Abstract
The membrane-bound serine protease CAP2/Tmprss4 has been previously identified in vitro as a positive regulator of the epithelial sodium channel (ENaC). To study its in vivo implication in ENaC-mediated sodium absorption, we generated a knockout mouse model for CAP2/Tmprss4. Mice deficient in CAP2/Tmprss4 were viable, fertile, and did not show any obvious histological abnormalities. Unexpectedly, when challenged with sodium-deficient diet, these mice did not develop any impairment in renal sodium handling as evidenced by normal plasma and urinary sodium and potassium electrolytes, as well as normal aldosterone levels. Despite minor alterations in ENaC mRNA expression, we found no evidence for altered proteolytic cleavage of ENaC subunits. In consequence, ENaC activity, as monitored by the amiloride-sensitive rectal potential difference (ΔPD), was not altered even under dietary sodium restriction. In summary, ENaC-mediated sodium balance is not affected by lack of CAP2/Tmprss4 expression and thus, does not seem to directly control ENaC expression and activity in vivo.
Introduction
The regulation of sodium balance throughout the body is important to maintain blood volume and blood pressure. In tight epithelia as in kidney and colon, aldosterone promotes sodium reabsorption through the amiloride-sensitive epithelial sodium channel ENaC [1]. This channel was initially identified in the colon of rats challenged with a low salt diet [2,3]. ENaC is composed of three subunits, Scnn1a, Scnn1b, and Scnn1g, sharing 30% homology with each other at the protein level [3].
One regulatory mechanism of ENaC-mediated sodium reabsorption is achieved through channel-activating proteases (CAPs) as e.g. CAP1 (Prss8 or prostasin), CAP2 (Tmprss4) and CAP3 (ST-14 or matriptase) [4–7]. All three are membrane-bound serine proteases that are able to significantly increase ENaC-mediated sodium transport by increasing the open probability (Po) of single channels [6–8] and/or activating a population of near-silent ENaC channels at the plasma membrane [9]. In vivo studies conducted on different mouse models for CAP1/Prss8 have shown that this protease is a regulator of ENaC in several epithelia where the two proteins are co-expressed. In lung, absence of CAP1/Prss8 leads to impaired lung fluid clearance mediated by ENaC, and to altered-adrenergic response which may impact on the resolution of pulmonary oedema after lung injury [10–12]. Colon-specific deletion of CAP1/Prss8 resulted in decreased amiloride-sensitive rectal potential difference (PD) upon either regular or low salt diet [13]. Decreased rectal PD was also observed in two spontaneous CAP1/Prss8 mutants, in frizzy mice harbouring a V170D transversion, and in frCR rats, that carry a G54-P57 deletion [14–17].
CAP2/Tmprss4, previously termed Tmprss3 [18] belongs to subfamily A of the S1 chymotrypsin family. Proteases of this family are characterized by the presence of a catalytic triad, composed of one histidine (H), one aspartate (D) and a serine (S), forming together a catalytic pocket that enables hydrolysis of target peptide bonds. CAP2/Tmprss4 is a type II transmembrane serine protease, and harbours a N-terminal transmembrane domain, one low-density lipoprotein (LDL) class A domain, one scavenger receptor cysteine-rich (SRCR) domain, the protease domain, and a short C-terminal tail [19,20]. While the physiological role of CAP2/Tmprss4 is largely unknown due to lack of a knockout model, CAP2/Tmprss4 was identified as involved in pathologies such as cancer, influenza infections and neurological disorders [21–23].
Experiments in Xenopus oocytes strongly supported the hypothesis that CAP2/Tmprss4 activates ENaC-mediated sodium current by cleaving the Scnn1g subunit at position R138 [24], previously identified as furin-consensus cleavage site [25,26], although the significance for final ENaC activation is still under debate [27].
In the present study, we aimed to investigate the in vivo physiological function of CAP2/Tmprss4 using constitutive knock-out mice. Our data indicate that ENaC-mediated sodium reabsorption is not regulated by CAP2/Tmprss4 arguing for a redundant protease network regulating sodium homeostasis.
Material and Methods
Animals and ethics statement
All experimental procedures and animal maintenance followed Swiss federal guidelines. This study has been reviewed and approved (authorization no. 1003.7 to EH) by the “Service de la consommation et des affaires vétérinaires” (SCAV) of the canton of Vaud, Switzerland. Animals were anaesthetized by intraperitoneal injection of 10μl per gram of body weight with a solution containing 10% of Rompun (Bayer) and 10% Ketanarkon (Streuli Pharma) diluted in water. If necessary, animals were sacrificed by cervical dislocation and bleeding. Animals were housed in rooms with controlled temperature and humidity levels and a 12h/12h light/dark cycle, and had free access to food and drinking water. Age-matched homozygous mutant (CAP2/Tmprss4 Δ/Δ, Δ/Δ, knockout, KO), heterozygous mutant (CAP2/Tmprss4 Δ/+, Δ/+, HET), and CAP2/Tmprss4 wildtype (CAP2/Tmprss4 +/+, +/+, WT) littermates were obtained by interbreeding mice heterozygous mutant for the CAP2/Tmprss4 Δ/+ allele. Genotyping of the 350bp floxed, 500bp knockout and 250bp wildtype allele was performed by PCR on genomic DNA using following primers [5’sense: s3, 5’-GGTCAGATGTAAAAGGTAGAC-3’; VR anti-sense: as3, 5’-CACACCAGCCCTGAATCATC-3’; and 3’-anti-sense: as2 5’-GCTAGGTTCCTTGTTCCTG-3’. PCR amplification was performed for 36 cycles for 1’ at 95°C, 1’ at 56°C and 1’ at 72°C. PCR products were visualized by ethidium bromide staining and run by electrophoresis on 2% agarose gel. Male and female animals (mice homozygous for Ren-1 c), if not stated elsewise, were used at the age of 3 to 6 months and fed with standard (0.17%) Na+ diet (ssniff, Spezialdiäten GmbH, Germany).
Generation of conditional and null mutant CAP2/Tmprss4 mice
To construct the CAP2/Tmprss4 replacement-type targeting vector, a 14kb genomic DNA contig (strain 129S5/SvEvBrd) spanning exon 6–13 was cloned into pREC-1 vector containing a HSV-TK cassette. A loxP site was inserted into the BstEII site upstream of exon 8 resulting in a 4.2kb 5’ homologous region containing exons 6 and 7 and the 1.8kb vital region harbouring exons 8 (histidine, H243) and 9 (aspartate, D288) of the catalytic triad. A 1.5kb BamHI/PvuI FRT-neo-FRT-lox cassette (pAT-FRT-K13; [28]) was introduced into the SpeI site, generating the 3.4kb SpeI/EcoRI 3’ homologous region containing exons 10–13. The targeting vector was linearized with SalI and electroporated into mouse embryonic stem (ES) cells (129S5/SvEvBrd) [29]. Briefly, G418- and ganciclovir-resistant colonies were expanded and screened by PCR using following primers: 3’ recombination: sense 5’-GGACATTGCCCTTGTTAAGCTG-3’ or sense: s1, 5’-TCGCCTTCTTGACGAGTTCTTC-3’ combined with antisense: as1, 5’-GTTTGTCATTGGTGCCGTGTG-3’. Targeted clones were further confirmed by Southern blot analysis using a 5’ external probe (523bp NdeI/PstI fragment) revealing 7.5kb wildtype and 9.6kb mutant (loxneo) alleles on SpeI/NheI-digested genomic DNA, and using a 3’-probe (530bp SphI/SacI fragment) detecting 7.5kb wildtype and 8.9kb mutant (loxneo) alleles, on BamHI-digested DNA as well as an internal probe (PCR-amplified neomycin fragment) which revealed a 4.7kb mutant band on EcoRI-digested genomic DNA. Following deletion of the neomycin cassette, the 5’ probe detected a 7.5kb (wildtype or floxed), a 9.6kb (loxneo) or a 5.6kb (knockout) fragment on SpeI/NheI-digested genomic DNA. Position of loxP sites was verified by PCR with loxP-specific primers (details available on request).
