Abstract
Marine sediments host a large population of diverse, heterotrophic, uncultured microorganisms with unknown physiologies that control carbon flow through organic matter decomposition. Recently, single-cell genomics uncovered new key players in these processes, such as the miscellaneous crenarchaeotal group. These widespread archaea encode putative intra- and extracellular proteases for the degradation of detrital proteins present in sediments. Here, we show that one of these enzymes is a self-compartmentalizing tetrameric aminopeptidase with a preference for cysteine and hydrophobic residues at the N terminus of the hydrolyzed peptide. The ability to perform detailed characterizations of enzymes from native subsurface microorganisms, without requiring that those organisms first be grown in pure culture, holds great promise for understanding key carbon transformations in the environment as well as identifying new enzymes for biomedical and biotechnological applications.—Michalska, K., Steen, A. D., Chhor, G., Endres, M., Webber, A. T., Bird, J., Lloyd, K. G., Joachimiak, A. New aminopeptidase from “microbial dark matter” archaeon.
Keywords: carbon cycle, marine sediments, single-cell genomics, detrital proteins
The vast majority of microorganisms in the environment have never been grown in the laboratory. They have therefore been referred to as microbial dark matter (MDM) (1) because it is difficult to study their physiology and determine their impact on ecosystems and major global elemental cycles. Each of these uncultured microorganisms harbors uncharacterized enzymes that have never been studied or tapped for their biotechnological and biomedical potential (2). Recently, the sequences of genomes from MDM have become available, by amplifying DNA from single cells (3) and assembling MDM genomes in silico from environmental metagenomes (4). However, the novelty of the information that can be gathered from these genomes is restricted because the annotation of individual genes relies on their sequence similarity to proteins that have been characterized previously in cultured organisms. This is a major drawback in the study of MDM genomes because it hinders the discovery of truly unique functions that do not exist in the cultured realm studied thus far.
Nevertheless, sequencing genomes from MDM projects has the potential to reveal unexpected functionalities. For example, some archaeal species encode predicted proteases that may be utilized for degradation of detrital proteins in marine sediments and can contribute to carbon and nitrogen cycling (5). To uncover the true biochemical functions of these enzymes, we have investigated recombinant proteins, including here as an example a putative protease [designated as bathyaminopeptidase MCG (miscellaneous crenarchaeotal group)-15 (BAP)] from Thaumarchaeota archaeon SCGC AB-539-E09. This uncultured microorganism is phylogenetically assigned to the MCG, which is widespread in marine sediments (5) and has recently been given the phylum name Candidatus Bathyarchaeota (6). BAP is of particular interest because no close homologs exist in cultured organisms, making the original annotation ambiguous. As indicated by the Basic Local Alignment Search Tool (BLAST), it shares sequence similarity with proteins annotated as 1) X-prolyl dipeptidyl aminopeptidase (PepX) from Caulobacter sp. K31 (S15 peptidase family, 51% sequence identity, EC 3.4.14.11); 2) cocaine esterase (CocE) from Janthinobacterium sp. HH01 (51% sequence identity, EC 3.1.1.84); and 3) α-amino acid ester hydrolase (AEH) from Xanthomonas citri (XcAEH; 51% sequence identity, EC 3.1.1.43); some close homologs are also annotated (most likely incorrectly) as glutaryl-7-ACA acylase, EC 3.5.1.93. Among the proteins that have been biochemically characterized, only the confirmed AEH holds its relatively high sequence identity. The other homologs with experimentally validated activities and structural characterization are in fact more distant relatives, with sequence identity of 27% for CocE (7) and 16% for PepX (8).
All of the above enzymes belong to the large family of serine hydrolases that have conserved catalytic residues, and very likely, they share a common mechanism. Namely, they utilize a classic Ser-His-Asp catalytic triad and a water molecule to carry out acylation and deacylation steps with the help of an oxyanion hole, which stabilizes tetrahedral intermediates (9). These enzymes can carry out both hydrolytic and synthetic reactions using a variety of substrates. Aminopeptidases progressively remove N-terminal residues from short peptides through peptide bond cleavage. Esterases can hydrolyze ester bonds but also carry out esterification and transesterification reactions. AEHs catalyze the synthesis of β-lactam antibiotics using an α-amino acid ester [such as d-phenylglycine (Phg) methyl ester] as a donor and a β-lactam as an acceptor (10). They also are able to hydrolyze product antibiotics, and some AEHs exhibit α-aminopeptidase activities (11). In this work, we collectively refer to them as S15-like family, and we follow standard nomenclature as used for serine proteases.
MATERIALS AND METHODS
Cloning to vector pMCSG73 and in vitro tobacco vein mottling virus protease cleavage of transcription termination/antitermination protein-BAP fusion
The DNA template for BAP was obtained and amplified from a single cell of T. archaeon SCGC AB-539-E09. The fragment coding for the BAP protein (residues Met1-Ser623) was amplified with KOD DNA Polymerase using conditions and reagents provided by EMD Millipore (Billerica, MA, USA). The following primers were used: 5′-TACTTCCAATCCAATGCCATGAAAAAACTCAGAGATGATTTCTCCGAAGA-3′, and 5′-TTATCCACTTCCAATGTTATGAAATAATATGTATCTCGATCGCTGTCG-3′. The PCR product was cloned according to the ligation-independent procedure (12, 13) into vector pMCSG73, which is a derivative of the pMCSG53 vector (14). Proteins expressed from vector pMCSG73 are produced as a C-terminal fusion to Escherichia coli transcription termination/antitermination protein (NusA) in the following protein construct: NusA-[tobacco vein mottling virus (TVMV) protease recognition site]-His6-Strep tag-[tobacco etch virus (TEV) protease recognition site]-(TARGET) (NusA-ETVRFQ/S-HHHHHH-WSHPQFEK-ENLYFQ/SNA-GENE SEQUENCE). The fusion is cleaved in vitro by TVMV protease overexpressed in a separate batch of bacteria. An auxiliary plasmid expressing TVMV (pMCSG-TVMV) was created by cloning of the TVMV gene into pMCSG7 (15) where a fragment between the START codon and the TEV recognition site had been deleted. The process of releasing NusA starts at sonication and is completed by the time of loading onto the Ni resin. The amount of TVMV protease overexpressed in 1 L growth medium is sufficient to cleave an NusA-target fusion from 10 L target-producing bacteria.
