Abstract
Docosahexaenoic acid (DHA) is a Δ4-desaturated C22 fatty acid and the limiting highly unsaturated fatty acid (HUFA) in neural tissue. The biosynthesis of Δ4-desaturated docosanoid fatty acids 22:6n-3 and 22:5n-6 are believed to proceed via a circuitous biochemical pathway requiring repeated use of a fatty acid desaturase 2 (FADS2) protein to perform Δ6 desaturation on C24 fatty acids in the endoplasmic reticulum followed by 1 round of β-oxidation in the peroxisomes. We demonstrate here that the FADS2 gene product can directly Δ4-desaturate 22:5n-3→22:6n-3 (DHA) and 22:4n-6→22:5n-6. Human MCF-7 cells lacking functional FADS2-mediated Δ6-desaturase were stably transformed with FADS2, FADS1, or empty vector. When incubated with 22:5n-3 or 22:4n-6, FADS2 stable cells produce 22:6n-3 or 22:5n-6, respectively. Similarly, FADS2 stable cells when incubated with d5-18:3n-3 show synthesis of d5-22:6n-3 with no labeling of 24:5n-3 or 24:6n-3 at 24 h. Further, both C24 fatty acids are shown to be products of the respective C22 fatty acids via elongation. Our results demonstrate that the FADS2 classical transcript mediates direct Δ4 desaturation to yield 22:6n-3 and 22:5n-6 in human cells, as has been widely shown previously for desaturation by fish and many other organisms.—Park, H. G., Park, W. J., Kothapalli, K. S. D., Brenna, J. T. The fatty acid desaturase 2 (FADS2) gene product catalyzes Δ4 desaturation to yield n-3 docosahexaenoic acid and n-6 docosapentaenoic acid in human cells.
Keywords: metabolism, nutrition, polyunsaturated fatty acids, Δ6 desaturation
The 22 carbon polyunsaturated fatty acids (PUFAs) 22:6n-3 and 22:5n-6 are ubiquitous throughout human tissue. They are particularly concentrated in neural tissue (1), where they play structural and signaling roles that mediate signal amplification and speed, synaptic function (2), and inflammatory signaling (3), and are nutritionally limiting for newborns, particularly premature infants (4). Humans can consume 22:6n-3 and 22:5n-6 from marine foods and can also biosynthesize them from the corresponding precursor fatty acids 18:3n-3 and 18:2n-6 and downstream intermediates by sequential desaturations and elongations (5). The desaturase biosynthetic activities are known to localize to the endoplasmic reticulum (ER) (6), though over the years data suggest that they operate in other organelles, chiefly the mitochondria (7) and nuclear membrane (8). The synthesis of 22:6n-3 is widely accepted to be limited in humans (9), with serious translational consequences. For instance, neural function development as reflected by the ontogeny of visual acuity is related to the content and duration of dietary 22:6n-3 provision in otherwise healthy preterm infants (10).
The presently accepted mammalian biochemical pathway for 22:6n-3 and 22:5n-6 biosynthesis worked out with rodents is presumed to operate via sequential desaturation and elongation on the ER with a final round of β-oxidation in the peroxisomes (px) demonstrated for rat liver and other tissues (11, 12), via pathway A:
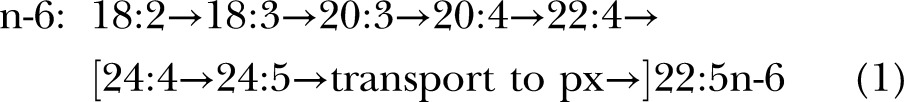 |
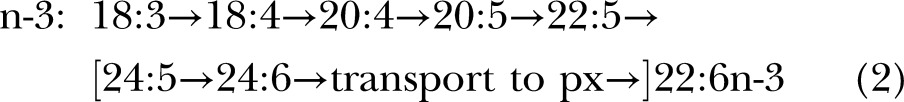 |
expressed as n:d where the steps that increment the double bond number (d) are desaturations (D) and those that increase the carbon number (n) are elongations (E).
Before 1991, the biosynthetic pathway for synthesis of 22:6n-3 and 22:5n-6 from C18 precursors was assumed to be via alternating desaturations and elongations terminating in Δ4 desaturation (13) via pathway B:
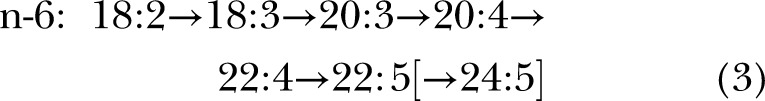 |
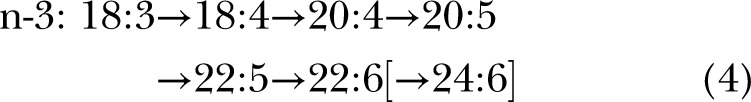 |
The difference in the 2 pathways lies after the first appearance of the C22 species. In pathway A, another round of elongation and Δ6 desaturation is followed by transport to the peroxisomes for a single round of β-oxidation to yield a net Δ4 desaturation (brackets in displays 1 and 2). In pathway B, Δ4 desaturation is a single step mediated by a Δ4-desaturase. Genes whose protein products perform the first 2 desaturations in both pathways are well established, specifically fatty acid desaturase 2 (FADS2, coding for a Δ6 and Δ8-desaturase) and FADS1 (Δ5-desaturase), which catalyze desaturation for both the n-6 and n-3 fatty acids at each step, although at differing rates (14, 15).
The entirety of pathway B was assumed to proceed at the ER, though numerous lines of evidence suggest that newly synthesized 22:6n-3 may be closely associated with the mitochondria (16, 17). Were 22:6n-3 and 22:5n-6 synthesized in mitochondria, one might expect isolated mitochondria to also synthesize 22:6n-3 or 22:5n-6; however, we are aware of no such reports, consistent with our measurements (unpublished data). Such observations are not definitive, however, because isolated mitochondria may function metabolically very differently than those in intact cells.
A serious difficulty of both pathways is the hypothesis that common enzymes are responsible for n-3 and n-6 transformations. For instance, the much attenuated 22:6n-3 in acyl-coA oxidase (Aox)-null mouse liver is accompanied by a 3-fold 22:5n-6 rise, with acyl-CoA oxidation presumed to be the final step in 22:6n-3 and 22:5n-6 synthesis (18); these results are paralleled by human neonatal adrenoleukodystrophy, showing a decrease in 22:6n-3 but an increase in 22:5n-6 (19). Two different “Δ6 desaturases” may explain these results if they are n-3 and n-6 specific; at present, candidates for the molecular intermediaries of these processes are the alternative transcripts of the FADS2 (20) and possibly FADS1 (21) or FADS3 (22) as modifiers of activity, though no evidence for this function in these pathways has yet emerged.
In humans and rodents, PUFA desaturation is mediated by proteins coded by the fatty acid desaturase (FADS) genes (human genome at 11q12-13.1). Before the widespread use of molecular techniques, the PUFA desaturases were assumed to be highly substrate specific. FADS gene products are now known to operate on numerous substrates. A single vertebrate gene in the rabbitfish codes for a protein that performs dual Δ5 and Δ4 desaturation to synthesize 22:6n-3 directly by pathway B, established by the appearance of 22:6n-3 before the appearance of 24:6n-3 in yeast cells transiently transfected with the fish Fad gene (23). Other Δ4 desaturation activities have long been established in marine microorganisms by similar methods (24).
We recently described novel functions of the proteins coded by the baboon FADS using yeast and mammalian cancer cells. FADS2 coding for a protein that performs Δ6 desaturation on numerous PUFA substrates also catalyzes Δ8 desaturation (14):
Similarly, we showed that FADS1 coding for Δ5-desaturase activity (e.g., 20:3n-6→20:4n-6) also catalyzes in MCF-7 human breast cancer cells Δ5 desaturation of 20:2 to yield sciadonic acid (25):
MCF-7 cells have no detectable Δ6-desaturase (FADS2) activity (26). FADS1-mediated desaturation to yield non-methylene-interrupted PUFAs apparently operates when FADS2 is absent either experimentally via gene ablation (27) or naturally in vivo, as in the cat, which has negligible Δ6-desaturase activity (28).
