SUMMARY
CodY and ScoC are Bacillus subtilis transcriptional regulators that control the expression of dozens of genes and operons. Using scoC-lacZ fusions and DNA-binding experiments, we show here that scoC is directly repressed by CodY. This effect creates multiple forms of cascade regulation. For instance, expression of the dtpT gene, which is directly and negatively controlled by ScoC and encodes a putative oligopeptide permease, was activated indirectly by CodY due to CodY-mediated repression of scoC. The opp operon, which encodes an oligopeptide permease that is essential for sporulation and genetic competence development, proved to be a direct target of repression by both ScoC and CodY, but was not significantly affected in codY or scoC single mutants. The combined actions of CodY and ScoC maintain opp repression when either one of the regulators loses activity, but limit the level of repression to that provided by one of the regulators acting alone. Under conditions of nitrogen limitation, repression by ScoC of dtpT and opp was partly prevented by TnrA. Thus, the functioning of ScoC is determined by other transcription factors via modulation of its expression or DNA binding.
Keywords: Bacillus subtilis, CodY, ScoC, TnrA, oligopeptide permease
INTRODUCTION
CodY, a global regulatory protein first identified in Bacillus subtilis (Slack et al., 1995), controls directly or indirectly the transcription of more than 200 genes, many of which encode metabolic pathways involved in nutrient acquisition by macromolecular degradation, nutrient transport, and intracellular catabolism, as well as utilization of nutrients for biosynthesis (Barbieri et al., 2015; Belitsky and Sonenshein, 2013; Brinsmade et al., 2014; Molle et al., 2003; Sonenshein, 2005; Sonenshein, 2007). Homologs of CodY are present in most other low G+C Gram-positive bacteria and have been shown to play a global role in metabolic regulation and in coordinating expression of virulence-associated and metabolic genes (Sonenshein, 2007) [(see also refs. (Zhang et al., 2014), (Roux et al., 2014) and references therein)].
The DNA-binding affinity of CodY from B. subtilis and most other species is increased by interaction with two types of ligands, the branched-chain amino acids [isoleucine, leucine, and valine (ILV)] (Guedon et al., 2001; Petranovic et al., 2004; Shivers and Sonenshein, 2004) and GTP (Brinsmade and Sonenshein, 2011; Handke et al., 2008; Molle et al., 2003; Ratnayake-Lecamwasam et al., 2001; Shivers and Sonenshein, 2004). Most genes regulated by CodY have in their regulatory or coding sequences sites of direct CodY binding (Belitsky and Sonenshein, 2013; Molle et al., 2003). In these cases, there are multiple mechanisms by which CodY alters gene expression (Belitsky, 2011): they include binding within or near a promoter site to activate or block RNA polymerase binding, competing with a positive regulator for binding near the promoter region, and binding within a coding sequence in order to act as a roadblock to RNA polymerase.
Interestingly, genome-wide analyses have revealed that some genes whose expression is affected by a codY null mutation do not have CodY-binding sites and that expression of some genes that have CodY-binding sites is not affected by codY mutations (Belitsky and Sonenshein, 2013; Molle et al., 2003). These findings could potentially be explained by complex regulatory schemes in which CodY regulates genes indirectly by controlling the synthesis of the direct regulators or in which CodY and other proteins regulate the same genes redundantly. In fact, established or potential targets of CodY regulation in B. subtilis include several genes encoding known or probable transcriptional regulators (Belitsky and Sonenshein, 2013; Brinsmade et al., 2014; Molle et al., 2003), creating the potential for regulatory cascades involving two or more transcriptional regulators. For instance, the negative effect of CodY on expression of ComK, an activator of genetic competence genes, has been previously described (Serror and Sonenshein, 1996a; Smits et al., 2007).
The B. subtilis scoC gene (formerly known as hpr or catA) codes for a transcriptional regulator that negatively controls expression of multiple genes, including some that encode extracellular proteases and oligopeptide permeases (Caldwell et al., 2001; Dod et al., 1978; Higerd et al., 1972; Kallio et al., 1991; Koide et al., 1999). In cells growing in rich media, many of these genes are expressed more strongly during the transition from exponential growth phase to stationary phase (Strauch and Hoch, 1993). ScoC is also involved in the regulation of sporulation (Dod and Balassa, 1978; Perego and Hoch, 1988).
ScoC is a 203-amino acid member of the MarR family of transcriptional regulators (Kallio et al., 1991; Perego and Hoch, 1988; Perera and Grove, 2010). It is a dimeric (or possibly tetrameric) protein containing a winged helix-turn-helix DNA-binding motif. The three-dimensional structure of ScoC (PDB 2FXA) resembles that of many other proteins of the MarR family despite the low level of sequence similarity (http://www.rcsb.org/pdb/explore/explore.do?structureId=2FXA). ScoC is longer by about 40 amino acids than most members of the MarR family; the role of this C-terminal extension in ScoC activity remains unknown. ScoC is present in most Bacillus species and in bacteria of several related genera.
Global analyses of gene expression have indicated that expression of the B. subtilis scoC gene is increased ~3-fold in a codY null mutant (http://www.genome.jp/kegg/expression/) (Brinsmade et al., 2014), although scoC was not detected as a direct CodY target in ChIP-to-chip experiments (Molle et al., 2003) and was not on the list of the strongest CodY-binding sites detected by IDAP-Seq (Belitsky and Sonenshein, 2013). However, a weaker CodY-binding site, just beyond the arbitrary cut-off that defines the strongest sites, was in fact detected in the regulatory region of scoC by IDAP-Seq [Dataset S2 of ref. (Belitsky and Sonenshein, 2013)].
In this work, we showed that ScoC expression is indeed under direct CodY control and that CodY-mediated repression of scoC leads to an underestimation of the potential repressive effect of ScoC under conditions when CodY is active. CodY appears to employ ScoC as part of a regulatory cascade to maintain or increase repression of certain genes under conditions in which CodY loses activity. Moreover, some genes are subject to direct negative control by both ScoC and CodY, which creates a feed-forward regulatory loop. In addition, the functioning of ScoC proved to be influenced by the global nitrogen metabolism regulator, TnrA, indicating that B. subtilis has evolved complex mechanisms for determining the level of expression of certain genes under a variety of nutritional states.
RESULTS
CodY-dependent regulation of the scoC gene
Using a transcriptional fusion, scoC561-lacZ, containing a 561-bp fragment that includes the entire intergenic region upstream of the scoC gene, the 3’ end of the upstream yhaH gene and 102 bp of the scoC coding region (Fig. 1A), we determined that expression of the fusion under conditions of maximal CodY activity in minimal (TSS) glucose-ammonium medium containing a mixture of 16 amino acids (hereafter referred to as the TSS + 16 aa medium) was 4.6-fold higher in the codY null mutant strain GB1041 than in the wild-type strain GB1039 (Table 1). This result is in accord with the results of genome-wide analyses of the CodY regulon (http://www.genome.jp/kegg/expression/) (Brinsmade et al., 2014).
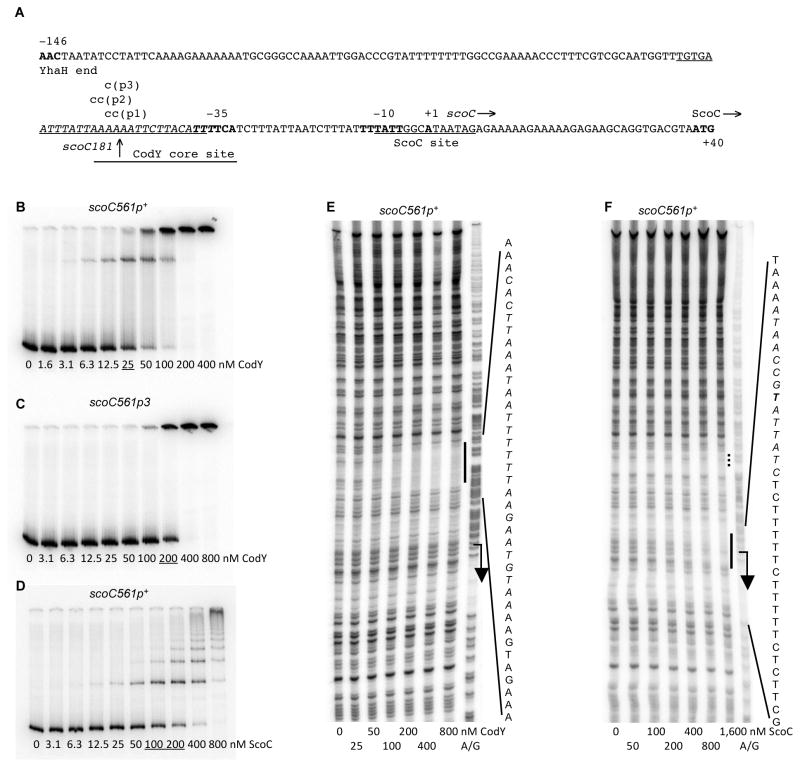
Fig. 1.
Binding of CodY and ScoC to the scoC regulatory region.
A. The sequence (5’ to 3’) of the coding (non-template) strand of the scoC regulatory region. Coordinates are reported with respect to the transcription start point (Abe et al., 2009). The upstream endpoints of inserts within the scoC561 and scoC350 fusions are at positions −422 and −211, respectively. The upstream boundary of the scoC181-lacZ fusion at position −42 is indicated by a vertical arrow. The downstream boundary for all fusions is at position +139. The likely translation initiation codon (ATG), −10 and −35 promoter regions, and transcription start point are in bold. The directions of transcription and translation are indicated by the horizontal arrows. The mutated nucleotides are shown in lowercase above the sequence. The sequences on the template strand that were protected by CodY or ScoC in DNase I footprinting experiments are underlined. The core CodY-binding site identified by IDAP-Seq (Fig. 2A) is shown by a horizontal line below the sequence. Two overlapping CodY-binding motifs with 4 and 5 mismatches to the consensus are italicized; two additional motifs with 4 and 5 mismatches to the consensus can be found in the promoter region but are not indicated.
B and C. Gel shift assays of CodY binding in the presence of 10 mM ILV to radioactively labeled scoC561p+ (B) or scoC561p3 (C) PCR fragments obtained with oligonucleotides oBB67 and oBB102. CodY concentrations used (nM of monomer) are reported below each lane; concentrations corresponding to the apparent KD for binding are underlined.
D. Gel shift assay of ScoC binding to the scoC561p+ PCR fragment. ScoC concentrations (nM of monomer) are reported below each lane; concentrations corresponding to the apparent KD for binding are underlined.
E and F. DNase I footprinting analysis of protein binding to the scoC regulatory region. The scoC561p+ DNA fragment labeled on the template strand and used in panel B was incubated with increasing amounts of purified (E) CodY in the presence of 10 mM ILV or (F) ScoC and then treated with DNase I. The protected areas are indicated by vertical lines and the corresponding sequences are reported; the protected nucleotides are italicized. The transcription start point and direction of transcription are shown by a bent arrow. Protein concentrations used (nM of monomer) are indicated below each lane. The A + G sequencing ladder of the template DNA strand is shown in the right lane in each case.
Table 1.
