Highlights
-
•
Mumps vaccines contain live attenuated viruses that are manufactured in cell substrates.
-
•
The production of the JL-CK vaccine in primary canine kidney cells is atypical.
-
•
Genetic changes introduced by different cell types have not been investigated.
-
•
JL-CK vaccine contains a unique mix of mumps viruses that have acquired a number of mutations.
-
•
Growth in cell or animal substrates dramatically alters the population dynamic of JL-CK.
Keywords: Mumps virus, Jeryl Lynn, Vaccines, Deep sequencing, Neurovirulence
Abstract
Mumps vaccines are live attenuated viruses. They are known to vary in effectiveness, degree of attenuation and adverse event profile. However, the underlying reasons are poorly understood. We studied two closely related mumps vaccines which originate from the same attenuated Jeryl Lynn-5 strain but have different efficacies. Jeryl Lynn-Canine Kidney (JL-CK), produced on primary canine kidney cells, is less effective than RIT4385, which is produced on chicken embryo fibroblasts. JL-CK and RIT4385 could be distinguished by a number of in vitro and in vivo properties. JL-CK produced heterogeneous, generally smaller plaques than RIT4385, but gave 100-fold higher titres when grown in cells and showed a higher degree of hydrocephalus formation in neonatal rat brains. Sanger sequencing of JL-CK identified 14 regions of heterogeneity throughout the genome. Plaque purification of JL-CK demonstrated the presence of five different Jeryl Lynn-5 variants encompassing the 14 mutations. One JL-CK mutation was associated with a small plaque phenotype, the effects of the others in vitro or in vivo were less clear. Only 4% of the JL-CK population corresponded to the parental Jeryl Lynn-5 strain. Next generation sequencing of JL-CK and virus before and after growth in cell lines or neonatal rat brains showed that propagation in vitro or in vivo altered the population dramatically. Our findings indicate that growth of JL-CK in primary canine kidney cells resulted in the selection of a mixture of mumps virus variants that have different biological properties compared to the parent Jeryl Lynn-5 virus. We also report three previously unknown heterogenic regions within the N gene of the RIT4385 vaccine.
1. Introduction
Mumps is caused by infection with a negative-strand RNA paramyxovirus. Disease is characterised by pain and swelling of the parotid gland(s) [1]. Mumps is preventable by vaccination with live mumps viruses (MuV) that have been attenuated by passage in various cell substrates [2–6]. MuV vaccine strains vary in protective efficacy, degree of attenuation [7] and adverse event profiles [8–11]. A correlate of protection in terms of neutralising antibody has not been established [12] and an animal model which mimics human disease has yet to be identified [13–16].
The Jeryl Lynn vaccine strain is most widely used [1]. Attenuated by serial passage in embryonated hens’ eggs and chicken embryo culture [17], it has been shown to be a mixture of two distinct but related MuVs, JL-2 and JL-5 [18–20]. The Jeryl Lynn-5 (JL-5) component was passaged in chicken embryo fibroblasts (CEFs) to produce RIT4385 [21], which has the same efficacy as the original Jeryl Lynn vaccine [12] and a seroconversion rate of ∼94% [21].
The Jeryl Lynn-Canine Kidney (JL-CK) vaccine is also derived from the Jeryl Lynn vaccine [22] but was passaged in primary canine kidney cells [4,23], which is atypical for MuV vaccine production. Seroconversion after JL-CK vaccination was only 75% 6–8 weeks post-vaccination [22], increasing to 82% after 35 months [4]. JL-CK has been associated with vaccine failure [24,25]. A large mumps outbreak occurred in the Czech Republic between January 2005 and June 2006 [26], despite high vaccination coverage with JL-CK, suggesting the vaccine may be over-attenuated [7].
MuV adaptation to cell substrates is well known and we hypothesised that propagation in primary canine kidney cells may have altered the virus. We compared the JL-CK and RIT4385 vaccine viruses to identify differences. We report genotypic and phenotypic variations between the two viruses, despite their shared origin. We identified mutations and distinct virus subpopulations within the JL-CK vaccine, which are likely a result of the virus’ passage in primary canine kidney cells. We highlight that propagation of the JL-CK vaccine virus, both in vitro and in vivo, alters the composition of the virus subpopulations, selecting certain MuV subpopulations over others. We also describe previously unknown mutations in the RIT4385 vaccine.
2. Materials and methods
2.1. Cell lines and viruses
African green monkey (Vero) and Madin Darby canine kidney (MDCK) cell lines were used as substrates for virus growth. JL-CK vaccine was provided by the Official Medicines Control Laboratory-Prague, Czech Republic. Mu90, a clinical mumps isolate passaged four times on Vero cells, and RIT4385 were archived material held at NIBSC. Infected cells were incubated at 35 °C, 5% CO2.
2.2. In vitro growth kinetics
Vero cells were inoculated, incubated and viable cells measured using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega). Vero or MDCK cells were inoculated in triplicate at a multiplicity of infection (MOI) of 0.0001, incubated and one plate removed daily for 7 days before clarification by centrifugation and titration on Vero cells.
2.3. Titration and plaque isolation of viruses
Viruses were titrated by plaque assay on Vero cells [27] with a 10 day incubation period before fixation/staining with methyl violet. Virus titres were calculated as plaque-forming-units (pfu) per ml. Plaque sizes were compared using ImageJ software [28,29]. JL-CK plaques (n = 81) were picked to a confluent monolayer of Vero cells and grown until cytopathic effect observed.
2.4. RNA extraction, RT-PCR and Sanger sequencing
Viral RNA was extracted using Trizol® LS reagent (Life Technologies). RT-PCR assays were performed using QIAGEN® One-Step RT-PCR Master mix (primers available upon request), amplicons purified by QIAquick® Gel Extraction and sequenced (Eurofins Genomics, GmBH). Sequencing data was analysed using Lasergene 8.0 – SeqMan (DNASTAR) and ClustalW (EMBL-EBI).