Using MCF-7 cancer cells stably expressing FADS2 and FADS1, we demonstrate that pathway B operates for both 22:6n-3 and 22:5n-6 in human cells. We also provide the first molecular evidence showing the localization of FADS2 in mitochondria.
MATERIALS AND METHODS
Cornell University (IACUC, protocol 02-105) and Texas Biomedical Research Institute (formerly the Southwest Foundation for Biomedical Research) institutional animal care and use committees approved studies on baboons.
RNA extraction, cDNA synthesis, and vector construction
Liver tissue from a 12-wk-old baboon neonate collected at necropsy in RNAlater was stored at −80°C. Total RNA was isolated with an RNeasy mini kit (Qiagen, Germantown, MD, USA). RNA quality and quantity was evaluated by NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). The resulting cDNA was stored at −20°C and was used as template for real-time PCR reactions. The protein coding region of baboon FADS1 (GenBank accession no. EF531577) and FADS2 (GenBank accession no. EU780003) were amplified and cloned into pcDNA3.1 expression vector (Life Technologies, Grand Island, NY, USA) containing cytomegalovirus promoter by using HindIII and XhoI restriction enzyme sites.
Mammalian cell culture and expression of FADS
MCF-7 human breast cancer cells (a kind gift of R. H. Liu, Cornell University) were grown in minimum essential medium α (MEM-α) (Life Technologies) containing 10% fetal bovine serum (FBS) and 10 mM [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; HEPES] buffer at 37°C with 5% CO2. To perform functional studies, MCF-7 cells stably expressing FADS1 and FADS2 were established, along with negative control empty vector cells. The stable cells were selected by using Geneticin (Life Technologies) as selection marker. The control, FADS1, and FADS2 stable MCF-7 cells were supplemented with albumin bound fatty acid substrates to investigate the functionality of the expressed FADS. Pulse studies were generally performed by adding substrate to medium for a short time, followed by nonsupplemented medium, whereas incubation studies were performed by adding substrate and not changing medium. Choice of pulse or incubation was made on the basis of preliminary experiments and were chosen to produce cells with moderate incorporation of substrate. Most studies used 1 × 106 cells; d5-18:3n-3 (Cambridge Isotope Laboratories, Cambridge, MA, USA) kinetics used 2 × 106 cells. Kinetic studies were carried out using 50 μM of 22:5n-6, 50 μM of 22:6n-3, 10 μM of 22:5n-3, or 100 μM of d5-18:3n-3 substrates. Cells after trypsinization were collected for fatty acid analysis at 0, 3, 6, 12, 18, and 24 h. For d5-18:3n-3 experiments, cells were collected at 24, 36, 48, 60, and 72 h. In a pulse kinetic experiment, 100 μM of 22:4n-6 was added to the medium for 1 h. After 1 h of incubation, the medium was removed, cells were washed with 1× PBS, and fresh MEM-α without FBS was added to the cells. Cells were collected through trypsinization at 1, 3, 6, 12, 18, and 24 h for fatty acid analysis. When cells were dosed with an n-3 PUFA, PUFAs were normalized to the sum of n-6 PUFAs with chain lengths of 20 to 24 carbons, and vice versa.
Fatty acid extraction and analysis
Collected stable cells were used for fatty acid extraction. After centrifugation, the supernatant was removed, and lipid extraction was carried out on the cell pellets. Fatty acid methyl esters (FAMEs) were prepared using a modified 1-step lipid extraction method (29, 30). Briefly, pelleted cells were incubated with an aqueous digesting/methylating reagent and an organic extraction reagent and FAMEs obtained directly. Methylated fatty acids were structurally identified by gas chromatography covalent adduct chemical ionization tandem mass spectrometry (31–36) by molecular weight and by diagnostic ion analysis in MS/MS because the FAME double bond position cannot be established by either electron or proton transfer chemical ionization MS. All were quantified by gas chromatography–flame ionization detection using an equal weight FAME standard mixture to verify response factors daily according to standard methods (37).
Subcellular localization and Immunoblotting
SK-N-SH cells were used for subcellular localization experiments and were cultured as described previously (21). Earlier we had shown localization of FADS1 to mitochondria and ER. The open reading frame (ORF) of FADS2 was cloned into the pEGFP-N1 vector driven by the cytomegalovirus promoter. At confluence, SK-N-SH cells were transfected with the constructed FADS2 and empty vector using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA). MitoTracker Red CMXRos or ER-tracker Blue-White DPX (Molecular Probes, Eugene, OR, USA; Invitrogen) organelle-specific stains were used to stain the organelles. Inverted Meta Confocal Microscope (LSM 510 META; Carl Zeiss GmbH, Jena, Germany) was used to visualize organelle-stained live cells. Mitochondrial isolation kit (Thermo Fisher Scientific) was used to isolate mitochondria from the transfected SK-N-SH cells. Western blot was carried out using green fluorescent protein (GFP) antibody (Abcam, Cambridge, MA, USA), cytochrome oxidase IV (COX IV, inner mitochondrial membrane-specific) antibody (Cell Signaling Technology, Danvers, MA, USA), protein disulfide isomerase (PDI, ER-specific) antibody (Cell Signaling Technology), and β-actin antibody as control (Roche, Basel, Switzerland). Immunoblots were visualized by using Li-Cor Odyssey infrared imaging system as directed by the manufacturer (Li-Cor Biosciences, Lincoln, NE, USA).
RESULTS
MCF-7 cells stably expressing FADS1 and FADS2, as well as empty vector control cells, were generated using a Geneticin-selective agent. The ORF sequence of primate (baboon, Papio sp.) FADS1 (EF531577) consists of 1335 bp, encoding a protein of 444 aa and a stop codon. Analysis and comparison of the amino acid sequence of baboon FADS1 shows 95% identity and 97% similarity with human FADS1 (NP_037534). Similarly, FADS2 (EU780003) also consists of 1335 bp ORF, encoding a protein of 444 aa and a stop codon showing 98% identity and 99% similarity with human FADS2 (NP_004256). The protein product of FADS1 catalyzes Δ5 desaturation, whereas the product of FADS2 catalyzes Δ6 desaturation and Δ8 desaturation (14).
24:5n-6 and 24:6n-3 are products of 22:5n-6 and 22:6n-3
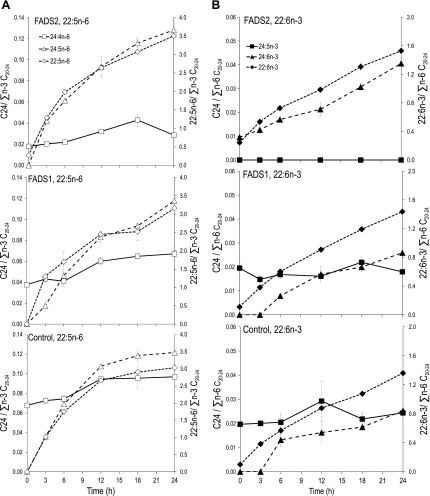
FADS2, FADS1, or control cells have no detectable 24:5n-6. Incubation of all 3 transformed cells with 22:5n-6 (50 μM) causes a time-dependent accumulation of 24:5n-6, demonstrating that 24:5n-6 is an elongation product of 22:5n-6 (Fig. 1A). Similarly, control and FADS1 cells have no detectable 24:6n-3; FADS2 cells have basal 24:6n-3. Incubation of all 3 cell types with 22:6n-3 (50 μM) causes time-dependent increases in 24:6n-3, demonstrating that 24:6n-3 is a product of 22:6n-3 (Fig. 1B).
Figure 1.