Expression of scoC-lacZ fusions
| Strain | Fusion promoter | Relevant genotype | β-Galactosidase activity | Fold regulation | ||
|---|---|---|---|---|---|---|
| codY/codY+ | scoC/scoC+ | abrB/abrB+ | ||||
| GB1039 | scoC561p+ | wild type | 52.0 | 4.6 | 1.3 | 0.95 |
| GB1041 | codY | 241.4 | 1.8 | |||
| GB1061 | scoC | 68.8 | 6.2 | |||
| GB1063 | codY scoC | 427.2 | 0.80 | |||
| BB3897 | abrB | 49.4 | ||||
| BB3907 | codY scoC abrB | 342.4 | ||||
| BB3890 | scoC350p+ | wild type | 48.6 | 4.7 | 1.8 | |
| BB3902 | codY | 228.4 | 2.0 | |||
| BB3901 | scoC | 87.1 | 5.1 | |||
| BB3908 | codY scoC | 445.4 | ||||
| BB3898 | scoC181p+ | wild type | 120.4 | 0.94 | 1.0 | |
| BB3904 | codY | 113.5 | 1.1 | |||
| BB3903 | scoC | 120.8 | 0.99 | |||
| BB3909 | codY scoC | 119.8 | ||||
| GB1045 | scoC561p1 | wild type | 125.5 | 0.61 | 1.8 | |
| GB1046 | codY | 76.7 | 3.3 | |||
| BB4001 | scoC | 236.1 | 1.1 | |||
| BB4002 | codY scoC | 255.3 | ||||
| GB1059 | scoC561p2 | wild type | 25.1 | 1.4 | 2.6 | |
| GB1060 | codY | 34.0 | 3.2 | |||
| BB4003 | scoC | 68.6 | 1.6 | |||
| BB4004 | codY scoC | 107.5 | ||||
| BB3919 | scoC561p3 | wild type | 8.64 | 0.55 | 1.2 | |
| BB3925 | codY | 4.71 | 2.3 | |||
| BB3920 | scoC | 10.7 | 1.0 | |||
| BB3926 | codY scoC | 10.8 | ||||
Cells were grown in TSS glucose-ammonium medium with a mixture of 16 amino acids. β-Galactosidase activity was assayed and expressed in Miller units. All values are averages of at least two experiments, and the standard errors of the mean did not exceed 30%.
Because it is common for transcriptional regulators to be autoregulated, we tested expression of the scoC561-lacZ fusion in a scoC null mutant. Indeed, expression of the fusion was increased 1.3- to 1.8-fold in codY+ and codY mutant strains, respectively, indicating a low level of negative autoregulation. The autorepression appears to increase in a codY mutant strain, consistent with the higher level of ScoC expression in this strain (Table 1). The negative autoregulation by ScoC should moderate the effect of scoC derepression in a codY mutant. In a codY scoC double mutant, expression of the scoC561-lacZ fusion was 8.2-fold higher than in a wild-type strain, consistent with the conclusion that effects of thecodY and scoC mutations are additive (Table 1, strain GB1063). Thus, the results show that the two proteins control scoC independently.
ScoC was reported to be under positive control by AbrB, an important transition state regulator (Perego and Hoch, 1988; Strauch and Hoch, 1993). However, little or no effect of an abrB null mutation on scoC561-lacZ expression was observed in TSS + 16 aa medium (Table 1).
The scoC350-lacZ fusion, containing a similar fragment of the scoC regulatory region, but truncated by 211 bp from the 5’ end, had nearly the same activity as the longer fusion in all strains tested, indicating that all regions important for expression and regulation were retained within the shorter fusion (Table 1, strain BB3890 and derivatives). However, a scoC181-lacZ fusion that lacks an additional 169 bp from the 5’ end and retains only 42 bp upstream of the scoC transcription start point (Fig. 1A), lost the ability to be regulated by either CodY or ScoC (Table 1, strain BB3898 and derivatives). Under fully derepressing conditions, in a scoC codY double mutant, the expression level of the scoC181-lacZ fusion was ~4-fold less than that of the other two fusions (Table 1), indicating that this fusion construct contains a scoC promoter with reduced activity, presumably because the fusion lacks the sequence just upstream of the −35 region (Fig. 1A).
In gel shift experiments, purified CodY bound to a DNA fragment containing the scoC regulatory region with moderate affinity (apparent KD 25 nM; the equilibrium dissociation constant was estimated as the protein concentration needed to shift 50% of DNA fragments under conditions of vast protein excess over DNA) (Fig. 1B). In general, non-specific binding of CodY in gel shift experiments is observed only at 400–800 nM CodY (Belitsky and Sonenshein, 2011a; Belitsky and Sonenshein, 2011b).
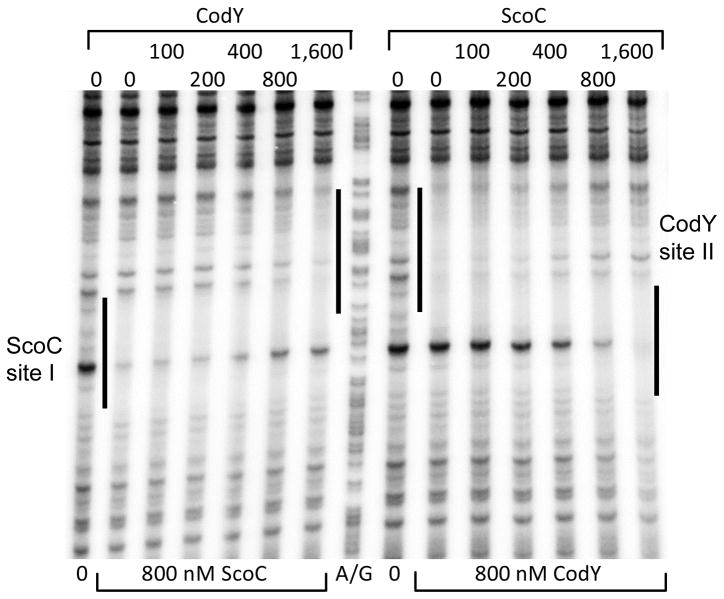
DNase I footprinting experiments showed that CodY protects a region from positions −58 to −31 with respect to the scoC transcription start point (Fig. 1A and 1E). This region corresponds very well to the core CodY-binding site, from positions −51 to −31, as determined by the global IDAP-Seq analysis (Fig. 1A and 2A)(Belitsky and Sonenshein, 2013); a core binding site is defined as a sequence that is limited by the 5’ and 3’ nucleotides that are essential for CodY binding. The scoC CodY-protected site includes two 15-bp sequences, motif I (from positions −53 to −39) and motif II (from positions −44 to −30), that have 4 and 5 mismatches, respectively, with respect to the CodY-binding consensus motif, AATTTTCWGAAAATT (Belitsky and Sonenshein, 2008; den Hengst et al., 2005; Guedon et al., 2005) (we use the terms “site” and “motif” to describe an experimentally determined location of CodY binding and a 15-bp sequence that is similar to the CodY-binding consensus, respectively) (Fig. 1A). The two motifs overlap each other by 6 bp as is common for many CodY-binding sites (Belitsky and Sonenshein, 2013; Wray and Fisher, 2011). A third CodY-binding motif with 5 mismatches with respect to the consensus is located at positions −65 to −51 and also overlaps the binding site determined by footprinting; a fourth motif with 4 mismatches almost fully overlaps motif I (from positions −45 to −31).
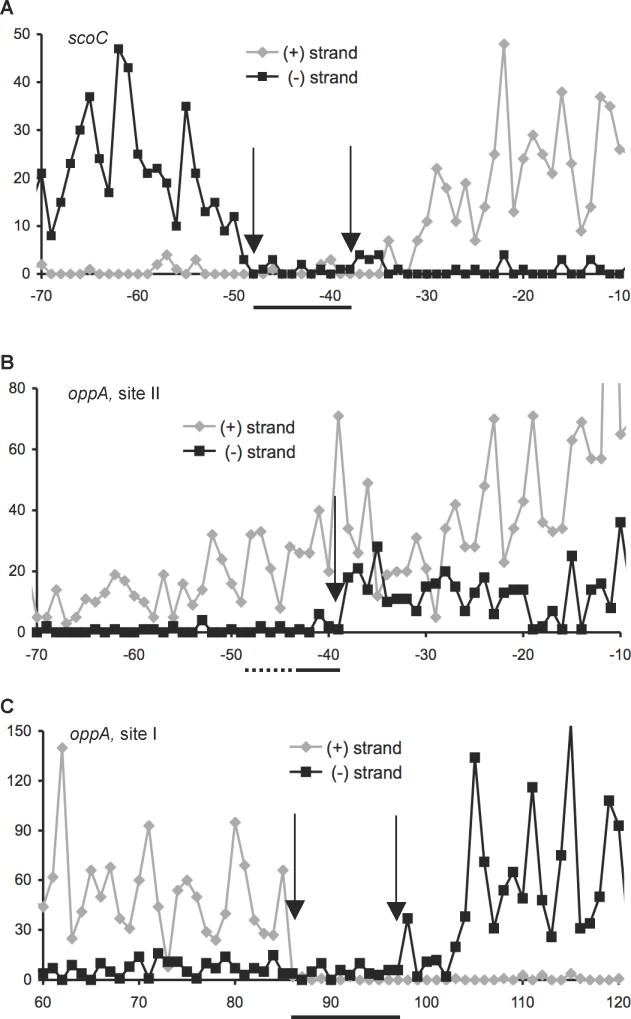
Fig. 2.
Coverage maps of CodY-binding sites in the (A) scoC and (B and C) oppA regulatory regions using strand-specific counting of 5’ nucleotides of Illumina sequencing reads obtained in IDAP-Seq experiments. The coverage of the (+) and (−) strands is shown in grey and black, respectively. Arrows indicate boundaries of coverage gaps corresponding to the boundaries of core binding sites at the CodY concentration used. The core site location is also indicated by horizontal lines below x-axes; the 5’ end of the oppA site II was not determined. Forty nM CodY was used for affinity purification of DNA fragments in presented experiments; the boundaries of the core sites, mentioned in the text, were deduced from multiple experiments using a range of CodY concentrations. Non-uniform coverage is likely due to varying position-specific efficiency of DNA shearing during sonication and to non-uniform size of DNA fragments. Coordinates are reported with respect to the transcription start points. The results shown here were deduced from data provided as supplementary material in a previous publication (Belitsky and Sonenshein, 2013) but were not previously analyzed.
Only low-affinity binding of purified ScoC (apparent KD 150 nM) to the scoC regulatory fragment could be observed in gel shift experiments (Fig. 1D). In our hands, despite the presence of a large excess of salmon sperm DNA in the binding buffer, ScoC had a propensity to bind non-specifically to several randomly chosen DNA fragments with an apparent KD of 200–400 nM (data not shown), making interpretation of scoC gel shift experiments difficult. Low-affinity binding of ScoC from positions −7 to +7 with respect to the scoC transcription start point was detected in DNase I footprinting assays (Fig. 1F). Therefore, it is likely that the low-level negative autoregulation of ScoC reflects a direct effect.
Mutagenesis of the scoC CodY-binding site
We introduced separately three single or double substitution mutations into the CodY-binding site of the scoC561-lacZ fusion in such a way as to reduce the similarity of one or both of the CodY-binding motifs I and II to the consensus (Fig. 1A). All three mutations abolished or strongly reduced the derepressing effect of a codY null mutation (Table 1). In fact, expression of the fusions containing the stronger p1 or p3 mutations in a codY mutant strain was lower than in a wild-type strain, probably indicating stronger negative autoregulation by the elevated level of ScoC (Table 1). (Note that in all these strains ScoC protein is expressed from a wild-type promoter). In gel shift experiments, a scoC fragment containing the p3 mutation interacted with CodY ~8-fold more weakly than did a corresponding wild-type fragment and did not form a complex with an intermediate mobility (Fig. 1C), confirming that the mutation severely impaired the interaction between CodY and the scoC promoter. Taken together, the in vivo and in vitro results indicate that CodY binding to the scoC promoter region is directly responsible for regulation.
The mutations also reduced the derepressed level of fusion expression in a codY scoC double mutant (Table 1), suggesting that all three mutations directly affect the intrinsic activity of the scoC promoter. This outcome is likely due to the positions of the mutations within the AT-rich sequence (a probable UP element) upstream of the −35 promoter region (Helmann, 1995; Meijer and Salas, 2004). However, we cannot explain why the single mutation (A-44C) in the p3 mutant promoter had a much stronger negative effect on scoC expression than did the double mutation (A-44C, A-43C) in the p1 mutant promoter.