2.5. Mass spectrometry (MS) and sequence analysis
JL-CK and RIT4385 were grown in Vero cells for 3 days, sucrose gradient purified, subjected to SDS-PAGE before staining with Colloidal Blue (Life Technologies). Gel bands within 76–52 kDa (JL-CK, n = 2; RIT4385, n = 3) were excised, washed and trypsin-digested. Tryptic peptides were extracted and subjected to online nano-LC/MS/MS analysis using a LTQ-Orbitrap mass spectrometer (Thermo Scientific, UK) [30]. Mass spectra were processed and database searched using Proteome Discoverer v.1.2 (Thermo Scientific) and UniProt FASTA database. A FASTA format of the JL-CK nucleoprotein with additional amino acids at the C-termini (HANTFPNNPNQNAQLQVGDWDEQITDMIKP/KLTPIATIPGQSSHS) was created.
2.6. cDNA synthesis, PCR and deep sequencing
Random hexamer cDNA was synthesised by Superscript III First Strand cDNA Synthesis (Life Technologies). Fusion primers were designed with universal tails (available upon request) to generate targeted amplicons using Phusion® High-Fidelity PCR Master mix (New England Biolabs). Amplicons were purified with Agencourt AMPure XP Beads (Beckman-Coulter) and quantified using Agilent High Sensitivity DNA assay. Purified amplicons were pooled in equimolar concentration and libraries prepared by Nextera® XT DNA Sample preparation Kit (Illumina), before analysis on an Illumina MiSeq Sequencer.
2.7. Neurovirulence testing
Neonatal rat pups were inoculated intracerebrally [31] with 100 pfu either JL-CK, RIT4385, Mu90 or JL-CK variants. After 25 days, brains were removed and fixed in 10% neutral-buffered formalin. Brains were processed and stained with haematoxylin/eosin (Propath UK Limited) [29]. Hydrocephalus severity was calculated as a percentage of the cross-sectional area of the ventricle from the area of the whole brain using ImageJ [28]. All animal procedures were performed in accordance with Institutional guidelines.
2.8. Statistical analyses
The in vitro growth data and in vivo neurovirulence data were analysed using Student's t test. The error bars represented the standard error of the mean for each data point with a p value <0.05 considered significant.
2.9. Accession numbers
The JL-CK variants have been designated GenBank® accession numbers. JL-CK1, JQ946550; JL-CK2, JQ946551; JL-CK3, JQ946552; JL-CK4, JQ946553 and JL-CK5, JQ946554.
3. Results
3.1. In vitro growth of JL-CK and RIT4385
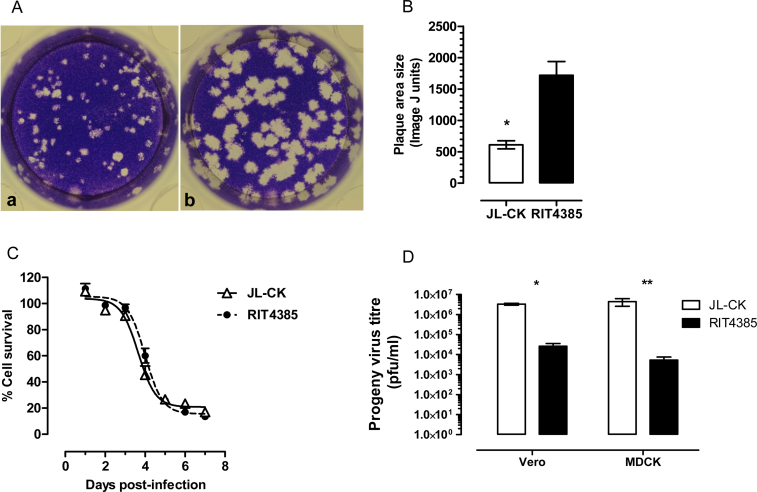
Titration of MuV is routinely performed on the interferon-deficient Vero cell line [27]. Plaques produced by JL-CK in Vero cells were at least three times smaller than those formed by RIT4385 (Fig. 1A and B). JL-CK and RIT4385 growth and virus yield was measured in both simian (Vero) and canine kidney (MDCK) cells. Fig. 1C demonstrates JL-CK and RIT4385 lysed Vero cells at similar rates, with 50% lysis observed at 3.65 or 3.99 days, respectively. By day 7, JL-CK produced 100-fold more progeny virus in Vero (p < 0.0001) and MDCK cells (p = 0.0365), Fig. 1D.
Fig. 1.
(A) Plaque morphology after infection of Vero cells. Panel a represents JL-CK and Panel b represents RIT4385. JL-CK produced a mixture of small and large plaques whereas RIT4385 formed a more homogeneous population of large plaques. (B) Measurement of plaque size of JL-CK and RIT4385 using ImageJ. The data shown is from JL-CK plaques (n = 25) and RIT4385 plaques (n = 33) (*p < 0.0001). (C) Growth kinetics of JL-CK and RIT4385 in Vero cells. JL-CK and RIT4385 were inoculated at MOI 0.0001 and lysed Vero cells at comparable rates. Viability of infected cells was expressed as the percentage of uninfected cells. Sigmoidal curves were fitted to the data points to calculate 50% cell lysis using GraphPad Prism 5 software (3.65 days for JL-CK and 3.99 days for RIT4385). The 95% confidence intervals overlapped (JL-CK, 3.89–4.42, RIT4385, 3.85–4.15 days). (D) JL-CK and RIT4385 progeny virus titres in Vero and MDCK cells. Infections were performed in two independent experiments each in triplicate, cells were infected at MOI 0.0001 and progeny viruses were titred in Vero cells after 7 days. JL-CK produced more progeny viruses on both Vero and MDCK cells (*p < 0.0001, **p = 0.0365), compared with RIT4385 on the same cell line (unpaired, two-tailed t-test).