Correctly targeted cells (clone #1) were injected into C57BL/6N blastocysts and germline chimeras were obtained. Breeding of CAP2/Tmprss4 loxneo/loxneo mice with Flp mice [30] allowed the excision of the neomycin cassette to generate mice carrying the CAP2/Tmprss4 lox (CAP2/Tmprss4 Lox) allele. Breeding with nestin-Cre mice [31] allowed to generate mice carrying the Δ (CAP2/Tmprss4 Δ CAP2/Tmprss4 KO, knockout, Tmprss4tm1.1Hum) allele.
RNA extraction and qRT-PCR
Organs were frozen in liquid nitrogen and stored at -80°C. Tissues were homogenized using TissueLyser (Qiagen, Valencia, CA), and mRNA was isolated using Qiagen RNeasy Mini Kit (Basel, Switzerland) according to the manufacturer’s instructions. cDNA synthesis was performed using 1.5μg of mRNA and reverse transcribed using PrimeScript RT reagent kit according to the manufacturer’s instructions (Takara Bio Inc Japan). Real-time PCR was performed using TaqMan Universal PCR Master Mix (Applied Biosystems) for CAP1/Prss8, CAP3/ST-14, Scnn1a, Scnn1b, Scnn1g, and furin, or Power SYBRgreen PCR Master Mix (Applied Biosystems) for CAP2/Tmprss4, and run using Applied Biosystems 7500 Fast (Carlsbad, CA). Each measurement was performed as duplicate. Quantification of fluorescence was normalized to β-actin for TaqMan reagents, and to mouse Gapdh for SYBRgreen reagents. Primer and probe sequences for CAP1/Prss8, CAP3/ST-14, Scnn1a, Scnn1b and Scnn1g have been described previously [13]. The primer sequences used for CAP2/Tmprss4 were: 5’-CTGCCTTGACTGTGGAAAG-3’ and 5’-GCTGCTTGTTGTACTGGATG-3’, and for furin: 5’-GCCGGAAAGTGAGCCATTC-3’, 5’-GGGTTCCACCAGGATTTCAA-3’ and 5’-FAM-TGCCATGGTGGCTCTGGCCC-BHQ1-3’.
SDS-PAGE and Western blot analysis
30μg of proteins were separated by SDS-PAGE on 10% acrylamide gels, and proteins were electrically transferred to PolyScreen PVDF hybridization transfer membranes (Perkin Elmer, Boston, MA). Membranes were incubated overnight at 4°C with primary rabbit antibody for Scnn1a (1:500), Scnn1b and Scnn1g (1:1000) [32], CAP2/Tmprss4 [33] (1:200) and β-actin (1:1000, Sigma-Aldrich) and for 1 hour with donkey anti-rabbit IgG HRP-conjugated secondary antibody (1:10000, Amersham, Burkinghampshire, UK) (all antibodies in TBS-Tween 1% and dried milk 2%). The signal was revealed using SuperSignal West Dura detection system (Pierce, Rockford, IL) and quantified using ImageStudioTM Lite program (LI-COR). Kidney extracts from inducible renal tubule-specific Scnn1a KO mice, generated by interbreeding of Scnn1a lox/lox mice [34] and Pax8::rtTA/LC1 mice [35], were used as control for Scnn1a-specific signals on Western blot (control non-doxycycline-induced animals [Ctrl WT], control doxycycline-induced animals [Ctrl KO]). The same strategy was applied for Scnn1b- and Scnn1g-specific bands [36]. The specificity of the primary antibody for CAP2/Tmprss4 has been described previously and extensively tested in vitro using the Xenopus oocyte expression system [33], and corroborated using protein extracts from CAP2/Tmprss4 knock-out mice that were used as control.
Histological analyses
Organs were fixed in 4% paraformaldehyde and processed for paraffin embedding. Following organs were taken for histological analyses: skin, kidney, colon, lung, heart, brain, eye, tongue, stomach, small intestine, spleen, spine, femur, testis, uterus, thymus, salivary gland, pancreas, and adrenal gland. 3μm sagittal sections were cut, prepared and stained with eosin and hematoxylin as previously described [17]. Sections were visualized by optical microscopy (Axioplan, Carl Zeiss Microscopy, Jena, Germany) and pictures were taken using an AxioCam HR microscope (Carl Zeiss Microscopy, Jena, Germany).
Measurement of physiological parameters
Mice were kept in standard cages with free access to food and water and fed with regular sodium (RS: 0.17% Na+) or sodium-deficient diet (<0.01% Na+) (ssniff, Spezialdiäten GmbH, Germany) for 21 consecutive days. At the end of the experiment, blood samples were collected. Plasma aldosterone levels were measured according to standard procedures using radioimmunoassay (RIA) (Coat-A-Count RIA kit, Siemens Medical Solutions Diagnostics, Ballerup, Denmark) [37]. Samples with values > 1200 pg/ml were further diluted using a serum pool with low aldosterone concentration (<50 pg/ml). Aldosterone concentration is indicated as pg/ml. Plasma electrolytes were analyzed using an Instrumentation Laboratory 943 Electrolyte Analyzer (UK).
Amiloride-sensitive rectal transepithelial potential difference measurements
Amiloride-sensitive transepithelial rectal potential difference (ΔPD) measurements were performed as described [38,17]. Briefly, amiloride-sensitive rectal ΔPD was measured in the morning and in the afternoon on two days the same week in anaesthetized animals. Rectal PD was monitored by a VCC600 electrometer (Physiologic instruments, San Diego, CA, USA) connected to a chart recorder. After stabilization of PD, saline solution was injected through the first barrel as control procedure and PD was recorded. Saline solution containing 25μmol/l amiloride was injected through the second barrel and PD was recorded. Potential difference was recorded before and after addition of amiloride as amiloride-sensitive ΔPD.
Statistical analysis
Results are presented as mean ± SEM. Throughout the study, and if not otherwise stated, data were analyzed by one-way ANOVA. P < 0.05 was considered statistically significant.