Cascade regulation of the dtpT (yclF) gene by CodY and ScoC
Most previous studies aimed at analyzing ScoC function were performed in amino acid-containing media (Caldwell et al., 2001; Dod et al., 1978; Koide et al., 1999), i.e., under growth conditions that allow CodY to be active and repress scoC. Repression of scoC by CodY may have caused an underestimation of the potential role of ScoC in gene regulation. To test the extent to which CodY modulates ScoC-mediated regulation, we sought a reporter gene that is regulated by ScoC but is not subject to direct CodY-mediated regulation.
The dtpT gene (also called yclF) is the gene most highly (6-fold) negatively regulated by ScoC during exponential growth in rich nutrient broth medium (Caldwell et al., 2001). It encodes a protein of uncharacterized function but with strong similarity to certain oligopeptide permeases (Doki et al., 2013; Kunji et al., 1993). Genome-wide analysis of CodY binding to B. subtilis chromosomal DNA failed to show any binding of CodY in the vicinity of dtpT either in vivo or in vitro (Belitsky and Sonenshein, 2013; Molle et al., 2003).
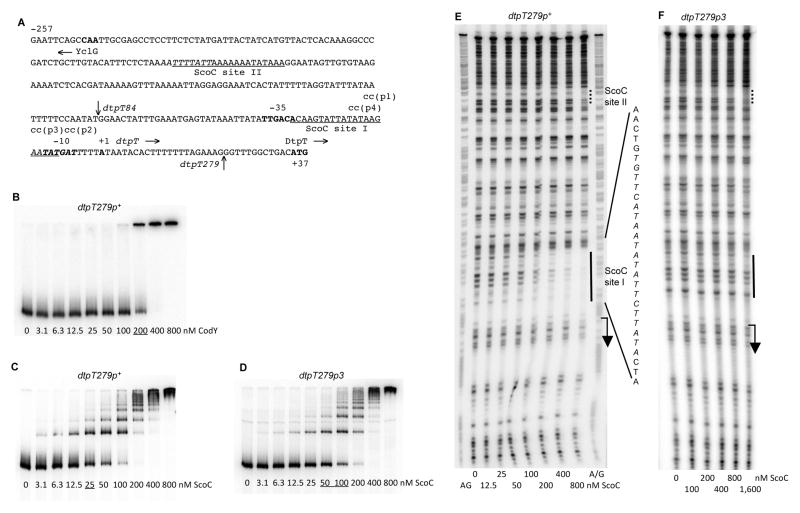
In gel shift experiments using a fragment containing the entire 279-bp intergenic region upstream of dtpT (Fig. 3A), CodY only bound at a concentration of 200 nM or higher, which is unlikely to be of physiological significance (Fig. 3B). By contrast, ScoC bound the dtpT regulatory region with an apparent KD of ~25 nM, i. e., with a 6- to 16-fold higher affinity than that for the regulatory regions of scoC and several other genes (Fig. 3C). This binding appears to be specific, although formation of multiple protein-DNA complexes may reflect a non-specific component of ScoC binding. DNase I footprinting experiments showed that ScoC protects a 21-bp region, site I, from positions −28 to −8 with respect to the transcription start point (see below), an appropriate location for repression (Fig. 3E). Another weak ScoC-binding site, site II, is located from positions −171 to −152 (Fig. 3E).
Fig. 3.
Binding of CodY and ScoC to the dtpT regulatory region.
A. The sequence (5’ to 3’) of the coding (non-template) strand of the dtpT regulatory region. Coordinates are reported with respect to the transcription start point as deduced from the mutational analysis (see text). The 5’ nucleotide of the sequence presented corresponds to the first nucleotide of the dtpT insert within the dtpT279-lacZ fusions. The vertical arrow above the sequence indicates the 5’ nucleotide of the dtpT84-lacZ fusion at position −62. The vertical arrow below the sequence indicates the junction point, at position +22, between the dtpT and lacZ sequences. The likely translation initiation codon (ATG), −10 and −35 promoter regions, and transcription start point are in bold. The directions of transcription and translation are indicated by the horizontal arrows. The mutated nucleotides are shown in lowercase above the sequence. The sequences protected by ScoC in DNase I footprinting experiments on the template strand of DNA are underlined. The ScoC-binding motifs are italicized.
B, C and D. Gel shift assay of protein binding to a radioactively labeled dtpT279p+ (B and C) or dtpT279p3 (D) PCR fragment, obtained with oligonucleotides oBB67 and oBB102. The proteins tested for binding were (B) CodY in the presence of 10 mM ILV or (C and D) ScoC. Protein concentrations (nM of monomer) are reported below each lane. The underlined concentrations indicate the apparent KD for binding.
E and F. DNase I footprinting analysis of ScoC binding to the dtpT regulatory region. The dtpT279p+ (E) or dtpT279p3 (F) DNA fragment labeled on the template strand and used in panels B or D, respectively, was incubated with increasing amounts of purified ScoC and then treated with DNase I. See the legend to Fig. 1E for additional details.
A dtpT279-lacZ fusion, containing the same promoter fragment, was created and shown to be derepressed 6.9-fold in a scoC mutant in TSS + 16 aa medium (Table 2, strains BB3928 and BB3929). Importantly, expression of the dtpT279-lacZ fusion in this medium, which provides for high CodY activity, was reduced a further 6.7-fold in a codY mutant (Table 2, strain BB3930). A double scoC codY mutant expressed the dtpT279-lacZ fusion at the same high level as did the single scoC mutant (Table 2, strain BB3931). These results indicate that CodY is a strong activator of dtpT, but exerts control indirectly through ScoC. This is fully consistent with our prediction that, in a codY mutant, ScoC expression should increase and provide a higher level of ScoC-mediated repression (superrepression). It also shows that the full potential of ScoC-mediated regulation (47-fold) can be revealed only when CodY is inactive. The maximal effect detected in microarray experiments was 21-fold, reflecting apparently the partial loss of activity by CodY in the stationary phase of growth (Caldwell et al., 2001).
Table 2.
Expression of dtpT-lacZ fusions
| Strain | Fusion promoter | Relevanta genotype | β-Galactosidase activity | Fold regulation | |
|---|---|---|---|---|---|
| codY/codY+ | scoC/scoC+ | ||||
| BB3928 | dtpT279p+ | wild type | 4.30 | 0.15 | 6.9 |
| BB3929 | scoC | 29.8 | 1.0 | ||
| BB3930 | codY | 0.64 | 47.3 | ||
| BB3931 | codY scoC | 30.3 | |||
| BB4057 | scoCp1 | 1.01 | 0.89 | ||
| BB4059 | codY scoCp1 | 0.90 | |||
| BB3954 | dtpT279p1 | wild type | 7.63 | 0.14 | 4.7 |
| BB3958 | scoC | 36.2 | 0.93 | ||
| BB3957 | codY | 1.08 | 31.3 | ||
| BB3959 | codY scoC | 33.8 | |||
| BB3970 | dtpT279p2 | wild type | 0.41 | 1.2 | |
| BB3973 | scoC | 0.48 | |||
| BB3971 | dtpT279p3 | wild type | 17.6 | 0.74 | 1.1 |
| BB3975 | scoC | 20.2 | 0.96 | ||
| BB3976 | codY | 13.0 | 1.5 | ||
| BB3982 | codY scoC | 19.3 | |||
| BB3978 | dtpT279p4 | wild type | 10.5 | 0.94 | 1.1 |
| BB3983 | scoC | 11.4 | 0.86 | ||
| BB3984 | codY | 9.91 | 1.0 | ||
| BB3987 | codY scoC | 9.93 | |||
| BB4047 | dtpT84p+ | wild type | 47.8 | 0.71 | 1.2 |
| BB4049 | scoC | 56.0 | 1.1 | ||
| BB4048 | codY | 33.8 | 1.9 | ||
| BB4050 | codY scoC | 62.9 | |||
Cells were grown and β-galactosidase activity was assayed as described in Table 1.
scoCp1 is a promoter mutation; other mutations are null.
Our results are in accord with and explain the 6-fold reduction in dtpT expression that was observed in a codY null mutant in the global RNA-Seq analysis of the CodY regulon (Brinsmade et al., 2014) [microarray analysis did not detect dtpT as a target of positive regulation by CodY (http://www.genome.jp/kegg/expression/)].
The positive effect of CodY was almost completely lost if the chromosomal scoC allele was engineered to be expressed from the CodY-insensitive scoCp1 promoter (Table 2, strains BB4057 and BB4059). Simultaneously, expression from the dptT promoter in codY+ cells was decreased 4.3-fold, apparently due to higher, unregulated expression of ScoC. These results prove that the CodY-mediated superrepression is indeed indirect and mediated through regulation of ScoC expression. As a corollary, the results confirm that, as predicted, the low-affinity CodY binding to the dtpT promoter detected in the gel shift experiments (Fig. 3B) has no physiological significance.
Mutational and deletion analysis of the dtpT ScoC-binding site
Three pairs of mutations were introduced separately into the strong dtpT ScoC-binding site I (Fig. 3A). The p1 version of the promoter retained almost fully the ability to be repressed by ScoC; however, the p3 and p4 mutant promoters lost all repression by ScoC (Table 2). Moreover, the mutations in the p3 and p4 promoters abolished the superrepression of dtpT expression observed in the codY mutant; such superrepression was not affected by the p1 promoter mutation (Table 2). In gel shift experiments, the affinity of the p3-containing regulatory region for ScoC was decreased ~3-fold (Fig. 3D)(other mutant regulatory regions were not tested). No binding of ScoC to the dtpTp3 fragment was detected by DNase footprinting (Fig. 3F). These results prove that the regulatory effect of ScoC is due to its direct interaction with the dtpT promoter.
Another pair of mutations, p2, located immediately downstream of ScoC-binding site I (Fig. 3A), almost completely wiped out expression of the dtpT279p2-lacZ fusion (Table 2). Considering that p2 affects the last two nucleotides of the sequence TATGAT, which is preceded at a distance of 17 bp by the sequence TTGACA, it is likely that the p2 version of the dtpT promoter has a defective −10 region (the two hexanucleotide sequences have one and zero mismatches, respectively, to the consensus −10 and −35 regions of σA-dependent promoters of most bacteria). For convenience, we assumed that the A nucleotide, located 5 bp downstream from the presumed −10 region is the transcription start point of the dtpT gene; this position is only 5 bp downstream from the transcription start point estimated by tiling DNA microarray experiments (Nicolas et al., 2012). Based on this assumption, the mutations in the p1, p3, and p4 promoters are located at positions −15 and −14, −12 and −11, and −18 and −17, with respect to the dtpT transcription start point, respectively. Therefore, it is not surprising that these three pairs of mutations also altered, to different extents (1.5- to 2.6-fold in a scoC single mutant strain), expression from the dtpT promoter.
Nine-bp sequences with one mismatch to the proposed ScoC-binding consensus, AATAnTATT (Kallio et al., 1991), are located from positions −173 to −165 and −12 to −4 with respect to the dtpT transcription start point and overlap the experimentally determined ScoC-binding sites (Fig. 3A). The p3 mutations are within the downstream 9-bp sequence, but the p4 mutations are located several nucleotides upstream.
To test the role of the weak upstream ScoC-binding site II in dtpT regulation, we constructed a dtpT84-lacZ fusion, which lacks this site due to the deletion of 195 nt from the 5’ end of the dtpT279-lacZ fusion. The dtpT84-lacZ fusion lost almost completely ScoC-mediated regulation (Table 2). We conclude that site II is also essential for efficient dtpT repression though it is located far upstream of the transcription start point. Similar roles of upstream ScoC-binding sites were previously reported for the aprE and nprE genes (Henner et al., 1988; Kallio et al., 1991; Ogura et al., 2004; Toma et al., 1986). A possible explanation would be that interaction between ScoC molecules bound to the two sites is required for efficient repression.