3.2. Isolation and sequence analysis of JL-CK virus variants
In order to identify genomic differences between the two viruses, we sequenced unpassaged JL-CK and aligned it against the published sequence of RIT4385. Thirteen positions throughout the JL-CK genome displayed polymorphic nucleotides, resulting in 10 predicted amino acid changes (nucleoprotein: E108D, V529A, Ter550Q which extends the nucleoprotein by 21 amino acids and incorporates L557P, phosphoprotein/V/I: N56T, matrix: C244R, fusion: P93S and F295S based on two changes in adjacent nucleotides, haemagglutinin-neuraminidase: E569K and large: D311G), one silent mutation (L542L) and one mutation in the leader sequence (G/Ant92). We found no polymorphisms in the gene encoding the small hydrophobic (SH) protein, which is highly variable between mumps strains [32]. This indicated the presence of multiple JL-5 viruses, consistent with the mixed plaque morphology in Fig. 1A. We confirmed that the elongated nucleoprotein incorporating Ter550Q is expressed at the protein level. Using mass spectrometry, we identified the C-terminus of the extended protein (LTPIATIPGQSSHS) in the JL-CK vaccine, but not in RIT4385, Table S1. The L557P mutation, which is located in the same peptide of the elongated nucleoprotein of variant JL-CK5 (PTPIATIPGQSSHS), was not detected due to the proline disrupting the trypsin digestion motif.
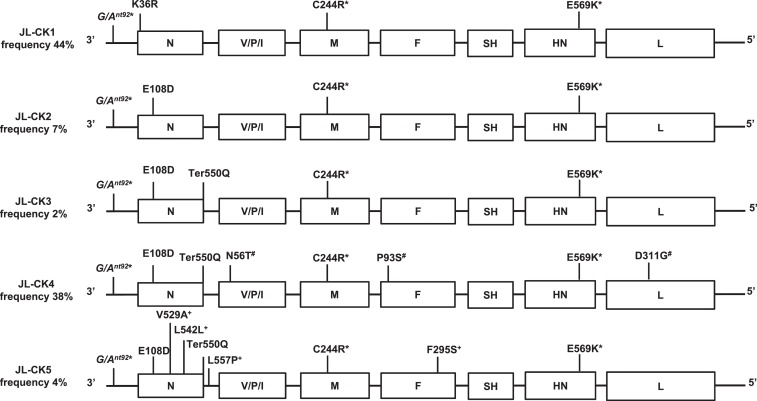
Plaque purification and sequencing the polymorphic regions in 81 plaques from JL-CK confirmed the presence of the mutations and also identified an additional mutation leading to a change in the nucleoprotein (K36R), Table S2. We identified five virus variants (JL-CK1, JL-CK2, JL-CK3, JL-CK4 and JL-CK5) plus a parent-like variant, which was identical to the published sequence of RIT4385 and found at only 4% frequency in the population. The predicted amino acid changes, silent mutation and mutation in the non-coding leader sequence in each variant, compared to the RIT4385 sequence are depicted in Fig. 2.
Fig. 2.
Schematic representation of the JL-CK virus variants plaque-purified from the JL-CK vaccine. The variants are numbered JL-CK1-5 in order of increasing number of mutations. Locations where the sequence differs from the published RIT4385 genome are indicated. Those positions on the genome which are linked are marked *(G/Ant92, C244R and E569K), #(N56T, P93S, D311G) and +(V529A, L542L, L557P, F295S). The K36R mutation was identified in plaques, it was not identified by Sanger sequencing of the unpassaged JL-CK vaccine. The Ter550Q mutation results in an extended nucleoprotein open reading frame, incorporating the L557P mutation. The ratio of the different subpopulations is expressed as a percentage of the 81 plaques isolated from Vero cells and Sanger sequenced.
The G/Ant92, C244R and E569K mutations are common to all variants. Each variant has in addition a variety of the other mutations. The parent-like virus represents only 4% of the population (not shown). Some mutations are unique to one variant; K36R in JL-CK1; N56T/P93S/D311G in JL-CK4 and V529A/L542L/L557P/F295S in JL-CK5. The E108D mutation is found in variants JL-CK2-5 and Ter550Q in variants JL-CK3-5 (Fig. 2).
3.3. Deep sequencing of JL-CK and RIT4385
To identify whether the plaque-purified virus populations are represented at similar ratios in unpassaged JL-CK, we performed amplicon-based deep sequencing of JL-CK and RIT4385 vaccines. The frequency of each mutation determined by plaque purification (JL-CK) and deep sequencing of unpassaged virus (JL-CK and RIT4385) is shown in Table 1. The mutations unique to JL-CK5 (V529A, L542L, L557P, F295S) all reduced from 26% (unpassaged) to 4% in plaques. The K36R mutation, unique to JL-CK1, increased from undetectable (Sanger) and 9.1% (deep sequencing) in unpassaged virus to 46% in plaques. JL-CK4 represented 38% of the plaques isolated and JL-CK4 mutations (N56T/P93S/D311G) occurred at similar frequencies in the unpassaged virus (40.5, 45.3 and 43.4%, respectively). E108D, found in all variants other than JL-CK1, was identified at 51% in plaques but at 86.6% in unpassaged virus. The G/Ant92/C244R/E569K mutations present in all JL-CK variants occurred as frequently in the unpassaged vaccine as in plaques, as did the N56T/P93S/D311G mutations (JL-CK4).
Table 1.