Protein expression and purification
The E. coli BL21-Gold(DE3) strain carrying the pMCSG73-BAP plasmid was grown in 1 L enriched M9 medium (16) at 37°C, shaking at 200 rpm until it reached an optical density at a wavelength of 600 nm (OD600) of 1.0. Inhibitory amino acids (25 mg each of l-valine, l-isoleucine, l-leucine, l-lysine, l-threonine, and l-phenylalanine) and 90 mg selenomethionine (SeMet) (Orion Enterprises, Wheeling, IL, USA) were added to the culture, which was then cooled to 4°C for 60 min. To induce protein expression, 0.5 mM isopropyl-β-d-thiogalactoside was added. The cells were incubated overnight at 18°C and then centrifuged for 10 min at 7880 g. The supernatant was removed, and the cells were resuspended in 40 ml lysis buffer [500 mM NaCl, 5% glycerol, 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 8.0), 20 mM imidazole, and 10 mM 2-ME]. To remove NusA, 3 ml TVMV protease-expressing cell suspension (OD600, ∼70) was added to the target cells, which were then disrupted by 5 min of sonication. The lysate was centrifuged for 1 h at 29,500 g to separate insoluble fractions, which were discarded. The protein present in the supernatant was purified using Ni-NTA affinity chromatography and the ÄKTAxpress system (GE Healthcare Biosciences, Pittsburgh, PA, USA) as described previously (17, 18). Subsequently, the N-terminal His6 tag was removed using recombinant His7-tagged TEV protease. In the next purification step, subtractive Ni-NTA affinity chromatography was applied to remove the protease, affinity tag, and any uncut protein. Native BAP was also expressed and purified analogously to the SeMet-labeled variant, with the exception that the growth media consisted of both M9 and Luria-Bertani. The pure proteins were concentrated in crystallization buffer [20 mM HEPES (pH 8.0), 250 mM NaCl, and 2 mM DTT] using an Amicon Ultra-15 concentrator (EMD Millipore). Protein concentrations were determined based on the absorbance at 280 nm measured on a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA) and their theoretic, sequence-deduced absorption coefficients.
Crystallization
The SeMet-labeled BAP protein was screened for crystallization conditions with the help of a Mosquito nanoliter liquid handler (TTP Labtech, Cambridge, MA, USA) using the sitting-drop vapor diffusion technique in 96 well CrystalQuick plates (Greiner Bio-One, Monroe, NC, USA). The crystallization was set up at 4°C using the MCSG 1–4 screens from Microlytic (Woburn, MA, USA). For each condition, 0.4 µl protein (at 50 mg/ml) and 0.4 µl crystallization formulation were mixed; the mixture was equilibrated against 140 µl of the crystallization solution in each reservoir well. For cocrystallization of native BAP (20 mg/ml) with dl-Phe, the protein was incubated with the ligand for 4 h at 4°C prior to plate setup. The crystals appeared under a number of conditions, 2 of which were ultimately used. Crystals X1 grew from a solution containing 0.2 M potassium thiocyanate and 20% (w/v) polyethylene glycol (PEG) 3350, whereas crystals X2 grew from 0.2 M sodium citrate, 20% (w/v) PEG 3350, and 2.8 mM dl-Phe.
Data collection
Prior to data collection, the crystals were briefly soaked in mother liquors supplemented with 15% glycerol for cryoprotection and immediately flash cooled in liquid nitrogen. The diffraction experiments were performed at the 19-Insertion Device (19-ID) beamline of the Structural Biology Center at the Advanced Photon Source (APS) at Argonne National Laboratory using the program SBCcollect (19). There were 2 diffraction data sets collected at 100 K near the selenium absorption peak. Crystals X1 (λ = 0.9792 Å) diffracted to 2.40 Å resolution, whereas the resolution of the X2 crystals (λ = 0.9793 Å) extended to 2.10 Å. The crystals are isomorphous and belong to the monoclinic P21 space group. The diffraction images were processed with the HKL-3000 suite (20). Intensities were converted to structure factor amplitudes in the CTRUNCATE program (21, 22) from the CCP4 package (23). The data collection and processing statistics are given in Table 1 (for X2 only; see below).
TABLE 1.
BAP data collection and refinement statistics
| Statistic | Value |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 118.4, 108.1, 120.4 |
| β (deg) | 95.1 |
| Temperature (K) | 100 |
| Radiation source | 19-ID |
| Wavelength (Å) | 0.9793 |
| Resolution (Å)a | 30.00–2.10 (2.14–2.10) |
| Unique reflections | 176,530 (8826) |
| Rmergeb | 0.137 (0.570) |
| 〈I〉/〈σI〉 | 9.0 (1.9) |
| Completeness (%) | 99.8 (99.8) |
| Redundancy | 3.2 (3.0) |
| Refinement | |
|---|---|
| Resolution (Å) | 29.69–2.10 |
| No. of reflections work/test set | 173,579/2166 |
| Rwork/Rfreec | 0.205/0.229 |
| No. of atom proteins/ligands/water | 19,666/130/1124 |
| Average B factor protein/ligands/water (Å2) | 24.4/36.2/23.8 |
| Rmsd | |
| Bond lengths (Å) | 0.008 |
| Bond angles (deg) | 0.73 |
| Ramachandran plot | |
| Most favored (%) | 95.9 |
| Outliers (%) | 0.25 |
| MolProbity score | 1.26 |
| Clash score | 1.39 |
Values in parentheses correspond to the highest-resolution shell.
Rmerge = ΣhΣj|Ihj − 〈Ih〉|/ΣhΣjIhj, where Ihj is the intensity of observation j of reflection h.
Rwork = Σh|Fo| − |Fc|/Σh|Fo| for all reflections, where Fo and Fc are observed and calculated structure factors, respectively. Rfree is calculated analogously for the test reflections, randomly selected, and excluded from the refinement.
Structure solution
Initial attempts to determine the structure of crystal X1 through the selenium-based single-wavelength anomalous diffraction method failed due to insufficient anomalous signal for successful experimental phasing. Therefore, the structure was solved by molecular replacement (MR) with the XcAEH [Protein Data Bank (PDB) entry 1MPX] (24), as a search model. The calculations were performed in the HKL-3000 package (20), which executes MOLREP (25) for MR search followed by automated model building in ARP/wARP (26). The preliminary model was further rebuilt in Coot (27) and refined in REFMAC (28). Then, the structure was refined against the diffraction data for crystal X2. Because the electron density did not reveal the presence of the phenylalanine ligand, the final model of the apo structure was obtained through alternating manual rebuilding and crystallographic refinement in BUSTER (29) against the X2 crystal data. For the refinement, 4 translation/libration/screw (TLS) groups were defined, each of them for a single protein chain. In the refined model, the following residues could be reliably modeled in the electron density map: Phe8-Glu391 and Glu397-Ser623 in chains A and D, and Asp7-Glu391 and His399-Ser623 in chains B and C. In addition to the protein molecules, 1124 water molecules have been identified, 15 glycerol molecules, and 4 PEG chains. An unassigned positive peak in the DFo-mFc electron density map is present near the catalytic Ser158 in chain D. The quality of the final structure was verified by the MolProbity server (30). The refinement statistics are shown in Table 1. The atomic coordinates and structure factors have been deposited in the PDB under accession code 4PF1.