Paradoxical regulation of genes controlled by both CodY and ScoC
The global analyses of CodY-binding sites revealed that several ScoC-repressed genes or operons, such as aprE, nprE, opp, and app, contain strong CodY-binding sites (Belitsky and Sonenshein, 2013; Molle et al., 2003). However, only one of the operons, app, was derepressed in a codY null mutant strain in DNA microarray or RNA-Seq experiments; expression of the three other genes/operons did not change significantly (Brinsmade et al., 2014; Molle et al., 2003). Based on the results presented above, we considered the possibility that increased repression of these genes by the elevated level of ScoC in a codY mutant masks the loss of direct CodY-mediated repression.
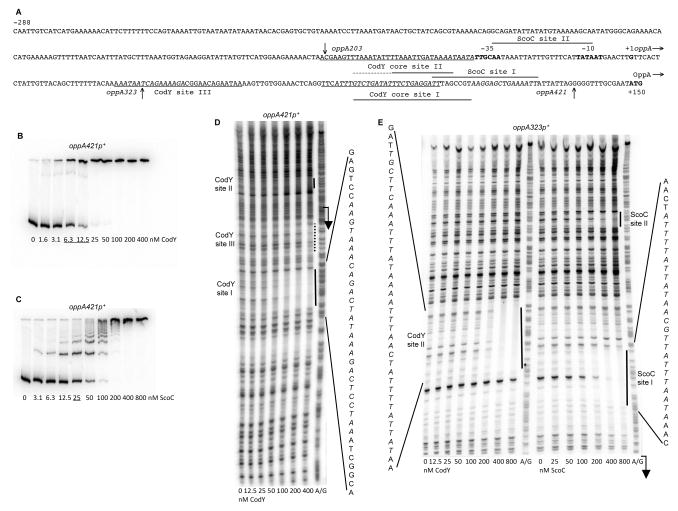
The oppABCDF operon (formerly known as spo0K) encodes an ABC-type oligopeptide permease (Perego et al., 1991; Rudner et al., 1991). It is responsible for transporting signaling oligopeptides that are essential for sporulation and genetic competence development (Lazazzera et al., 1997; Perego, 1997; Solomon et al., 1995). In gel shift experiments, CodY bound a fragment containing ~0.4 kb upstream of the oppA coding region with high affinity (apparent KD 8 nM) (Fig. 4A and 4B). DNase I footprinting experiments showed that CodY strongly protects a region (site I) (Fig. 4A and 4D) from positions +78 to +104 with respect to the transcription start point (Irnov et al., 2010). A weaker CodY-binding region (site II) was detected by footprinting at positions −70 to −36 and a third, even weaker, region (site III) at positions +31 to +59 (Fig. 4D and 4E). A shorter 323-bp oppA fragment was used for footprinting in Fig. 4E to resolve better the upstream binding site II; it follows that binding to site II, at least in vitro, is independent of binding to other sites. The first two sites correlated very well with the core binding sites determined by IDAP-Seq (from positions +85 to +105 and upstream of position −41); the upstream boundary of site II and location of the weak third site could not be determined by IDAP-Seq due to the signal interference from the strongest binding site I (Fig. 2B, 2C and 4A) (Belitsky and Sonenshein, 2013).
Fig. 4.
Binding of CodY and ScoC to the oppA regulatory region.
A. The sequence (5’ to 3’) of the coding (non-template) strand of the oppA regulatory region. Coordinates are reported with respect to the transcription start point (Irnov et al., 2010). The 5’ nucleotide of the sequence presented corresponds to the first nucleotide of the oppA insert within the oppA421- and oppA323-lacZ fusions. The vertical arrow above the sequence indicates the 5’ nucleotide of the oppA203-lacZ fusion at position −69. The vertical arrows below the sequence indicate the junction points, at positions +36 and +134, between the oppA and lacZ sequences for the oppA323 or oppA421 and oppA203 fusions, respectively. The likely translation initiation codon (ATG), −10 and −35 promoter regions, and transcription start point are in bold. The directions of transcription and translation are indicated by the horizontal arrows. The sequences protected by CodY in DNase I footprinting experiments on the template strand of DNA are underlined. The core CodY-binding sites I and II identified by IDAP-Seq (Fig. 2B and 2C) and ScoC-binding sites determined by footprinting are shown by horizontal lines below the sequence; the 5’ boundary of the core site II is unknown. The CodY-binding motifs overlapping sites I and II and the ScoC-binding motif within site I are italicized; other CodY-binding motifs with 4 and 5 mismatches to the consensus can be found in the promoter region and are not shown.
B and C. Gel shift assays of protein binding to a radioactively labeled oppA421p+ PCR fragment, obtained with oligonucleotides oBB67 and oBB102. The proteins tested were (B) CodY in the presence of 10 mM ILV and (C) ScoC. Protein concentrations used (nM of monomer) are reported below each lane; concentrations corresponding to the apparent KD for binding are underlined.
D and E. DNase I footprinting analysis of protein binding to the oppA regulatory region. The oppA421p+ (D) or oppA323p+ (E) DNA fragment, obtained with oligonucleotides oBB67 and oBB102 and labeled on the template strand, was incubated with increasing amounts of (D and E) CodY in the presence of 10 mM ILV or (E) ScoC and then treated with DNase I. See the legend to Fig. 1E for additional details.
The oppA regulatory region contains four 15-bp sequences (each with four mismatches) that resemble the consensus CodY-binding motif. Two motifs, overlapping by 6 bp, are located at positions +81 to +95 and +90 to +104 with respect to the transcription start point; another sequence is located at positions +32 to +46 (Fig. 4A). These two locations correspond very well to the strongest (site I) and weakest (site III) CodY-binding sites detected by footprinting. No candidate CodY-binding motif with fewer than six mismatches could be detected for the upstream binding site of intermediate strength (site II).
Binding of ScoC to the oppA regulatory region has never been assayed. In gel shift experiments, ScoC bound the oppA promoter region with an affinity similar to that of the dtpT gene (apparent KD≈25 nM) (Fig. 4C). In DNase I footprinting experiments, ScoC protected two regions, sites II and I, from positions −179 to −157 and −45 to −22 with respect to the oppA transcription start point, respectively (Fig. 4E). We conclude that the negative effect of ScoC on the oppA promoter is direct. A 9-bp sequence with no mismatches to the proposed ScoC-binding consensus is located from positions −42 to −34 with respect to the oppA transcription start point and within the experimentally determined ScoC-binding site I; no such sequence with ≤2 mismatches to the consensus can be found at the location of site II.
All CodY-binding sites and ScoC-binding site I are located at positions appropriate for repression of oppA transcription through interference with either initiation or elongation of transcription. The downstream ScoC-binding site I partially overlaps the upstream CodY-binding site II (Fig. 4A and 4E). As a result, each protein in increasing concentrations was able to displace the other from oppA DNA (Fig. 5).
Fig. 5.
Competition between CodY and ScoC at oppA CodY-binding site II and ScoC-binding site I. The oppA323p+ DNA fragment used in Fig. 4E was preincubated for 16 min with or without 800 nM ScoC or 800 nM CodY in the presence of 10 mM ILV, then incubated for additional 16 min with increasing amounts of either CodY or ScoC, and finally treated with DNase I. The corresponding A+G sequencing ladder is shown in the middle. The protected areas are indicated by vertical lines. Protein concentrations (nanomolar of monomer) used in the first and the second stages of the incubation are indicated below or above each lane, respectively.
A transcriptional oppA421-lacZ fusion, containing the 421-bp oppA promoter region used for DNA binding, was constructed. Despite strong binding of CodY in vitro, no significant effect of a codY mutation on expression of the fusion was detected (Table 3), consistent with the results of global analyses of the CodY regulon (Brinsmade et al., 2014; Molle et al., 2003). A transcriptional oppA735-lacZ fusion, containing the entire long intergenic region upstream of oppA was tested previously [Table S1 of Ref. (Belitsky and Sonenshein, 2013)]. The level of its expression was almost identical to that of the oppA421-lacZ fusion both in wild-type and codY mutant strains.
Table 3.
Expression of oppA-lacZ fusions
| Strain | Fusion promoter | Binding sites within the promoter | Relevanta genotype | β-Galactosidase activity | Fold regulation | |
|---|---|---|---|---|---|---|
| codY/codY+ | scoC/scoC+ | |||||
| BB3626 | oppA421p+ | CodY: I, II, III ScoC: I, II |
wild type | 51.0 | 0.86 | 2.6 |
| BB3628 | codY | 43.9 | 15.5 | |||
| BB3874 | scoC | 132.5 | 5.1 | |||
| BB3878 | codY scoC | 679.0 | ||||
| BB4059 | scoCp1 | 15.4 | 6.3 | |||
| BB4060 | codY scoCp1 | 96.6 | ||||
| BB3993 | oppA323p+ | CodY: II ScoC: I, II |
wild type | 251.8 | 0.30 | 1.3 |
| BB3995 | codY | 74.3 | 9.9 | |||
| BB3996 | scoC | 315.0 | 2.3 | |||
| BB3999 | codY scoC | 735.9 | ||||
| BB4029 | oppA203p+ | CodY: I, II, III ScoC: I |
wild type | 172.0 | 4.5 | 1.1 |
| BB4030 | codY | 775.4 | 1.2 | |||
| BB4031 | scoC | 181.8 | 5.0 | |||
| BB4032 | codY scoC | 915.0 | ||||
Cells were grown and β-galactosidase activity was assayed as described in Table 1.
scoCp1 is a promoter mutation; other mutations are null.
In accord with a previous report (Koide et al., 1999), expression of the oppA421-lacZ fusion increased about 2.6-fold in a scoC mutant (Table 3). Importantly, in a codY scoC double mutant, expression from the oppA promoter increased a further 5.1-fold (Table 3). We conclude that CodY is indeed a direct repressor of the opp operon but the effect of a codY mutation can be observed only in the absence of ScoC. This result supports the prediction that increased expression of ScoC, resulting from the absence of CodY, can compensate for the loss of CodY-mediated repression. Thus, paradoxically, in addition to being a direct negative regulator of the opp operon, CodY acts as an indirect positive regulator by repressing the synthesis of the second negative regulator, ScoC.
The negative, 6.3-fold, effect of CodY was also revealed in a scoCp1 strain in which the chromosomal scoC allele cannot be repressed by CodY (Table 3, strains BB4058 and BB4060). In codY+ scoCp1 cells, expression from the oppA promoter was 3.3-fold lower than in codY+ scoC+ cells, apparently due to higher, unregulated expression of ScoC (Table 3). In fact, these results may underestimate the role of ScoC since the scoCp1 mutation appears to reduce expression from the scoC promoter (Table 1); indeed, the higher expression from the oppA promoter in codY scoCp1 cells, compared with codY scoC+ cells (Table 3), apparently results from the lower level of ScoC expressed from the scoCp1 allele.
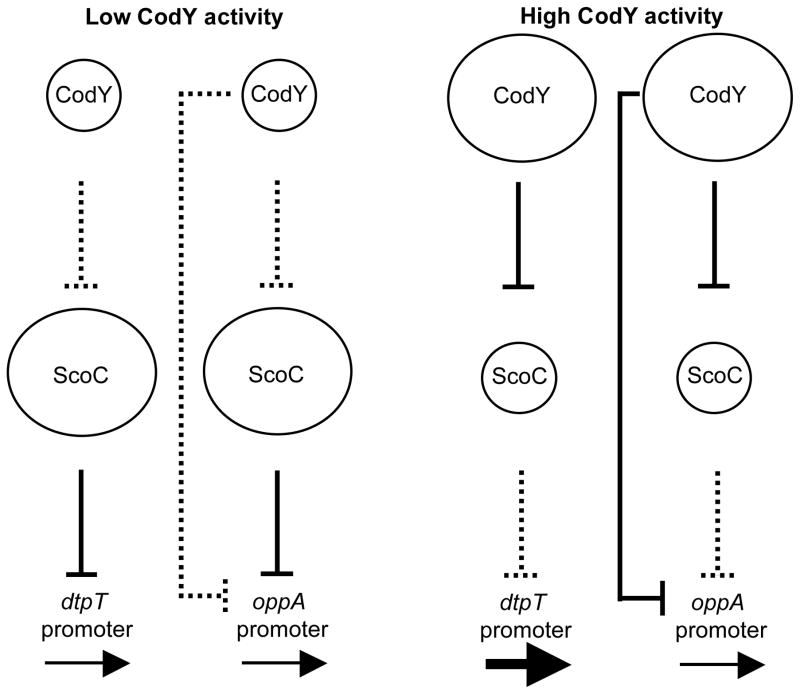
The model summarizing the functional interaction between CodY and ScoC at the oppA promoter and its comparison to the regulation of the dtpT promoter is presented in Fig. 6. Competition between CodY and ScoC at the level of DNA binding is also likely to contribute to the regulation of the oppA promoter.