Mutation frequencies identified by deep sequencing of JL-CK and RIT4385 compared to Sanger sequencing of JL-CK plaques. Unpassaged virus and virus isolated from JL-CK plaques, 10 days post-infection (d.p.i.) are shown in the top section. Virus in Vero and MDCK cell lysates, 3 d.p.i. are compared in the middle section. Mutation frequencies identified by deep sequencing of neonatal rat brains 4 d.p.i. with JL-CK virus, are shown in the lower section.
| Substrate | Virus | Mutation frequency (%) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Leader | K36R | E108D | V529A | L542L | Ter550Q | L557P | N56T | C244R | P93S | F295S | E569K | D311G | ||
| Unpassaged | JL-CK | 96.7 | 9.1 | 86.6 | 27 | 26.2 | 74.2 | 26.5 | 40.5 | 97.5 | 45.3 | 27.3 | 96.9 | 43.4 |
| RIT4385 | n.d. | n.d. | n.d. | 6.2 | 16.6 | n.d. | 21.3c | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |
| Vero Plaques# |
JL-CK | 98a | 46b | 51 | 4 | 4 | 44 | 4 | 38 | 96 | 38 | 4 | 96 | 38 |
| Vero* 3 d.p.i. |
JL-CK | 94.5 | 27.0 | 70.5 | 10.4 | 9.5 | 48.0 | 9.7 | 42.2 | 95.6 | 39.9 | 11.0 | 95.8 | 35.1 |
| RIT4385 | n.d. | n.d. | n.d. | 11.6 | 21.5 | n.d. | 30.8 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |
| MDCK* 3 d.p.i. |
JL-CK | 84.1 | 18.8 | 68.3 | 23.3 | 23.0 | 53.1 | 23.2 | 31.7 | 89.5 | 31.0 | 26.3 | 89.7 | 29.5 |
| RIT4385 | n.d. | n.d. | n.d. | 2.9 | 31.6 | n.d. | 33.1 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |
| RNV1 | JL-CK | 100 | 69.6 | 33.4 | 1.3 | 1.3 | 26.6 | 1.2 | 25.7 | 99.9 | 27.2 | 1.2 | 99.9 | 26.0 |
| RNV2 | 100 | 93.9 | 5.4 | n.d. | n.d. | 18.4 | n.d. | 5.1 | 100 | 5.8 | n.d. | 100 | 5.5 | |
| RNV3 | 100 | 94.3 | 6.6 | n.d. | n.d. | 3.1 | n.d. | 3.0 | 100 | 3.8 | 0.3 | 99.9 | 3.5 | |
81 plaques were sequenced except a 49 plaques and b 79 plaques due to lack of material, c Mutation in non-coding region, * data for Vero and MDCK lysates are the mean frequencies detected in 2 independent deep sequencing assays, n.d. not detected, d.p.i days post infection.
We also found three of the mutations in the RIT4385 vaccine (V529A, L542L and L557P at 6.2%, 16.6% or 21.3% respectively), Table 1. We did not detect Ter550Q in RIT4385.
3.4. Variant population dynamics after in vitro passage
We investigated changes to the virus subpopulations after passage in Vero and MDCK cells. Deep sequencing was performed on cell lysates 3 days post-infection (d.p.i.) with JL-CK or RIT4385, the point at which cytopathic effect was evident, which confirmed the Sanger sequencing plaque data (10 d.p.i.), Table 1. K36R increased from 9.1% in unpassaged virus to 27% in Vero and 18.8% in MDCK cells. The JL-CK4 variant appeared consistent, with N56T/P93S/D311G occurring at 42.2%, 39.9% and 35%, in Vero lysates and 31.7%, 31%, and 29.5% in MDCK lysates, although direct linkage cannot be confirmed, it suggests selection of JL-CK1 and JL-CK4 in Vero cells. All other mutations reduced in frequency from unpassaged virus and were in line with the plaque frequencies shown in the top section of Table 1.
The frequencies of mutations observed in MDCK cell lysates were more in line with unpassaged JL-CK virus, suggesting the canine cell line supports growth of all variants. The mutations identified in unpassaged RIT4385, were also detected in RIT4385-infected Vero or MDCK cells, at similar levels to the unpassaged virus, indicating that RIT4385 is a more stable virus mixture compared to JL-CK.
3.5. In vitro growth of JL-CK variants
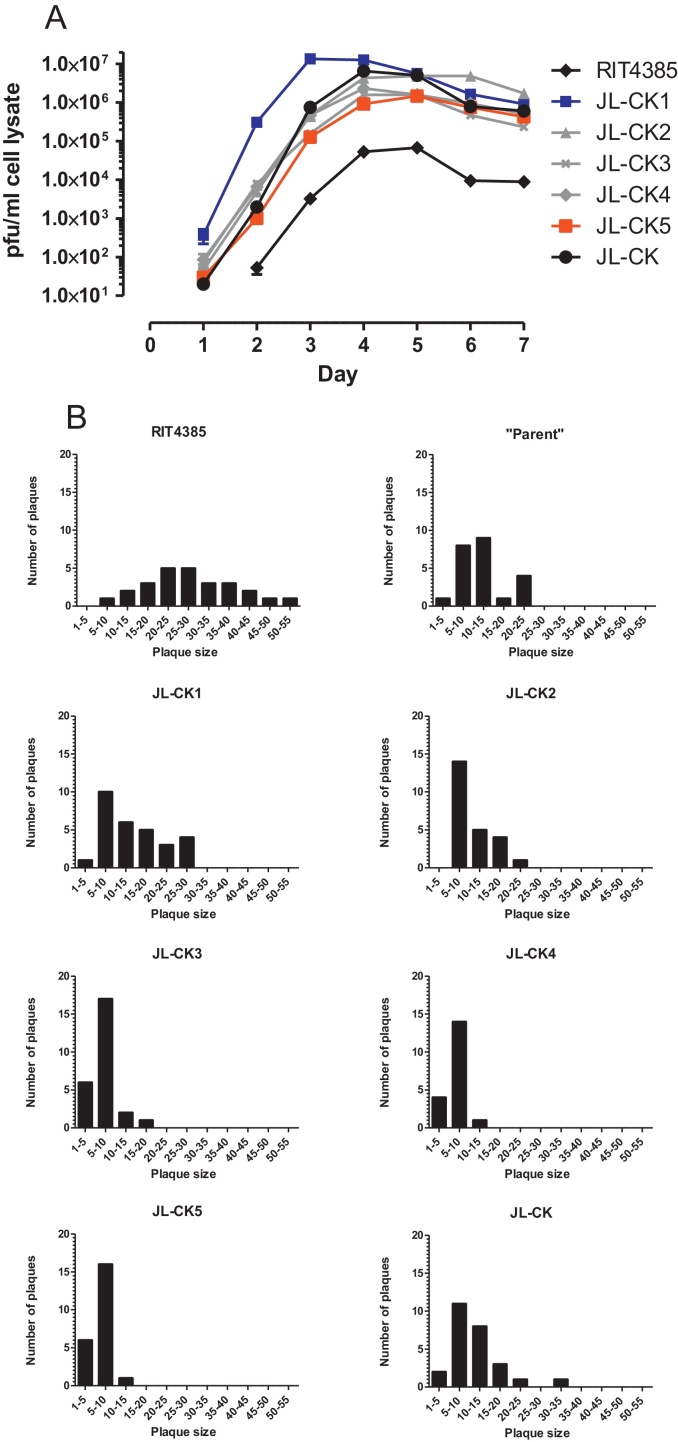
To compare the growth of the individual variants with JL-CK and RIT4385, we propagated each virus in Vero cells over 7 days. Fig. 3A shows JL-CK5 grew slowest, reaching maximum titre (1.46 × 106 pfu/ml) at 5 d.p.i, whilst JL-CK1 produced the highest virus titre (1.35 × 107 pfu/ml), in the shortest time (3 d.p.i.). All variants and the JL-CK vaccine produced higher titres of virus than RIT4385. We titrated the JL-CK variants to identify if the smaller plaque phenotype could be attributed to one particular mutation. Plaque analysis depicted in Fig. 3B showed that JL-CK3, JL-CK4 and JL-CK5 all displayed the smaller plaque phenotype, suggesting that Ter550Q, which is common to all of these variants, may play a role in this phenotype.