Enzymatic assays
The peptidase activity was determined directly, using fluorogenic substrate analogs bearing 7-amido-4-methylcoumarin (AMC) or 4-methylumbelliferone linked to an amino acid or a peptide chain at its C terminus or with dipeptides, or indirectly, via a competition assay. The AMC-based experiment requires the AMC molecule to be released from the substrate. Thus, for peptides containing ≥2 residues, the detection of AMC would indicate that the enzyme is an endopeptidase, a progressive aminopeptidase that sequentially cleaves off N-terminal residues until AMC is reached, or, much less probably, a nonspecific carboxypeptidase.
In the AMC-based assays, AMC substrate hydrolysis rates were measured at 500 μM substrate concentration (saturating) at 25°C. A total of 6.5 μl substrate, dissolved in DMSO, was added to 245 μl buffer [100 mM sodium citrate, 200 mM sodium phosphate, and 30 mM KCl (pH 6.85)] containing 4.35 μg enzyme ml−1. Fluorescence (excitation, 360 nm; emission, 445 nm) was monitored over 10 min in a BioTek Cytation 3 plate reader (BioTek Instruments, Winooski, VT, USA) and calibrated with AMC standards. Hydrolysis rates in triplicate samples with the enzyme were compared to no-enzyme controls to assess the importance of uncatalyzed substrate hydrolysis.
The competitive inhibition experiment used 2000 μM inhibitor and 200 μM l-Phe-AMC. Each reaction contained 10 μl substrate dissolved in DMSO, 20 μl inhibitor dissolved in Q-H2O, and 970 μl citrate/phosphate buffer containing 1.74 μg enzyme ml−1, mixed in 1 ml methacrylate cuvettes and held at 21°C. Fluorescence was monitored with a Promega QuantiFluor ST solid state fluorimeter (Madison, WI, USA) set to the UV channel. The inhibition constant KI was calculated as
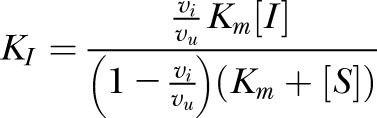 |
where Km is the Michaelis constant, vi is reaction velocity in the presence of the inhibitor, vu is velocity in the uninhibited control, [I] is inhibitor concentration, and [S] is substrate concentration.
Temperature sensitivity was assessed using l-Leu-AMC at 500 mM. The incubation temperature was slowly increased from 0 to 46°C. Maximum velocity Vmax was calculated as the hydrolysis rate of fluorescence production between each set of measurements, and optimal temperature Topt was calculated based on a nonlinear least-squares fit of a normal function to the temperature-activity data.
For thin-layer chromatography (TLC)-based transferase assay (Supplemental Fig. S1), substrates were dissolved in 50 mM HEPES/Na buffer (pH 6.8 or 7.6; data not shown) and 2.5% DMSO to yield 100 mM stock solutions. 6-Aminopenicillanic acid (6-APA) solution also contained 150 mM NaOH. Reactions were carried out at room temperature for 1.5 h in buffer 1 [50 mM HEPES/Na buffer (pH 6.8) and 2.5% DMSO] or buffer 2 [50 mM HEPES/Na buffer (pH 7.6) and 2.5% DMSO] with final concentrations of substrates 15 mM and enzyme 0.55 mg/ml. Silica gel-coated TLC plates loaded with 1 μl reaction mixture were eluted in a mobile phase consisting of n-butanol:acetic acid:water in a 3:1:1 ratio. The plates were dried, briefly soaked in ninhydrin stain (0.1 g ninhydrin, 0.5 ml acetic acid, and 100 ml acetone), dried, and heated to develop spots.
Hydrolysis of l-Phe-l-Leu was measured using a distinct TLC-based assay. Here, solutions of the dipeptide (10 mM in the citrate/phosphate buffer described above and 1% DMSO) were incubated with 0.435 μg/ml BAP for 3 h at 30°C. Products were separated by TLC. Silica gel-coated TLC plates loaded with 0.5 μl reaction mixture were eluted in a mobile phase consisting of n-butanol:acetic acid:water in a 4:1:1 ratio. The plates were dried, sprayed with ninhydrin, and heated to develop.
There were 2 experiments performed to assess the effect of oxygen on BAP. First, activity of BAP (0.435 μg/ml), which had been stored frozen in oxic citrate/phosphate buffer (pH 6.85) for ∼3 mo (including ∼3 h thawed), was assessed in the presence or absence of 10 mM tris(2-carboxyethyl)phosphine (TCEP). Stored BAP was thawed, mixed with a TCEP solution to a final concentration of 10 mM TCEP, and incubated with l-Phe-AMC, l-Leu-AMC, or l-Leu-l-Leu-AMC. Second, fresh BAP [stored with 20 mM HEPES (pH 8.0), 250 mM NaCl, and 2 mM DTT] was thawed, diluted into citrate/phosphate buffer (0.435 μg/ml final concentration enzyme), and the experiment described above was repeated.
Phylogenetic analysis
BLAST 2.2.28 was used to search databases representing 135 large metagenomes (>109 bp) from integrated microbial genomes (IMGs) (31) and MG-RAST (32) encompassing a wide range of commensal and environmental samples, for homologs of the crystallized protein. In addition, all S15 homologs identified by annotations with Pfam PF02129 were obtained from all cultured microorganisms in the IMG database. All BLAST hits from environmental metagenomes with E values < 10−100 were aligned along with the sequence of the crystallized protein and Pfam hits from cultured microorganisms using MUSCLE (33). These sequences were imported into ARB (www.arb-home.de) and used to build a maximum likelihood tree using randomized axelerated maximum likelihood (RAxML). Only 4 sequences from a marine sediment metagenome grouped with the sequence of the crystallized protein. Metatranscriptomic hits were identified by applying Pfam searches on metatranscriptomes downloaded from MG-RAST using InterProScan 5.3-46.0 (34) to search the Pfam database with an E value cutoff of 1 × 10−5. These were also imported into ARB and aligned manually. Because these are short reads, they were not included in the final tree.
