Abstract
Signal transducers and activators of transcription (STAT) belongs to a family of latent cytoplasmic factors that can be activated by tyrosine phosphorylation by members of the Jak tyrosine kinase family in response to a variety of cytokines and growth factors. Activated STATs form dimers and translocate into nucleus to induce expression of critical genes essential for normal cellular events. In the past several years, significant progress has been made in the characterization of STAT acetylation, which is dependent on the balance between histone deacetylases (HDACs) and histone acetyltransferases (HATs) such as CBP/p300. Acetylation of STAT1, STAT2, STAT3, STAT5b and STAT6 has been identified. This review will highlight acetylation on the modulation of STAT activation.
1. Introduction
Although phosphorylation is a crucial posttranslational mechanism that regulates the activities of numerous proteins, there are many others including acetylation, methylation, ubiquitination, sumoylation, isgylation that are involved in protein modification. Acetylation has been studied widely in the recent years.
The phenomenon of histone acetylation in the eukaryotic cell has been known for many years. It occurs widely among eukaryotes, and various histone acetyltransferases (HATs) have been isolated and partially characterized since the early 1970s. Co-translational Nα-terminal acetylation is one of the most frequent protein modifications, occurring on approximately 85% of eukaryotic proteins [1]. A less common, but perhaps more important, form of protein acetylation takes place on the ε-amino group of lysines. HATs function enzymatically by transferring an acetyl group from acetyl-coenzyme A (acetyl-CoA) to the ε-amino group of certain lysine side chains within a histone’s basic N-terminal tail region [2, 3].
Each of these HAT enzymes generally belongs to one of two categories: type A, located in the nucleus and acetylate nucleosomal histones within chromatin in the nucleus, or type B, located in the cytoplasm, acetylates newly synthesized free histones in the cytoplasm for transport into the nucleus, where they may be deacetylated and incorporated into chromatin [4, 5]. The best-understood set of acetyltransferases is the GNAT (Gcn5-related N-acetyltransferase) superfamily which includes GCN5, PCAF, Hat1, Elp3, Hpa2, and other acetyltransferases and important for transcriptional initiation. Another group of evolutionarily related proteins that are known to be HATs is the MYST family, MOZ, Ybf2/Sas3, Sas2, and Tip60 yeast Esa1, Drosophila MOF, and human HBO1 and MORF are included in this family. After the discovery of histone acetylation by GCN5 and PCAF, the critical role of acetyltransferases in transcriptional regulation was also demonstrated by the fact that a pair of well-characterized coactivators of p300 and its close homolog CBP (CREB-binding protein) [3, 4]. Both of them contain a bromodomain and are often found within the same complexes and have the broadcast substrate for histones and non-histones proteins.
Acetylation is a highly reversible process, which is regulated by histone deacetylases (HDACs). HDACs are classified into four groups based on their homology to yeast histone deacetylases: Class I (HDAC1, 2, 3 and 8) are related to yeast RPD3 gene and are mostly located in the nuclei; Class II (HDAC4, 5, 6, 7, 9 and 10) are related to yeast Hda1 gene and are primarily located in the cytoplasm but can shuttle to nucleus; Class III (SIRT1–7), also known as the sirtuins, are related to the Sir2 gene and Class IV (HDAC11) has a conserved domain in the catalytic regions of both Class I and Class II enzymes [6]. The balance between acetylation and deacetylation is the important modulation mechanism for many cellular functions in diverse proteins. Here we will focus primarily on acetylation and transcriptional regulation of signal transducers and activators of transcription (STAT).
2. STAT structure, function and signaling pathway
The principal signaling route of cytokines was discovered over a decade ago, and its basic mechanisms are currently well understood [7]. Ligand-induced receptor dimerization activates receptor-associated tyrosine kinases, the Janus kinases (JAKs), and initiates tyrosine phosphorylation of intracellular signaling proteins, including latent cytoplasmic transcription factors: STATs [8, 9].
STATs comprise a family of seven structurally and functionally related proteins: STAT1, STAT2, STAT3, STAT4, STAT5a/b, and STAT6 [9]. STATs share structurally and functionally conserved domains. This includes the amino-terminal domain (NH2), the coiled-coiled domain (CCD), the DNA binding domain (DBD), the linker domain and the SH2/tyrosine activation domain. In contrast, the carboxy-terminal transcriptional activation domain (TAD) is quite divergent and contributes to STAT specificity (Figure 1). As the most highly conserved STAT domain, SH2 domains play an important role in signaling through their capacity to bind to specific phosphotyrosine motifs [10]. STATs are critical mediators of functional responses and specificity in cytokine signaling. At the receptor complex, STATs become phosphorylated on a conserved tyrosine residue, which induces their dimerization, nuclear translocation, and DNA binding, and leads to the induction of cytokine-responsive genes [11, 12]. After termination of the signal, STATs will translocate back to the cytoplasm from nucleus. STATs have been shown to play a role in development, cell growth and differentiation, proliferation, immune responses, cell survival and apoptosis [13].
Figure 1.

Structure of STAT proteins with the most important conserved domains. P is phosphorylation sites.
Different STATs are activated by distinct groups of cytokines. Interferon-γ is a potent activator of STAT-1; the interleukin-6 (IL-6) family members including IL-6, leukemia inhibitory factor (LIF) primarily activate STAT-3; STAT2 is almost uniquely tyrosine phosphorylated and activated in the presence of IFNα/β; STAT4 is predominantly activated in response to IL-12 and STAT6 is primarily activated by IL-4 and the highly related cytokine IL-13. Unlike most of the STATs, the two highly related STAT5 proteins are activated in response to a variety of cytokines as well as tyrosine kinase receptors. Therefore, it was assumed that STAT5a/b would have very basic functions in regulating cell growth [11].
3. Acetylation and its role in regulation of kinase, transcriptional activity and protein stability
3.1. Transcriptional activity
Transcriptional activators can be generally defined as proteins that bind to specific sites on the promoter DNA and bring about increased transcription of specific genes through interactions with other proteins. Eukaryotic transcription is a highly regulated process, and acetylation is now known to play a major role in this regulation nearly 50 years ago [14]. Specifically, acetyltransferase enzymes that act on particular lysine side chains of histones and other proteins are intimately involved in transcriptional activation. By modifying chromatin proteins and transcription-related factors, these acetylates are believed to regulate the transcription of many genes.