Kinetics of C22→C24 PUFAs. A) 22:5n-6 incubations. FADS2, FADS1, and control stable MCF-7 cells were incubated with 22:5n-6 at 50 μM and sampled at 3, 6, 12, 18, and 24 h. In all cells, 24:5n-6 starts to appear at 3 h and increases to 24 h, rising in parallel with 22:5n-6 (right y axis) showing that 24:5n-6 is a product of 22:5n-6. B) 22:6n-3 incubations. FADS2, FADS1, and control stable MCF-7 cells were incubated with 22:6n-3 at 50 μM. Analogous to (A), the time-dependent increase in 24:6n-3 is seen in all 3 cell types, showing that 24:6n-3 is a product of 22:6n-3.
The results of Fig. 1 show that accumulation of newly synthesized 22:5n-6 and 22:6n-3 eventually results in the synthesis of 24:5n-6 and 24:6n-3, respectively. Demonstration of direct Δ4-desaturase-mediated synthesis of 22:6n-3 and 22:5n-6 from 22:5n-3 and 22:4n-6, respectively, requires appearance of the C22 fatty acids before detection of C24 fatty acids.
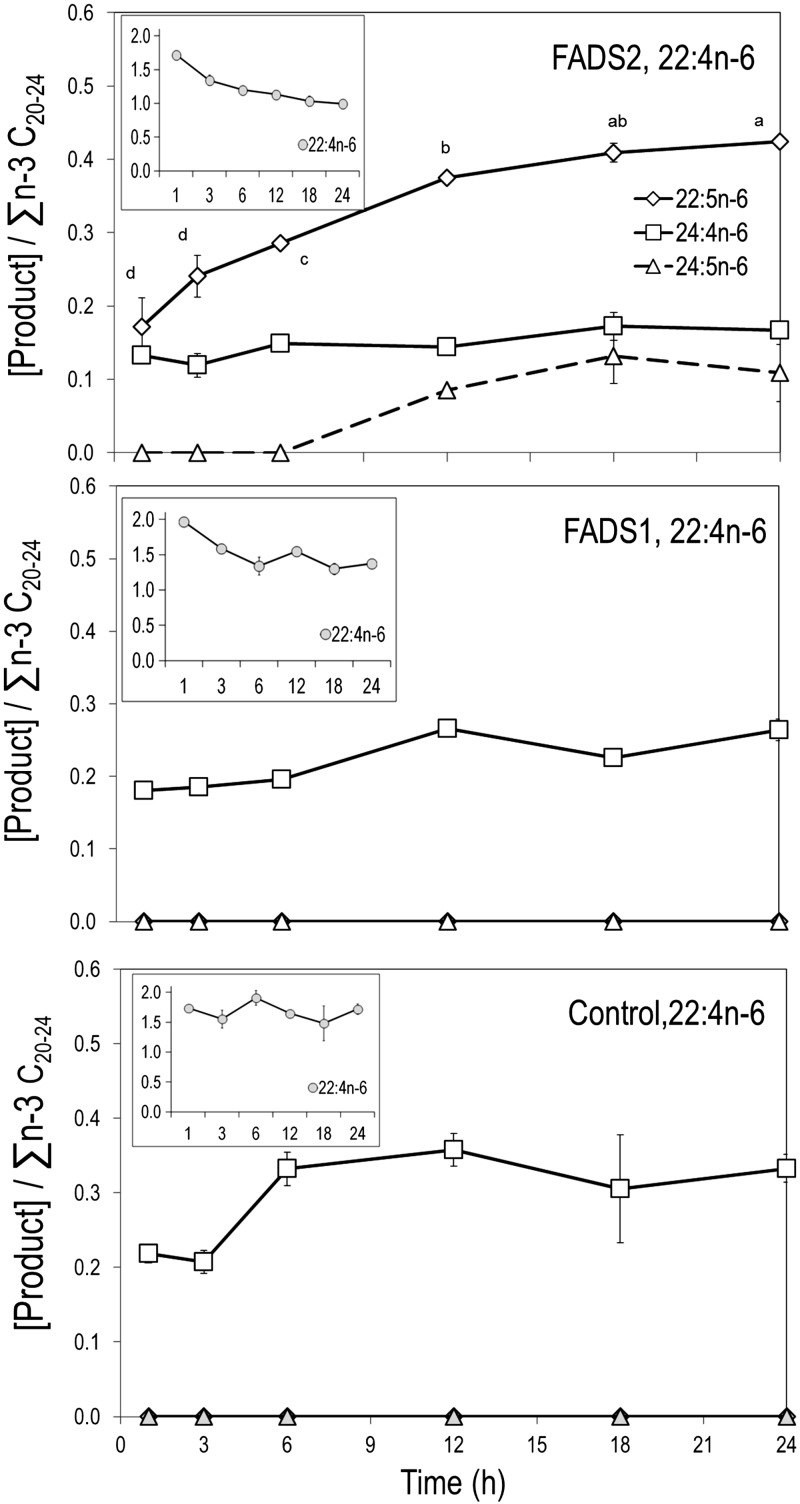
22:4n-6→22:5n-6 in FADS2 cells before 24:5n-6 is detectable
Substrate 22:4n-6 was applied to medium as a pulse (100 μM, 1 h), then replaced with MEM-α (10% FBS) with no substrate and incubated up to 24 h to allow cells to operate on 22:4n-6 that had been taken up. Control and FADS1 cells show no activity toward 22:4n-6; however, both cells slowly elongate 22:4n-6 to 24:4n-6. In contrast, FADS2 cells show synthesis of 22:5n-6; 24:5n-6 appears later, only at and after 12 h (Fig. 2).
Figure 2.
22:4n-6 incubations. FADS2, FADS1, and control stable MCF-7 cells were incubated with 22:4n-6 at 100 μM for 1 h, then incubated with medium. FADS2 cells synthesize 22:5n-6; putative intermediate 24:5n-6 appears only at 12 h. Both FADS1 and control cells elongate 22:4n-6 to 24:4n-6; no synthesis of 22:5n-6 is detected. Points labeled with different letters are significantly different at P < 0.05.