Fig. 3.
(A) Progeny virus titration on Vero cells after a 7 day timecourse assay on Vero cells. Viruses were inoculated at a MOI 0.0001. Progeny viruses were titrated in triplicate and plotted as pfu/ml of cell lysate. (B) Measurement of plaque size of JL-CK, RIT4385 and JL-CK variants using ImageJ. The “Parent” panel is the isolated parent-like variant which has the identical sequence to RIT4385. The data plotted is the number of plaques with corresponding plaque size, measured in ImageJ units.
3.6. Neurovirulence potential of JL-CK and variants
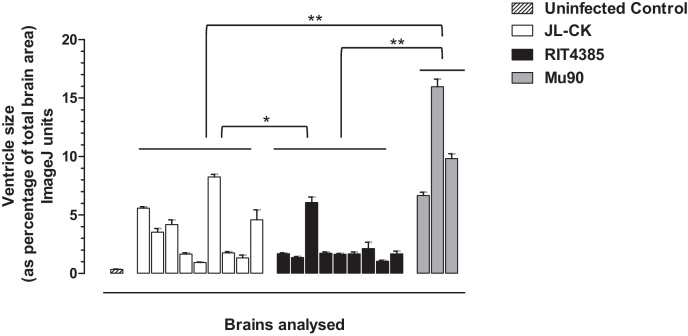
The neonatal rat model has been proposed for neurovirulence safety testing of MuV [29,33]. The neurovirulence potential of JL-CK, RIT4385 and Mu90 was compared. Fig. 4 shows the results from individual rat brains. On average, JL-CK has higher neurovirulence potential than RIT4385 (p = 0.0035), but when compared to Mu90, a neurovirulent MuV [29], the hydrocephalus observed in JL-CK was significantly less (p < 0.001). We observed variation between individual animals.
Fig. 4.
Neurovirulence scoring from neonatal rat brains 25 days after inoculation with JL-CK, RIT4385 or Mu90. Neurovirulence calculated by measuring the cross-sectional areas of the ventricles expressed as the percentage of the cross-sectional area of the whole brain. The data shown is from nine brains from each group (except Mu90, n = 3), with measurements taken from four sections of each brain. JL-CK showed a significant increase in neurovirulence score compared to RIT4385 (*p = 0.0035 unpaired two-tailed t-test). As a control, the neurovirulent Mu90 strain cause significantly higher scores than JL-CK, RIT4385 or uninfected control (**p < 0.0001 unpaired two-tailed t-test for all).
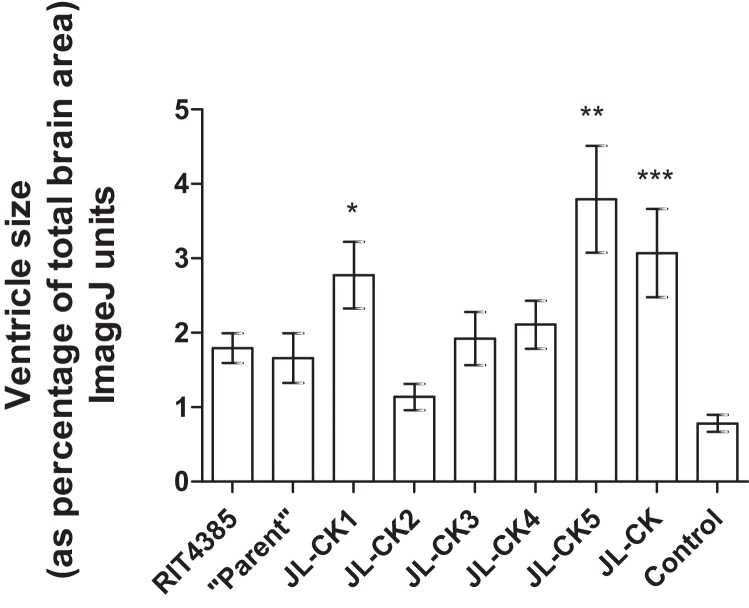
Fig. 5 shows the neurovirulence potential of the isolated JL-CK variants, confirming the difference observed between JL-CK and RIT4385 shown in Fig. 4, albeit slightly less in this instance (p = 0.0478). JL-CK1 and JL-CK5 produced increased ventricle sizes compared to RIT4385 (*p = 0.0406 and **p = 0.0100 respectively), suggesting they may have increased neurovirulence potential. As expected, the parent-like virus behaved like RIT4385, as did JL-CK3 (p = 0.1055) and JL-CK4 (p = 0.162). JL-CK2 showed significantly lower neurovirulence potential compared to both RIT4385 (p = 0.0181) and JL-CK (p = 0.0032).