The acetylation of histones increases the accessibility of nucleosomal DNA to transcription factors, relieving transcriptional repression and correlating with the potential for transcriptional activity in vivo [15]. The characterization of several histone acetyltransferases, including the transcription coactivator p300/CBP, the human GCN5 homolog PCAF (p300/CBP-associated factor), and TAFII250 [16], has provided a potential explanation for the relationship between histone acetylation and transcriptional activation. In addition to histones, however, other components of the basal transcription machinery might be acetylated by these enzymes and directly affect transcription.
There are many transcription factors that are regulated by acetylation. Of them, p53, NF-κB, STATs, YY1, c-Myc are well-known. Transcription factor NF-κB plays a central role in inflammation, immune responses, cell survival, differentiation, apoptosis and proliferation. Phosphorylated p65 preferentially interacts with p300/CBP, resulting in p65 acetylation at multiple sites. Acetylation of K221 and K310 is associated with an increased transcription of NF-κB target genes, and is required for the full activity of p65 [17].
The tumor suppressor p53 is a key player in cellular signaling and stress responses and regulate the expression of genes contributing to cell cycle arrest, senescence and apoptosis [18]. Tumor suppressor and sequence-specific DNA binding transcription factor p53 is the symbol of non-histone targets of HAT acetylation [19]. p53 is acetylated by p300/CBP at multiple lysine residues at the C-terminal DNA binding regulatory domain, which activates its sequence-specific DNA binding activity and, consequently, increases activation of its target genes. The p53 protein can be acetylated by distinct acetyltransferases at different lysines: K120, K164, K320, K370, K372, K373, K381, K382 and K386 [20]. A report suggested that acetylation of the C terminus of p53 by p300 is not necessary for binding or promoter activation [21], further studies have convinced that p300/CBP-mediated acetyltransferase activity indeed is critical for p53-dependent function [22].
STAT1, STAT2, STAT3, STAT5 and STAT6 have also been shown to cooperate with CBP/p300 in transcriptional activation [4]. We will discuss this issue in detail in the latter part of this review.
3.2. Protein stability
It is no doubt that acetylation and deacetylation may have roles in cellular processes independent of transcription.
Ubiquitination is another post-translational modification which is to target ubiquitin labeled proteins for degradation and is involved in the regulation of numerous cellular events, ranging from progression through the cell cycle, transcription, and DNA repair, to antigen presentation and apoptosis [23]. The ubiquitin-proteasomal pathway involves a cascade of reactions that is catalyzed by ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (Ubc) (E2); and finally, ubiquitin ligase (E3) catalyzes the transfer of ubiquitin to the ε-amino group of lysine residues in the substrate protein. The polyubiquitinated proteins are then recognized and degraded by the 26S proteasome. Other types of ubiquitin modifications, such as monoubiquitination or multiubiquitination, do not result in protein degradation but instead alter the activity of the modified protein [24].
The primary similarity between protein ubiquitination and acetylation is based on the nature of the modified amino acid, which in both cases is a lysine. The cross-talk between these two protein lysine modifications is a critical regulatory mechanism controlling vital cellular functions. One of the emerging concepts is that acetylation status regulates protein stability by lysine competition. Lysine residues are targets for both acetylation and ubiquitination, and one of these modifications seems to prevent the other. deacetylation by HDACs in many cases is a prerequisite for subsequent ubiquitination. Conversely, acetylation may protect a protein from ubiquitination and degradation. For example, lysine residues acetylated in p53 overlap with those that are ubiquitinated, p53 acetylation serves to promote protein stability [25]. Unacetylated lysines then are targets for ubiquitination catalyzed by Mdm2, which ultimately leads to the destruction of p53.
Increasing evidence indicates that the interplay between the two lysine modifications goes beyond a simple competition mechanism and involves a more complex functional interaction between different actors of these signaling pathways. In these cases, lysine acetylation governs protein stability essentially by modulating protein – protein interaction. It’s also making it possible that acetylation modulates protein degradation. For example, the acetylation of a specific lysine would create a binding site for the recruitment of an E3-containing complex, which would then ubiquitinate the target protein [26]. In contrast, a given acetylated lysine may attract a partner, which would then mask other lysines and protect them against the activity of E3s [27]. Lysine acetylation may lead to a complex dissociation and would thereby render their components accessible to the action of protein degradation machinery [28, 29]. In addition to acetylation directly blocking ubiquitination at specific residues, it may also attenuate ubiquitination of other unacetylated lysine residues by inducing a protein conformational change [30].
Three HATs, CBP, p300 and TAF1, could directly be involved in the control of protein ubiquitination [31]. Tip60 is known to form a complex with at least two proteins with E3 activity, Mdm2 and Pihr2. Mdm2–Tip60 interaction has been shown to mediate Tip60 ubiquitination and degradation [32], while Tip60 stabilizes Pihr2, consequently forming a stable complex with an altered subcellular localization and containing both HAT and E3 activities [33].
3.3. Others
The association of nuclear receptors with their co-activator ACTR is inhibited by acetylation [34] was an example that acetylation could modulate protein and protein interactions. Also, acetylation could affect DNA binding. In the case of DNA-binding transcription factors, p53, E2F1, EKLF and GATA1, the acetylation sites located directly adjacent to the DNA-binding domain and acetylation results in stimulation of DNA binding [19, 35, 36]. On the contrary, the acetylated lysines within HMGI(Y) transcription factor fall within the DNA-binding domain and result in disruption of DNA binding [37].
4. The role of acetylation in regulation of STAT proteins
The first STAT protein shown to undergo acetylation was STAT6 [38]. Acetylation of other STAT such as STAT1, STAT2, STAT3, STAT5b has been recently characterized [39–44] (Table 1 and 2). Here we discuss the role of acetylation in the regulation of STAT proteins one by one in detail.
Table 1.
Acetylation-mediated modulation of STAT proteins
Table 2.