Fig. 6.
A model of regulation of the dtpT and oppA promoters by the combined actions of CodY and ScoC. The sizes of the circles reflect the relative amount of the active form of each protein. The solid vertical lines indicate relatively strong effects on transcription. Dotted lines indicate relatively weak effects on transcription. The boldness of the horizontal arrows indicates the relative strength of transcription of the target genes.
Both CodY and ScoC contributed to repression of the opp operon in a wild-type strain under our growth conditions. The high, derepressed level of ScoC in a codY mutant strain caused more efficient opp repression (15.5-fold) than that provided by the CodY-repressed level of ScoC (2.6-fold) or by CodY (5.1-fold, as detected in a scoC mutant strain). The combined effect of the two regulators in a wild-type strain was less than a sum of their maximal effects, reflecting their complex roles in opp regulation (Table 3).
To test the role of different CodY-binding sites in oppA regulation, we constructed an oppA323-lacZ fusion, which lacks sites I and III due to the deletion of 98 nt from the 3’ end of the oppA421-lacZ fusion. The oppA323-lacZ fusion lost half of CodY-mediated regulation (2.3-fold versus 5.1-fold) (Table 3). We conclude that both downstream and upstream CodY-binding sites contribute to oppA regulation.
To test the roles of ScoC-binding sites in oppA regulation, we constructed an oppA203-lacZ fusion, which lacks the upstream site II due to the deletion of 218 nt from the 5’ end of the oppA421-lacZ fusion. The oppA203-lacZ fusion lost completely ScoC-mediated regulation (Table 3), though in vitro ScoC was still able to bind to the oppA203 fragment (data not shown). We conclude that, as for other genes, the far upstream ScoC-binding site II is essential for oppA repression.
Role of TnrA in ScoC-mediated dtpT and oppA regulation
To determine whether the activity of ScoC varies under different growth conditions, we altered the composition of the medium, using the ScoC-regulated dtpT279p+-lacZ fusion as a reporter. In a wild-type strain in TSS glucose-ammonium medium without added amino acids, CodY has low activity (Belitsky and Sonenshein, 2008; Slack et al., 1995) and, therefore, transcription of the scoC gene was derepressed (Table 4, strain GB1039); as a result, the dtpTp+-lacZ fusion was expressed at the same low level as in a codY mutant strain in TSS + 16 aa medium (Tables 2 and 5, strain BB3928). However, if glutamate, a poor nitrogen source, was substituted for ammonium as the sole nitrogen source in TSS medium, expression from the scoC promoter remained largely unchanged (Table 4), but expression of the dtpTp+-lacZ fusion increased 4.7-fold (Table 5). As expected in the absence of other exogenous amino acids, a codY null mutation had only a small effect on dtpT expression either in glucose-ammonium or in glucose-glutamate medium (Table 5, strain BB3930).
Table 4.
Role of TnrA in expression of the scoC561p+-lacZ fusion
| Strain | Relevant genotype | Nitrogen source | β-Galactosidase activity | Fold regulation | |
|---|---|---|---|---|---|
| Glt/NH4+ | tnrA+/tnrA | ||||
| GB1039 | wild type | NH4+ | 180.8 | 0.88 | |
| Glt | 131.4 | 0.73 | 0.59 | ||
| BB3927 | tnrA | NH4+ | 205.1 | ||
| Glt | 223.3 | 1.1 | |||
| GB1063 | codY scoC | NH4+ | 633.8 | 1.1 | |
| Glt | 849.5 | 1.3 | 1.0 | ||
| BB3967 | codY scoC tnrA | NH4+ | 587.3 | ||
| Glt | 809.3 | 1.4 | |||
Cells were grown in TSS glucose medium with 0.2% NH4Cl or 0.2% glutamate as sole nitrogen source and β-galactosidase activity was assayed as described in Table 1.
Table 5.
Role of TnrA in expression of dtpT-lacZ fusions
| Strain | Fusion promoter | Relevant genotype | Nitrogen source | β-Galactosidase activity | Fold regulation | |
|---|---|---|---|---|---|---|
| Glt/NH4+ | tnrA+/tnrA | |||||
| BB3928 | dtpT279p+ | wild type | NH4+ | 0.69 | 1.6 | |
| Glt | 3.26 | 4.7 | 7.8 | |||
| BB3930 | codY | NH4+ | 0.42 | 1.2 | ||
| Glt | 2.28 | 5.4 | 6.7 | |||
| BB3992 | tnrA | NH4+ | 0.43 | |||
| Glt | 0.42 | 1.0 | ||||
| BB3960 | codY tnrA | NH4+ | 0.34 | |||
| Glt | 0.34 | 1.0 | ||||
| BB3929 | scoC | NH4+ | 61.7 | 0.96 | ||
| Glt | 57.1 | 0.93 | 1.0 | |||
| BB3969 | scoC tnrA | NH4+ | 64.6 | |||
| Glt | 56.4 | 0.87 | ||||
| BB3971 | dtpT279p3 | wild type | NH4+ | 24.5 | ||
| Glt | 26.0 | 1.1 | ||||
Cells were grown and β-galactosidase activity was assayed as described in Table 4.
Because TnrA, a transcriptional regulator of many nitrogen metabolism genes in B. subtilis, is known to be activated in glucose-glutamate medium (Fisher, 1999; Yoshida et al., 2003), we tested expression of the dtpTp+-lacZ fusion in a tnrA null mutant strain. Indeed, in the absence of TnrA, expression from the dtpTp+ promoter was reduced in glucose-glutamate medium to the level seen in glucose-ammonium medium, indicating that glutamate-mediated activation of the dtpTp+-lacZ fusion is due to altered TnrA activity (Table 5, strains BB3992 and BB3960). Interestingly, no effect of glutamate or a tnrA mutation on the derepressed level of dtpT expression was observed in a scoC mutant (Table 5, strains BB3929 and BB3969). Similarly, no effect of glutamate was detected on expression of the dtpTp3-lacZ fusion containing a mutation in the ScoC-binding site (Table 5, strain BB3971). Thus, TnrA is not able to activate the dtpT promoter by itself, indicating that the role of TnrA is to interfere with ScoC-mediated repression.
In contrast to the dtpT gene, which was not previously known to be regulated by TnrA, opp expression has been shown to be under direct positive TnrA control (Yoshida et al., 2003). In accord with this result, in TSS glucose-glutamate medium, expression of the oppA421-lacZ fusion was 3.7-fold higher in tnrA+ cells compared to that in tnrA mutant cells (Table 6). However, no effect of a tnrA mutation was observed in a scoC mutant strain, indicating that oppA activation by TnrA, like that of the dtpT gene, is apparently due to interference with repression by ScoC (Table 6). In both cases, TnrA reduced the ability of ScoC to repress the dtpT or oppA promoter only partially, because the level of expression of the corresponding fusions in TSS glucose-glutamate medium in a tnrA mutant did not reach that seen in scoC mutant cells (Tables 5 and 6).
Table 6.
Role of TnrA in expression of the oppA421-lacZ fusion
| Strain | Relevant genotype | Nitrogen source | β-galactosidase activity | Fold regulation | |
|---|---|---|---|---|---|
| Glt/NH4+ | tnrA+/tnrA | ||||
| BB3626 | wild type | NH4+ | 53.5 | 0.71 | |
| Glt | 166.3 | 3.1 | 3.7 | ||
| BB3979 | tnrA | NH4+ | 75.4 | ||
| Glt | 44.8 | 0.59 | |||
| BB3874 | scoC | NH4+ | 657.7 | 0.98 | |
| Glt | 621.6 | 0.95 | 1.0 | ||
| BB3980 | scoC tnrA | NH4+ | 669.3 | ||
| Glt | 596.2 | 0.89 | |||
Cells were grown and β-galactosidase activity was assayed as described in Table 4.
It was reported that scoC may be a target of direct repression by TnrA (Abe et al., 2009). We have tested the effect of TnrA on scoC expression in glucose-glutamate medium, which supports high activity of TnrA, but it was very small if any (Table 4). Therefore, it is unlikely that the positive effect of TnrA on dtpT or oppA expression is mediated through reduced scoC expression.
DISCUSSION
Functional interaction between CodY and ScoC
The results presented here establish that B. subtilis ScoC expression is under direct, negative control by CodY. The Bacillus anthracis scoC gene (BA1045) also appears to be repressed by CodY (van Schaik et al., 2009). Therefore, ScoC-regulated genes, such as dtpT, are potentially subject to cascade regulation that depends on the level of CodY activity (Fig. 6). Under growth conditions in which CodY activity is high, CodY should act as an indirect positive regulator of ScoC-repressed genes by reducing the level of ScoC if those genes do not require other factors for expression under the growth condition used and if the relatively low level of ScoC in wild-type cells is not already sufficient for maximal repression. In addition to dtpT, several other genes and operons, such as rbs and yppF, that are negatively regulated by ScoC (Caldwell et al., 2001)(Caldwell et al., personal communication) have been catalogued as genes positively regulated by CodY (Brinsmade et al., 2014) but lack any associated CodY-binding sites (Belitsky and Sonenshein, 2013). It is likely that for these genes and possibly other targets of ScoC the effect of CodY is indirect and mediated through ScoC.
Our results also describe a second, seemingly paradoxical form of CodY-ScoC regulatory interaction seen for genes that are negatively regulated by both CodY and ScoC. For such genes and operons, e.g., opp, CodY and ScoC form a feed-forward regulatory loop, an arrangement in which two regulatory proteins control expression of the same target gene and one of the regulators controls expression of the other (Alon, 2007; Mangan and Alon, 2003)(Fig. 6). In this case, when CodY activity decreases due to nutrient exhaustion, an increase in expression of the target gene will only occur if the increased expression of ScoC is insufficient to maintain repression; in other cases, it might lead to enough accumulation of ScoC to decrease expression even further. Thus, the extent to which any target gene responds to this feed-forward loop depends on the independent efficiencies of repression of that gene by CodY and ScoC. Depending on the parameters of interaction between each protein and DNA, feed-forward loops may affect the kinetics of the target gene response when the upstream regulator gains or loses activity (Mangan and Alon, 2003; Wall et al., 2005).
In the case of the opp operon, the direct negative and indirect positive effects of CodY are complemented by the competition with ScoC at CodY-binding site II and appear to balance each other almost perfectly; consequently, no increase in expression was seen when the codY gene was inactivated. Only weak opp repression by ScoC was achieved in codY+ cells due to low scoC expression. Both codY and scoC genes had to be inactivated simultaneously in order to fully derepress opp transcription under the steady-state growth conditions used. Thus, the combined actions of CodY and ScoC serve to maintain a significant level of opp repression even when one of the regulators is not active or not expressed but also avoid overly strong repression of the operon by fully active regulators.