Fig. 5.
Neurovirulence scoring from neonatal rat brains 25 days after inoculation with JL-CK, RIT4385 or JL-CK variants. The data shown is from six brains from each group, with measurements taken from four sections of each brain. An increase in neurovirulence compared to RIT4385 was observed for JL-CK1 (*p = 0.0406) and JL-CK5 (**p = 0.0100). JL-CK displayed an increase in neurovirulence potential compared to the parent-like variant isolated from JL-CK (***p = 0.0440).
3.7. Variant population dynamics after in vivo passage
Given the mixed population present in JL-CK and the difference in the neurovirulence scores of each variant it was of interest to examine variation of the virus subpopulations after growth in rat brains. Deep sequencing was performed on three brains 4 d.p.i. with JL-CK virus, Table 1, lower section. Compared with the inoculate (JL-CK) or the plaque data, the frequency of most of the mutations decreased. The frequencies of G/Ant92, C244R and E569K mutations, present in all JL-CK variants, remained mostly unchanged. However, K36R in the nucleoprotein was significantly more prominent after growth of the virus in vivo with a 9.6-fold increase, suggesting that JL-CK1 predominantly grew in the rat brain. JL-CK4 was present but at lower frequency than in Vero cells (∼3.5-fold decrease). The results obtained from each animal were largely consistent, although the difference was less prominent in one animal (RNV1). Growth of JL-CK vaccine in vivo preferentially selected JL-CK1. The JL-CK5 variant did not appear to grow effectively despite being one of the most virulent variants when tested individually (Fig. 5).
4. Discussion
JL-CK and RIT4385 are two related mumps vaccine viruses that provide different levels of protection against MuV. The JL-CK vaccine was thought to be equivalent to RIT4385 and has been used in some countries [26,34]. However, little is known about the passage history of JL-CK and its production on primary canine kidney cells raised additional concerns. When reports emerged about a potential failure of the vaccine to protect against mumps [24,26], we hypothesised that it differs from the RIT4385 strain. MuV is known to adapt to many cell lines and different growth conditions can alter the ratio of Jeryl Lynn MuV subpopulations [19,20,35]. Unpublished data from our lab and others [27,36] suggested that Vero cells may alter MuVs. The objective of this study was to identify the differences and to compare the two vaccines at the genomic level and in in vitro and in vivo biological assays. Our data show that both vaccines derive from the same Jeryl Lynn-5 strain, but plaques formed by JL-CK were small compared to those of RIT4385 and whilst their growth displayed similar kinetics, the yield from JL-CK was far higher. Deep sequencing suggested not all variants in the JL-CK vaccine produced plaques equally well in Vero cells. We identified selection of a subpopulation within JL-CK (JL-CK1), confirmed by plaque sequencing, which also grew faster in Vero cells compared to the other variants.
Sequencing of JL-CK revealed 14 polymorphisms throughout the genome which have not been reported before. One is in the non-coding leader (G/Ant92), a second is silent (L542L), all others result in changes to the predicted amino acid sequence, mostly in the nucleoprotein (K36R, E108D, V529A, Ter550Q, L557P). All other proteins contained at least one mutation, except SH which was identical to RIT4385. Three of the mutations (V529A, L542L and L557P) were also found in RIT4385 at a lower level, although L557P was in a non-coding region.
JL-CK contains a more mixed population than RIT4385, confirmed by plaque isolation, with mutation frequencies consistent with sequence heterogeneity and deep sequencing results. Mutation K36R was identified at low frequency by deep sequencing of unpassaged JL-CK vaccine, but accumulated during passage in Vero cells, in two independent assays after 3 or 10 days incubation, confirming previous findings of selective pressure from Vero cells [27,36]. Variant populations were less affected by passage on MDCK cells, suggesting a key effect from the species of cell used. RIT4385 was more genetically stable on both Vero and MDCK cell lines. We did not assess virus growth or population dynamic within other cell lines, and it is possible that additional phenotypes may be observed.
The nucleoprotein contains between one (JL-CK1) and five (JL-CK5) mutations. Ter550Q occurs in JL-CK3, JL-CK4 and JL-CK5, extending the nucleoprotein by 21 amino acids, confirmed by mass spectrometry. Plaque isolates containing Ter550Q correlated with a small plaque phenotype, although a definitive link would require further studies using recombinant MuVs. The nucleoprotein is required for transcription, replication and virus assembly, and antigenic sites have been found in its C-terminus [37,38]. It is possible that elongation of the nucleoprotein may affect these functions.
JL-CK was more virulent than RIT4385 in the neonatal rat model causing a higher degree of hydrocephalus, albeit not as virulent as the wild-type Mu90. We observed variation between animals, which has been described for this model before [39]. Group sizes were kept low in order to minimise animal usage. The differences we observed between RIT4385 and Mu90 have been previously reported [29]. However, in previous studies, differences between closely related strains had been hard to detect, including the Jeryl Lynn vaccine (containing JL-5 and JL-2) which was indistinguishable from RIT4385 [29].
Changes in viral genetic heterogeneity may impact on MuV neurovirulence [40]. Interestingly, compared to JL-CK, the two variants which caused similar levels of hydrocephalus were JL-CK1 containing the fewest mutations and JL-CK5 containing the most, suggesting the requirement of a combination of mutations for a neurovirulence phenotype. Studies using recombinant MuVs have shown that combinations of genes can alter neurovirulence in neonatal rats [41].