Results
Generation of CAP2/Tmprss4 constitutive knockout mice
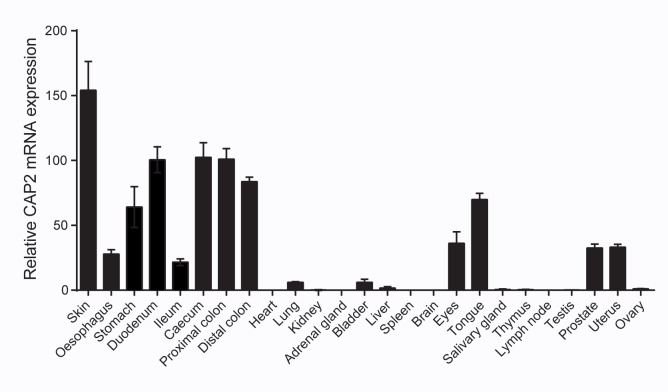
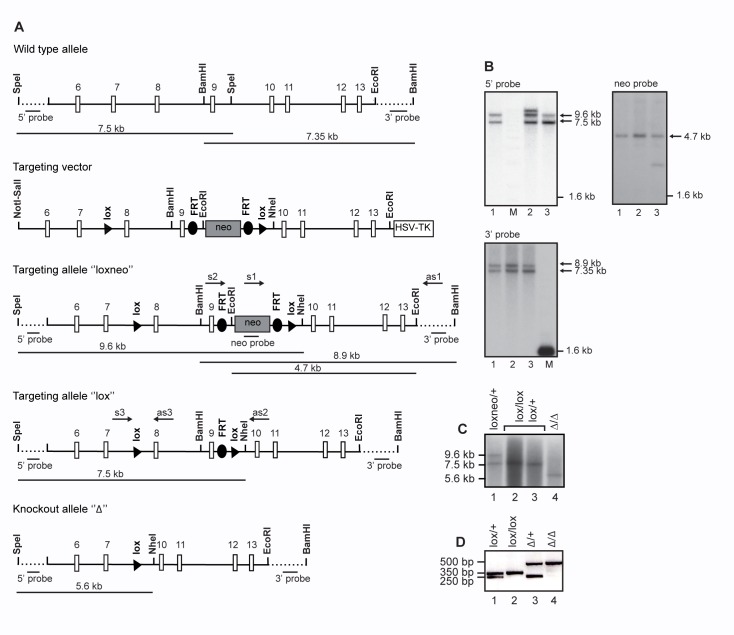
CAP2/Tmprss4, as analysed by quantitative RT-PCR analysis, shows high expression in epithelia like skin and whole digestive tract including duodenum and distal colon, moderate expression in eye, prostate and uterus, low expression in lung, bladder and liver and no detectable expression in whole organs such as heart, kidney, and testis (Fig 1). Homologous recombination in mouse embryonic stem (ES) cells was performed to position loxP sides around exons 8 and 9 of the CAP2/Tmprss4 gene locus containing the histidine and the aspartate of the catalytic triad (Fig 2A). Southern blot analyses confirmed correct targeting of ES cell clone #1, which was chosen to generate germline chimeras (Fig 2B). CAP2/Tmprss4 loxneo/+ mice were mated with Cre- or Flp-deleter mouse strains [31,30], and floxed CAP2/Tmprss4 (CAP2/Tmprss4 lox/+, CAP2/Tmprss4 loxlox), CAP2/Tmprss4 heterozygous mutant (CAP2/Tmprss4 Δ/+) and knockout (CAP2/Tmprss4 Δ/Δ) mice were obtained as evidenced by Southern blot (Fig 2C) and DNA-based PCR analyses (Fig 2D).
Fig 1. Distribution of wildtype CAP2/Tmprss4 mRNA transcript expression.
CAP2/Tmprss4 mRNA expression profile in WT mice (n = 4) in various organs as indicated; individual values were normalized to β-actin.
Fig 2. Inactivation of the CAP2/Tmprss4 gene locus.
(A) Scheme of the wild-type allele, the targeting vector, and the targeted CAP2/Tmprss4 loxneo allele following homologous recombination, and the CAP2/Tmprss4 lox and the CAP2/Tmprss4 Δ allele following breeding with Flp- and Cre-deleter mice, respectively. Relevant restriction enzymes for cloning and diagnosis of targeted ES cell clones are shown. Exons 8 and 9 and the neomycin cassette (flanked by frt sites) are flanked by loxP sites. 5’ and 3’ probes as well as PCR primers used for ES cell screening and mouse genotyping are indicated. (B) Southern blot analyses of targeted ES cell clones using the external 5’probe (upper left panel) following digestion with SpeI and NheI, the neo probe (upper right panel) following EcoRI digestion, and the external 3’probe following digestion with BamH1; note that clone #2 and #3 harbour additional recombination and integration events as evidenced by Southern blot analyses using the 5’ and neo probe, respectively. (C) Southern blot analysis of CAP2/Tmprss4 loxneo/+, CAP2/Tmprss4 lox/lox and/or CAP2/Tmprss4 lox/+ and CAP2/Tmprss Δ/Δ mice using the 5’ probe following SpeI/NheI digestion. (D) PCR-based genotyping of mice harbouring the wild type (+, 250bp, lane 1 and 3), lox alleles (lox, 350bp, lane 2) and knockout alleles (Δ, 500bp, lane 3 and 4).
CAP2/Tmprss4 knockout mice do not show an obvious phenotype
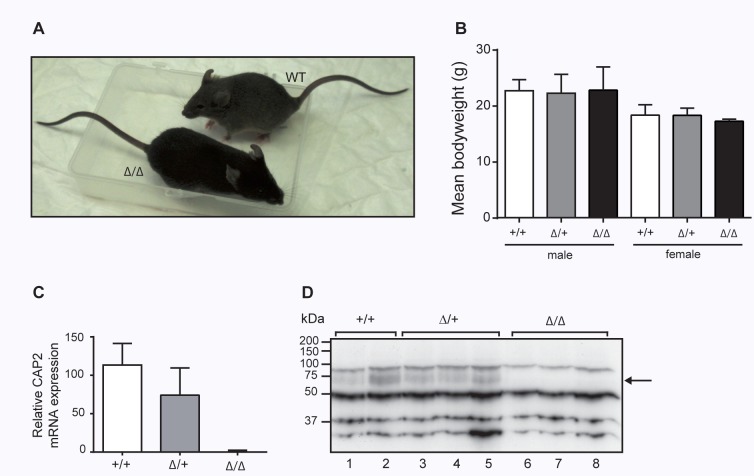
Following interbreeding of heterozygotes, CAP2/Tmprss4 wildtype (CAP2/Tmprss4 +/+) heterozygous mutant (CAP2/Tmprss4 Δ/+) and homozygous mutant (CAP2/Tmprss4 Δ/Δ) mice were born according to Mendelian ratio (272 pups: +/+, n = 92; Δ/+, n = 131; Δ/Δ, n = 49; P <0.1). CAP2/Tmprss4 knockout mice appeared healthy and were not affected in body weight (Fig 3A and 3B). CAP2/Tmprss4 knockout mice completely lacked mRNA transcript and protein expression as evidenced by qRT-PCR and Western blot analysis, while heterozygous CAP2/Tmprss4 mice showed intermediate expression levels (Fig 3C and 3D). Histopathology of skin, kidney, colon and lung from knockout mice did not reveal any deviation from wildtype or heterozygous mice (Fig 4). Analysis of 16 additional organs revealed no differences either (data not shown).
Fig 3. Phenotype of CAP2/Tmprss4-deficient mice.