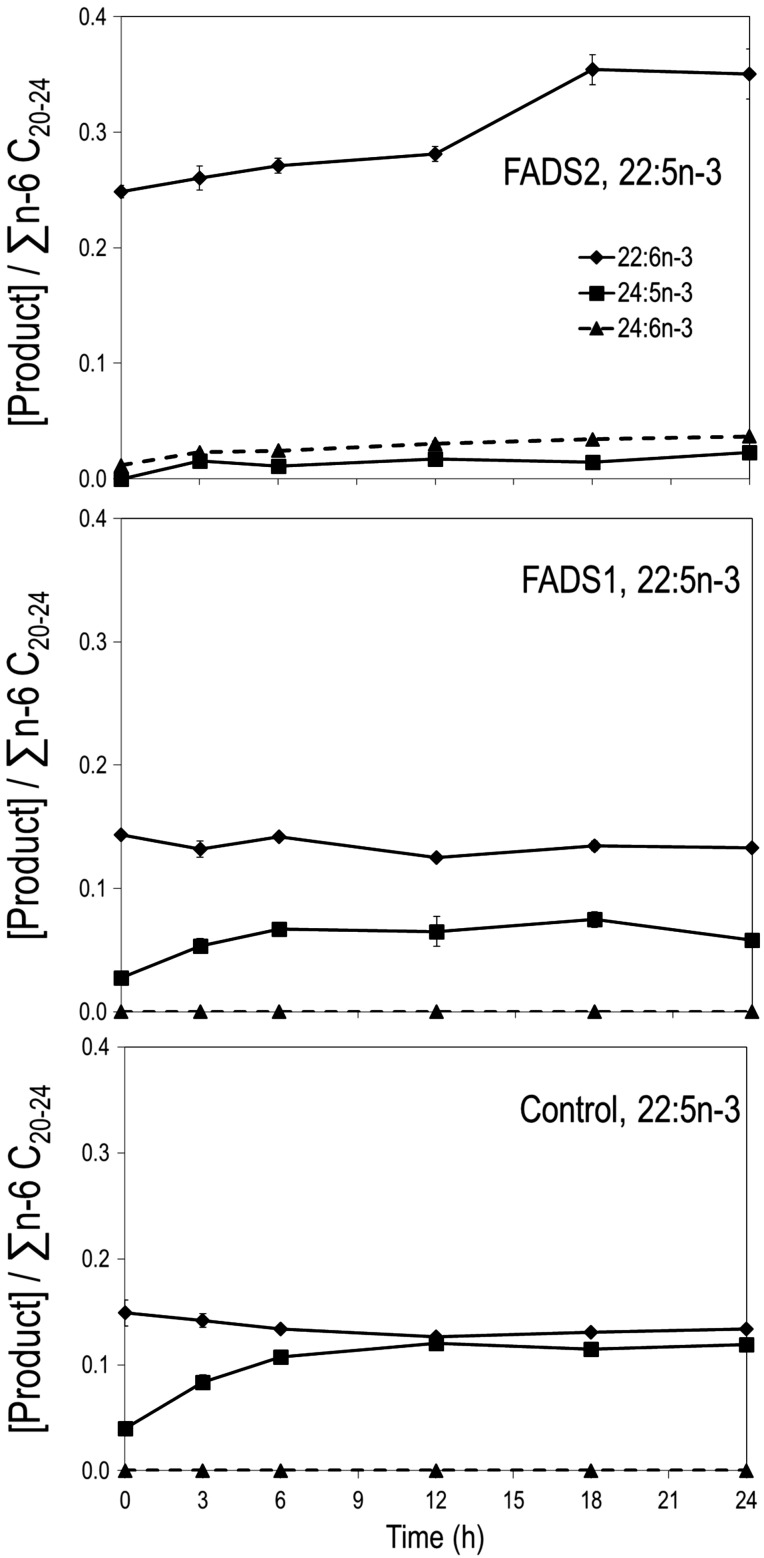
22:5n-3 metabolism
Cells were incubated with 22:5n-3 (10 μM) for up to 24 h. FADS2 cells show an increase in 22:6n-3 to 24 h, while control and FADS1 cells show no discernable change in 22:6n-3 (Fig. 3). At baseline, FADS2 cells have no detectable 24:5n-3 and trace 24:6n-3; both rise and plateau at 3 h. In contrast, control and FADS1 cells have no detectable 24:6n-3 at any time point, while 24:5n-3 rises to a plateau by 12 h.
Figure 3.
22:5n-3 incubations. FADS2, FADS1, and control stable MCF-7 cells incubated with 22:5n-3 at 10 μM and sampled at 3, 6, 12, 18, and 24 h. FADS2 cells show an increase in 22:6n-3 to 24 h, whereas 24:6n-3 plateaus at 3 h. Both FADS1 and control cells elongate 22:5n-3 to 24:5n-3, which plateaus by 12 h; no change in 22:6n-3 is evident.
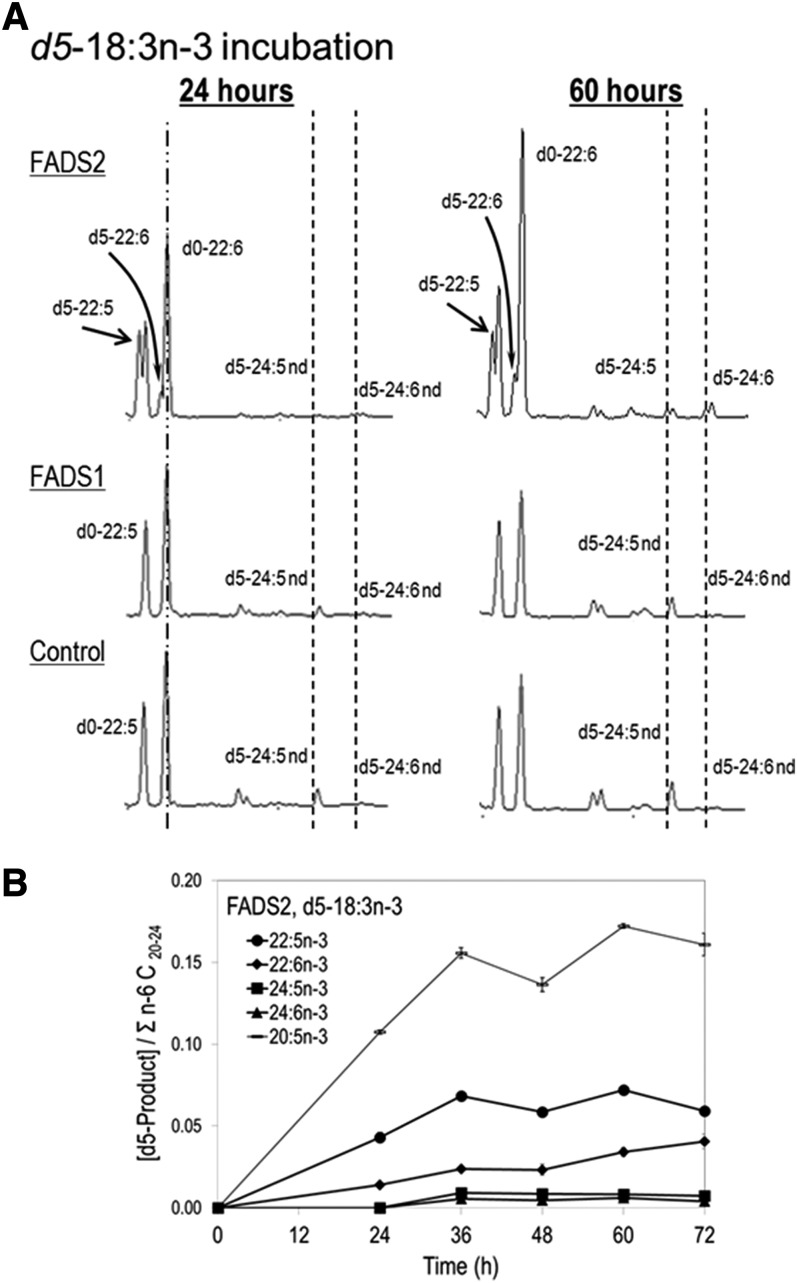
d5-18:3→d5-22:6n-3 prior to detection of d5-24:5n-3 or d5-24:6n-3
Synthesis of 22:6n-3 was investigated with isotopically labeled d5-18:3n-3 (Fig. 4). Incubation of control or FADS1 cells with d5-18:3n-3 results in no labeled C22 products or other n-3 intermediates 24 h. Incubation of FADS2 cells with d5-18:3n-3 for 24 h shows labeling in all n-3 intermediates shown in pathway B, including synthesis of d5-22:6n-3 but no labeling of 24:5n-3 or 24:6n-3. Both d5-24:5n-3 and d5-24:6n-3 are detected at 60 h and as early as 36 h (Fig. 4A). Kinetic plots show that 20:5n-3 and 22:5n-3 accumulate from the first time point at 24 h and then plateaus between 36 and 72 h, while 22:6n-3 continues to rise even at 72 h. Both 24:5n-3 and 24:6n-3 plateau between 36 and 72 h (Fig. 4B).
Figure 4.
d5-18:3n-3 metabolism. A) FADS2, FADS1, and control stable MCF-7 cells were incubated with 100 μM of d5-18:3n-3 and sampled at 24 and 60 h. No labeled intermediates or C22 products are seen in FADS1 and control cells. FADS2 cells show labeling in all n-3 intermediates and 22:6n-3; no labeled C24 products were detected at 24 h. B) d5-18:3n-3 kinetics for FADS2 cells. C24 products (d5-24:5n-3 and d5-24:6n-3) are detected at 36 h and plateau between 36 and 72 h. C22 (d5-22:6n-3) continues to rise at 72 h.