More generally, the presence of CodY and ScoC in the same cell allows different kinds of outcomes when CodY loses activity due to nutrient exhaustion. Genes directly repressed solely by CodY should show increased expression. Genes directly repressed by ScoC, but not by CodY, should have reduced expression due to the increased level of ScoC, whereas genes controlled negatively and directly by both CodY and ScoC can have varying responses depending on the concentrations of the active forms of the proteins needed to bind to the regulatory regions of the genes in question and the strength of the corresponding binding sites.
In addition to ScoC, synthesis of another important regulator, ComK, as well as the ComS protein, which controls ComK degradation (Hamoen et al., 1995), were shown to be directly repressed by CodY in B. subtilis (Serror and Sonenshein, 1996a; Smits et al., 2007). ComK is a master regulator of genetic competence and its expression is under the control of several additional transcriptional regulators (Hamoen et al., 2003a; Hamoen et al., 2003b); however, none of them is known to be regulated by CodY.
The sinIR operon was reported to be under direct, negative ScoC control (Kallio et al., 1991; Shafikhani et al., 2002). SinR is a master regulator of biofilm formation in B. subtilis whose activity is inhibited by SinI (Bai et al., 1993; Kearns et al., 2005). This finding implies that at least some SinR targets (Chu et al., 2006) may be subject to a two-step cascade regulation mediated by CodY, ScoC, and SinR, but, since sinI and sinR are both repressed by ScoC, it remains unclear whether the contribution of ScoC to SinR activity is negative or positive.
Regulation of ScoC activity
We have detected a low level (two- to three-fold or less) of negative autoregulation by ScoC that apparently serves to moderate the effect of CodY-mediated regulation on expression of the scoC gene. That is, expression of scoC increases when CodY activity decreases, but the maximum extent of ScoC accumulation is limited by its self-repression.
CodY is the only DNA-binding regulator known to affect expression of ScoC directly. Expression of scoC is moderately increased in nutrient broth medium in a spo0A mutant strain, which is consistent with activation of scoC by AbrB, an important transition state regulator (Perego and Hoch, 1988)(B. R. Belitsky and A. L. Sonenshein, data not shown). However, we did not observe any significant effect of an abrB null mutation on scoC-lacZ expression under our growth conditions and attempts to demonstrate specific binding of AbrB to the scoC regulatory region have been unsuccessful (Strauch et al., 1989). A global study of sites that are bound by AbrB in vivo also failed to detect scoC as a target (Chumsakul et al., 2010). SalA and multicopy SenS have also been reported to negatively affect expression of scoC, but the molecular mechanisms of action of these proteins are not known (Kawachi et al., 2005; Ogura et al., 2004).
Although many transcriptional regulators of the MarR-family respond to low molecular-weight effectors (Perera and Grove, 2010; Wilkinson and Grove, 2006), no effectors of ScoC activity are known. Expression of the ScoC-regulated dtpT gene varies to a limited extent (less than 1.7-fold) over a very wide range of growth conditions (Nicolas et al., 2012), indicating that activity of ScoC was not drastically altered in those experiments. Although we found that dtpT expression is activated in glucose-glutamate medium, this effect is due to TnrA-mediated interference with ScoC functioning. Thus, the activity of ScoC appears to be determined principally, if not solely, by modulation of its expression or DNA-binding by other transcription factors.
In other studies, ScoC was reported to be phosphorylated by the protein arginine kinase McsB at the Arg3 position (Elsholz et al., 2012). In our hands, neither an mcsB null mutation nor a null mutation in the ywlE gene, encoding protein arginine phosphatase, affected expression of the dtpTp+-lacZ fusion used as a reporter for ScoC activity in a codY+ or codY mutant strain (data not shown).
Oligopeptide permeases as targets of complex transcriptional regulation
Though the function of the DtpT protein has never been established experimentally, its sequence strongly suggests that DtpT is an oligopeptide permease. Three other peptide permeases of the ABC transporter type and encoded by the dpp-ykf, opp, and app operons are present in B. subtilis (Barbe et al., 2009). Interestingly, three of the four oligopeptide permeases, DtpT, Opp, and App, have been shown to be negatively regulated by ScoC in this work and previously (Caldwell et al., 2001; Koide et al., 1999). Dpp, Opp, and App oligopeptide permeases are also direct targets of repression by CodY (Belitsky and Sonenshein, 2013; Molle et al., 2003; Serror and Sonenshein, 1996b; Slack et al., 1995)(this work). Thus, ScoC and CodY cooperate in the regulation of B. subtilis oligopeptide uptake either by binding to the promoter regions or through CodY-mediated repression of ScoC or both. Expression of the dpp-ykf and app operons is also known to be negatively regulated by AbrB; in contrast, the opp operon appears to be under positive regulation of AbrB (Koide et al., 1999; Slack et al., 1991).
As noted above, CodY serves as an indirect positive regulator of the ScoC-repressed dtpT gene and opp operon. Interestingly, under growth conditions when CodY is inactive, another transcription regulator, TnrA, which is active only under conditions of nitrogen limitation, acts as a positive regulator of the same two oligopeptide permeases via interference with ScoC action. This role of TnrA is consistent with its main function, which is to increase the supply of nitrogen-containing nutrients under conditions of nitrogen limitation (Fisher, 1999). The TnrA-binding sites of the dtpT and oppA genes have been identified recently by a ChIP-on-chip experiment (Mirouze et al., 2015). They do not overlap the ScoC-binding sites of these genes, and the mechanism(s) of interaction between TnrA and ScoC remains unknown. Because scoC expression was reported to be negatively affected by TnrA (Abe et al., 2009), we cannot exclude that TnrA, like CodY, affects dtpT and oppA expression both directly and indirectly.
In summary, our results indicate that ScoC’s function as a transcriptional repressor may be determined by the activities of two other transcriptional regulators, CodY and TnrA. Both CodY and ScoC act as direct repressors of the opp operon, forming a rare feed-forward regulatory loop. It also follows that a complete picture of the ScoC regulon can be revealed only when both CodY and TnrA are inactive. Although in most previous experiments the growth media used contained an excess of amino acids, thereby activating CodY and inactivating TnrA, in some experiments amino acids could have been partially exhausted and the level of ScoC would have increased due to CodY inactivation. Whether the resulting concentration of ScoC would have reached the level needed for efficient binding to all of its targets remains unknown, as does the full impact of ScoC on gene expression.
Experimental procedures
Bacterial strains and culture media
The B. subtilis strains constructed and used in this study were all derivatives of strain SMY (Zeigler et al., 2008) and are described in Table 7 or in the text. Escherichia coli strain JM107 (Yanisch-Perron et al., 1985) was used for isolation of plasmids. Bacterial growth in TSS 0.5% glucose medium supplemented with 0.2% NH4Cl or sodium glutamate or DS nutrient broth medium was as described (Belitsky and Sonenshein, 2011b).
DNA manipulations
Methods for common DNA manipulations, transformation, and sequence analysis were as previously described (Belitsky and Sonenshein, 1998). All oligonucleotides used in this work are described in Table 8. Chromosomal DNA of B. subtilis strain SMY or plasmids constructed in this work were used as templates for PCR. All cloned PCR-generated fragments were verified by sequencing.
Table 8.
Oligonucleotides used in this work
| Name | Sequencea | Specificity |
|---|---|---|
| Flanking forward primers | ||
| oBB67 | 5’-GCTTCTAAGTCTTATTTCC | erm (pHK23) |
| oGB26 | 5’-GGACTCTAGAGCACCTTCCTCAGGAAAGC | scoC |
| oBB576 | 5’-TCGTTGAATTCATGAAAACATCAACC | oppA735/421/323 |
| oBB697 | 5’-AATATGAATTCAGCCAATTGC | dtpT279 |
| oBB711 | 5’-GAAAAGAATTCGAAGTTTAAATATTTTAAATTG | oppA203 |
| oBB712 | 5’-TTTTCGAATTCGGAACTATTTGAAATGAG | dtpT84 |
| Flanking reverse primers | ||
| oBB102 | 5’-CACCTTTTCCCTATATAAAAGC | lacZ (pHK23) |
| oGB28 | 5’-CGGGAAAGCTTATCCTTCTCGATCGATTTCC | scoC |
| oBB577 | 5’-CGCAAAGATCTAATAATAATTTTCAGCTC | oppA735/421/203 |
| oBB698 | 5’-GCCAAAAGCTTTCTAAAAAAAGTGTA | dtpT |
| oBB701 | 5’-GCCAAAAGCTTTCTAAAAAAAGTGTA TTATAAAAATCATATTCggATATAATAC |
dtpTp1 |
| oBB702 | 5’-GCCAAAAGCTTTCTAAAAAAAGTGTA TTATAAAAATCAggTTCTTATATAA |
dtpTp2 |
| oBB703 | 5’-GCCAAAAGCTTTCTAAAAAAAGTGTA TTATAAAAATCATAggCTTATATAATAC |
dtpTp3 |
| oBB704 | 5’-GCCAAAAGCTTTCTAAAAAAAGTGTA TTATAAAAATCATATTCTTAggTAATAC |
dtpTp4 |
| oBB708 | 5’-TCTTTAGATCTTATTTTTGTAAAAAAGCTG | oppA323 |
| Internal mutagenic forward primers | ||
| oGB38 | 5’-GGTTTGTGAATTTATTAAccAATTCTTACAT | scoCp1 |
| oGB40 | 5’-GGTTTGTGAATTTATTccAAAATTCTTACAT | scoCp2 |
| oBB696 | 5’-GGTTTGTGAATTTATTAAcAAATTCTTACAT | scoCp3 |
| Internal mutagenic reverse primers | ||
| oGB39 | 5’-ATGTAAGAATTggTTAATAAATTCACAAACC | scoCp1 |
| oGB41 | 5’-ATGTAAGAATTTTggAATAAATTCACAAACC | scoCp2 |
| oBB695 | 5’-ATGTAAGAATTTgTTAATAAATTCACAAACC | scoCp3 |
The altered nucleotides conferring down mutations in the CodY- or ScoC-binding site are in lowercase. The restriction sites are underlined.
Construction of transcriptional fusions
Plasmid pGB15 (scoC561p+-lacZ) was created by cloning the XbaI- and HindIII-treated PCR product, containing the entire scoC regulatory region, in an integrative plasmid pHK23 (erm) (Belitsky and Sonenshein, 2008). The 0.57-kb scoC PCR product was synthesized with oGB26 and oGB28 as primers; the resulting fusion is identical to one described earlier (Kawachi et al., 2005). Plasmids pBB1812 (scoC350p+-lacZ) or pBB1813 (scoC181p+-lacZ), containing the same scoC regulatory region but truncated from the 5’ end, were constructed in a similar way by cloning the 3’ part of the same PCR fragment after its digestion with BstYI and HindIII or ApoI and HindIII in pHK23, treated with BamHI and HindIII or EcoRI and HindIII, respectively.
Plasmid pBB1819 (dtpT279p+-lacZ) was created by cloning the EcoRI- and HindIII-treated PCR product, containing the entire dtpT regulatory region, in pHK23. The 0.29-kb dtpT PCR product was synthesized with oBB697 and oBB698 as direct and reverse primers, respectively. Plasmid pBB1833 (dtpT84p+-lacZ), containing the dtpT regulatory region truncated from the 5’ end, was constructed as pBB1819, but using oBB712 as the direct PCR primer. Plasmid pBB1741 (oppA735p+-lacZ) was created by cloning the EcoRI- and BglII-treated PCR product, containing the entire oppA regulatory region, in pHK23. The 0.74-kb oppA PCR product was synthesized with oBB576 and oBB577 as direct and reverse primers, respectively. Plasmid pBB1742 (oppA421p+-lacZ), containing the oppA regulatory region truncated from the 5’ end, was constructed in a similar way by cloning the 0.42-kb 3’ part of the same PCR fragment after its digestion with MfeI and BglII. Plasmid pBB1827 (oppA323p+-lacZ), containing the oppA regulatory region truncated from the 3’ end, was constructed as pBB1742, but using oBB708 as the reverse PCR primer. Plasmid pBB1831 (oppA203p+-lacZ), containing the oppA regulatory region truncated further from the 5’ end, was constructed as pBB1741, but using oBB711 as the direct PCR primer.