(A) Representative pictures of 3 months old (male) CAP2/Tmprss4 wildtype (WT) and CAP2/Tmprss4 knockout (KO) littermates. (B) Mean body weight (g) of 3-month-old male and female wildtype (WT, n = 6), heterozygous mutant (HET, n = 11 and n = 9, respectively), and knockout (KO, n = 6 and n = 5, respectively) mice. (C) Relative CAP2/Tmprss4 mRNA transcript expression in colon from CAP2/Tmprss4 WT, CAP2/Tmprss4 HET and CAP2/Tmprss4 KO mice (n = 6 mice per group); β-actin is used as internal control. (D) Representative immunoblot showing the presence of a 70kDa CAP2/Tmprss4-specific band in colon extracts from CAP2/Tmprss4 WT (lane 1 and 2), CAP2/Tmprss4 HET (lane 3–5) mice and absence in CAP2/Tmprss4 KO (lane 6–8) mice; arrow indicates the size of the expected but absent CAP2/Tmprss4-specific band in knockouts.
Fig 4. Histopathological analysis in ENaC-expressing organs from CAP2/Tmprss4 knockout mice.
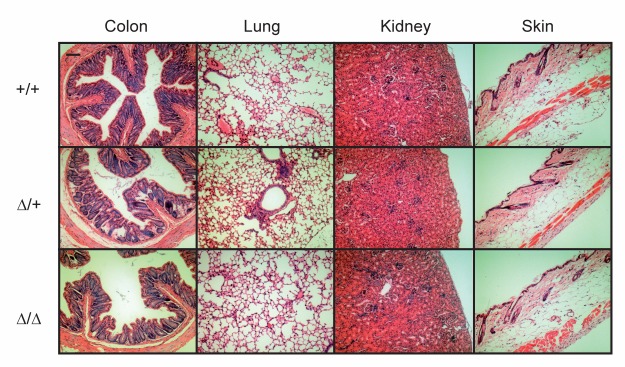
Representative H&E stained section of colon, lung, kidney and skin from CAP2/Tmprss4 wildtype (WT), heterozygous mutant (HET) and knockout (KO) mice; n = 2 females and 2 males for each group and genotype; bar indicates 100μm.
CAP2/Tmprss4 deletion does not affect ENaC expression and activity
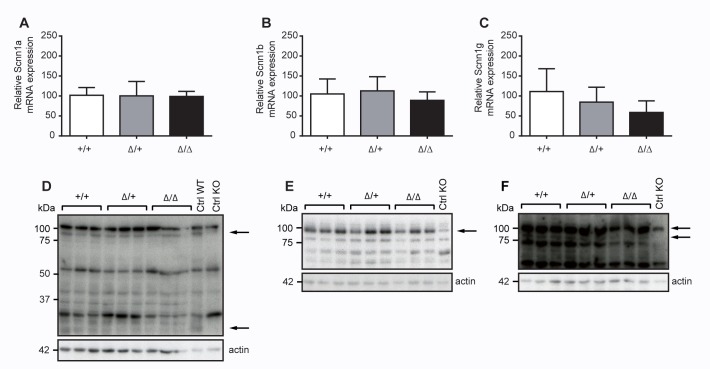
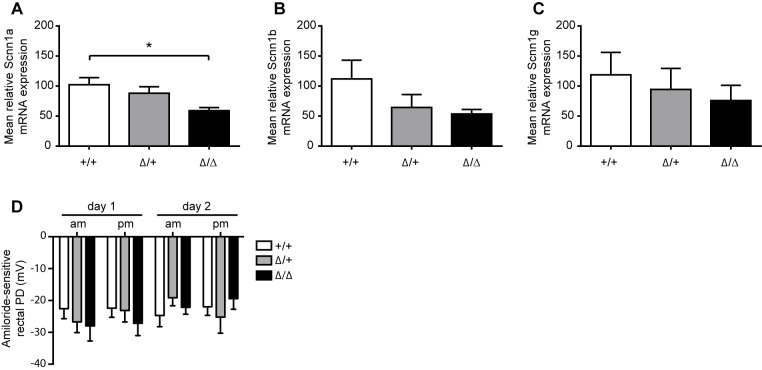
Since channel-activating proteases like CAP2/Tmprss4 are supposed to activate the amiloride-sensitive epithelial sodium channel, thereby possibly affecting the whole net sodium balance, we analysed furthermore renal mRNA transcript expression levels of ENaC subunits in CAP2/Tmprss4 knockout mice. Here, we could not find any changes between Scnn1a, Scnn1b and Scnn1g expression levels (Fig 5A, 5B and 5C). When we analysed ENaC subunit protein expression levels, not only the full-length Scnn1a, Scnn1b or Scnn1g proteins were equally expressed among the different CAP2/Tmprss4 genotypes, but cleaved Scnn1a (32kDa) and Scnn1g (70kDa) ENaC proteins were equally present and expressed (Fig 5D, 5E and 5F; data not shown). This also coincides with plasma sodium and potassium, and plasma aldosterone levels that were not significantly different among the genotypes upon regular sodium diet (Table 1) indicating that ENaC expression and activity is not affected in kidney. Furthermore, we could not observe any functional redundancy in renal mRNA transcript expression levels among CAP1/Prss8 and CAP3/ST-14 (Table 1).
Fig 5. ENaC mRNA transcript and protein expression in kidneys from CAP2/Tmprss4 wildtype (WT), heterozygous mutant (HET) and knockout (KO) mice under regular sodium diet.
(A-C) Relative mRNA transcript and (D-F) ENaC and β-actin protein expression in kidneys of (A) Scnn1a in CAP2/Tmprss4 wildtype (WT, n = 6), heterozygous mutant (HET, n = 7) and knockout (KO, n = 5) mice, (B) Scnn1b in CAP2/Tmprss4 wildtype (WT, n = 6), heterozygous mutant (HET, n = 7) and knockout (KO, n = 5) mice, and (C) Scnn1g in CAP2/Tmprss4 wildtype (WT, n = 6), heterozygous mutant (HET, n = 5) and knockout (KO, n = 5) mice; β-actin was used as internal control. Representative immunoblots of (D) Scnn1a, (E) Scnn1b and (F) Scnn1g and its corresponding β-actin protein expression in CAP2/Tmprss4 wildtype (WT), heterozygous mutant (HET) and knockout (KO) mice; kidney extracts from Scnn1 wildtype (WT) and knockout (KO) mice were used as positive and negative control respectively; arrows indicate the full-length and the corresponding cleaved ENaC fragments.