Acetylation sites and domain in STAT proteins.
| STATs | Acetylation sites | Domain | References |
|---|---|---|---|
| STAT1 | K410 and K413 | DNA-binding domain (DBD) | [39] |
| STAT2 | K390 | DNA-binding domain (DBD) | [40] |
| STAT3 | K685 | SH2 | [41, 42] |
| STAT3 | K49, K87 | NH2-terminus | [43] |
| STAT5b | K694 and K701 | SH2 | [44] |
| STAT5b | K359 | DNA-binding domain | [44] |
| STAT6 | Amino acids 750–788 in the carboxyl terminus | [72] |
4.1. The role of acetylation in regulation of STAT3
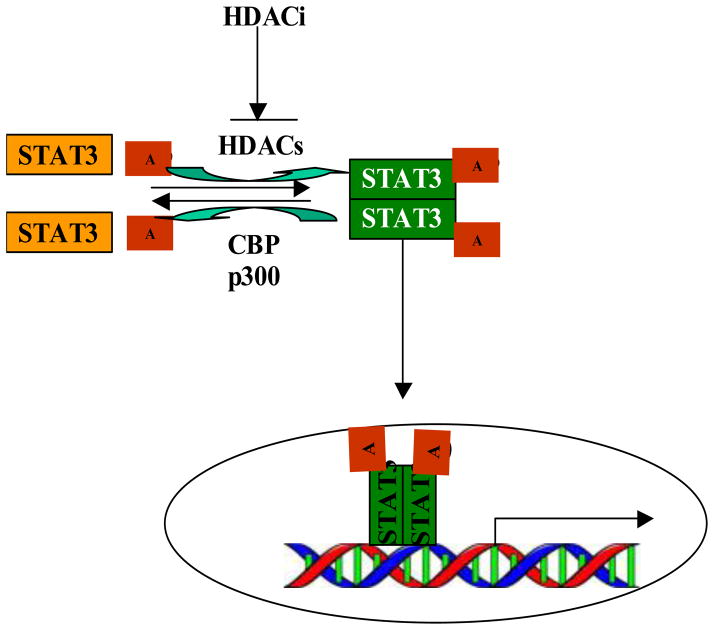
STAT3, one of the downstream effectors of cytokine signal transduction pathways, is a latent cytoplasmic transcription factor and was originally identified as an acute-phase response factor and activated by IL-6. In response to stimuli, tyrosine phosphorylation activates STAT3 and this phosphorylation is required for STAT3 dimerization. The nuclear translocation of STAT3 and its subsequent activation of target genes depend on dimerization. However, some studies have suggested the existence of phosphorylation-independent regulation of STAT3 dimerization [45, 46]. Recent studies have identified reversible acetylation as a modulator of STAT3 dimerization [41, 42]. CBP/p300 has been reported as a transcriptional coactivator that regulates STAT3 activity in vivo [47]. Upon cytokine treatment, CBP/p300 that adds acetyl groups to amino acids acetylates STAT3 on a single amino acid residue, lysine 685. Acetylation enhances both DNA binding, transactivation activity and nuclear localization (Figure 2). A STAT3 acetylation site mutant which carrys an arginine instead of a lysine residue at this position could be phosphorylated on tyrosine and serine and move to the nucleus, has impaired dimerization ability, and consequently has impaired cytokine-stimulated DNA binding and transcriptional activation capability. Deacetylation by HDAC3, and to a lesser extent, HDAC1 and HDAC2 (removes acetyl groups from amino acids), inhibits transcription of STAT3 target genes [41]). On the contrary, HDAC inhibitors (HDACi) can induces or increases STAT3 acetylation. Acetylation of STAT3 is a cytokine-induced post-translational modification that, like tyrosine phosphorylation and serine phosphorylation, is critical for STAT3 to attain its full transcriptional modulation (Figure 2. These results suggest a possible alternative mechanism for transducing intracellular signals within the Jak-STAT pathway or at least to compliment its tyrosine phosphorylation pathway and provide fresh insights into how an important family of DNA binding proteins might regulate gene expression, which may apply for other STAT family members.
Figure 2.
Regulation of STAT3 dimerization by acetylation. CBP/p300 induces STAT3 acetylation, which is required for its dimerization and subsequent nuclear translocation and gene transcription. On the contrary, HDACs promote STAT3 deacetylation.
Ray et al [43] reported that additional two Lys residues at amino acids 49 and 87 in the STAT3 NH2 terminus are also acetylated by p300. Comparing with STAT3 K685R, STAT3 K49R/K87R had no effect on inducible DNA binding, but blocked p300-mediated acetyl (Ac)-STAT3 formation and abrogated IL-6-induced human angiotensinogen (hAGT) gene activation in hepatocytes. Although STAT3 K49R/K87R rapidly translocated into the nucleus, it did not bind p300 and had delayed cytoplasmic redistribution. They also observed that STAT3 interacts with histone deacetylases (HDACs), specifically HDAC1, that down-regulate IL-6-induced hAGT transactivation [43]. Thus, IL-6-induced target gene activation requires p300-mediated STAT3 acetylation, and HDACs are involved in the termination of STAT3 action. These studies indicate the acetylation-deacetylation reaction as a novel signaling mechanism controlling the IL-6-STAT3 pathway in the hepatic APR. In addition, acetylation status of the STAT3 NH(2)-terminal domain regulates its interaction with p300 [48] and STAT3 NH(2)-terminal domain plays an important role in the IL-6 signaling pathway by interacting with the p300 bromodomain, thereby stabilizing some assembly.
Recently, Nie and colleagues also identified K679, which is highly conserved within the STAT family, as well as K707 and K709 as novel acetylated sites in STAT3. These three new lysine residues are all in the vicinity of the Y705 of STAT3, indicating that these sites may be pertinent to STAT3 phosphorylation. Arginine substitution of all four lysines (K679/685/707 to R) reduced STAT3 phosphorylation more strongly than the K685R mutant alone, which indicates a regulatory importance of multiple acetylation sites [49]
Acetylation of STAT3 has involved in many signaling pathway. Nadiminty et al [50] demonstrated that active but not latent STAT3, was expressed in numerous types of human cancers and involved in cell proliferation and survival, as well as induced p100 processing to p52 by activation of IKKα and subsequent phosphorylation of p100. The STAT3-mediated p100 processing to p52 requires activation of STAT3 by the acetyltransferase activity CBP/p300. STAT3-K685R blocked STAT3-mediated p100 processing to p52. Furthermore, overexpression of p52 protected cells from apoptotic cell death. Thus, activation of the processing of p100 to p52 by STAT3 may represent one of the common pathways used by cancer cells to survive and escape therapy.