RESULTS
BAP is unique to MDM
To investigate genomic BAP abundance, we analyzed 135 environmental metagenomes and 15,000 genomes of cultured organisms. This search revealed highly similar (84–96% at the amino acid level) sequences only from estuarine sediment in North Carolina, United States (Fig. 1). These metagenome sequences share perfect matches to the residues that appear to define the signature motif for BAP-like enzymes, as discussed below (Supplemental Fig. S2). The existence of these genes in uncultured microorganisms in such distantly located marine sediments (Denmark and United States) suggests that further exploration of marine sediments will reveal that BAP is present in marine sediments worldwide because MCGs are broadly distributed (5). Many other marine sediment metagenomes also contained homologous proteins (Supplemental Table S1; BLAST hits of E < 10−100), although they were not as closely matched as those from the North Carolina estuary. These include environments as varied as the Arctic Ocean, Antarctica, Baltic Sea, and Peru Margin. We also found homologs of BAP in marine sediment metatranscriptomes (Supplemental Table S2) (35), suggesting that this protein is not just a genomic feature but is actually expressed in the environment.
Figure 1.
Phylogenetic placement of BAP relative to homologs from cultured organisms. RAxML amino acid tree of BAP homologs in cultured organisms (black), marine sediment metagenomes, and BAP protein characterized functionally and structurally in this study (red) is presented. Sequences that have been evaluated as proteins are in bold.
Enzymatic activity
BAP was originally annotated as a putative protease and/or esterase. To validate these predictions, we assessed BAP’s activity either directly using fluorogenic substrate analogs or a peptide or indirectly through a competition assay (see Supplemental Data). In a standardized assay using 0.44 μg/ml enzyme solution, BAP is an aminopeptidase most efficiently hydrolyzing the amide bond of l-configured dipeptide proxies with cysteine or a hydrophobic residue at the N terminus (Table 2). The activity is decreased in the presence of divalent cations, with Mg2+ having a more pronounced effect than Ca2+, regardless of the substrate used. Among the tested S1 residues, l-Cys is the most preferred (Kml-Cys-AMC = 80 ± 17 μM), followed by l-Ala, l-Phe, l-Leu, l-Tyr, l-Trp, l-Ile, and Gly. l-Arg and l-Phg are much poorer substrates. Among longer peptides, the only tripeptide proxy detectably hydrolyzed under the same experimental conditions is l-Leu-l-Leu-AMC. This substrate seems to be progressively hydrolyzed (first, the l-Leu-l-Leu peptide bond is cut, and then the l-Leu-AMC amide bond). Hydrolysis of Gly-l-Pro-AMC is only detectable with longer incubations (∼3 h) and higher enzyme concentrations, most likely due to poor performance of l-Pro in the P1 site, but also, the l-Pro mismatch in the P1′ site cannot be excluded. Similar conditions are necessary to observe hydrolysis of tetrapeptide proxies Gly-Gly-l-Arg-AMC and l-Ala-l-Ala-l-Phe-AMC, even though all residues in these substrates are acceptable in the S1 position and should allow progressive cleavage. Thus, either these proxies are too large to access BAP’s active site, or the enzyme is an aminopeptidase with specific requirements for the P1′, P2′, and P3′ sites. However, the fact that l-Phe-l-Leu peptide is readily hydrolyzed (Supplemental Fig. S2) with efficiency comparable to that against l-Cys-AMC suggests that peptide size rather than residue identity at the P1 or P1′ positions is a discriminating factor.
TABLE 2.
Relative aminopeptidase activity of BAP against a set of peptide proxies
| Substrate | Normalized activity | Activity se |
|---|---|---|
| l-Cys-AMC | 1.00 × 10+0 | 3.30 × 10−2 |
| l-Ala-AMC | 4.50 × 10−1 | 8.28 × 10−3 |
| l-Phe-AMC | 3.24 × 10−1 | 3.72 × 10−3 |
| l-Leu-AMC | 1.24 × 10−1 | 2.37 × 10−3 |
| l-Tyr-AMC | 1.04 × 10−1 | 1.47 × 10−3 |
| l-Trp-AMC | 4.42 × 10−2 | 4.87 × 10−4 |
| l-Ile-AMC | 2.46 × 10−2 | 7.12 × 10−4 |
| Gly-AMC | 2.29 × 10−2 | 6.57 × 10−4 |
| l-Leu-l-Leu-AMC | 1.17 × 10−2 | 7.62 × 10−3 |
| l-Cysteine-AMC | 1.11 × 10−2 | 5.80 × 10−4 |
| d-Phe-AMC | 6.56 × 10−3 | 3.48 × 10−4 |
| l-Arg-AMC | 3.95 × 10−3 | 1.74 × 10−3 |
| l-Phg-AMC | 2.07 × 10−3 | 7.04 × 10−4 |
| Z-l-Phe-l-Arg-AMC | 1.99 × 10−3 | 1.39 × 10−3 |
| l-Ala-l-Ala-l-Phe-AMC | n.d. | n.d. |
| d-Phg-AMC | n.d. | n.d. |
| Gly-l-Pro-AMC | n.d. | n.d. |
| Pyr-AMC | n.d. | n.d. |
| Z-l-Phe-l-Val-l-Arg-AMC | n.d. | n.d. |
| Gly-Gly-l-Arg-AMC | n.d. | n.d. |
n.d., not determined.
d-configured substrates are poorly recognized by BAP. The cleavage of d-Phe-AMC could be detected at a much lower rate (relv0l-Cys-AMC = 0.65 ± 0.03%). The binding of other d-configured derivatives of phenylalanine and leucine (esters and amides) was also observed, but affinity of the inhibitor was considerably lower for d than for l stereoisomers (Supplemental Table S3). Contrary to previous findings for AEHs, d-Phg derivatives are poorly recognized—cleavage of d-Phg-AMC was not detected under standard conditions; only TLC analysis showed slow hydrolysis of d-Phg-OMe (Supplemental Fig. S2). In addition, no significant difference is observed between d-Phg-OMe and its l enantiomer. Therefore, BAP is unlikely to function as an esterase that uses such a donor in a synthetic reaction.
The l-Phe-AMC inhibition assay with 10-fold molar excess of amide (−NH2) and methyl ester (−OMe) derivatives of l-Phe shows the importance of the S1′ residue, with preference for the more hydrophobic −AMC moiety over −OMe (relv0l-Phe-AMC = 18 ± 1%) and −NH2 (relv0l-Phe-AMC = 49 ± 2%). Similarly, stronger binding of l-Leu methyl ester is observed than l-Leu amide.
Esterase activity was confirmed for BAP with methyl esters of l-Leu, l-Cys (data not shown), l-Phe, and d-Phg (Supplemental Fig. S2). Ampicillin was also hydrolyzed with release of d-Phg (Supplemental Fig. S2). However, no transferase reaction could be observed with either l-Phe-OMe or d-Phg-OMe as a donor and 6-APA as an acceptor.