Table 1. Physiological parameters under regular salt diet or sodium-deficient diet.
| Parameters | Regular salt diet | ||
| WT | Δ/+ | Δ/Δ | |
| n | 6 | 7 | 5 |
| Body weight (g) | 22.70±0.77 | 22.46±1.59 | 20.49±1.77 |
| Plasma aldosterone (pg/ml) | 358.5±156.8 | 353.9±126.3 | 586.6±304.4 |
| Plasma sodium (mmol) | 143.15±1.43 | 145.18±0.66 | 143.65±0.60 |
| Plasma potassium (mmol) | 5.37±0.28 | 5.32±0.22 | 5.32±0.11 |
| CAP1, relative mRNA expression (% of control) | 100±18.37 | 97.96±14.78 | 99.44±17.10 |
| CAP3, relative mRNA expression (% of control) | 100±10.16 | 84.15±6.70 | 84.80±5.13 |
| Furin, relative mRNA expression (% of control) | n.d. | n.d. | n.d. |
| Sodium-deficient diet | |||
| WT | Δ/+ | Δ/Δ | |
| n | 5 | 5 | 4 |
| Body weight (g) | 25.75±1.36 | 25.58±1.48 | 23.44±2.44 |
| Plasma aldosterone (pg/ml) | 803.3±203.6 | 474.3±157.9 | 723.7±300.6 |
| Plasma sodium (mmol) | 152.20±1.83 | 155.94±1.97 | 156.28±3.34 |
| Plasma potassium (mmol) | 4.55±0.31 | 4.79±0.14 | 4.31±0.29 |
| CAP1, relative mRNA expression (% of control) | 100±9.96 | 119.09±10.06 | 72.11±4.56 |
| CAP3, relative mRNA expression (% of control) | 100±4.72 | 113.10±10.39 | 74.93±13.08 |
| Furin, relative mRNA expression (% of control) | 100±2.97 | 123.76±17.44 | 144.04±8.02 |
Physiological parameters of CAP2/Tmprss4 wildtype (WT), heterozygous mutant (HET, Δ/+) and knockout (KO, Δ/Δ) mice under regular sodium or sodium-deficient diets. Data are presented as mean ± SEM.
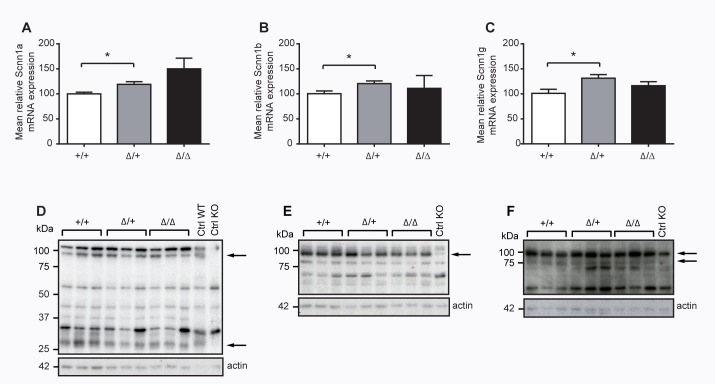
When challenging CAP2/Tmprss4 knockout mice with sodium-deficient diet, we unveiled no alteration in plasma sodium and potassium or plasma aldosterone levels (Table 1). Similarly, mRNA transcript expression levels of ENaC subunits did not differ between knockout and wildtype mice, even though we found a significant difference of ENaC subunit expression between wildtype and heterozygous mutant CAP2/Tmprss4 mice (Fig 6A, 6B and 6C). However, this difference could not be confirmed on protein levels and we found no difference in protein expression for full-length Scnn1a, Scnn1b and Scnn1g and cleaved Scnn1a and Scnn1g subunits (Fig 6D, 6E and 6F; data not shown). In colon, we found a significantly decreased mRNA transcript expression of Scnn1a in CAP2/Tmprss4 knockout mice under sodium restriction, while mRNA transcript expression of Scnn1b and Scnn1g did not significantly differ (Fig 7A, 7B and 7C). We finally tested in vivo ENaC activity upon sodium-deficiency in distal colon and determined the amiloride-sensitive rectal potential difference (ΔPDamil). This did not reveal an effect of CAP2/Tmprss4-deficiency on ENaC activity (Fig 7D) demonstrating that CAP2/Tmprss4 is not required for in vivo colonic ENaC activity.
Fig 6. ENaC mRNA transcript and protein expression in kidneys from CAP2/Tmprss4 wildtype (WT), heterozygous mutant (HET) and knockout (KO) mice under sodium-deficient diet.
(A-C) Relative mRNA transcript and (D-F) protein expression of (A) Scnn1a, (B) Scnn1b and (C) Scnn1g from CAP2/Tmprss4 wildtype (WT), heterozygous mutant (HET), and knockout (KO) mice; n = 4 for each group and genotype; β-actin was used as internal control. Representative immunoblots of (D) Scnn1a, (E) Scnn1b and (F) Scnn1g and its corresponding β-actin protein expression from CAP2/Tmprss4 wildtype (WT), heterozygous mutant (HET) and knockout (KO) mice (n = 5 for each group and genotype); kidney extracts from Scnn1 wildtype (WT) and knockout (KO) mice were used as positive and negative control respectively; arrows indicate the full-length and the corresponding cleaved ENaC fragments; * P< 0.05).
Fig 7. ENaC mRNA transcript expression and activity in colon from CAP2/Tmprss4 mice under sodium-deficient diet.
(A-C) Relative mRNA transcript expression of (A) Scnn1a, (B) Scnn1b and (C) Scnn1g from CAP2/Tmprss4 wildtype (WT, n = 4), heterozygous mutant (HET, n = 5), and knockout (KO, n = 4) mice; *P< 0.05); β-actin was used as internal control. (D) Morning and afternoon amiloride-sensitive rectal potential difference (PD) measurements at 10-12am and 4-6pm of two consecutive days in Tmprss4 wildtype (WT), heterozygous mutant (HET) and knockout (KO) mice; n = 4 for each group and genotype.
Discussion
In the present study, we generated constitutive knockout mice for CAP2/Tmprss4, targeting exons 8 and 9 that contain two out of three amino acids (histidine and aspartate) of the catalytic triad (Fig 2). Disruption of the CAP2/Tmprss4 gene locus and CAP2/Tmprss4-deficiency was verified at the genomic, mRNA transcript and protein expression level (Figs. 2 and 3).
The knockout mice seemed healthy and we detected no obvious effects on embryonic development or after birth (Fig 3), in contrary to the phenotype described for CAP2/Tmprss4 knockdown experiments in zebrafish embryos which exhibit severe defects in tissue development and cell differentiation including disturbed skeletal muscle formation, decelerated heartbeat, a degenerated vascular system, and impaired epidermal keratinocytes [39]. This strongly suggests a functional redundancy among serine proteases in the mammalian system, although we could not reveal any upregulation of other channel-activating proteases, such as CAP1/Prss8 (prostasin), CAP3/ST-14 (matriptase) or furin (Table 1). Absence of the channel-activating proteases such as CAP1/Prss8 leads to embryonic lethality due to placental failure [40]. Skin-specific conditional knockout of CAP1/Prss8 and a constitutive knockout of CAP3/ST-14 result in early postnatal lethality due to severe impaired skin barrier function [41,42].
Although target substrates of CAP2/Tmprss4 are largely unknown, in vitro experiments in Xenopus oocytes identified the amiloride-epithelial sodium channel ENaC as potential downstream target [7,24]. In vitro, in presence of CAP2/Tmprss4, the open probability (Po) of the amiloride-sensitive ENaC channel is significantly increased and can be blocked by preincubation of Xenopus oocytes [7] with aprotinin, an inhibitor of serine proteases [4,6]. Thereby, catalytic activity seems to be required since the mutation of the serine (S385) of the catalytic triad in CAP2/Tmprss4 completely inhibits ENaC activation in vitro [33]. Previously, in vitro experiments pointed to an implication of CAP2/Tmprss4 in Scnn1g and Scnn1a cleavage, and several putative cleavage sites including the Scnn1g furin (R138) site were reported to significantly reduce ENaC-mediated sodium current [24,43]. Moreover, a recent study confirmed that Scnn1g is processed proteolytically in human kidney [44]. Although CAP2/Tmprss4 mRNA expression was low in kidney, the protein was previously identified in a mouse cortical collecting duct cell line (mpkCCDC14) [7]. mRNA expression was confirmed in the same cell line but could not be detected in whole kidney, suggesting a low and localized expression of CAP2/Tmprss4 in kidney [7]. We thus concentrated on ENaC-expressing organs for histopathology, such as skin, lung, kidney and colon, but could not detect any alterations in CAP2/Tmprss4 knockout or heterozygous mice (Fig 4). We expected cleavage changes in Scnn1a and Scnn1g, but did not detect differences of the potentially cleaved 32kDa Scnn1a and 70kDa Scnn1g fragments in CAP2/Tmprss4 knockout mice, strongly suggesting that in vivo cleavage of ENaC is independent of CAP2/Tmprss4 (Fig 5).