Ohbayashi et al [51] found that leukemia inhibitory factor (LIF) or IL-6 induced acetylation of STAT3 at Lys-685 in 293T and Hep3B cells. Moreover, acetylation of STAT3 at Lys-685 was suppressed by PI3K inhibitor, LY294002, or a dominant negative Akt. Their further studies indicated that endogenous STAT3 is acetylated at Lys-685 by LIF or IL-6 through PI3K/Akt activation.
Through SirT1 mediates deacetylation of key STAT3 lysine sites, STAT3 phosphorylation and its function in the liver is tightly regulated by the nutritional status of an animal. The connection between acetylation and phosphorylation of STAT3 implies that STAT3 may have an important role in other cellular processes [49]. Our recent studies showed that induction of STAT3 acetylation by TSA is accompanied by its dephosphorylation and inhibition of renal fibroblast activation and proliferation as well as renal fibrogenesis in a muirne model of unilateral ureteral obstruction, suggesting that acetylation of STAT3 may suppress its phosphorylation and functional consequences [52]. In support of our observation, recent studies have also indicated that treatment of diffuse large B-cell lymphoma (DLBLC) with a HDAC inhibitor, LBH589, leads to STAT3 hyperacetylation and dephosphorylatinon, and inhibiting the transcription of STAT3-responsive anti-apoptotic genes and the survival of DLBCL cells [53]. In addition, HDAC inhibitors-induced STAT3 dephosphorylation may also mechanically be associated with its negative regulation on its upstream activator. In this context, Na et al., found that blocking class I HDACs with MS-275 results in inhibition of EGFR expression and its phosphorylation and subsequent dephosphorylation of STAT3 [54].
One of targeted genes of STAT3 is cyclin D1, and temperospatial expression and location of cyclinD1 is associated with different biological functions. In the early phase of its expression cyclin D1 is localized mostly in the cytoplasm and influences cell migration. However, during the late phase of its expression, cyclin D1 is translocated to the nucleus, which is required for cell proliferation. Interestingly, STAT-3 is tyrosine-phosphorylated in both early and late phases, but the late phase requires NFATc1·(Nuclear Factor of Activated T cells c1) mediated acetylation. NFATc1·STAT-3 complex binding to the cyclin D1 promoter facilitates cyclin D1 expression and proliferation [55].
In addition to the cross-talk between acetylation and phosphorylation, STAT3 acetylation also cross-communicate with other post-modifications. Lee et al, recently reported [56] that acetylated STAT3 is crucial for methylation of tumor-suppressor gene promoters (i.e. estrogen receptor-α gene) through upregulating expression of DNA methyltransferase 1 (DNMT1) and interaction of it. Since inactivation of the ERα gene via methylation strongly correlates with poor prognosis as well as an aggressive phenotype in cancers [57, 58], reactivation of ERα gene expression in tumors by inhibition of its methylation by targeting STAT3 acetylation may have therapeutic application. In vitro studies have shown that genetically disrupting STAT3 acetylation at Lys685 can reduce tumor growth, which is accompanied by demethylation and reactivation of several tumor-suppressor genes including ERα.
A recent study showed that IFN stimulation can induce STAT3 phosphorylation and DNA binding without triggering transcription of a STAT3-dependent reporter and endogenous genes, such as SOCS3 and c-FOS [59]. Impairment of the STAT3-dependent gene transcription can be restored by inhibition of HDAC1 and HDAC2, suggesting the importance of HDACs-mediated deacetylation in interfering with transcriptional activities. Further studies identify the Sin3a complex as a repressor of STAT3 transciptional activity. Sin3a can directly interact with STAT3 and promotes its deacetylation and subsequently reduces STAT3 binding to SOCS3 promoter and suppresses its transcriptional regulation of this gene [4].
4.2. The role of acetylation in regulation of STAT1
Investigation into IFNs signaling pathways led to the discovery of STAT1 and STAT2, the first members of STAT family. IFN-γ activates STAT1 homodimers, which initiates transcription of GAS-driven genes. IFN-α and IFN-β (i.e. type I IFNs) induce formation of STAT1 homodimers and STAT1/STAT2 heterodimers. The latter complex can induce transcription of interferon-sensitive response element (ISRE)-driven genes [10].
Krämer [39] described that STAT1 is an acetylated protein and demonstrated how the acetylation of STAT1 regulates NF-κB activity and leads to cell apoptosis. STAT1 acetylation depends on the balance between STAT1-associated histone deacetylases (HDACs) and histone acetyltransferases (HATs) such as CBP. Remarkably, both inhibitors of HDACs and IFN-α alter this equilibrium and induce STAT1 acetylation. CBP-mediated acetylation of STAT1 depends on the ε-amino group of lysine residues K410 and K413, which belong to the surface-exposed DNA-binding domain (DBD) common to all STATs. Experiments with STAT1 mutants mimicking either constitutively acetylated or nonacetylated states show that only acetylated STAT1 is able to interact with NF-κB p65. As a consequence, p65 DNA binding, nuclear localization, and expression of anti-apoptotic NF-κB target genes decrease. Here, STAT1 functions as a molecular switch and make STAT1 and NF-κB binding possible and then enhances NF-κB signaling. In addition, acetylation of the STAT1 protein may downregulate iNOS expression in proinflammatory states [60] by decreasing STAT1 binding to the iNOS promoter with resultant inhibition of IFN-γ-mediated iNOS expression.
Krämer et al [61] showed there is a phosphorylation-acetylation switch which regulates STAT1 signaling. This group found that acetylation of STAT1 counteracts IFN-induced STAT1 phosphorylation, nuclear translocation, DNA binding, and target gene expression. Biochemical and genetic experiments altering the HAT/HDAC activity ratio and STAT1 mutants reveal that a phospho-acetyl switch regulates STAT1 signaling via CBP, HDAC3, and the T-cell protein tyrosine phosphates (TCP45). Strikingly, inhibition of STAT1 signaling via CBP-mediated acetylation is distinct from the functions of this HAT in transcriptional activation. STAT1 acetylation induces binding of TCP45, which catalyzes dephosphorylation and latency of STAT1. These findings reveal a new layer of physiologically relevant STAT1 regulation and suggest that a previously unidentified balance between phosphorylation and acetylation affects cytokine signaling. However, this view was challenged by a very recent report showing that the treatment of cells with HDAC inhibitor results in the undiminished phosphorylation of STAT1[62]. A given explanation is that lys410 and lys413 are embedded within the nuclear localization sequence (NLS) of STAT1. Acetylation of these lysines would lead to precluded nuclear accumulation through neutralizing their positive charge since their positive charge are required for STAT1 to associate with nuclear translocation. As such, the phosphorylation-acetylation switch model in regulating STAT1 signaling is needed to be further verified.