B. subtilis strains carrying various lacZ fusions at the amyE locus (Table 7) were isolated after transforming strain BB2511 (amyE::spc lacA) with the appropriate plasmids, by selecting for resistance to erythromycin conferred by the plasmids, and screening for loss of the spectinomycin-resistance marker, which indicated a double crossover, homologous recombination event. Strain BB2511 and all its derivatives have very low endogenous β-galactosidase activity due to a null mutation in the lacA gene [Daniel, 1997 #33].
Table 7.
B. subtilis strains used
| Strain | Genotype | Source or referencea |
|---|---|---|
| SMY | prototroph | (Zeigler et al., 2008) |
| JH12586 | ΔabrB::cat trpC2 pheA1 | (Perego et al., 1988) |
| JH14272 | ΔamyE::[aph Φ (opp-lacZ)] ΔscoC::cat trpC2 pheA1 | (Koide et al., 1999) |
| BB278 | tnrA62::Tn917 | (Belitsky et al., 2000) |
| BB382 | abrB::cat | SMY x DNA(JH12586) |
| BB383 | abrB::(cat::neo) | BB382 x pCm::Nm (Steinmetz and Richter, 1994) |
| BB1043 | codY::(erm::spc) | (Barbieri et al., 2015) |
| BB1888 | lacA::tet | (Belitsky and Sonenshein, 2008) |
| BB2511 | ΔamyE::spc lacA::tet | (Belitsky and Sonenshein, 2008) |
| GB1039 | ΔamyE::[erm Φ (scoC561p+-lacZ)] lacA::tet | BB2511 x pGB15 |
| GB1045 | ΔamyE::[erm Φ (scoC561p1-lacZ)] lacA::tet | BB2511 x pGB18 |
| GB1059 | ΔamyE::[erm Φ (scoC561p2-lacZ)] lacA::tet | BB2511 x pGB22 |
| BB3543 | tnrA62::(Tn917::neo) | BB278 x p917::Nm (Steinmetz and Richter, 1994) |
| BB3890 | ΔamyE::[erm Φ (scoC350p+-lacZ)] lacA::tet | BB2511 x pBB1812 |
| BB3898 | ΔamyE::[erm Φ (scoC181p+-lacZ)] lacA::tet | BB2511 x pBB1813 |
| BB3919 | ΔamyE::[erm Φ (scoC561p3-lacZ)] lacA::tet | BB2511 x pBB1817 |
| BB3928 | ΔamyE::[erm Φ (dtpT279p+-lacZ)] lacA::tet | BB2511 x pBB1819 |
| BB3929 | ΔamyE::[erm Φ (dtpT279p1-lacZ)] lacA::tet | BB2511 x pBB1821 |
| BB3970 | ΔamyE::[erm Φ (dtpT279p2-lacZ)] lacA::tet | BB2511 x pBB1822 |
| BB3971 | ΔamyE::[erm Φ (dtpT279p3-lacZ)] lacA::tet | BB2511 x pBB1823 |
| BB3978 | ΔamyE::[erm Φ (dtpT279p4-lacZ)] lacA::tet | BB2511 x pBB1825 |
| BB3626 | ΔamyE::[erm Φ (oppA421-lacZ)] lacA::tet | BB2511 x pBB1742 |
| BB3993 | ΔamyE::[erm Φ (oppA323-lacZ)] lacA::tet | BB2511 x pBB1827 |
| BB4029 | ΔamyE::[erm Φ (oppA203-lacZ)] lacA::tet | BB2511 x pBB1831 |
| BB4042 | scoCp1 lacA::tet | BB1888 x pBB1830 |
| BB4047 | ΔamyE::[erm Φ (dtpT84p+-lacZ)] lacA::tet | BB2511 x pBB1833 |
The symbol × indicates transformation by plasmid or chromosomal DNA.
Mutations in the CodY- and ScoC-binding sites
Plasmids pGB18 (scoC561p1-lacZ), pGB22 (scoC561p2-lacZ), and pBB1817 (scoC561p3-lacZ), containing 1-bp or 2-bp substitution mutations in the CodY-binding site, were constructed as described for pGB15 using fragments generated by two-step overlapping PCR. In the first step, a product containing the 5’ part of the scoC regulatory region was synthesized by using oligonucleotide oGB26 as the forward primer and mutagenic oligonucleotide oGB39, oGB41 or oBB695 as the reverse primer. A product containing the 3’ part of the scoC regulatory region was synthesized by using mutagenic oligonucleotides oGB38, oGB40 or oBB696 as the forward primer and oligonucleotide oGB28 as the reverse primer. The PCR products were used in a second, splicing step of PCR mutagenesis as overlapping templates to generate a modified fragment containing the entire scoC regulatory region; oligonucleotides oGB26 and oGB28 served as the forward and reverse PCR primers, respectively.
Plasmids pBB1821 (dtpTp1-lacZ), pBB1822 (dtpTp2-lacZ), pBB1823 (dtpTp3-lacZ), and pBB1825 (dtpTp4-lacZ), containing 2-bp substitution mutations in the ScoC-binding site, were constructed as described for pBB1819, using mutagenic oligonucleotides oBB701, oBB702, oBB703 or oBB704 as the reverse PCR primer, respectively.
Construction of the scoCp1 mutant
The 0.64-kb XbaI-SalI PCR fragment of pGB18 (scoC561p1-lacZ), containing the entire scoC insert, was cloned in the integrative plasmid pBB1579 (neo bgaB) (Belitsky and Sonenshein, 2011a). The resulting plasmid, pBB1830, was introduced by a single-crossover homologous recombination event into the scoC chromosomal locus of strain BB1888 (lacA). White Neos colonies indicating excision of pBB1830 from the chromosome were searched for on plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside), the colored substrate of bgaB-encoded β-galactosidase. The presence of the scoCp1 mutation in strain BB4042 was confirmed by sequencing of the chromosomal scoC allele.
Purification of CodY and ScoC
CodY-His5 was purified to near homogeneity as described previously (Belitsky and 6-ScoC was purified to near homogeneity in the same manner after induction plasmid-containing E. coli BL21(DE3) pLysS cells (Studier and Moffatt, 1986) with 0.1% IPTG for 4 hours. Elution from the Ni2+-affinity column (His·Bind resin; Novagen) was with a buffer containing 385 mM imidazole. The plasmid pRSETA-scoC, containing the scoC open reading frame cloned between the BamHI and EcoRI sites of pRSET A (Life Technologies, Thermo Fisher Scientific), was obtained from U. Gerth. The resulting tagged protein contained the peptide MRGSHHHHHHGMASMTGGQQMGRDLYDDDDKDRWGS as its N-terminal extension.
Labeling of DNA fragments
The PCR products containing the regulatory region of the scoC or dtpT or oppA gene were synthesized using vector-specific oligonucleotides oBB67 and oBB102 as the forward and reverse primer, respectively. The reverse primer for each PCR reaction (which would prime synthesis of the template strand of the PCR product) was labeled using T4 polynucleotide kinase and [γ-32P]ATP. oBB67 starts 96 bp or 112 bp upstream of the XbaI or EcoRI site used for cloning, respectively, and oBB102 starts 36 bp or 65 bp downstream of the HindIII or BglII site that served as a junction between the promoters and the lacZ part of the fusions, respectively.
Gel shift assays and DNase I protection experiments
Incubation of CodY or ScoC with the 32P-labeled promoter fragments was performed in a binding buffer containing 20 mM Tris-Cl (pH 8.0) - 50 mM KCl - 2 mM MgCl2 - 5% glycerol - 0.5 mM EDTA - 1 mM DTT - 0.05% Nonidet P-40 - 25 μg/ml sonicated salmon sperm DNA. Samples (11 μl) containing varying amounts of proteins and less than 1 fmole of DNA were incubated for 16 min at room temperature and separated on 8% nondenaturing 50 mM Tris - 384 mM glycine - 1 mM EDTA polyacrylamide gels in 35 mM Hepes - 43 mM imidazole buffer. In some experiments, 10 mM ILV were present in the incubation mixture and gel buffers.
For DNase I protection experiments, samples containing 20–40 fmoles of labeled DNA were incubated with proteins as described above. One μl of the binding buffer containing 0.1–0.2 U RQ1 DNase I (Promega), 10 mM MgCl2 and 20 mM CaCl2 was then added, followed by addition, after 1 min, of 4 μl of 20 mM EDTA-95% formamide dye solution and subsequent heating of the samples at 80°C for 5 min. The samples were loaded without further purification on 7 M urea - 6% polyacrylamide DNA sequencing gels. The G+A sequencing ladder, generated according to a published procedure by boiling the appropriate samples of labeled DNA for 20 min (Liu and Hong, 1998), served to locate precisely the protected region.
The gels were dried, and the radioactive bands were detected and quantified using storage screens, an Applied Biosystems PhosphorImager, and ImageQuant software (GE Healthcare).
Enzyme assays
β-Galactosidase specific activity was determined as described previously (Belitsky and Sonenshein, 1998).
Acknowledgments
This paper is dedicated to the memory of Mark Strauch, who was the first to characterize CodY-responsive expression of the scoC gene. We are grateful to L. Handke, who performed initial footprinting experiments of CodY binding to the scoC promoter, and M. Perego and U. Gerth for the gift of strains. This work was supported by a research grant (GM042219) from the U. S. National Institute of General Medical Sciences to A. L. Sonenshein. The content is solely the responsibility of the authors and does not represent the official view of the National Institute of Health or NIGMS. G. Barbieri was supported by the University of Pavia PostDoc Fund and AA-FAR 2012-13.