It has been reported that dietary salt restriction promotes both cleavage and release of an imbedded inhibitory tract from the Scnn1g subunit, that could account for the increased Na+ absorption observed in rats on low Na+ diet [45–47]. When lowering dietary salt intake, ENaC activity is enhanced upon increased aldosterone secretion to preserve sodium homeostasis [48]. Even though significant increase of mRNA transcript levels for all three ENaC subunits was detected in kidney of heterozygous mutant, but not in knockout CAP2/Tmprss4 mice, protein levels for full-length and cleaved ENaC subunit forms were unchanged between genotypes. Body weight, plasma aldosterone, sodium and potassium were not changed (Fig 6 and Table 1), and no obvious compensation was detected when measuring mRNA levels for CAP1/Prss8, CAP3/ST-14 or furin (Table 1).
We cannot exclude that other proteases recently identified as in vitro potent ENaC activators, as trypsin IV [49] trypsin I [49] meprin β [50] or cathepsin B [51] might be implicated in in vivo ENaC activation. As CAP2/Tmprss4 mRNA expression level was high in wildtype colon, we investigated whether ENaC mRNA transcript expression could be affected in this organ (Fig 7). Although the significant decrease in mRNA transcript expression might be indicative for reduced colonic ENaC activity, monitoring of ENaC activity by amiloride-sensitive PD showed no difference between genotypes (Fig 7D). It has been shown that the activity of ENaC is not proportional to the amount of expressed ENaC protein levels, and that de novo synthesis of ENaC subunits might play an important role in channel regulation [52]. The detected protein pool (Figs. 5 and 6) represents the cytoplasmic as well as the plasma membrane pool of total proteins. Even when ENaC is located at the plasma membrane, the channel can remain silent and not active [9]. The unaltered ENaC activity is thus consistent with the measured physiological parameters such as plasma sodium and potassium, plasma aldosterone and the Scnn1a and Scnn1g protein cleavage pattern that was not altered in the CAP2/Tmprss4 knockout mice on sodium-deficient diet (Fig 6 and Table 1). CAP1/Prss8, in contrary, is implicated in in vivo activation of ENaC in colon as mutations in CAP1/Prss8 in frizzy mice and frCR rats [17], and the colon-specific CAP1/Prss8 knockout led to significant reduced amiloride-sensitive rectal PD and consequently to 2–3 times elevated plasma aldosterone levels to compensate fecal ENaC-mediated sodium loss via the activation of the renin-angiotensin-aldosterone (RAAS) system [13].
Target substrate specificity of CAP2/Tmprss4 under physiological conditions is still largely unknown, and no other target substrate than ENaC has so far been proposed in this area of research. Its implication in pathophysiological processes, however, becomes more evident. CAP2/Tmprss4 was found mutated in a new form of pediatric neurodegenerative disorder, termed Autosomal Recessive Cerebral Atrophy (ARCA), where a point mutation in the gene (c.995C>T) leads to severe CNS degeneration [23]. A role of CAP2/Tmprss4 in influenza virus spreading was proposed, mediated through proteolytic cleavage of the viral protein hemagglutinin (HA), although virus specificity has not been identified so far due to lack of a suitable knockout model [53,22,54]. Upregulation of CAP2/Tmprss4 is observed in various cancer types originating from pancreas, lung, breast, colon and stomach [18,55–60], and was found associated with poor prognosis in patients [59–63].
In conclusion, in this study, we generated and analysed CAP2/Tmprss4 knockout mice and demonstrate that the protease CAP2/Tmprss4 is not required for in vivo ENaC-mediated sodium regulation. We propose that these knockout mice can be used to determine the target substrate specificity and its further implication in physiological and pathophysiological processes.
Acknowledgments
We thank Bernard Rossier for continuous support on the project. We are grateful to Jean-Christophe Stehle and the mouse histology platform (University of Lausanne). We are also thankful for the help provided by the Transgenic Animal Facility (TAF, University of Lausanne) for the generation of the mouse line.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the Swiss National Science Foundation (http://www.snf.ch/fr/Pages/default.aspx) grant number 31003A_144198/1, to AK, DA, AMM, SM and EH, the National Center for Competence in Research (NCCR) Kidney Control of Homeostasis (Kidney.CH) (http://www.nccr-kidney.ch) to AK, AMM, JB, SM and EH, and Fondation Leducq, grant number 26077648 (https://www.fondationleducq.org) to AK, DA, AMM, SM and EH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rossier BC, Staub O, Hummler E. Genetic dissection of sodium and potassium transport along the aldosterone-sensitive distal nephron: importance in the control of blood pressure and hypertension. FEBS Lett. 2013;587: 1929–1941. 10.1016/j.febslet.2013.05.013 [DOI] [PubMed] [Google Scholar]
- 2. Canessa CM, Horisberger JD, Rossier BC. Epithelial sodium channel related to proteins involved in neurodegeneration. Nature 1993;361: 467–470. [DOI] [PubMed] [Google Scholar]
- 3. Canessa CM, Schild L, Buell G, Thorens B, Gautschi I, Horisberger JD, et al. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature 1994;367: 463–467. [DOI] [PubMed] [Google Scholar]
- 4. Vallet V, Chraibi A, Gaeggeler HP, Horisberger JD, Rossier BC. An epithelial serine protease activates the amiloride-sensitive sodium channel. Nature 1997;389: 607–610. [DOI] [PubMed] [Google Scholar]
- 5. Vallet V, Horisberger JD, Rossier BC. Epithelial sodium channel regulatory proteins identified by function expression cloning. Kidney Int. 1998;54: 109–114. [DOI] [PubMed] [Google Scholar]
- 6. Vuagniaux G, Vallet V, Fowler Jaeger N, Pfister C, Bens M, Farman N, et al. Activation of the amiloride-sensitive epithelial sodium channel by the serine protease mCAP1 expressed in a mouse cortical collecting duct cell line. J. Am. Soc. Nephrol. 2000;11: 828–834. [DOI] [PubMed] [Google Scholar]
- 7. Vuagniaux G, Vallet V, Fowler Jaeger N, Hummler E, Rossier BC. Synergistic activation of ENaC by three membrane-bound channel-activating serine proteases (mCAP1, mCAP2, and mCAP3) and serum- and glucocorticoid-regulated kinase (Sgk1) in Xenopus oocytes. J. Gen. Physiol. 2002;120: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vallet V, Pfister C, Loffing J, Rossier BC. Cell-surface expression of the channel activating protease xCAP-1 is required for activation of ENaC in the Xenopus oocyte. J. Am. Soc. Nephrol. 2002;13: 588–594. [DOI] [PubMed] [Google Scholar]
