel disease locus on chromosome 3p26: linkage, transmission/disequilibrium and partitioning of linkage. Hum Mol Genet. 2002 Oct 1;11(21):2599–606. doi: 10.1093/hmg/11.21.2599. [DOI] [PubMed] [Google Scholar]
- 22.Leonard EJ, Skeel AH. Isolation of macrophage stimulating protein (MSP) from human serum. Exp Cell Res. 1978 Jun;114(1):117–26. doi: 10.1016/0014-4827(78)90043-5. [DOI] [PubMed] [Google Scholar]
- 23.Leonard EJ, Skeel A. A serum protein that stimulates macrophage movement, chemotaxis and spreading. Exp Cell Res. 1976 Oct 15;102(2):434–8. doi: 10.1016/0014-4827(76)90065-3. [DOI] [PubMed] [Google Scholar]
- 24.Morrison AC, Wilson CB, Ray M, Correll PH. Macrophage-stimulating protein, the ligand for the stem cell-derived tyrosine kinase/RON receptor tyrosine kinase, inhibits IL-12 production by primary peritoneal macrophages stimulated with IFN-gamma and lipopolysaccharide. J Immunol. 2004 Feb 1;172(3):1825–32. doi: 10.4049/jimmunol.172.3.1825. [DOI] [PubMed] [Google Scholar]
- 25.Chen YQ, Fisher JH, Wang MH. Activation of the RON receptor tyrosine kinase inhibits inducible nitric oxide synthase (iNOS) expression by murine peritoneal exudate macrophages: phosphatidylinositol-3 kinase is required for RON-mediated inhibition of iNOS expression. J Immunol. 1998 Nov 1;161(9):4950–9. [PubMed] [Google Scholar]
- 26.Zhou YQ, Chen YQ, Fisher JH, Wang MH. Activation of the RON receptor tyrosine kinase by macrophage-stimulating protein inhibits inducible cyclooxygenase-2 expression in murine macrophages. J Biol Chem. 2002 Oct 11;277(41):38104–10. doi: 10.1074/jbc.M206167200. [DOI] [PubMed] [Google Scholar]
- 27.Liu QP, Fruit K, Ward J, Correll PH. Negative regulation of macrophage activation in response to IFN-gamma and lipopolysaccharide by the STK/RON receptor tyrosine kinase. J Immunol. 1999 Dec 15;163(12):6606–13. [PubMed] [Google Scholar]
- 28.Tello-Ruiz MK, Curley C, DelMonte T, Giallourakis C, Kirby A, Miller K, et al. Haplotype-based association analysis of 56 functional candidate genes in the IBD6 locus on chromosome 19. Eur J Hum Genet. 2006 Jun;14(6):780–90. doi: 10.1038/sj.ejhg.5201612. [DOI] [PubMed] [Google Scholar]
- 29.van Bodegraven AA, Curley CR, Hunt KA, Monsuur AJ, Linskens RK, Onnie CM, et al. Genetic variation in myosin IXB is associated with ulcerative colitis. Gastroenterology. 2006 Dec;131(6):1768–74. doi: 10.1053/j.gastro.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 30.De Jager PL, Franchimont D, Waliszewska A, Bitton A, Cohen A, Langelier D, et al. The role of the Toll receptor pathway in susceptibility to inflammatory bowel diseases. Genes Immun. 2007 Jul;8(5):387–97. doi: 10.1038/sj.gene.6364398. [DOI] [PubMed] [Google Scholar]
- 31.Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170:2–6. doi: 10.3109/00365528909091339. discussion 16–9. [DOI] [PubMed] [Google Scholar]
- 32.Podolsky DK. Inflammatory bowel disease (1) N Engl J Med. 1991 Sep 26;325(13):928–37. doi: 10.1056/NEJM199109263251306. [DOI] [PubMed] [Google Scholar]
- 33.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005 Nov;37(11):1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 34.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 35.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007 Jun 7;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han S, Stuart LA, Degen SJ. Characterization of the DNF15S2 locus on human chromosome 3: identification of a gene coding for four kringle domains with homology to hepatocyte growth factor. Biochemistry. 1991 Oct 8;30(40):9768–80. doi: 10.1021/bi00104a029. [DOI] [PubMed] [Google Scholar]
- 37.Yoshimura T, Yuhki N, Wang MH, Skeel A, Leonard EJ. Cloning, sequencing, and expression of human macrophage stimulating protein (MSP, MST1) confirms MSP as a member of the family of kringle proteins and locates the MSP gene on chromosome 3. J Biol Chem. 1993 Jul 25;268(21):15461–8. [PubMed] [Google Scholar]
- 38.Wang MH, Gonias SL, Skeel A, Wolf BB, Yoshimura T, Leonard EJ. Proteolytic activation of single-chain precursor macrophage-stimulating protein by nerve growth factor-gamma and epidermal growth factor-binding protein, members of the kallikrein family. J Biol Chem. 1994 May 13;269(19):13806–10. [PubMed] [Google Scholar]
- 39.Wang MH, Skeel A, Leonard EJ. Proteolytic cleavage and activation of pro-macrophage-stimulating protein by resident peritoneal macrophage membrane proteases. J Clin Invest. 1996 Feb 1;97(3):720–7. doi: 10.1172/JCI118470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwama A, Wang MH, Yamaguchi N, Ohno N, Okano K, Sudo T, et al. Terminal differentiation of murine resident peritoneal macrophages is characterized by expression of the STK protein tyrosine kinase, a receptor for macrophage-stimulating protein. Blood. 1995 Nov 1;86(9):3394–403. [PubMed] [Google Scholar]