All these data suggest that the enzyme can hydrolyze both amide and ester bonds and is promiscuous in accepting different substrates. BAP is also the first microbial enzyme to show high specificity for l-Cys. The preference for l-Cys and activity toward l-Phe-, l-Tyr-, and l-Trp-containing substrates indicate that BAP might be responsible for securing cysteine and aromatic amino acids. Such a function would be highly beneficial for the cell in a nutrient-restricted environment because synthesis of aromatic amino acids requires substantial energy resources, and cysteine is an important source of sulfur. Interestingly, cysteine is a catalytic residue in the majority of archaeal MDM peptidases proposed to process detrital proteins (5). Because terminal cysteine residues are stable only in anoxic environments, it appears that BAP has adapted to life in such an ecosystem; it loses activity on a timescale of days in the presence of oxygen (Supplemental Fig. S3) and cannot efficiently utilize oxidized cysteine (Table 2).
Crystal structure
To gain further insight into the molecular basis of BAP function, we performed crystallographic studies. The crystal structure of BAP was determined by an MR method with characterized XcAEH as a template [PDB entry 1MPX, sequence identity 51% (36)]. The atomic model was refined against 2.10 Å resolution data (Table 1). The asymmetric unit contains 4 nearly identical protein molecules. Analogously to their structurally closest bacterial cousins, the individual BAP chains associate into a homotetramer composed of 2 dimers. The assembly adopts a hollow sphere shape with an external diameter of ∼110 Å and 222-point group symmetry (Fig. 2). The channel runs across the tetramer and can be accessed through 2 identical, elongated orifices with approximate dimensions 21 × 9 Å. Comparisons with other AEHs located 4 independent active sites that face the interior of the homotetramer (see below). This arrangement poses significant restrictions on the size of acceptable substrates, which could only reach the catalytic pockets through those relatively narrow openings. Such architecture is consistent with a self-compartmentalizing hydrolase (37). The monomer, nearly identical to 2 other AEHs available in the PDB [root-mean-sd (rmsd) 1.00 Å for 597 residues aligned with XcAEH (Fig. 3A), and 0.96 Å for 581 residues aligned with Acetobacter turbidans homolog, AtAEH, PDB entry 2B9V (38)], consists of 3 domains and an N-terminal protrusion that plays an important role in the formation of the primary dimer unit (Fig. 2). The N-terminal domain, with the classic α/β-hydrolase fold, is followed by the cap domain and the jelly roll-like C-terminal domain.
Figure 2.
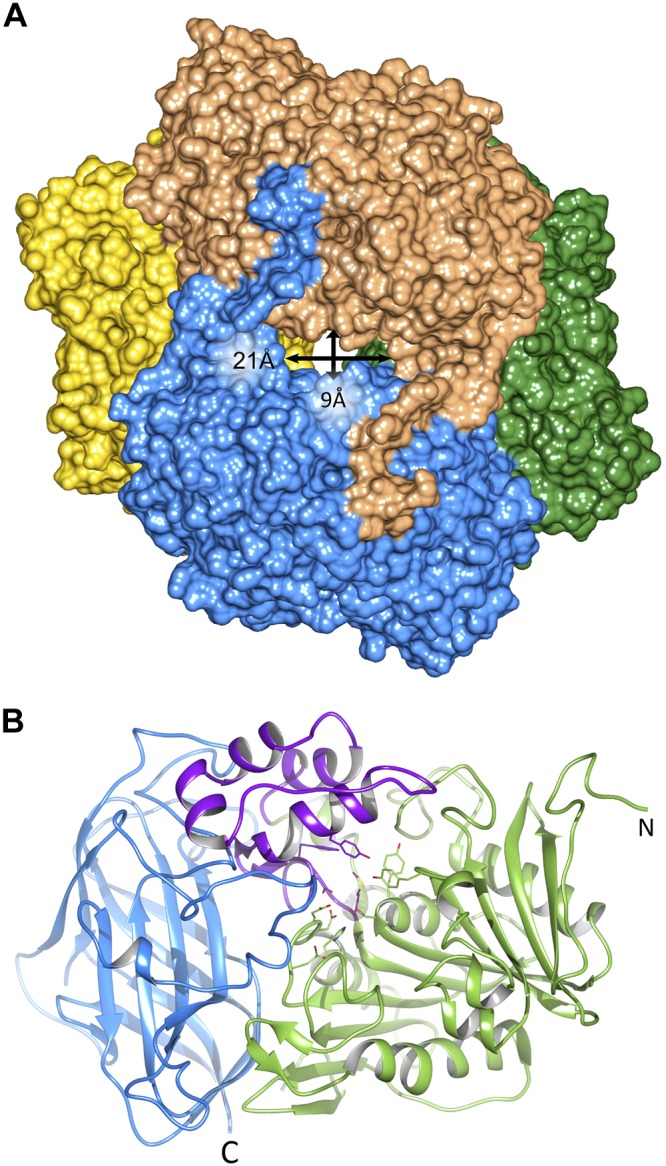
Structure of BAP. A) A surface representation of the BAP homotetramer. B) A ribbon diagram of the BAP monomer (chain C) with individual domains colored green (N-terminal domain; “N”), purple (cap domain), and blue (C-terminal domain; “C”). The key residues in the active site are shown in a stick representation.
Figure 3.