Subcellular localization
Front-end desaturases are known to be ER proteins. We recently showed that the FADS1 classic isoform localizes to mitochondria (21). To examine the subcellular localization of FADS2 (444 aa), we used the GFP tagging system, and to localize organelles, SK-N-SH neuroblastoma cells transiently transfected with constructed vectors were stained with MitoTracker Red and ER-tracker Blue-White DPX. Imaging analysis shows FADS2 localizing to ER and to mitochondria (Fig. 5A, B). To verify the mitochondria localization, we isolated intact mitochondria, and Western blot analysis was carried out using GFP, COX IV, PDI and β-actin antibodies. We detected a 73 kDa band (46 kDa FADS2; 27 kDa GFP) for FADS2. A 27 kDa band alone corresponding to GFP was seen in empty vector control. Clear bands were observed using COX IV and β-actin antibodies (Fig. 5C). No visible bands were seen using PDI antibody, ruling out ER contamination from the mitochondrial isolates and showing FADS2 localizing to mitochondria.
Figure 5.
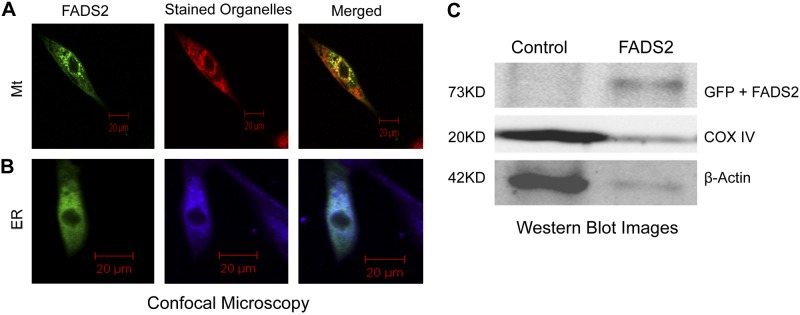
FADS2 subcellular localization. SK-N-SH cells transfected with control or FADS2 were stained with MitoTracker Red (Mt) or ER-tracker Blue-White DPX (ER), then visualized with an inverted meta confocal microscope (A, B). C) Western blot analysis using GFP, COX IV, and β-actin antibodies. No visible bands were seen with PDI antibody.
DISCUSSION
Until 1991, the biosynthesis of biologically active 22:5n-6 and 22:6n-3 PUFAs in all organisms was thought to occur via Δ4 desaturation. However, difficulties in demonstrating Δ4 desaturation activity collected from microsomes of rats fed minimal fat diets led to the proposal of a coupled microsomal–peroxisomal pathway (11). Classic biochemical radiotracer evidence has been presented in numerous studies in support of this pathway in tissue, primarily from rats. Fractionated organelles, cells, or an intact animal are treated with specially labeled 14C or 3H PUFA precursors, and products are analyzed by HPLC or other methods with radioactivity detection without localization of the radioactivity within the product (11, 38–40). For instance, the first article to introduce this pathway incubated rat liver microsomes or primary hepatocytes with 1-14C-18:2, 1-14C-22:5, 3-14C-24:5 or 3-14C-24:6 showed flow of label into 22:6 when incubated with cells but not microsomes (11). Addition of a peroxisome-enriched fraction led to incorporation of label into 22:6n-3 and 22:5n-6, respectively, leading to the “coupled microsomal–peroxisomal” pathway (pathway A). However, a well-established alternative pathway that leads to labeling of all PUFAs provided only a single labeled fatty acid is β-oxidation in both the mitochondria and the peroxisomes to yield labeled acetate (14C-2:0), followed by incorporation of label into downstream products at the elongation steps. Indeed, many reports offering data that show flow of label from, for instance, 3-14C-24:6 → 14C-22:6n-3 or other labeled substrates, show in the same chromatograms label also appearing in 16:0, 18:0, and/or 18:1, a process that demonstrates unequivocally that partial β-oxidation extensively labels acetate pools (38, 39). Cultured rat hippocampal neurons appear to incorporate more label from [1-14C]22:5n-3 and [3-14C]24:5n-3 into 16:0 and 18:0 than into 22:6 at 72 h incubation, while [3-14C]24:6n-3 appears to be completely consumed and its label appears only in 22:6n-3 (41). To our knowledge, no studies ran controls to rule out the possibility that, for instance, 3-14C-24:6→14C-2:0 and 18:3 + 14C-2:0→→→ 1- or 3-14C-22:6 was the major process delivering label to end products. We are also not aware of double label studies (e.g., 3H, 14C) showing that the ratio of label is similar in the precursor or products, as was done in early studies that demonstrated direct chain elongation and desaturation (42), or β-oxidation in peroxisomes (43). Finally, no studies comparing 1-14C-24:6 to 3-14C-24:6 labeling of product fatty acids have been reported; the C24 fatty acid labeled in the 1 position could have been synthesized by then-available methods (11). Our labeling studies are not vulnerable to this alternative because conversion of the d5 moiety to acetate would result in the d3 COOH-CD3, which, upon incorporation into an elongated fatty acid, would retain only 2 deuteriums, resulting in a d2-labeled fatty acid product.
Numerous data are not consistent with the microsomal–peroxisomal pathway, reviewed elsewhere (16, 17). First, Aox knockout mice simultaneously increase 22:5n-6 and decrease 22:6n-3 (18). A single peroxisomal Aox as the sole final step operating in a single compartment in synthesis of both 22:5n-6 and 22:6n-3 cannot yield this result. Second, treatments that enhance peroxisomes do not increase 22:6n-3 and 22:5n-6; they usually decrease them. For instance, fenofibrate, a peroxisome proliferator–activated receptor α agonist, also increases hepatic Fads1 and Fads2 mRNA by about 3-fold; however, 22:6n-3 and 20:5n-3 decrease by 15 and 27%, respectively (44). In these same animals, hepatic de novo fatty acid synthesis increase dramatically (150–600%). Third, in vivo mouse data suggest alternative channeled pathways for 22:5n-6 biosynthesis from 18:2n-6 or 20:3n-6 (45). Fourth, X-linked adrenoleukodystrophy or adrenomyeloneuropathy patients having defective peroxisomal β-oxidation of very long chain fatty acids show normal 22:6n-3 levels (19).
Synthesis of labeled 22:6n-3 from labeled [1-14C]18:3 or [1-14C]20:5 appears to be impaired in human fibroblasts with defects in mitochondrial fatty acid transport genes CPT1, CACT, and CPT2 (46). Radioactivity found in 22:6n-3 considered as a percentage of total radioactivity detected in all intermediates is reduced by 47, 41, and 31% in cells with specific defects in the mitochondrial fatty acid transport proteins CPT1, CACT, and CPT2 compared to controls. Moreover, the block appears to be in the metabolism of 22:5n-3, as it is greater by 12 to 27% than controls. Whether this defect is in desaturation or elongation is not evident.