- 9. Caldwell RA, Boucher RC, Stutts MJ. Serine protease activation of near-silent epithelial Na+ channels. Am. J. Physiol. Cell Physiol. 2004;286: 190–194. [DOI] [PubMed] [Google Scholar]
- 10. Planès C, Leyvraz C, Uchida T, Apostolova Angelova M, Vuagniaux G, Hummler E, et al. In vitro and in vivo regulation of transepithelial lung alveolar sodium transport by serine proteases. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005;288: L1099–L1109. [DOI] [PubMed] [Google Scholar]
- 11. Planès C, Randrianarison NH, Charles RP, Frateschi S, Cluzeaud F, Vuagniaux G, et al. ENaC-mediated alveolar fluid clearance and lung fluid balance depend on the channel-activating protease 1. EMBO Mol. Med. 2010;2: 26–37. 10.1002/emmm.200900050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Donaldson SH, Hirsh A, Li DC, Holloway G, Chao J, Boucher R, et al. Regulation of the epithelial sodium channel by serine proteases in human airways. J. Biol. Chem. 2002;277: 8838–8345. [DOI] [PubMed] [Google Scholar]
- 13. Malsure S, Wang Q, Charles RP, Sergi C, Perrier R, Christensen BM, et al. Colon-specific deletion of epithelial sodium channel causes sodium loss and aldosterone resistance. J. Am. Soc. Nephrol. 2014;25: 1453–1464. 10.1681/ASN.2013090936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falconer DS, Snell GD. Two new hair mutants, rough and frizzy. J. Hered. 1952;43: 53–57. [Google Scholar]
- 15. Panteleyev AA, Christiano AM. The Charles River "Hairless" rat mutation is distinct from the Hairless mouse allele. Comp. Med. 2001;51: 49–55. [PubMed] [Google Scholar]
- 16. Spacek DV, Perez AF, Ferranti KM, Wu LKL, Moy DM, Magnan DR, et al. The mouse frizzy (fr) and rat "hairless" (frCR) mutations are natural variants of protease serine S1 family member 8 (Prss8). Exp. Dermatol. 2010;19: 527–532. 10.1111/j.1600-0625.2009.01054.x [DOI] [PubMed] [Google Scholar]
- 17. Frateschi S, Keppner A, Malsure S, Iwaszkiewicz J, Sergi C, Mérillat AM, et al. Mutations of the serine protease CAP1/Prss8 lead to reduced embryonic viability, skin defects, and decreased ENaC activity. Am. J. Pathol. 2012;181: 605–615. 10.1016/j.ajpath.2012.05.007 [DOI] [PubMed] [Google Scholar]
- 18. Wallrapp C, Hähnel S, Müller-Pillasch F, Burghardt B, Iwamura T, Ruthenbürger M, et al. A novel transmembrane serine protease (TMPRSS3) overepressed in pancreatic cancer. Cancer Res. 2000;60: 2602–2606. [PubMed] [Google Scholar]
- 19. Hooper JD, Clements JA, Quigley JP, Antalis TM. Type II transmembrane serine proteases: insights into an emerging class of cell surface proteolytic enzymes. J. Biol. Chem. 2001;276: 857–860. [DOI] [PubMed] [Google Scholar]
- 20. Bugge TH, Antalis TM, Wu Q. Type II Transmembrane serine proteases. J. Biol. Chem. 2009;284: 23177–23181. 10.1074/jbc.R109.021006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim S, Lee JW. Membrane Proteins Involved in Epithelial-Mesenchymal Transition and Tumor Invasion: Studies on TMPRSS4 and TM4SF5. Genomics Inform. 2014;12: 12–20. 10.5808/GI.2014.12.1.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bertram S, Glowacka I, Blazejewska P, Soilleux E, Allen P, Danisch S, et al. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of Influenza virus in Caco-2 cells. J. Virol. 2010;84: 10016–10025. 10.1128/JVI.00239-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lahiry P, Racacho L, Wang J, Robinson JF, Gloor GB, Rupar CA, et al. A mutation in the serine protease TMPRSS4 in a novel pediatric neurodegenerative disorder. Orphanet J. Rare Dis. 2013;8: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garcia-Caballero A, Dang Y, He H, Stutts MJ. ENaC proteolytic regulation by channel-activating protease 2. J. Gen. Physiol. 2008;132: 521–535. 10.1085/jgp.200810030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hughey RP, Mueller GM, Bruns JB, Kinlough CL, Poland PA, Harkleroad KL, et al. Maturation of the epithelial Na+ channel involves proteolytic processing of the alpha- and gamma-subunits. J. Biol. Chem. 2003;272: 37073–37082. [DOI] [PubMed] [Google Scholar]
- 26. Hughey RP, Bruns JB, Kinlough CL, Harkleroad KL, Tong Q, Carttino MD, et al. Epithelial sodium channels are activated by furin-dependent proteolysis. J. Biol. Chem. 2004;279: 18111–18114. [DOI] [PubMed] [Google Scholar]
- 27. Harris M, Garcia-Caballero A, Stutts MJ, Firsov D, Rossier BC. Preferential assembly of epithelial sodium channel (ENaC) subunits in Xenopus oocytes: role of furin-mediated endogenous proteolysis. J. Biol. Chem. 2008;283: 7455–7463. 10.1074/jbc.M707399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, et al. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 2001;414: 768–773. [DOI] [PubMed] [Google Scholar]
- 29. Porret A, Mérillat AM, Guichard S, Beermann F, Hummler E. Tissue-specific transgenic and knockout mice. Methods Mol. Biol. 2006;337: 185–205. [DOI] [PubMed] [Google Scholar]
- 30. Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25: 139–140. [DOI] [PubMed] [Google Scholar]
- 31. Buchholz F, Refaeli Y, Trumpp A, Bishop JM. Inducible chromosomal translocation of AML1 and ETO genes through Cre/loxP-mediated recombination in the mouse. EMBO Rep. 2000;1: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rubera I, Loffing J, Palmer LG, Frindt G, Fowler Jaeger N, Sauter D, et al. Collecting duct-specific gene inactivation of alphaENaC in the mouse kidney does not impair sodium and potassium balance. J. Clin. Invest. 2003;112: 554–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andreasen D, Vuagniaux G, Fowler Jaeger N, Hummler E, Rossier BC. Activation of epithelial sodium channels by mouse channel activating proteases (mCAP) expressed in Xenopus oocytes requires catalytic activity of mCAP3 and mCAP2 but not mCAP1. J. Am. Soc. Nephrol. 2006;17: 968–976. [DOI] [PubMed] [Google Scholar]
- 34. Hummler E, Mérillat AM, Rubera I, Rossier BC, Beermann F. Conditional gene targeting of the Scnn1a (alphaENaC) gene locus. Genesis 2002;32: 169–172. [DOI] [PubMed] [Google Scholar]
- 35. Traykova-Brauch M, Schönig K, Greiner O, Miloud T, Jauch A, Bode M, et al. An efficient and versatile system for acute and chronic modulation of renal tubular function in transgenic mice. Nat. Med. 2008;14: 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mérillat AM, Charles RP, Porret A, Maillard M, Rossier BC, Beermann F, et al. Conditional gene targeting of the ENaC subunit genes Scnn1b and Scnn1g. Am. J. Physiol. Renal Physiol. 2009;296: 249–256. [DOI] [PubMed] [Google Scholar]