References
- Abe S, Yasumura A, Tanaka T. Regulation of Bacillus subtilis aprE expression by glnA through inhibition of scoC and sigma(D)-dependent degR expression. J Bacteriol. 2009;191:3050–3058. doi: 10.1128/JB.00049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Bai U, Mandic-Mulec I, Smith I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 1993;7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, Vallenet D, Wang TZ, Moszer I, Medigue C, Danchin A. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology. 2009;155:1758–1775. doi: 10.1099/mic.0.027839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri G, Voigt B, Albrecht D, Hecker M, Albertini AM, Sonenshein AL, Ferrari E, Belitsky BR. CodY regulates expression of the Bacillus subtilis extracellular proteases Vpr and Mpr. J Bacteriol. 2015;197:1423–1432. doi: 10.1128/JB.02588-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR. Indirect repression by Bacillus subtilis CodY via displacement of the activator of the proline utilization operon. J Mol Biol. 2011;413:321–336. doi: 10.1016/j.jmb.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Sonenshein AL. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J Bacteriol. 1998;180:6298–6305. doi: 10.1128/jb.180.23.6298-6305.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Sonenshein AL. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J Bacteriol. 2008;190:1224–1236. doi: 10.1128/JB.01780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Sonenshein AL. CodY-mediated regulation of guanosine uptake in Bacillus subtilis. J Bacteriol. 2011a;193:6276–6287. doi: 10.1128/JB.05899-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Sonenshein AL. Contributions of multiple binding sites and effector-independent binding to CodY-mediated regulation in Bacillus subtilis. J Bacteriol. 2011b;193:473–484. doi: 10.1128/JB.01151-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Sonenshein AL. Genome-wide identification of Bacillus subtilis CodY-binding sites at single-nucleotide resolution. Proc Natl Acad Sci U S A. 2013;110:7026–7031. doi: 10.1073/pnas.1300428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Wray LV, Jr, Fisher SH, Bohannon DE, Sonenshein AL. Role of TnrA in nitrogen source-dependent repression of Bacillus subtilis glutamate synthase gene expression. J Bacteriol. 2000;182:5939–5947. doi: 10.1128/jb.182.21.5939-5947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinsmade SR, Alexander EL, Livny J, Stettner AI, Segre D, Rhee KY, Sonenshein AL. Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY. Proc Natl Acad Sci USA. 2014;111:8227–8232. doi: 10.1073/pnas.1321308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinsmade SR, Sonenshein AL. Dissecting complex metabolic integration provides direct genetic evidence for CodY activation by guanine nucleotides. J Bacteriol. 2011;193:5637–5648. doi: 10.1128/JB.05510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell R, Sapolsky R, Weyler W, Maile RR, Causey SC, Ferrari E. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis. J Bacteriol. 2001;183:7329–7340. doi: 10.1128/JB.183.24.7329-7340.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- Chumsakul O, Takahashi H, Oshima T, Hishimoto T, Kanaya S, Ogasawara N, Ishikawa S. Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 2010;39:414–428. doi: 10.1093/nar/gkq780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hengst CD, van Hijum SA, Geurts JM, Nauta A, Kok J, Kuipers OP. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J Biol Chem. 2005;280:34332–34342. doi: 10.1074/jbc.M502349200. [DOI] [PubMed] [Google Scholar]
- Dod B, Balassa G. Spore control (sco) mutations in Bacillus subtilisIII Regulation of extracellular protease synthesis in the spore control mutations scoC. Molec Gen Genet. 1978;163:57–63. [Google Scholar]
- Dod B, Balassa G, Raulet E, Jeannoda V. Spore control (sco) mutations in Bacillus subtilisII Sporulation and the production of extracellular protease and a-amylase by scoC mutants. Molec Gen Genet. 1978;163:45–56. [Google Scholar]
- Doki S, Kato HE, Solcan N, Iwaki M, Koyama M, Hattori M, Iwase N, Tsukazaki T, Sugita Y, Kandori H, Newstead S, Ishitani R, Nureki O. Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT. Proc Natl Acad Sci USA. 2013;110:11343–11348. doi: 10.1073/pnas.1301079110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsholz AK, Turgay K, Michalik S, Hessling B, Gronau K, Oertel D, Mader U, Bernhardt J, Becher D, Hecker M, Gerth U. Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc Natl Acad Sci USA. 2012;109:7451–7456. doi: 10.1073/pnas.1117483109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SH. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol Microbiol. 1999;32:223–232. doi: 10.1046/j.1365-2958.1999.01333.x. [DOI] [PubMed] [Google Scholar]
- Guedon E, Serror P, Ehrlich SD, Renault P, Delorme C. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol Microbiol. 2001;40:1227–1239. doi: 10.1046/j.1365-2958.2001.02470.x. [DOI] [PubMed] [Google Scholar]
- Guedon E, Sperandio B, Pons N, Ehrlich SD, Renault P. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology. 2005;151:3895–3909. doi: 10.1099/mic.0.28186-0. [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Eshuis H, Jongbloed J, Venema G, van Sinderen D. A small gene, designated comS, located within the coding region of the fourth amino acid-activation domain of srfA, is required for competence development in Bacillus subtilis. Mol Microbiol. 1995;15:55–63. doi: 10.1111/j.1365-2958.1995.tb02220.x. [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Kausche D, Marahiel MA, van Sinderen D, Venema G, Serror P. The Bacillus subtilis transition state regulator AbrB binds to the -35 promoter region of comK. FEMS Microbiol Lett. 2003a;218:299–304. doi: 10.1111/j.1574-6968.2003.tb11532.x. [DOI] [PubMed] [Google Scholar]
- Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003b;149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- Handke LD, Shivers RP, Sonenshein AL. Interaction of Bacillus subtilis CodY with GTP. J Bacteriol. 2008;190:798–806. doi: 10.1128/JB.01115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucl Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner DJ, Ferrari E, Perego M, Hoch JA. Location of the targets of the hpr-97, sacU32(Hy), and sacQ36(Hy) mutations in upstream regions of the subtilisin promoter. J Bacteriol. 1988;170:296–300. doi: 10.1128/jb.170.1.296-300.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higerd TB, Hoch JA, Spizizen J. Hyperprotease-producing mutants of Bacillus subtilis. J Bacteriol. 1972;112:1026–1028. doi: 10.1128/jb.112.2.1026-1028.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irnov I, Sharma CM, Vogel J, Winkler WC. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 2010;38:6637–6651. doi: 10.1093/nar/gkq454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio PT, Fagelson JE, Hoch JA, Strauch MA. The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J Biol Chem. 1991;266:13411–13417. [PubMed] [Google Scholar]
- Kawachi E, Abe S, Tanaka T. Inhibition of Bacillus subtilis scoC expression by multicopy senS. J Bacteriol. 2005;187:8526–8530. doi: 10.1128/JB.187.24.8526-8530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Koide A, Perego M, Hoch JA. ScoC regulates peptide transport and sporulation initiation in Bacillus subtilis. J Bacteriol. 1999;181:4114–4117. doi: 10.1128/jb.181.13.4114-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunji ER, Smid EJ, Plapp R, Poolman B, Konings WN. Di-tripeptides and oligopeptides are taken up via distinct transport mechanisms in Lactococcus lactis. J Bacteriol. 1993;175:2052–2059. doi: 10.1128/jb.175.7.2052-2059.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera BA, Solomon JM, Grossman AD. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell. 1997;89:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Liu ST, Hong GF. Three-minute G + A specific reaction for DNA sequencing. Anal Biochem. 1998;255:158–159. doi: 10.1006/abio.1997.2457. [DOI] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer WJJ, Salas M. Relevance of UP elements for three strong Bacillus subtilis phage phi29 promoters. Nucl Acids Res. 2004;32:1166–1176. doi: 10.1093/nar/gkh290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze N, Bidnenko E, Noirot P, Auger S. Genome-wide mapping of TnrA-binding sites provides new insights into the TnrA regulon in Bacillus subtilis. MicrobiologyOpen. 2015 doi: 10.1002/mbo3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol. 2003;185:1911–1922. doi: 10.1128/JB.185.6.1911-1922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Haertig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RAT, Nannapaneni P, Noone D, Pohl S, Rinn B, Rugheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stulke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Volker U, Bessieres P, Noirot P. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- Ogura M, Matsuzawa A, Yoshikawa H, Tanaka T. Bacillus subtilis SalA (YbaL) negatively regulates expression of scoC, which encodes the repressor for the alkaline exoprotease gene, aprE. J Bacteriol. 2004;186:3056–3064. doi: 10.1128/JB.186.10.3056-3064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci U S A. 1997;94:8612–8617. doi: 10.1073/pnas.94.16.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M, Higgins CF, Pearce SR, Gallagher MP, Hoch JA. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol Microbiol. 1991;5:173–185. doi: 10.1111/j.1365-2958.1991.tb01838.x. [DOI] [PubMed] [Google Scholar]
- Perego M, Hoch JA. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J Bacteriol. 1988;170:2560–2567. doi: 10.1128/jb.170.6.2560-2567.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M, Spiegelman GB, Hoch JA. Structure of the gene for the transition state regulator, abrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- Perera IC, Grove A. Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J Mol Cell Biol. 2010;2:243–254. doi: 10.1093/jmcb/mjq021. [DOI] [PubMed] [Google Scholar]
- Petranovic D, Guedon E, Sperandio B, Delorme C, Ehrlich D, Renault P. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol Microbiol. 2004;53:613–621. doi: 10.1111/j.1365-2958.2004.04136.x. [DOI] [PubMed] [Google Scholar]
- Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 2001;15:1093–1103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Todd DA, Velazquez JV, Cech NB, Sonenshein AL. CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J Bacteriol. 2014;196:1184–1196. doi: 10.1128/JB.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, LeDeaux JR, Ireton K, Grossman AD. The spo0K locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J Bacteriol. 1991;173:1388–1398. doi: 10.1128/jb.173.4.1388-1398.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serror P, Sonenshein AL. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J Bacteriol. 1996a;178:5910–5915. doi: 10.1128/jb.178.20.5910-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serror P, Sonenshein AL. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol Microbiol. 1996b;20:843–852. doi: 10.1111/j.1365-2958.1996.tb02522.x. [DOI] [PubMed] [Google Scholar]
- Shafikhani SH, Mandic-Mulec I, Strauch MA, Smith I, Leighton T. Postexponential regulation of sin operon expression in Bacillus subtilis. J Bacteriol. 2002;184:564–571. doi: 10.1128/JB.184.2.564-571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivers RP, Sonenshein AL. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol. 2004;53:599–611. doi: 10.1111/j.1365-2958.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Mueller JP, Strauch MA, Mathiopoulos C, Sonenshein AL. Transcriptional regulation of a Bacillus subtilis dipeptide transport operon. Mol Microbiol. 1991;5:1915–1925. doi: 10.1111/j.1365-2958.1991.tb00815.x. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Serror P, Joyce E, Sonenshein AL. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol Microbiol. 1995;15:689–702. doi: 10.1111/j.1365-2958.1995.tb02378.x. [DOI] [PubMed] [Google Scholar]
- Smits WK, Hoa TT, Hamoen LW, Kuipers OP, Dubnau D. Antirepression as a second mechanism of transcriptional activation by a minor groove binding protein. Mol Microbiol. 2007;64:368–381. doi: 10.1111/j.1365-2958.2007.05662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon JM, Magnuson R, Srivastava A, Grossman AD. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev. 1995;9:547–558. doi: 10.1101/gad.9.5.547. [DOI] [PubMed] [Google Scholar]
- Sonenshein AL. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol. 2005;8:203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Sonenshein AL. Control of key metabolic intersections in Bacillus subtilis. Nat Rev Microbiol. 2007;5:917–927. doi: 10.1038/nrmicro1772. [DOI] [PubMed] [Google Scholar]
- Steinmetz M, Richter R. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene. 1994;142:79–83. doi: 10.1016/0378-1119(94)90358-1. [DOI] [PubMed] [Google Scholar]
- Strauch MA, Hoch JA. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol Microbiol. 1993;7:337–342. doi: 10.1111/j.1365-2958.1993.tb01125.x. [DOI] [PubMed] [Google Scholar]
- Strauch MA, Spiegelman GB, Perego M, Johnson WC, Burbulys D, Hoch JA. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989;8:1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Toma S, Del Bue M, Pirola A, Grandi G. nprR1 and nprR2 regulatory regions for neutral protease expression in Bacillus subtilis. J Bacteriol. 1986;167:740–743. doi: 10.1128/jb.167.2.740-743.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik W, Chateau A, Dillies MA, Coppee JY, Sonenshein AL, Fouet A. The global regulator CodY regulates toxin gene expression in Bacillus anthracis and is required for full virulence. Infect Immun. 2009;77:4437–4445. doi: 10.1128/IAI.00716-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall ME, Dunlop MJ, Hlavacek WS. Multiple functions of a feed-forward-loop gene circuit. J Mol Biol. 2005;349:501–514. doi: 10.1016/j.jmb.2005.04.022. [DOI] [PubMed] [Google Scholar]
- Wilkinson SP, Grove A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr Issues Mol Biol. 2006;8:51–62. [PubMed] [Google Scholar]
- Wray LV, Jr, Fisher SH. Bacillus subtilis CodY operators contain overlapping CodY binding sites. J Bacteriol. 2011;193:4841–4848. doi: 10.1128/JB.05258-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Yamaguchi H, Kinehara M, Ohki YH, Nakaura Y, Fujita Y. Identification of additional TnrA-regulated genes of Bacillus subtilis associated with a TnrA box. Mol Microbiol. 2003;49:157–165. doi: 10.1046/j.1365-2958.2003.03567.x. [DOI] [PubMed] [Google Scholar]
- Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Dahlsten E, Korkeala H, Lindstrom M. Positive regulation of botulinum neurotoxin gene expression by CodY in Clostridium botulinum ATCC 3502. Appl Environ Microbiol. 2014;80:7651–7658. doi: 10.1128/AEM.02838-14. [DOI] [PMC free article] [PubMed] [Google Scholar]