Elongation of very long chain fatty acids protein 2 (Elovl2) and 5 (Elovl5) both mediate elongation of PUFAs. Mice with ablated Elovl2 (47) and Elovl5 (48) both have reduced docosahexaenoic acid (DHA) levels. Very recent evidence shows that ablation of Elovl2 results in simultaneous reduction in 22:6n-3 and 22:5n-6, and increase in 22:5n-3 and 22:4n-6, with no strong effect on C18 or C20 PUFA levels, all consistent with pathway A (47). A plausible possibility is that ablation of Elovl2 influences Fads2 expression or may influence a coupled or channeled pathway for which evidence has been presented (18). Both human ELOVL2 and mouse Elovl2 elongate C20 and C22 PUFAs, though the human enzyme does not operate on C18 PUFAs (49) and plausibly may differ from the human/primate enzyme in other ways; the difference in substrate specificity may translate into a difference in regulation or participation in the PUFA pathways. The present results are not inconsistent with a major role for ELOVL2 in synthesis of C24 PUFAs for Δ6 desaturation, as they do not address whether that pathway operates. It is curious, however, that the concentration of C24 intermediates are unaffected in Elovl2-null mice, since 24:5n-3 and 24:4n-6 are the products of elongation initiated by Elovl2, and 24:6n-3 and 24:5n-6 must be transported from ER to peroxisomes and therefore cannot be part of a channeled pathway in a single organelle. Absent input from elongated 22:4n-6 and 22:5n-3, the peroxisomal pathway might be expected to convert all available 24:5n-3 and 24:4n-6 to 22:6n-3 and 22:5n-6, respectively, reducing C24 highly unsaturated fatty acids (HUFAs) to nil since pathway A requires rapid β-oxidation to support the very low C24 levels compared to products. Plausible also is that both pathways operate as redundant systems for 22:6n-3 synthesis for physiologic conditions and/or tissues with high 22:6n-3 demand (e.g., perinatal brain).
Human ELOVL2 specifically elongates C20 and C22 PUFAs to their respective C22 and C24 metabolites, and this gene may mediate synthesis of 22:6n-3 via 24:6n-3 (49). More recent data indicate that ablation of Elovl2 is responsible for loss of fertility in male mice due to near-complete elimination of elongation of 20:4n-6, present at near-normal levels, to 22:4n-6 and presumably subsequent elongation to at least 30:5n-6 in testes, which also involves Elovl4 for the longer chain members of the family (50).
Enzymatic Δ4 desaturation activity is common across phyla. Heterologous expression of the full-length Thraustochytrium Fad4 desaturase into yeast and brassica resulted in the production of 22:6n-3 via Δ4 desaturation (24). A functional Δ4 Fads gene has been isolated from the freshwater organism Euglena gracilis (51), marine algae (Pavlova lutheri) (52), and a parasitic (trypanosomes) protozoan (53). The first functional Δ4 Fads isolated from a vertebrate (rabbitfish) is a bifunctional enzyme that has both Δ5/Δ4 activities (23), and subsequently, functional Δ4 Fads have been described in other fish species (Solea senegalensis, Chirostoma estor) (54, 55), including a carnivorous freshwater species (Channa striata) (56). Human FADS2 is a multifunctional enzyme with known substrate specificities for 16, 18, 20, and 24 carbon fatty acids (14), though no previous reports show FADS2 specificity for 22 carbon PUFAs. Our data support the existence of functional human/primate Δ4 FADS catalyzed by FADS2 with substrate specificity for 22 carbon PUFAs. Thus, human/primate FADS2 catalyzes Δ6, Δ8, and Δ4 activities and can be considered an even-numbered desaturase. The Δ6-desaturase multiple substrate use is fully consistent with the apparent attenuation of 22:6n-3 synthesis by 18:2n-6, which has been explained by competition for Δ6 desaturation of 24:5n-3→24:6n-3 (57); in pathway B, competition for action of the protein product of FADS2, Δ6 desaturation of 18:2n-6→18:3n-6 versus Δ4 desaturation of 22:5n-3→22:6n-3, would also operate. Notably, the existence of Δ4 desaturation pathway in human cells does not rule out the coupled microsomal–peroxisomal pathway; however, because controls were never presented for recycling of 14C via, for instance, 14C-24:5→14C-2:0; 14C-2:0+18:3n-3→22:6n-3, we consider that pathway to not be unequivocally demonstrated.
Our kinetic data do not address the intracellular site or sites where 22:6n-3/22:5n-6 are synthesized. Previous suggestive data implies that 22:6n-3 synthesis may occur in the mitochondria. Using the GFP tagging system and Western blot analysis, we show the first molecular evidence of FADS2 localization to the mitochondria. On the basis of the baboon FADS2 ORF (GenBank accession no. NM_001145087), a protein molecular weight calculator (58) predicted a protein size of 52.31 kDa. However, our Western blot analysis shows a smaller product of 46 kDa. We previously reported a shorter product for FADS1 using a FADS1 antibody (21), and similar observations have also made by others using a FADS3 antibody (59). This observed shorter product may be due to protein degradation or cleavage. FADS2 is known to code for ER proteins; however, mitochondrial Δ6-desaturase (FADS2) activity has been reported more than once (7, 60).
The Δ6-desaturase activity was 2-fold higher in a mouse liver mitochondrial fraction compared to the microsomal fraction (7). Primary rat hepatocytes, when incubated with radiolabel [14C] 18:3n-3, showed 20% of the total radiolabel recovery in the 22:6 phosphatidyl choline fraction of the mitochondria, compared to 5% in microsomes (60). A carnitine-dependent channeled mitochondrial pathway was proposed as the primary route for longer chain PUFA (20:4n-6, 22:5n-6, and 22:6n-3) biosynthesis. Our biochemical results demonstrate a FADS2-mediated Δ4-desaturase pathway in human cells, and the observation of FADS2 and FADS1 in mitochondria support a role in long-chain PUFA biosynthesis, not related to oxidation/retroconversion of a 24:6n-3 intermediate.
In conclusion, we provide first unequivocal molecular evidence showing a mammalian gene product introducing a double bond at the Δ4 position, demonstrating a FADS2 pathway for endogenous biosynthesis of 22:6n-3 and 22:5n-6. Considered in the context of previous data, the evidence supports trifunctional FADS (FADS2 classical transcript with Δ6, Δ8, and Δ4 activities) in mammals, analogous to bifunctionality demonstrated previously for vertebrate fish. We also demonstrate the first molecular evidence of FADS2 localization in mitochondria, supporting that organelle’s involvement in long-chain polyunsaturated fatty acid synthesis. The present data do not rule out a coupled microsomal–peroxisomal pathway but rather support further study of the relative importance of putatively redundant biosynthetic pathways.