JL-CK1 contains G/Ant92, C244R, E569K and K36R, which increased 9.6-fold during in vivo growth in rat brains, suggesting a role for these mutations in the neurovirulence phenotype observed. The majority of the other mutations reduced in frequency indicating a change in the virus subpopulation in vivo. The results for the neurovirulence studies and deep sequencing of the rat brains provide conflicting data, with previous studies suggesting a correlation [41], although other biological factors may play a role. JL-CK5 produces a high neurovirulence score but deep sequencing suggests this virus does not replicate as effectively as the other variants in vivo. These findings are the subject of further study. We found variation between animals infected with the same virus, with variant frequencies showing the same trend between animals but to varying degrees. In particular, the Ter550Q mutation (common to JL-CK3, 4 and 5) decreased in frequency compared to JL-CK (74.2%), in each animal the frequency was substantially different, RNV1 26.6%, RNV2 18.4% and RNV3 3.1%. This may imply preferential replication of JL-CK4 over JL-CK3 and JL-CK5 within the rat brain and may explain the variation seen within groups of animals in the neurovirulence study.
Passage of MuV through cell lines is the basis for attenuation. The genetic differences between JL-CK and RIT4385 are presumably a reflection of the different passage history of these viruses. The high heterogeneity of JL-CK was unexpected. Our findings show that passaging JL-CK in vitro or in vivo affects the phenotype of the viruses, with preferential selection of some virus populations over others, within a short time. The selection effects of the human host on the virus are unknown and only few studies have investigated what happens to mumps vaccine viruses after immunisation. For example, we have previously reported genomic variations in clinical MuV isolates following vaccination with the L-Zagreb vaccine [11].
This study confirms the genetic instability of MuV and adds to the current evidence concerning the unstable nature of the MuV genome. It also emphasises the importance of choosing suitable cell substrates for mumps vaccine manufacture. Genetic drift during virus propagation has the potential to alter vaccine efficacy or safety. Deep sequencing may be a useful tool to assess the genetic heterogeneity between different batches, and variant frequency may be a useful indicator of manufacturing consistency. However, further interpretation of the results is hampered by the fact that the impact of variations on vaccinated populations is not yet understood. Our study brings to the forefront the fact that still very little is known about MuV pathogenesis and that the development of an effective animal model still proves to be elusive.
Conflicts of interest
The authors declare no conflicts of interest.
Author contributions
Conceived and designed the study: SMC, EV, PM, SS. Performed the experiments: SMC, JXW. Analysed the data: SMC, JXW. Contributed to the writing of the manuscript: SMC, JXW, PM, SS. Approved the manuscript: SMC, JXW, EV, PM, SS.
Acknowledgements
We thank Shaun Baker, Maureen Bentley, Charlotte Crofts, Graham Crossland, Rose Curran, Valdemar Gomes, Alan Haynes and Edward Mee for technical assistance, Ruth Harvey (NIBSC) for critical reading of the manuscript and Mike Skinner (Imperial College London) for helpful discussion. This study is independent research which, in part, was funded by the Department of Health Policy Research Programme (NIBSC Regulatory Science Research Unit, 044/0069). The views expressed in this publication are those of the author(s) and not necessarily those of the Department of Health.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vaccine.2015.06.109.
Contributor Information
Sarah M. Connaughton, Email: Sarah.Gilliland@nibsc.org.
Jun X. Wheeler, Email: Jun.Wheeler@nibsc.org.
Philip Minor, Email: Philip.Minor@nibsc.org.
Silke Schepelmann, Email: Silke.Schepelmann@nibsc.org.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Carbone K.M., Rubin S. Mumps virus. In: Knipe D.M., Howley P.M., editors. Fields virology. 5th ed. Lippincott Williams & Wilkins; 2007. pp. 1527–1550. [Google Scholar]
- 2.WHO . WHO; 2007. Weekly epidemiology record; pp. 49–60. [Google Scholar]
- 3.Beck M., Welsz-Malecek R., Mesko-Prejac M., Radman V., Juzbasic M., Rajninger-Miholic M. Mumps vaccine L-Zagreb, prepared in chick fibroblasts. I. Production and field trials. J Biol Stand. 1989;17:85–90. doi: 10.1016/0092-1157(89)90031-0. [DOI] [PubMed] [Google Scholar]
- 4.Fedova D., Bruckova M., Plesnik V., Slonim D., Sejda J., Svandova E. Detection of postvaccination mumps virus antibody by neutralization test, enzyme-linked immunosorbent assay and sensitive hemagglutination inhibition test. J Hyg Epidemiol Microbiol Immunol. 1987;31:409–422. [PubMed] [Google Scholar]
- 5.Odisseev H., Gacheva N. Vaccinoprophylaxis of mumps using mumps vaccine, strain Sofia 6, in Bulgaria. Vaccine. 1994;12:1251–1254. doi: 10.1016/s0264-410x(94)80047-4. [DOI] [PubMed] [Google Scholar]
- 6.Sassani A., Mirchamsy H., Shafyi A., Ahourai P., Razavi J., Gholami M.R. Development of a new live attenuated mumps virus vaccine in human diploid cells. Biologicals. 1991;19:203–211. doi: 10.1016/1045-1056(91)90036-j. [DOI] [PubMed] [Google Scholar]
- 7.Goh K.T. Resurgence of mumps in Singapore caused by the Rubini mumps virus vaccine strain. Lancet. 1999;354:1355–1356. doi: 10.1016/s0140-6736(99)02853-6. [DOI] [PubMed] [Google Scholar]