4.3. The role of acetylation in regulation of STAT2
STAT2 is activated by type I IFNs and represents a unique member of the STAT family. It is the only STAT that is not known to bind GAS elements following activation. STAT2 acts as an adaptor to recruit STAT1 to IFNαR2 for activation which associates with p48 to form the interferon-stimulated gene factor 3 (ISGF3) complexes. It is also well known that STAT2 plays a critical role in promoting the antiviral immune response [10, 63].
Recently, more research was conducted on STAT2 acetylation. Génin et al., [64] showed that the trichostatin A (TSA), a deacetylase inhibitor, dependent inhibition of ISGF3 is related to impaired nuclear accumulation of STAT2 and suggest that an acetylation/deacetylatoin mechanism participates in the regulation of cellular distribution and function of STAT2 in IFN-alpha/beta signaling. Paulson et al., show that STAT2 recruits histone acetyltransferases (HAT) through its transactivation domain, resulting in localized transient acetylation of histones. GCN5, but not p300/CBP or PCAF, is required for STAT2 function [47].
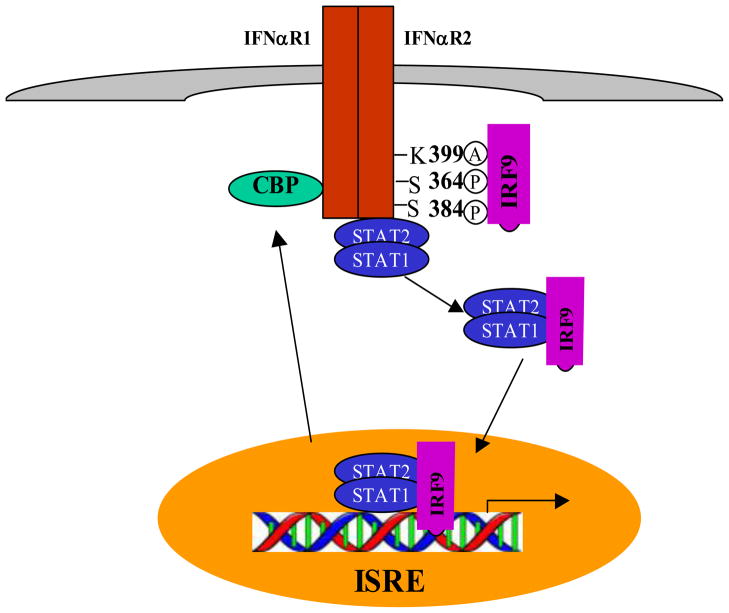
Type I IFNs (IFNα/β)-induced formation of the ISCF3 complex (STAT1/STA2/ interferon regulatory factor 9 (IRF9)) is critical for the induction of anti-viral gene expression and innate cellular immunity [65]. Tang et al [40] recently reported IFNα receptors recruit cytoplasmic CREB-binding protein (CBP) upon type 1 interferon (IFNα) stimulation, which acetylates IFNαR2 on Lys399 by binding to IFNαR2 within the region where two adjacent proline boxes bear phospho-Ser364 and phospho-Ser384. The process of acetylation in turn serves as the docking site for interferon regulatory factor 9 (IRF9). IRF9 interacts with the acetyl-Lys399 motif by means of its IRF homology2 (IH2) domain, leading to formation of the ISGF3 complex that includes IRF9, STAT1, and STAT2. All three components are acetylated by CBP. Acetylation within the DNA-binding domain (DBD) of both IRF9 and STAT2 is critical for the ISGF3 complex activation and its associated antiviral gene regulation. STAT2:STAT1 interaction was mainly regulated by STAT2 acetylation on Lys390, which dissociates DBD of STAT2 from STAT1, a step required for the active ISGF3 complex formation. Such significant roles for CBP-mediated acetylation of IFNαR, STAT1, STAT2, and IRF9 in IFN signaling enable acetylation as a positive regulator of STAT1 signaling (Figure 3). These results have significant demonstration concerning the important role of acetylation in cytokine receptor signal transduction.
Figure 3.
Acetylation of IFNαR-ISGF3 pathway is required for the antiviral activity of IFNα. Upon stimulation with IFNα, CBP is translocated from the nucleus to the cytosol and induces IFNαR acetylation on Lys 399. IRF9 interacts with this acetylation site leads to formation of the ISGF3 complex, which includes IRF9, STAT1 and STAT2. This complex is then translocated to the nucleus where it binds DNA and drives gene expression.
4.4. The role of acetylation in regulation of STAT5
The two highly related STAT5 proteins, STAT5a and STAT5b, are activated in response to a variety of cytokines as well as tyrosine kinase receptors, for example, IL-2 family, IL-3 family, growth hormone, prolactin, Epo and Tpo [10, 11, 66]. They share ~96% identity at the protein level and only diverse at their carboxy termini and have crucial roles in development, particularly in haematopoiesis [66, 67]. STAT5 was originally identified as a Prolactin-induced “mammary gland factor”, which is responsible for mediating ligand prolactin signaling and is required for mammary gland development and function [63, 68]. It would be interesting to study how acetylation modulates prolactin and its downstream STAT5. Recently, Ma et al., provides the evidence that acetylation does modulate STAT5b dimerization upon prolactin (PRL) stimulation [44].
Prolactin receptor (PRLR) is an essential type I cytokine receptor, which is involved in mammary gland development during pregnancy and lactation. Upon ligand binding, the membrane proximal regions of the PRLR cytoplasmic loop are brought into defined proximity to ensure activation of the protein kinase JAK2, which then phosphorylated PRLR on multiple tyrosine sites along the loop and the loop-associated STAT5a, and STAT5b on a conserved tyrosine residue within the C-terminal SH2-dimerization domain. Subsequently, tyrosine-phosphorylated STAT5 dissociates from the receptor and forms a transcriptional active dimer, and then translocates into the nucleus, where it regulates gene expression associated with the functions of the ligand PRL [69]. Cytokine-activated receptors undergo extracellular domain dimerization, which is necessary to activate intracellular signaling pathways. While the modulation of cytoplasmic loop dimerization is still unclear.