Supplementary Material
Acknowledgments
This work was supported by U.S. National Institutes of Health (NIH) Grant R01 AT007003 from the National Center for Complementary and Integrative Health (formerly the National Center for Complementary and Alternative Medicine) and the Office of Dietary Supplements. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Glossary
- Aox
acyl-CoA oxidase
- COX IV
cytochrome oxidase IV
- DHA
docosahexaenoic acid
- Elovl2
elongation of very long chain fatty acids protein 2
- ER
endoplasmic reticulum
- FADS
fatty acid desaturase
- FAME
fatty acid methyl ester
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- MEM-α
minimum essential medium α
- ORF
open reading frame
- PDI
protein disulfide isomerase
- PUFA
polyunsaturated fatty acid
- px
peroxisome
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Diau G. Y., Hsieh A. T., Sarkadi-Nagy E. A., Wijendran V., Nathanielsz P. W., Brenna J. T. (2005) The influence of long chain polyunsaturate supplementation on docosahexaenoic acid and arachidonic acid in baboon neonate central nervous system. BMC Med. 3, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breckenreidge W. C., Gombos G., Morgan I. G. (1971) The docosahexaenoic acid of the phospholipids of synaptic membranes, vesicles and mitochondria. Brain Res. 33, 581–583 [DOI] [PubMed] [Google Scholar]
- 3.Serhan C. N., Chiang N., Van Dyke T. E. (2008) Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol. 8, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lapillonne A., Groh-Wargo S., Gonzalez C. H., Uauy R. (2013) Lipid needs of preterm infants: updated recommendations. J. Pediatr. 162(3, Suppl)S37–S47 [DOI] [PubMed] [Google Scholar]
- 5.Brenna J. T. (2002) Efficiency of conversion of alpha-linolenic acid to long chain n-3 fatty acids in man. Curr. Opin. Clin. Nutr. Metab. Care 5, 127–132 [DOI] [PubMed] [Google Scholar]
- 6.Brenner R. R. (2003) Hormonal modulation of delta6 and delta5 desaturases: case of diabetes. Prostaglandins Leukot. Essent. Fatty Acids 68, 151–162 [DOI] [PubMed] [Google Scholar]
- 7.Hughes S., York D. A. (1985) Hepatic delta 6-desaturase activity in lean and genetically obese ob/ob mice. Biochem. J. 225, 307–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ves-Losada A., Brenner R. R. (1995) Fatty acid delta 5 desaturation in rat liver cell nuclei. Mol. Cell. Biochem. 142, 163–170 [DOI] [PubMed] [Google Scholar]
- 9.Brenna J. T., Salem N. Jr., Sinclair A. J., Cunnane S. C.; International Society for the Study of Fatty Acids and Lipids, ISSFAL (2009) alpha-Linolenic acid supplementation and conversion to n-3 long-chain polyunsaturated fatty acids in humans. Prostaglandins Leukot. Essent. Fatty Acids 80, 85–91 [DOI] [PubMed] [Google Scholar]
- 10.Morale S. E., Hoffman D. R., Castañeda Y. S., Wheaton D. H., Burns R. A., Birch E. E. (2005) Duration of long-chain polyunsaturated fatty acids availability in the diet and visual acuity. Early Hum. Dev. 81, 197–203 [DOI] [PubMed] [Google Scholar]
- 11.Voss A., Reinhart M., Sankarappa S., Sprecher H. (1991) The metabolism of 7,10,13,16,19-docosapentaenoic acid to 4,7,10,13,16,19-docosahexaenoic acid in rat liver is independent of a 4-desaturase. J. Biol. Chem. 266, 19995–20000 [PubMed] [Google Scholar]
- 12.Mohammed B. S., Luthria D. L., Bakousheva S. P., Sprecher H. (1997) Regulation of the biosynthesis of 4,7,10,13,16-docosapentaenoic acid. Biochem. J. 326, 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Innis S. M. (1991) Essential fatty acids in growth and development. Prog. Lipid Res. 30, 39–103 [DOI] [PubMed] [Google Scholar]
- 14.Park W. J., Kothapalli K. S., Lawrence P., Tyburczy C., Brenna J. T. (2009) An alternate pathway to long-chain polyunsaturates: the FADS2 gene product Delta8-desaturates 20:2n-6 and 20:3n-3. J. Lipid Res. 50, 1195–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Su H. M., Brenna J. T. (1998) Simultaneous measurement of desaturase activities using stable isotope tracers or a nontracer method. Anal. Biochem. 261, 43–50 [DOI] [PubMed] [Google Scholar]
- 16.Infante J. P., Huszagh V. A. (1997) On the molecular etiology of decreased arachidonic (20:4n-6), docosapentaenoic (22:5n-6) and docosahexaenoic (22:6n-3) acids in Zellweger syndrome and other peroxisomal disorders. Mol. Cell. Biochem. 168, 101–115 [DOI] [PubMed] [Google Scholar]
- 17.Infante J. P., Huszagh V. A. (1998) Analysis of the putative role of 24-carbon polyunsaturated fatty acids in the biosynthesis of docosapentaenoic (22:5n-6) and docosahexaenoic (22:6n-3) acids. FEBS Lett. 431, 1–6 [DOI] [PubMed] [Google Scholar]
- 18.Infante J. P., Tschanz C. L., Shaw N., Michaud A. L., Lawrence P., Brenna J. T. (2002) Straight-chain acyl-CoA oxidase knockout mouse accumulates extremely long chain fatty acids from alpha-linolenic acid: evidence for runaway carousel-type enzyme kinetics in peroxisomal beta-oxidation diseases. Mol. Genet. Metab. 75, 108–119 [DOI] [PubMed] [Google Scholar]
- 19.Martinez M. (1992) Abnormal profiles of polyunsaturated fatty acids in the brain, liver, kidney and retina of patients with peroxisomal disorders. Brain Res. 583, 171–182 [DOI] [PubMed] [Google Scholar]
- 20.Park W. J., Reardon H. T., Tyburczy C., Kothapalli K. S., Brenna J. T. (2010) Alternative splicing generates a novel FADS2 alternative transcript in baboons. Mol. Biol. Rep. 37, 2403–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park W. J., Kothapalli K. S., Reardon H. T., Lawrence P., Qian S. B., Brenna J. T. (2012) A novel FADS1 isoform potentiates FADS2-mediated production of eicosanoid precursor fatty acids. J. Lipid Res. 53, 1502–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park W. J., Kothapalli K. S., Reardon H. T., Kim L. Y., Brenna J. T. (2009) Novel fatty acid desaturase 3 (FADS3) transcripts generated by alternative splicing. Gene 446, 28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y., Monroig O., Zhang L., Wang S., Zheng X., Dick J. R., You C., Tocher D. R. (2010) Vertebrate fatty acyl desaturase with Δ4 activity. Proc. Natl. Acad. Sci. USA 107, 16840–16845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu X., Hong H., MacKenzie S. L. (2001) Identification of a Delta 4 fatty acid desaturase from Thraustochytrium sp. involved in the biosynthesis of docosahexanoic acid by heterologous expression in Saccharomyces cerevisiae and Brassica juncea. J. Biol. Chem. 276, 31561–31566 [DOI] [PubMed] [Google Scholar]
- 25.Park W. J., Kothapalli K. S., Lawrence P., Brenna J. T. (2011) FADS2 function loss at the cancer hotspot 11q13 locus diverts lipid signaling precursor synthesis to unusual eicosanoid fatty acids. PLoS ONE 6, e28186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grammatikos S. I., Subbaiah P. V., Victor T. A., Miller W. M. (1994) n-3 and n-6 fatty acid processing and growth effects in neoplastic and non-cancerous human mammary epithelial cell lines. Br. J. Cancer 70, 219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stroud C. K., Nara T. Y., Roqueta-Rivera M., Radlowski E. C., Lawrence P., Zhang Y., Cho B. H., Segre M., Hess R. A., Brenna J. T., Haschek W. M., Nakamura M. T. (2009) Disruption of FADS2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J. Lipid Res. 50, 1870–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trevizan L., de Mello Kessler A., Brenna J. T., Lawrence P., Waldron M. K., Bauer J. E. (2012) Maintenance of arachidonic acid and evidence of Δ5 desaturation in cats fed γ-linolenic and linoleic acid enriched diets. Lipids 47, 413–423 [DOI] [PubMed] [Google Scholar]
- 29.Garcés R., Mancha M. (1993) One-step lipid extraction and fatty acid methyl esters preparation from fresh plant tissues. Anal. Biochem. 211, 139–143 [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y., Nijland M., Miller M., Ford S., Nathanielsz P. W., Brenna J. T. (2008) The influence of maternal early to mid-gestation nutrient restriction on long chain polyunsaturated fatty acids in fetal sheep. Lipids 43, 525–531 [DOI] [PubMed] [Google Scholar]