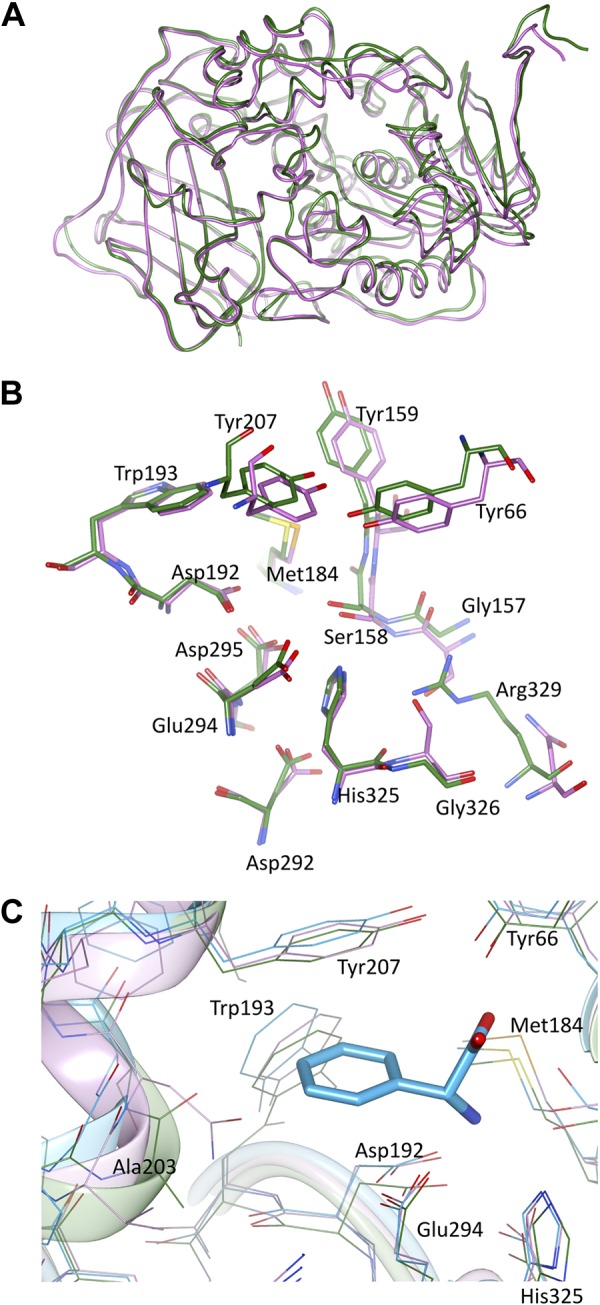
Comparison of BAP and AEHs. A) Superposition of the BAP monomer (green; this work) with XcAEH (pink; PDB entry 1MPX). B) Superposition of active sites: BAP is in green, and XcAEH is shown in pink. C) Superposition of active sites: BAP is shown in green, XcAEH in pink, and AtAEH in blue (PDB entry 2B4K). For reference, the d-Phg ligand from the AtAEH structure is shown in a stick representation.
BAP active site
The active site is formed at the domains’ interface and can be divided into 2 subcavities. The first one is relatively wide and faces the solvent channel. Deeper inside the pocket, the second subcavity is present. At the boundary between the subcavities, the N-terminal domain carries a catalytic triad consisting of Ser158, His325, and Asp292 (Fig. 3B and Supplemental Fig. S4). By analogy to other AEHs, this region also provides residues that are believed to create an oxyanion hole during catalysis: Tyr159 and Tyr66. In the former residue, the main-chain amide group is presumably involved in the stabilization of the tetrahedral intermediate, whereas Tyr66 interacts with the substrate molecule through its hydroxyl group.
The deep subpocket (acyl binding) closely resembles its equivalent in AEHs. This pocket has been shown to accommodate a d-Phg molecule in AtAEH [PDB entry 2B4K (38)] and, thus, corresponds to the P1 site. One side of this surface is lined up with several hydrophobic residues (Met184, Trp193, Tyr207, and Ala203) that interact with the side chain of d-Phg in AtAEH and very likely with small side chains of the BAP α-amino acid derivative substrates (esters or amides) (see below) (Fig. 3C). The second side of the acyl-binding subpocket exposes a constellation of conserved acidic residues: Asp192, Glu294, and Asp295. This negatively charged cluster is used to anchor an α-amino group of the substrate moiety [as in the structure of AtAEH (PDB entry 2B4K)], and it defines enzyme specificity at the P1 site as well as optimum pH. In fact, the pH optimum for BAP is 6.9. Given that the pKa of the N-terminal amino group is ∼7.8 [for phenylalanyl peptide (39)], it implies that the α-amino group of the peptide has to be protonated for the most efficient binding of the N-terminal residue into the strongly acidic P1 site. Compounds that do not bear an α-amino group, such as methylumbelliferyl-butyrate, are not recognized as substrates. These features define BAP as an aminopeptidase. Besides strictly conserved elements in the P1 site, there is also a protein fragment within the cap domain (helix αG in XcAEH) that adopts a slightly different main-chain conformation and has a variable amino acid composition. As a consequence, in BAP, unlike in XcAEH or AtAEH, a carbonyl group of Ala203 is facing the lumen (Fig. 3C). This feature may be responsible for the shift in substrate preference from Phg-based compounds toward Cys, Ala, or Phe derivatives. As the superposition with AtAEH/d-Phg shows, the aromatic ring of Phg would be too close to the C = O moiety. The longer side chain of the Phe ligand, on the other hand, most likely moves to avoid an unfavorable contact with Ala203. An equivalent region of AEHs is possibly responsible for discrimination between Phg-based substrates and its hydroxy-Phg analogs.
The channel-facing subcavity is much larger than the acyl-binding pocket. This space typically accommodates a leaving fragment of the hydrolyzed peptide (in peptidases) or a β-lactam ring (in esterases/transferases). The available data, based on modeling studies with antibiotics, predict very few contacts with the leaving group (24, 39), which guarantees broad substrate specificity with respect to the β-lactam moiety. The major one appears to be stacking interactions between the β-lactam ring and the Tyr66 equivalent—a dual-function residue that is also proposed to form an oxyanion hole (see above). The leaving group-binding pocket is more variable in terms of its shape, volume, and amino acid sequence than the acyl-binding region. The most striking difference between BAP and AEHs is the presence of Gly157 and Gly326 instead of 2 serine residues in the corresponding positions of XcAEH (Ser173 and Ser284) and AtAEH (Ser204 and Ser371; Fig. 3B). Gly326 is neighboring catalytic His325 and introduces the peptide bond switch. The altered main-chain conformation is stabilized by another mutation, namely Arg329 substituting Asn344 (in XcAEH numbering). The arginine guanidinium group forms a hydrogen bond with the carbonyl group of the switched peptide bond. Notably, the Arg329 side chain occupies space used by the serine residues in AEHs. Additional differences between BAP and AEHs are observed within the region contributed by the C-terminal domain, which may interact with S1 residues bearing longer side chains. These modifications are sufficiently large to suggest a difference in BAP and AEH function. These residues are conserved in BAPs found in species broadly distributed in marine sediments (Supplemental Fig. S1).
DISCUSSION
BAP is a true example of an enzyme found only in the uncultured MDM because sequenced cultured organisms do not contain a close homolog. This widespread enzyme may be an important factor in marine biology; yet, its function has been only assigned based on sequence comparison. Here, we have shown that a hypothesis generated from a single-cell genome of a deeply branching uncultured archaeon can be experimentally tested, and detailed characterizations of enzymes from MDM can be performed, without requiring that those organisms are first grown in pure culture. Recombinant BAP shows hydrolase activities, including that of a self-compartmentalizing α-aminopeptidase. In this capacity, it processes small peptides and peptide proxies with preference for cysteine and hydrophobic amino acids. The α-aminopeptidase activity is consistent with the hypothesis that widely distributed Bathyarchaeota and similar archaea hydrolyze detrital proteins in marine sediments. They use a set of secreted proteases to degrade proteins to short peptides that are imported across the membrane using peptide transporters. These molecules are further degraded into individual amino acids by intracellular peptidases; one of which is BAP. BAP and other peptidases have been identified in all 4 archaeal single-cell genomes derived from the same sediment sample. Interestingly, the temperature dependence of BAP (optimum, ∼28.8 ± 0.8°C) is consistent with mild psychrotrophy (40). This suggests that it is adapted to the temperatures of ocean sediments where MCG functions to provide an advantage. The α-aminopeptidase may also provide a housekeeping function and, together with other peptidases, is involved in recycling of intracellular archaeal proteins. The other BAP activities may be relevant to unidentified cellular processes.