Ma et al.,[44] reported that in PRL-treated cells, PRLR, undergoes cytoplasmic loop dimerization, which is independent of extracellular dimerization but tightly regulated by acetylation. Cytoplasmic loop-dimerized PRLR then activates STAT5b, which is also acetylated by CBP and undergoes acetylation-dependent dimerization.
STAT5 is the principal signaling factor recruited by the PRLR cytoplasmic loop for activation. Ma et al.,[44] confirmed the acetylation sites of STAT5b by the use of Mass spectrometry and site-directed mutagenesis that K694 and K701 within the SH2-dimerization domain and K359 within the DNA-binding domain were major acetyl-lysine sites of STAT5b protein. Specific antibodies clearly detected STAT5b acetylation induction on these three lysine sites by PRL in T47D cells. STAT5b was preferentially acetylated by CBP and NAM and, to a lesser extent; TSA enhanced CBP-mediated STAT5b acetylation. Coimmunoprecipitation analysis in STAT5-null PC3 cells confirmed that STAT5b with K694R substitution or, to a lesser extent, K701R substitution impaired STAT5b dimerization, whereas STAT5b with K359R substitution showed a minimal but repeatable positive effect on STAT5b dimerization. STAT5b with K701R substitution showed a minor effect on STAT5b transcriptional activation, whereas STAT5b with K359R or K694R substitution or K694/K701R double mutation apparently attenuated STAT5b transcriptional activity. These results indicate that acetylation on multiple lysine sites within different domains coordinately regulates transcriptionally active STAT5b dimer formation.
To elucidate the significance of cytokine receptor dimerization for signal transduction, Ma et al., [44] examined STAT5b recruitment and activation by PRLR K1–15R. Although PRLR K1–15R mutant still associated with STAT5b, Y699 phosphorylation and K694 acetylation were abolished. Both TPRLR and PRLR K1–15R mutant recruited JAK2 constitutively and responded to ligand stimulation for JAK2 autophosphorylation. These findings confirmed that cytokine receptor dimerization is needed for STAT activation but not for STAT or JAK docking. PRLR-T539A mutant inhibited STAT5b activation, whereas PRLR-T539D mutant showed normal STAT5b activation, supporting the finding that CBP can associate with PRLR on phospho-T539, leading to both PRLR and STAT5b activation. These results clearly demonstrate that CBP-catalyzed acetylation plays a critical role from PRLR to its downstream STAT5b activation.
STAT5 acetylation is subjected to regulation by multiple HDACs. Interestingly, loss of HDAC9, but not HDAC6 or sirtuin-1 (Sirt1), was associated with stabilization of the acetylated form of STAT5 and promoted its transcriptional activity in regulatory T cells [70]..In lymphocytes, STAT5 signaling acetylation can also be regulated by a sumoylation/acetylation-phosphorylation switch. Sumoylation and aceylation have opposing effects on the tyrosine phosphorylation of STAT5: while accumulation of SUM2-modifed STAT5 reduces its tyrosine phosphorylation, acetylation of STAT5A at lysine 696 is essential for its activation [71]. In addition, both acetylation and SUMOylation can occur on the same lysine residue in STAT5, SUMOylation of this lysine in STAT5 blocks its acetylation [71]. These results suggest a critical role of SUMOylation in the regulation of STAT5 acetylation.
4.5. The role of acetylation in regulation of STAT6
Interleukin-4 (IL-4) binding to its cell surface receptor leads to tyrosine phosphorylation of the intracellular part of the IL-4 receptor and to an activation of Janus kinases 1 and 3 [69]. These kinases may be involved in phosphorylation of signal transducer and activator of transcription 6 (STAT6) [69]. After phosphorylation, STAT6 dimerizes and translocates to the nucleus, where it may bind to a particular sequence elements in the promoter of IL-4-responsible genes.
The first study on CBP or p300 and STAT6 was reported by McDonald et al in 1999 [72]. CBP and p300 were both found to cooperate with STAT6 for induction of STAT6-dependent transcription. This cooperation is not due to acetylation of STAT6 but depends on the presence of a carboxyl-terminal region of STAT6. Both STAT6 molecules lacking this region and point mutations within this region affect transcription by STAT6 in response to IL-4, identifying a motif that appears to be required for transcription, possibly through functional cooperation with CBP/p300. McDonald also pointed out that although STAT6 can be acetylated both by CBP and p300, it’s not a direct substrate of acetylation substrate of p300 in in vitro studies
Another related research is on STAT6 and 15-lipoxygenase-1 (15-LOX-1). Shankaranarayanan et al., [38]demonstrated that acetylation of STAT6 by CBP/p300 is required for transcriptional activation of the 15-LOX-1 gene. IL-4 induces expression of reticulocyte-type 15-LOX-1 in various mammalian cells via the Janus kinase/STAT6 signaling system. They found that IL-4 up-regulated the CBP/p300, which is responsible for acetylation of nuclear histones and STAT6. The acetylation of both proteins appears to be essential for the IL-4-induced signal transduction cascade, because inhibition of CBP/p300 by the viral wild-type E1A oncoprotein abrogated acetylation of both histones and STAT6 and strongly suppressed transcriptional activation of the 15-LOX-1 gene. This data suggests that acetylation of STAT6 as well as nuclear histones are involved in transcriptional activation of the 15-LOX-1 gene. Hence, STAT6 acetylation and phosphorylation are required for translational activation of 15-LOX-1 gene.
Unlike other STAT1-3 and STAT5, The acetylation sites STAT6 have not been identified yet, thus the biological consequence of its site-specific lysine acetylation are not known at present.
5. Concluding remarks
Tyrosine phosphorylation has long been considered to play the central role in signal transduction for many cytokines in mammalian cells. Increasing evidence indicates that acetylation of STAT is critically involved in IFNs and prolactin initiated intracellular signaling. Acetylation of STAT depends on the balance between its associated HDACs and the HAT CBP/p300 [39–42, 44]. Research on STAT acetylation has broadened our understanding of cytokine signaling and subsequent gene regulation. Acetylated STAT can function as positive or negative factors to regulate various aspects of STAT signaling. Further, the crosstalk between STAT acetylation and phosphorylation, ubiquitination, methylation, sumorylation will alter STAT protein stability and transcriptional activities. Defining the mechanisms by which cytokine stimulation controls STAT acetylation and how acetylation regulates STAT function will be the key to further understanding of STAT signaling mechanisms.