- 31.Lawrence P., Brenna J. T. (2006) Acetonitrile covalent adduct chemical ionization mass spectrometry for double bond localization in non-methylene-interrupted polyene fatty acid methyl esters. Anal. Chem. 78, 1312–1317 [DOI] [PubMed] [Google Scholar]
- 32.Michaud A. L., Diau G. Y., Abril R., Brenna J. T. (2002) Double bond localization in minor homoallylic fatty acid methyl esters using acetonitrile chemical ionization tandem mass spectrometry. Anal. Biochem. 307, 348–360 [DOI] [PubMed] [Google Scholar]
- 33.Michaud A. L., Lawrence P., Adlof R., Brenna J. T. (2005) On the formation of conjugated linoleic acid diagnostic ions with acetonitrile chemical ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 19, 363–368 [DOI] [PubMed] [Google Scholar]
- 34.Michaud A. L., Yurawecz M. P., Delmonte P., Corl B. A., Bauman D. E., Brenna J. T. (2003) Identification and characterization of conjugated fatty acid methyl esters of mixed double bond geometry by acetonitrile chemical ionization tandem mass spectrometry. Anal. Chem. 75, 4925–4930 [DOI] [PubMed] [Google Scholar]
- 35.Van Pelt C. K., Brenna J. T. (1999) Acetonitrile chemical ionization tandem mass spectrometry to locate double bonds in polyunsaturated fatty acid methyl esters. Anal. Chem. 71, 1981–1989 [DOI] [PubMed] [Google Scholar]
- 36.Van Pelt C. K., Carpenter B. K., Brenna J. T. (1999) Studies of structure and mechanism in acetonitrile chemical ionization tandem mass spectrometry of polyunsaturated fatty acid methyl esters. J. Am. Soc. Mass Spectrom. 10, 1253–1262 [DOI] [PubMed] [Google Scholar]
- 37.Brenna J. T. (2013) Fatty acid analysis by high resolution gas chromatography and mass spectrometry for clinical and experimental applications. Curr. Opin. Clin. Nutr. Metab. Care 16, 548–554 [DOI] [PubMed] [Google Scholar]
- 38.Wang N., Anderson R. E. (1993) Synthesis of docosahexaenoic acid by retina and retinal pigment epithelium. Biochemistry 32, 13703–13709 [DOI] [PubMed] [Google Scholar]
- 39.Moore S. A., Hurt E., Yoder E., Sprecher H., Spector A. A. (1995) Docosahexaenoic acid synthesis in human skin fibroblasts involves peroxisomal retroconversion of tetracosahexaenoic acid. J. Lipid Res. 36, 2433–2443 [PubMed] [Google Scholar]
- 40.Su H. M., Moser A. B., Moser H. W., Watkins P. A. (2001) Peroxisomal straight-chain Acyl-CoA oxidase and D-bifunctional protein are essential for the retroconversion step in docosahexaenoic acid synthesis. J. Biol. Chem. 276, 38115–38120 [DOI] [PubMed] [Google Scholar]
- 41.Kaduce T. L., Chen Y., Hell J. W., Spector A. A. (2008) Docosahexaenoic acid synthesis from n-3 fatty acid precursors in rat hippocampal neurons. J. Neurochem. 105, 1525–1535 [DOI] [PubMed] [Google Scholar]
- 42.Stoffel W. (1966) [On biosynthesis and biological degradation of highly unsaturated fatty acids]. Naturwissenschaften 53, 621–630 [DOI] [PubMed] [Google Scholar]
- 43.Stoffel W., Eker, Assad H., Sprecher H. (1970) Enzymatic studies on the mechanism of the retroconversion of C22-polyenoic fatty acids to their C20-homologues. Hoppe Seylers Z. Physiol. Chem. 351, 1545–1554 [DOI] [PubMed] [Google Scholar]
- 44.Oosterveer M. H., Grefhorst A., van Dijk T. H., Havinga R., Staels B., Kuipers F., Groen A. K., Reijngoud D. J. (2009) Fenofibrate simultaneously induces hepatic fatty acid oxidation, synthesis, and elongation in mice. J. Biol. Chem. 284, 34036–34044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y. H., Salem N. Jr (2005) In vivo conversion of 18- and 20-C essential fatty acids in rats using the multiple simultaneous stable isotope method. J. Lipid Res. 46, 1962–1973 [DOI] [PubMed] [Google Scholar]
- 46.Ferdinandusse S., Denis S., Mooijer P. A., Zhang Z., Reddy J. K., Spector A. A., Wanders R. J. (2001) Identification of the peroxisomal beta-oxidation enzymes involved in the biosynthesis of docosahexaenoic acid. J. Lipid Res. 42, 1987–1995 [PubMed] [Google Scholar]
- 47.Pauter A. M., Olsson P., Asadi A., Herslöf B., Csikasz R. I., Zadravec D., Jacobsson A. (2014) Elovl2 ablation demonstrates that systemic DHA is endogenously produced and is essential for lipid homeostasis in mice. J. Lipid Res. 55, 718–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moon Y. A., Hammer R. E., Horton J. D. (2009) Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. J. Lipid Res. 50, 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leonard A. E., Kelder B., Bobik E. G., Chuang L. T., Lewis C. J., Kopchick J. J., Mukerji P., Huang Y. S. (2002) Identification and expression of mammalian long-chain PUFA elongation enzymes. Lipids 37, 733–740 [DOI] [PubMed] [Google Scholar]
- 50.Zadravec D., Tvrdik P., Guillou H., Haslam R., Kobayashi T., Napier J. A., Capecchi M. R., Jacobsson A. (2011) ELOVL2 controls the level of n-6 28:5 and 30:5 fatty acids in testis, a prerequisite for male fertility and sperm maturation in mice. J. Lipid Res. 52, 245–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer A., Cirpus P., Ott C., Schlecker R., Zähringer U., Heinz E. (2003) Biosynthesis of docosahexaenoic acid in Euglena gracilis: biochemical and molecular evidence for the involvement of a Delta4-fatty acyl group desaturase. Biochemistry 42, 9779–9788 [DOI] [PubMed] [Google Scholar]
- 52.Tonon T., Harvey D., Larson T. R., Graham I. A. (2003) Identification of a very long chain polyunsaturated fatty acid Delta4-desaturase from the microalga Pavlova lutheri. FEBS Lett. 553, 440–444 [DOI] [PubMed] [Google Scholar]
- 53.Tripodi K. E., Buttigliero L. V., Altabe S. G., Uttaro A. D. (2006) Functional characterization of front-end desaturases from trypanosomatids depicts the first polyunsaturated fatty acid biosynthetic pathway from a parasitic protozoan. FEBS J. 273, 271–280 [DOI] [PubMed] [Google Scholar]
- 54.Morais S., Castanheira F., Martinez-Rubio L., Conceição L. E., Tocher D. R. (2012) Long chain polyunsaturated fatty acid synthesis in a marine vertebrate: ontogenetic and nutritional regulation of a fatty acyl desaturase with Δ4 activity. Biochim. Biophys. Acta 1821, 660–671 [DOI] [PubMed] [Google Scholar]
- 55.Fonseca-Madrigal J., Navarro J. C., Hontoria F., Tocher D. R., Martínez-Palacios C. A., Monroig O. (2014) Diversification of substrate specificities in teleostei Fads2: characterization of Δ4 and Δ6Δ5 desaturases of Chirostoma estor. J. Lipid Res. 55, 1408–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kuah M., Jaya-Ram A., Shu-Chien A. C. (2015) The capacity for long-chain polyunsaturated fatty acid synthesis in a carnivorous vertebrate: functional characterisation and nutritional regulation of a Fads2 fatty acyl desaturase with Delta4 activity and an Elovl5 elongase in striped snakehead (Channa striata). Biochim. Biophys. Acta 1851, 248–260 [DOI] [PubMed] [Google Scholar]
- 57.Gibson R. A., Neumann M. A., Lien E. L., Boyd K. A., Tu W. C. (2013) Docosahexaenoic acid synthesis from alpha-linolenic acid is inhibited by diets high in polyunsaturated fatty acids. Prostaglandins Leukot. Essent. Fatty Acids 88, 139–146 [DOI] [PubMed] [Google Scholar]
- 58.Anonymous. (2014) Protein molecular weight calculator. http://www.sciencegateway.org/tools/proteinmw.htm.
- 59.Pédrono F., Blanchard H., Kloareg M., D’andréa S., Daval S., Rioux V., Legrand P. (2010) The fatty acid desaturase 3 gene encodes for different FADS3 protein isoforms in mammalian tissues. J. Lipid Res. 51, 472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jakobsson A., Ericsson J., Dallner G. (1990) Metabolism of fatty acids and their incorporation into phospholipids of the mitochondria and endoplasmic reticulum in isolated hepatocytes determined by isolation of fluorescence derivatives. Biochim. Biophys. Acta 1046, 277–287 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.