Molecular characterization of MDM-encoded proteins holds great promise for understanding key nutrient transformations in the environment, including very hard to access and study marine sediments. The complex protein/peptide degradation pathway that links archaea with carbon, nitrogen, and sulfur cycling requires a thorough investigation. These organisms are ubiquitous and abundant in marine sediments and therefore likely influence global geochemical cycles. Small changes in these ocean environments may change the enzyme activities and affect remineralization processes. Although sequence analysis can fuel some assumptions about microbial life, prediction of specificity, catalysis, turnover rates, thermal stability, and kinetics solely from the primary structure is still out of reach and, in some cases, may lead to false conclusions. For example, recognition of a mesophilic character and a different substrate preference of BAP than that of characterized bacterial AEHs is a good illustration of discoveries that can be easily missed in sequence-based assumptions. Our analysis demonstrates that the initial annotation of MDM genomes can be further refined and expanded by molecular and structural biology to discover novel functions that are not found in the cultured microorganisms studied thus far. Thorough characterization is also inescapable in the exploration of the enzymatic space for novel industrial and biomedical catalysts, including proteases.
Supplementary Material
Acknowledgments
The authors thank Dr. Robert Jedrzejczak (Argonne National Laboratory) for discussion of cloning strategy, Dr. Gyorgy Babnigg (Argonne National Laboratory) for help in designing the cloning construct, Katlyn Fayman (Argonne National Laboratory) for help with protein purification, members of the Structural Biology Center at Argonne National Laboratory for their help with data collection at the 19-Insertion Device Beamline, Dr. Steven Wilhelm (University of Tennessee Department of Microbiology) for provision of lab space to A.D.S., and Dr. B. B. Jørgensen and the staff of the Center for Geomicrobiology at Aarhus University (Aarhus, Denmark) for providing amplified genomic deoxyribonucleic acid. This work was supported by the following funds: U.S. National Institutes of Health, National Institute of General Medical Sciences Grant GM094585 (to A.J.); the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357 (to A.J.); and Center for Dark Energy Biosphere Investigations Grants 157595 (to K.G.L.) and 36202823 (to A.D.S.). This work is Center for Dark Energy Biosphere Investigation Contribution 268. The submitted manuscript has been created by UChicago Argonne, Limited Liability Company, Operator of Argonne National Laboratory (“Argonne”). Argonne, a U.S. Department of Energy Office of Science laboratory, is operated under Contract No. DE-AC02-06CH11357. The U.S. Government retains for itself, and others acting on its behalf, a paid-up nonexclusive, irrevocable worldwide license in said article to reproduce, prepare derivative works, distribute copies to the public, and perform publicly and display publicly, by or on behalf of the Government.
Glossary
- 6-APA
6-aminopenicillanic acid
- 19-ID
19-Insertion Device
- AEH
α-amino acid ester hydrolase
- AMC
7-amido-4-methylcoumarin
- APS
Advanced Photon Source
- BAP
bathyaminopeptidase miscellaneous crenarchaeotal group-15
- BLAST
Basic Local Alignment Search Tool
- CocE
cocaine esterase
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- IMG
integrated microbial genome
- KI
inhibition constant
- Km
Michaelis constant
- MCG
miscellaneous crenarchaeotal group
- MDM
microbial dark matter
- MR
molecular replacement
- NusA
transcription termination/antitermination protein
- OD600
optical density at a wavelength of 600 nm
- PDB
Protein Data Bank
- PEG
polyethylene glycol
- PepX
X-prolyl dipeptidyl aminopeptidase
- Phg
phenylglycine
- RAxML
randomized axelerated maximum likelihood
- rmsd
root-mean-sd
- SeMet
selenomethionine
- TCEP
tris(2-carboxyethyl)phosphine
- TEV
tobacco etch virus
- TLC
thin-layer chromatography
- Topt
optimal temperature
- TVMV
tobacco vein mottling virus
- Vmax
maximum velocity
- XcAEH
α-amino acid ester hydrolase from Xanthomonas citri
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Rinke C., Schwientek P., Sczyrba A., Ivanova N. N., Anderson I. J., Cheng J. F., Darling A., Malfatti S., Swan B. K., Gies E. A., Dodsworth J. A., Hedlund B. P., Tsiamis G., Sievert S. M., Liu W. T., Eisen J. A., Hallam S. J., Kyrpides N. C., Stepanauskas R., Rubin E. M., Hugenholtz P., Woyke T. (2013) Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 [DOI] [PubMed] [Google Scholar]
- 2.Hartmann E. M., Durighello E., Pible O., Nogales B., Beltrametti F., Bosch R., Christie-Oleza J. A., Armengaud J. (2014) Proteomics meets blue biotechnology: a wealth of novelties and opportunities. Mar. Genomics 17, 35–42 [DOI] [PubMed] [Google Scholar]
- 3.Stepanauskas R., Sieracki M. E. (2007) Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc. Natl. Acad. Sci. USA 104, 9052–9057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrighton K. C., Thomas B. C., Sharon I., Miller C. S., Castelle C. J., VerBerkmoes N. C., Wilkins M. J., Hettich R. L., Lipton M. S., Williams K. H., Long P. E., Banfield J. F. (2012) Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337, 1661–1665 [DOI] [PubMed] [Google Scholar]
- 5.Lloyd K. G., Schreiber L., Petersen D. G., Kjeldsen K. U., Lever M. A., Steen A. D., Stepanauskas R., Richter M., Kleindienst S., Lenk S., Schramm A., Jørgensen B. B. (2013) Predominant archaea in marine sediments degrade detrital proteins. Nature 496, 215–218 [DOI] [PubMed] [Google Scholar]
- 6.Meng J., Xu J., Qin D., He Y., Xiao X., Wang F. (2014) Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J. 8, 650–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen N. A., Turner J. M., Stevens J., Rosser S. J., Basran A., Lerner R. A., Bruce N. C., Wilson I. A. (2002) Crystal structure of a bacterial cocaine esterase. Nat. Struct. Biol. 9, 17–21 [DOI] [PubMed] [Google Scholar]