- 37. Christensen BM, Perrier R, Wang Q, Zuber AM, Maillard M, Mordasini D, et al. Sodium and potassium balance depends on αENaC expression in connecting tubule. J. Am. Soc. Nephrol. 2010;21: 1942–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Q, Horisberger JD, Maillard M, Brunner HR, Rossier BC, Burnier M. Salt- and angiotensin II-dependent variations in amiloride-sensitive rectal potential difference in mice. Clin. Exp. Pharmacol. Physiol. 2000;27: 60–66. [DOI] [PubMed] [Google Scholar]
- 39. Ohler A, Becker-Pauly C. Morpholino knockdown of the ubiquitously expressed transmembrane serine protease TMPRSS4a in zebrafish embryos exhibits severe defects in organogenesis and cell adhesion. Biol. Chem. 2011;392: 653–664. [DOI] [PubMed] [Google Scholar]
- 40. Hummler E, Dousse A, Rieder A, Stehle JC, Rubera I, Osterheld MC, et al. The channel-activating protease CAP1/Prss8 is required for placental labyrinth maturation. PLoS One. 2013;8: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Leyvraz C, Charles RP, Rubera I, Guitard M, Rotman S, Breiden B, et al. The epidermal barrier function is dependent on the serine protease CAP1/Prss8. J. Cell Biol. 2005;170: 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. List K, Haudernschild CC, Szabo R, Chen W, Wahl SM, Swaim W, et al. Matriptase/MT-SP1 is required for postnatal survival, epidermal barrier function, hair follicle development, and thymic homeostasis. Oncogene 2002;21: 3765–3779. [DOI] [PubMed] [Google Scholar]
- 43. Passero CJ, Mueller GM, Myerburg MM, Carattino MD, Hughey RP, Kleyman TR. TMPRSS4-dependent activation of the epithelial sodium channel requires cleavage of the γ-subunit distal to the furin cleavage site. Am. J. Physiol. Renal Physiol. 2012;302: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zachar RM, Skjødt K, Marcussen N, Walter S, Toft A, Nielsen MR, et al. The epithelial sodium channel γ-subunit is processed proteolytically in human kidney. J. Am. Soc. Nephrol. 2015;26: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carattino MD, Mueller GM, Palmer LG, Frindt G, Rued AC, Hughey RP, et al. Prostasin interacts with the epithelial Na+ channel and facilitates cleavage of the γ-subunit by a second protease. Am. J. Physiol. Renal Physiol. 2014;307: 1080–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bruns JB, Carattino MD, Sheng S, Maarouf AB, Weisz OA, Pilewski JM, et al. Epithelial Na+ channels are fully activated by furin- and prostasin-dependent release of an inhibitory peptide from the gamma-subunit. J. Biol. Chem. 2007;282: 6153–6160. [DOI] [PubMed] [Google Scholar]
- 47. Carattino MD, Hughey RP, Kleyman TR. Proteolytic processing of the epithelial sodium channel gamma subunit has a dominant role in channel activation. J. Biol. Chem. 2008;283: 25290–25295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Loffing J, Korbmacher C. Regulated sodium transport in the renal connecting tubule (CNT) via the epithelial sodium channel (ENaC). Pflugers Arch. 2009;458: 111–135. 10.1007/s00424-009-0656-0 [DOI] [PubMed] [Google Scholar]
- 49. Haerteis S, Krappitz A, Krappitz M, Murphy JE, Bertog M, Krueger B, et al. Proteolytic activation of the human epithelial sodium channel by trypsin IV and trypsin I involves distinct cleavage sites. J. Biol. Chem. 2014;289: 19067–19078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Garcia-Caballero A, Ishmael SS, Dang Y, Gillie D, Bond JS, Milgram SL, et al. Activation of the epithelial sodium channel by the metalloprotease meprin β subunit. Channels (Austin) 2011;5: 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tan CD, Hobbs C, Sameni M, Sloane BF, Stutts MJ, Tarran R. Cathepsin B contributes to Na+ hyperabsorption in cystic fibrosis airway epithelial cultures. J. Physiol. 2014;592: 5251–5268. 10.1113/jphysiol.2013.267286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. May A, Puoti A, Gaeggeler HP, Horisberger JD, Rossier BC. Early effect of aldosterone on the rate of synthesis of the epithelial sodium channel alpha subunit in A6 renal cells. J. Am. Soc. Nephrol. 1997;8: 1813–1822. [DOI] [PubMed] [Google Scholar]
- 53. Chaipan C, Kobasa D, Bertram S, Glowacka I, Steffen I, Tsegaye TS, et al. Proteolytic activation of the 1918 Influenza virus hemagglutinin. J. Virol. 2009;83: 3200–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bahgat MM, Blazejewska P, Schughart K. Inhibition of lung serine proteases in mice: a potentially new approach to control influenza infection. Virol. J. 2011;8: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Larzabal L, Nguewa PA, Pio R, Blanco D, Sanchez B, Rodriguez MJ, et al. Overexpression of TMPRSS4 in non-small cell lung cancer is associated with poor prognosis in patients with squamous histology. Br. J. Cancer. 2011;105: 1608–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nguyen TH, Weber W, Havari E, Connors T, Bagley RG, McLaren R, et al. Expression of TMPRSS4 in non-small cell lung cancer and its modulation by hypoxia. Int. J. Oncol. 2012;41: 829–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cheng D, Kong H, Li Y. TMPRSS4 as a poor prognostic factor for triple-negative breast cancer. Int. J. Mol. Sci. 2013;14: 14659–14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Huang A, Zhou H, Zhao H, Quan Y, Feng B, Zheng M. High expression level of TMPRSS4 predicts adverse outcomes of colorectal cancer patients. Med. Oncol. 2013;30 [DOI] [PubMed] [Google Scholar]
- 59. Huang A, Zhou H, Zhao H, Quan Y, Feng B, Zheng M. TMPRSS4 correlates with colorectal cancer pathological stage and regulates cell proliferation and self-renewal ability. Cancer Biol. Ther. 2014;15: 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Luo ZY, Wang YY, Zhao ZS, Li B, Chen JF. The expression of TMPRSS4 and Erk1 correlates with metastasis and poor prognosis in Chinese patients with gastric cancer. PLoS One 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dai W, Zhou Q, Xu Z, Zhang E. Expression of TMPRSS4 in patients with salivary adenoid cystic carcinoma: correlation with clinicopathological features and prognosis. Med. Oncol. 2013;30. [DOI] [PubMed] [Google Scholar]
- 62. Wu XY, Zhang L, Zhang KM, Zhang MH, Ruan TY, Liu CY, et al. Clinical implication of TMPRSS4 expression in human gallbladder cancer. Tumour Biol. 2014;35: 5481–5486. 10.1007/s13277-014-1716-4 [DOI] [PubMed] [Google Scholar]
- 63. Sheng H, Shen W, Zeng J, Xi L, Deng L. Prognostic significance of TMPRSS4 in gastric cancer. Neoplasma 2014;61: 213–217. 10.4149/neo_2014_027 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.