- 41.Waltz SE, Eaton L, Toney-Earley K, Hess KA, Peace BE, Ihlendorf JR, et al. Ron-mediated cytoplasmic signaling is dispensable for viability but is required to limit inflammatory responses. J Clin Invest. 2001 Aug;108(4):567–76. doi: 10.1172/JCI11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Correll PH, Iwama A, Tondat S, Mayrhofer G, Suda T, Bernstein A. Deregulated inflammatory response in mice lacking the STK/RON receptor tyrosine kinase. Genes Funct. 1997 Feb;1(1):69–83. doi: 10.1046/j.1365-4624.1997.00009.x. [DOI] [PubMed] [Google Scholar]
- 43.Carafoli F, Chirgadze DY, Blundell TL, Gherardi E. Crystal structure of the beta-chain of human hepatocyte growth factor-like/macrophage stimulating protein. Febs J. 2005 Nov;272(22):5799–807. doi: 10.1111/j.1742-4658.2005.04968.x. [DOI] [PubMed] [Google Scholar]
- 44.Danilkovitch A, Miller M, Leonard EJ. Interaction of macrophage-stimulating protein with its receptor. Residues critical for beta chain binding and evidence for independent alpha chain binding. J Biol Chem. 1999 Oct 15;274(42):29937–43. doi: 10.1074/jbc.274.42.29937. [DOI] [PubMed] [Google Scholar]
- 45.Kirchhofer D, Yao X, Peek M, Eigenbrot C, Lipari MT, Billeci KL, et al. Structural and functional basis of the serine protease-like hepatocyte growth factor beta-chain in Met binding and signaling. J Biol Chem. 2004 Sep 17;279(38):39915–24. doi: 10.1074/jbc.M404795200. [DOI] [PubMed] [Google Scholar]
- 46.Stamos J, Lazarus RA, Yao X, Kirchhofer D, Wiesmann C. Crystal structure of the HGF beta-chain in complex with the Sema domain of the Met receptor. Embo J. 2004 Jun 16;23(12):2325–35. doi: 10.1038/sj.emboj.7600243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raelson JV, Little RD, Ruether A, Fournier H, Paquin B, Van Eerdewegh P, et al. Genome-wide association study for Crohn’s disease in the Quebec Founder Population identifies multiple validated disease loci. Proc Natl Acad Sci U S A. 2007 Sep 11;104(37):14747–52. doi: 10.1073/pnas.0706645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schreiber S, Nikolaus S, Hampe J. Activation of nuclear factor kappa B inflammatory bowel disease. Gut. 1998 Apr;42(4):477–84. doi: 10.1136/gut.42.4.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura H, Miura S, Shigematsu T, Ohkubo N, Tsuzuki Y, Kurose I, et al. Increased nitric oxide production and inducible nitric oxide synthase activity in colonic mucosa of patients with active ulcerative colitis and Crohn’s disease. Dig Dis Sci. 1997 May;42(5):1047–54. doi: 10.1023/a:1018849405922. [DOI] [PubMed] [Google Scholar]
- 50.Omata F, Birkenbach M, Matsuzaki S, Christ AD, Blumberg RS. The expression of IL-12 p40 and its homologue, Epstein-Barr virus-induced gene 3, in inflammatory bowel disease. Inflamm Bowel Dis. 2001 Aug;7(3):215–20. doi: 10.1097/00054725-200108000-00006. [DOI] [PubMed] [Google Scholar]
- 51.Smith PD, Ochsenbauer-Jambor C, Smythies LE. Intestinal macrophages: unique effector cells of the innate immune system. Immunol Rev. 2005 Aug;206:149–59. doi: 10.1111/j.0105-2896.2005.00288.x. [DOI] [PubMed] [Google Scholar]
- 52.Smythies LE, Sellers M, Clements RH, Mosteller-Barnum M, Meng G, Benjamin WH, et al. Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J Clin Invest. 2005 Jan;115(1):66–75. doi: 10.1172/JCI19229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, et al. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999 Jan;10(1):39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 54.Fuss IJ, Becker C, Yang Z, Groden C, Hornung RL, Heller F, et al. Both IL-12p70 and IL-23 are synthesized during active Crohn’s disease and are down-regulated by treatment with anti-IL-12 p40 monoclonal antibody. Inflamm Bowel Dis. 2006 Jan;12(1):9–15. doi: 10.1097/01.mib.0000194183.92671.b6. [DOI] [PubMed] [Google Scholar]
- 55.Becker C, Wirtz S, Blessing M, Pirhonen J, Strand D, Bechthold O, et al. Constitutive p40 promoter activation and IL-23 production in the terminal ileum mediated by dendritic cells. J Clin Invest. 2003 Sep;112(5):693–706. doi: 10.1172/JCI17464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, McKenzie BS, et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006 Oct 30;203(11):2473–83. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kullberg MC, Jankovic D, Feng CG, Hue S, Gorelick PL, McKenzie BS, et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J Exp Med. 2006 Oct 30;203(11):2485–94. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.