- 8.Rigolet P., Mechin I., Delage M. M., Chich J. F. (2002) The structural basis for catalysis and specificity of the X-prolyl dipeptidyl aminopeptidase from Lactococcus lactis. Structure 10, 1383–1394 [DOI] [PubMed] [Google Scholar]
- 9.Hedstrom L. (2002) Serine protease mechanism and specificity. Chem. Rev. 102, 4501–4524 [DOI] [PubMed] [Google Scholar]
- 10.Kurochkina V. B., Sklyarenko A. V., Berezina O. V., Yarotskii S. V. (2013) Alpha-amino acid ester hydrolases: properties and applications. Appl. Biochem. Microbiol. 49, 672–694 [Google Scholar]
- 11.Krest’ianova I. N., Uvarov N. N., Rudenskaia G. N., Tsibanov V. V., Vasil’eva L. I., Stepanov V. M. (1990) [Intracellular aminopeptidase from Xanthomonas rubrilineans, hydrolyzing alpha-amino acid esters and cefalexin]. Biokhimiia 55, 2226–2238 [PubMed] [Google Scholar]
- 12.Aslanidis C., de Jong P. J. (1990) Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18, 6069–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eschenfeldt W. H., Lucy S., Millard C. S., Joachimiak A., Mark I. D. (2009) A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eschenfeldt W. H., Makowska-Grzyska M., Stols L., Donnelly M. I., Jedrzejczak R., Joachimiak A. (2013) New LIC vectors for production of proteins from genes containing rare codons. J. Struct. Funct. Genomics 14, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stols L., Gu M., Dieckman L., Raffen R., Collart F. R., Donnelly M. I. (2002) A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr. Purif. 25, 8–15 [DOI] [PubMed] [Google Scholar]
- 16.Stols L., Millard C. S., Dementieva I., Donnelly M. I. (2004) Production of selenomethionine-labeled proteins in two-liter plastic bottles for structure determination. J. Struct. Funct. Genomics 5, 95–102 [DOI] [PubMed] [Google Scholar]
- 17.Kim Y., Babnigg G., Jedrzejczak R., Eschenfeldt W. H., Li H., Maltseva N., Hatzos-Skintges C., Gu M., Makowska-Grzyska M., Wu R., An H., Chhor G., Joachimiak A. (2011) High-throughput protein purification and quality assessment for crystallization. Methods 55, 12–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y., Dementieva I., Zhou M., Wu R., Lezondra L., Quartey P., Joachimiak G., Korolev O., Li H., Joachimiak A. (2004) Automation of protein purification for structural genomics. J. Struct. Funct. Genomics 5, 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenbaum G., Alkire R. W., Evans G., Rotella F. J., Lazarski K., Zhang R. G., Ginell S. L., Duke N., Naday I., Lazarz J., Molitsky M. J., Keefe L., Gonczy J., Rock L., Sanishvili R., Walsh M. A., Westbrook E., Joachimiak A. (2006) The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J. Synchrotron Radiat. 13, 30–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 21.French G. S., Wilson K. S. (1978) Treatment of negative intensity observations. Acta Crystallogr. A 34, 517–525 [Google Scholar]
- 22.Padilla J. E., Yeates T. O. (2003) A statistic for local intensity differences: robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr. D Biol. Crystallogr. 59, 1124–1130 [DOI] [PubMed] [Google Scholar]
- 23.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barends T. R., Polderman-Tijmes J. J., Jekel P. A., Hensgens C. M., de Vries E. J., Janssen D. B., Dijkstra B. W. (2003) The sequence and crystal structure of the alpha-amino acid ester hydrolase from Xanthomonas citri define a new family of beta-lactam antibiotic acylases. J. Biol. Chem. 278, 23076–23084 [DOI] [PubMed] [Google Scholar]
- 25.Vagin A., Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 26.Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28.Winn M. D., Murshudov G. N., Papiz M. Z. (2003) Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 [DOI] [PubMed] [Google Scholar]
- 29.Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. (2011) BUSTER version 2.10.0, Global Phasing Limited, Cambridge, United Kingdom [Google Scholar]
- 30.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markowitz V. M., Korzeniewski F., Palaniappan K., Szeto E., Werner G., Padki A., Zhao X., Dubchak I., Hugenholtz P., Anderson I., Lykidis A., Mavromatis K., Ivanova N., Kyrpides N. C. (2006) The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34, D344–D348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer F., Paarmann D., D’Souza M., Olson R., Glass E. M., Kubal M., Paczian T., Rodriguez A., Stevens R., Wilke A., Wilkening J., Edwards R. A. (2008) The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar R. C. (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones P., Binns D., Chang H. Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., Pesseat S., Quinn A. F., Sangrador-Vegas A., Scheremetjew M., Yong S. Y., Lopez R., Hunter S. (2014) InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orsi W. D., Edgcomb V. P., Christman G. D., Biddle J. F. (2013) Gene expression in the deep biosphere. Nature 499, 205–208 [DOI] [PubMed] [Google Scholar]
- 36.Barends T. R., Dijkstra B. W. (2003) Acetobacter turbidans alpha-amino acid ester hydrolase: merohedral twinning in P21 obscured by pseudo-translational NCS. Acta Crystallogr. D Biol. Crystallogr. 59, 2237–2241 [DOI] [PubMed] [Google Scholar]
- 37.Lupas A., Flanagan J. M., Tamura T., Baumeister W. (1997) Self-compartmentalizing proteases. Trends Biochem. Sci. 22, 399–404 [DOI] [PubMed] [Google Scholar]
- 38.Barends T. R., Polderman-Tijmes J. J., Jekel P. A., Williams C., Wybenga G., Janssen D. B., Dijkstra B. W. (2006) Acetobacter turbidans alpha-amino acid ester hydrolase: how a single mutation improves an antibiotic-producing enzyme. J. Biol. Chem. 281, 5804–5810 [DOI] [PubMed] [Google Scholar]
- 39.Wallis M. (1973) The pKa values of the α-amino groups of peptides derived from the N-terminus of bovine growth hormone. Biochim. Biophys. Acta 310, 388–397 [DOI] [PubMed] [Google Scholar]
- 40.Zarubina S. A., Uporov I. V., Fedorchuk E. A., Fedorchuk V. V., Sklyarenko A. V., Yarotsky S. V., Tishkov V. I. (2013) 3D structure modeling of alpha-amino acid ester hydrolase from Xanthomonas rubrilineans. Acta Naturae 5, 62–70 [PMC free article] [PubMed] [Google Scholar]
- 41.Feller G., Gerday C. (2003) Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1, 200–208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