Highlights.
Tyrosine phosphorylation has long been considered to play the central role in signal transduction for many cytokines in mammalian cells. Increasing evidence indicates that acetylation of STAT is critically involved in IFNs and prolactin initiated intracellular signaling. Acetylation of STAT depends on the balance between its associated HDACs and the HAT CBP/p300. Research on STAT acetylation has broadened our understanding of cytokine signaling and subsequent gene regulation. Acetylated STAT can function as positive or negative factors to regulate various aspects of STAT signaling. Further, the crosstalk between STAT acetylation and phosphorylation, ubiquitination, methylation, sumorylation can alter STAT protein stability and transcriptional activities/p300. Acetylation of STAT1, STAT2, STAT3, STAT5b and STAT6 has been identified. This review have highlighted acetylation on the modulation of STAT activation.
Acknowledgments
This work was supported by grants from the National Institutes of Health (DK-085065 to SZ), the national nature science foundation of China (81270778 to SZ), and Pudong New District Foundation of China (PWZxk2010-02).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Polevoda B, Sherman F. J Biol Chem. 2000;275(47):36479–36482. doi: 10.1074/jbc.R000023200. [DOI] [PubMed] [Google Scholar]
- 2.Wieczorek M, Ginter T, Brand P, Heinzel T, Kramer OH. Cytokine Growth Factor Rev. 23(6):293–305. doi: 10.1016/j.cytogfr.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 3.Sterner DE, Berger SL. Microbiol Mol Biol Rev. 2000;64(2):435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Icardi L, De Bosscher K, Tavernier J. Cytokine Growth Factor Rev. 23(6):283–291. doi: 10.1016/j.cytogfr.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Cell. 1996;84(6):843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 6.Pang M, Zhuang S. J Pharmacol Exp Ther. 335(2):266–272. doi: 10.1124/jpet.110.168385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horvath CM, Darnell JE. Curr Opin Cell Biol. 1997;9(2):233–239. doi: 10.1016/s0955-0674(97)80067-1. [DOI] [PubMed] [Google Scholar]
- 8.Leonard WJ, O’Shea JJ. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 9.Darnell JE., Jr Science. 1997;277(5332):1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 10.Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. Gene. 2002;285(1–2):1–24. doi: 10.1016/s0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 11.Ihle JN. Curr Opin Cell Biol. 2001;13(2):211–217. doi: 10.1016/s0955-0674(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 12.Leonard WJ. Int J Hematol. 2001;73(3):271–277. doi: 10.1007/BF02981951. [DOI] [PubMed] [Google Scholar]
- 13.Levy DE, Darnell JE., Jr Nat Rev Mol Cell Biol. 2002;3(9):651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 14.Kouzarides T. Embo J. 2000;19(6):1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vitolo JM, Thiriet C, Hayes JJ. Mol Cell Biol. 2000;20(6):2167–2175. doi: 10.1128/mcb.20.6.2167-2175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. Nature. 1996;382(6589):319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 17.Chen LF, Greene WC. J Mol Med (Berl) 2003;81(9):549–557. doi: 10.1007/s00109-003-0469-0. [DOI] [PubMed] [Google Scholar]
- 18.Vousden KH. J Cell Sci. 2006;119(Pt 24):5015–5020. doi: 10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- 19.Gu W, Roeder RG. Cell. 1997;90(4):595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 20.Li AG, Piluso LG, Cai X, Gadd BJ, Ladurner AG, Liu X. Mol Cell. 2007;28(3):408–421. doi: 10.1016/j.molcel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Espinosa JM, Emerson BM. Mol Cell. 2001;8(1):57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- 22.Gu W, Luo J, Brooks CL, Nikolaev AY, Li M. Novartis Found Symp. 2004;259:197–205. discussion 205–197, 223–195. [PubMed] [Google Scholar]
- 23.Kaiser P, Tagwerker C. Methods Enzymol. 2005;399:243–248. doi: 10.1016/S0076-6879(05)99016-2. [DOI] [PubMed] [Google Scholar]
- 24.Hicke L. Nat Rev Mol Cell Biol. 2001;2(3):195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 25.Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E, Yao TP. Embo J. 2002;21(22):6236–6245. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH, Yoo MA, Song EJ, Lee KJ, Kim KW. Cell. 2002;111(5):709–720. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh JK, Chan FS, O’Connor DJ, Mittnacht S, Zhong S, Lu X. Mol Cell. 1999;3(2):181–193. doi: 10.1016/s1097-2765(00)80309-3. [DOI] [PubMed] [Google Scholar]
- 28.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Nature. 2002;419(6905):411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 29.Nimmanapalli R, Fuino L, Bali P, Gasparetto M, Glozak M, Tao J, Moscinski L, Smith C, Wu J, Jove R, Atadja P, Bhalla K. Cancer Res. 2003;63(16):5126–5135. [PubMed] [Google Scholar]
- 30.Li M, Luo J, Brooks CL, Gu W. J Biol Chem. 2002;277(52):50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 31.Col E, Caron C, Chable-Bessia C, Legube G, Gazzeri S, Komatsu Y, Yoshida M, Benkirane M, Trouche D, Khochbin S. Embo J. 2005;24(14):2634–2645. doi: 10.1038/sj.emboj.7600734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemercier C, Legube G, Caron C, Louwagie M, Garin J, Trouche D, Khochbin S. J Biol Chem. 2003;278(7):4713–4718. doi: 10.1074/jbc.M211811200. [DOI] [PubMed] [Google Scholar]
- 33.Halkidou K, Logan IR, Cook S, Neal DE, Robson CN. Nucleic Acids Res. 2004;32(5):1654–1665. doi: 10.1093/nar/gkh296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Cell. 1999;98(5):675–686. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 35.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Nature. 1998;396(6711):594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Bieker JJ. Proc Natl Acad Sci U S A. 1998;95(17):9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Mol Cell. 1998;2(4):457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 38.Shankaranarayanan P, Chaitidis P, Kuhn H, Nigam S. J Biol Chem. 2001;276(46):42753–42760. doi: 10.1074/jbc.M102626200. [DOI] [PubMed] [Google Scholar]
- 39.Kramer OH, Baus D, Knauer SK, Stein S, Jager E, Stauber RH, Grez M, Pfitzner E, Heinzel T. Genes Dev. 2006;20(4):473–485. doi: 10.1101/gad.364306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang X, Gao JS, Guan YJ, McLane KE, Yuan ZL, Ramratnam B, Chin YE. Cell. 2007;131(1):93–105. doi: 10.1016/j.cell.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 41.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Science. 2005;307(5707):269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 42.Wang R, Cherukuri P, Luo J. J Biol Chem. 2005;280(12):11528–11534. doi: 10.1074/jbc.M413930200. [DOI] [PubMed] [Google Scholar]
- 43.Ray S, Boldogh I, Brasier AR. Gastroenterology. 2005;129(5):1616–1632. doi: 10.1053/j.gastro.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 44.Ma L, Gao JS, Guan Y, Shi X, Zhang H, Ayrapetov MK, Zhang Z, Xu L, Hyun YM, Kim M, Zhuang S, Chin YE. Proc Natl Acad Sci U S A. 107(45):19314–19319. doi: 10.1073/pnas.1010253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braunstein J, Brutsaert S, Olson R, Schindler C. J Biol Chem. 2003;278(36):34133–34140. doi: 10.1074/jbc.M304531200. [DOI] [PubMed] [Google Scholar]
- 46.Kretzschmar AK, Dinger MC, Henze C, Brocke-Heidrich K, Horn F. Biochem J. 2004;377(Pt 2):289–297. doi: 10.1042/BJ20030708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paulson M, Pisharody S, Pan L, Guadagno S, Mui AL, Levy DE. J Biol Chem. 1999;274(36):25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 48.Hou T, Ray S, Lee C, Brasier AR. J Biol Chem. 2008;283(45):30725–30734. doi: 10.1074/jbc.M805941200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, Gao Q. Nat Cell Biol. 2009;11(4):492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nadiminty N, Lou W, Lee SO, Lin X, Trump DL, Gao AC. Proc Natl Acad Sci U S A. 2006;103(19):7264–7269. doi: 10.1073/pnas.0509808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ohbayashi N, Ikeda O, Taira N, Yamamoto Y, Muromoto R, Sekine Y, Sugiyama K, Honjoh T, Matsuda T. Biol Pharm Bull. 2007;30(10):1860–1864. doi: 10.1248/bpb.30.1860. [DOI] [PubMed] [Google Scholar]
- 52.Pang M, Kothapally J, Mao H, Tolbert E, Ponnusamy M, Chin YE, Zhuang S. Am J Physiol Renal Physiol. 2009;297(4):F996–F1005. doi: 10.1152/ajprenal.00282.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gupta M, Han JJ, Stenson M, Wellik L, Witzig TE. Leukemia. 26(6):1356–1364. doi: 10.1038/leu.2011.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu N, He S, Ma L, Ponnusamy M, Tang J, Tolbert E, Bayliss G, Zhao TC, Yan H, Zhuang S. PLoS One. 8(1):e54001. doi: 10.1371/journal.pone.0054001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kundumani-Sridharan V, Van Quyen D, Subramani J, Singh NK, Chin YE, Rao GN. J Biol Chem. 287(27):22463–22482. doi: 10.1074/jbc.M112.362996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee H, Zhang P, Herrmann A, Yang C, Xin H, Wang Z, Hoon DS, Forman SJ, Jove R, Riggs AD, Yu H. Proc Natl Acad Sci U S A. 109(20):7765–7769. doi: 10.1073/pnas.1205132109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mori T, Martinez SR, O’Day SJ, Morton DL, Umetani N, Kitago M, Tanemura A, Nguyen SL, Tran AN, Wang HJ, Hoon DS. Cancer Res. 2006;66(13):6692–6698. doi: 10.1158/0008-5472.CAN-06-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brinkman JA, El-Ashry D. J Mammary Gland Biol Neoplasia. 2009;14(1):67–78. doi: 10.1007/s10911-009-9113-0. [DOI] [PubMed] [Google Scholar]
- 59.Icardi L, Mori R, Gesellchen V, Eyckerman S, De Cauwer L, Verhelst J, Vercauteren K, Saelens X, Meuleman P, Leroux-Roels G, De Bosscher K, Boutros M, Tavernier J. Proc Natl Acad Sci U S A. 109(30):12058–12063. doi: 10.1073/pnas.1206458109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo L, Guo H, Gao C, Mi Z, Russell WB, Kuo PC. Surgery. 2007;142(2):156–162. doi: 10.1016/j.surg.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kramer OH, Heinzel T. Mol Cell Endocrinol. 315(1–2):40–48. doi: 10.1016/j.mce.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 62.Antunes F, Marg A, Vinkemeier U. Mol Cell Biol. 31(14):3029–3037. doi: 10.1128/MCB.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horvath CM. Trends Biochem Sci. 2000;25(10):496–502. doi: 10.1016/s0968-0004(00)01624-8. [DOI] [PubMed] [Google Scholar]
- 64.Genin P, Morin P, Civas A. J Virol. 2003;77(12):7113–7119. doi: 10.1128/JVI.77.12.7113-7119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kiu H, Nicholson SE. Growth Factors. 30(2):88–106. doi: 10.3109/08977194.2012.660936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schindler C, Strehlow I. Adv Pharmacol. 2000;47:113–174. doi: 10.1016/s1054-3589(08)60111-8. [DOI] [PubMed] [Google Scholar]
- 67.Reich NC, Liu L. Nat Rev Immunol. 2006;6(8):602–612. doi: 10.1038/nri1885. [DOI] [PubMed] [Google Scholar]
- 68.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Genes Dev. 1997;11(2):179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 69.Ihle JN, Kerr IM. Trends Genet. 1995;11(2):69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- 70.Beier UH, Wang L, Han R, Akimova T, Liu Y, Hancock WW. Sci Signal. 5(229):ra45. doi: 10.1126/scisignal.2002873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Van Nguyen T, Angkasekwinai P, Dou H, Lin FM, Lu LS, Cheng J, Chin YE, Dong C, Yeh ET. Mol Cell. 45(2):210–221. doi: 10.1016/j.molcel.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McDonald C, Reich NC. J Interferon Cytokine Res. 1999;19(7):711–722. doi: 10.1089/107999099313550. [DOI] [PubMed] [Google Scholar]