- 8.da Cunha S.S., Rodrigues L.C., Barreto M.L., Dourado I. Outbreak of aseptic meningitis and mumps after mass vaccination with MMR vaccine using the Leningrad-Zagreb mumps strain. Vaccine. 2002;20:1106–1112. doi: 10.1016/s0264-410x(01)00438-8. [DOI] [PubMed] [Google Scholar]
- 9.Kaic B., Gjenero-Margan I., Aleraj B., Ljubin-Sternak S., Vilibic-Cavlek T., Kilvain S. Transmission of the L-Zagreb mumps vaccine virus, Croatia, 2005–2008. Euro Surveill. 2008;13 [PubMed] [Google Scholar]
- 10.Brown E.G., Wright K.E. Genetic studies on a mumps vaccine strain associated with meningitis. Rev Med Virol. 1998;8:129–142. doi: 10.1002/(sici)1099-1654(199807/09)8:3<129::aid-rmv213>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 11.Gilliland S.M., Jenkins A., Parker L., Somdach N., Pattamadilok S., Incomserb P. Vaccine-related mumps infections in Thailand and the identification of a novel mutation in the mumps fusion protein. Biologicals: J Int Assoc Biol Standardiz. 2013;41:84–87. doi: 10.1016/j.biologicals.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Plotkin S.A., Rubin S.A. Mumps vaccine. In: Plotkin S.A., Orenstein W.A., Offit P.A., editors. Vaccines. 5th ed. Saunders; 2008. pp. 435–465. [Google Scholar]
- 13.Parker L., Gilliland S.M., Minor P., Schepelmann S. Assessment of the ferret as an in vivo model for mumps virus infection. J Gen Virol. 2013;94:1200–1205. doi: 10.1099/vir.0.052449-0. [DOI] [PubMed] [Google Scholar]
- 14.Tsurudome M., Yamada A., Hishiyama M., Ito Y. Replication of mumps virus in mouse: transient replication in lung and potential of systemic infection. Arch Virol. 1987;97:167–179. doi: 10.1007/BF01314419. [DOI] [PubMed] [Google Scholar]
- 15.Houard S., Varsanyi T.M., Milican F., Norrby E., Bollen A. Protection of hamsters against experimental mumps virus (MuV) infection by antibodies raised against the MuV surface glycoproteins expressed from recombinant vaccinia virus vectors. J Gen Virol. 1995;76(Pt 2):421–423. doi: 10.1099/0022-1317-76-2-421. [DOI] [PubMed] [Google Scholar]
- 16.Johnson C.D., Goodpasture E.W. An investigation of the etiology of mumps. J Exp Med. 1934;59:1–19. doi: 10.1084/jem.59.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buynak E.B., Hilleman M.R. Live attenuated mumps virus vaccine. 1. Vaccine development. Proc Soc Exp Biol Med. 1966;123:768–775. doi: 10.3181/00379727-123-31599. [DOI] [PubMed] [Google Scholar]
- 18.Afzal M.A., Pickford A.R., Forsey T., Heath A.B., Minor P.D. The Jeryl Lynn vaccine strain of mumps virus is a mixture of two distinct isolates. J Gen Virol. 1993;74(Pt 5):917–920. doi: 10.1099/0022-1317-74-5-917. [DOI] [PubMed] [Google Scholar]
- 19.Chambers P., Rima B.K., Duprex W.P. Molecular differences between two Jeryl Lynn mumps virus vaccine component strains, JL5 and JL2. J Gen Virol. 2009;90:2973–2981. doi: 10.1099/vir.0.013946-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amexis G., Rubin S., Chizhikov V., Pelloquin F., Carbone K., Chumakov K. Sequence diversity of Jeryl Lynn strain of mumps virus: quantitative mutant analysis for vaccine quality control. Virology. 2002;300:171–179. doi: 10.1006/viro.2002.1499. [DOI] [PubMed] [Google Scholar]
- 21.Usonis V., Bakasenas V., Chitour K., Clemens R. Comparative study of reactogenicity and immunogenicity of new and established measles, mumps and rubella vaccines in healthy children. Infection. 1998;26:222–226. doi: 10.1007/BF02962367. [DOI] [PubMed] [Google Scholar]
- 22.Plesnik V., Fedova D., Sejda J., Slonim D., Cervenka P., Walderova O. Results of vaccination with Czechoslovakian vaccine against parotitis. Cesk Pediatr. 1984;39:540–546. [PubMed] [Google Scholar]
- 23.Limberkova R., Lexova P. Genotyping results, laboratory diagnosis, and epidemiology of the mumps virus circulating in the Czech Republic in 2012. Epidemiol, Mikrobiologie, Imunologie: Casopis Spolecnosti Pro Epidemiologii a Mikrobiologii Ceske Lekarske Spolecnosti JE Purkyne. 2014;63:36–42. [PubMed] [Google Scholar]
- 24.Mrazova M., Smelhausova M., Sestakova Z., Svandova E., Benes C. The 2001 serological survey in the Czech Republic – mumps. Cent Eur J Public Health. 2003;11(Suppl.):S50–S53. [PubMed] [Google Scholar]
- 25.Stepanova V., Pliskova L., Kosina P., Splino M., Forstl M., Bolehovska R. Mumps – a reemerging infection? The current incidence of mumps in the East Bohemian region in the Czech Republic. Epidemiologie, Mikrobiologie, Imunologie: Casopis Spolecnosti Pro Epidemiologii a Mikrobiologii Ceske Lekarske Spolecnosti JE Purkyne. 2006;55:127–135. [PubMed] [Google Scholar]
- 26.Boxall N., Kubinyiova M., Prikazsky V., Benes C., Castkova J. An increase in the number of mumps cases in the Czech Republic, 2005–2006. Euro Surveill. 2008:13. [PubMed] [Google Scholar]
- 27.Afzal M.A., Dussupt V., Minor P.D., Pipkin P.A., Fleck R., Hockley D.J. Assessment of mumps virus growth on various continuous cell lines by virological, immunological, molecular and morphological investigations. J Virol Methods. 2005;126:149–156. doi: 10.1016/j.jviromet.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 28.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rubin S.A., Afzal M.A., Powell C.L., Bentley M.L., Auda G.R., Taffs R.E. The rat-based neurovirulence safety test for the assessment of mumps virus neurovirulence in humans: an international collaborative study. J Infect Dis. 2005;191:1123–1128. doi: 10.1086/428098. [DOI] [PubMed] [Google Scholar]
- 30.Chen R., Vendrell I., Chen C.P., Cash D., O’Toole K.G., Williams S.A. Proteomic analysis of rat plasma following transient focal cerebral ischemia. Biomark Med. 2011;5:837–846. doi: 10.2217/bmm.11.89. [DOI] [PubMed] [Google Scholar]
- 31.Rubin S.A., Pletnikov M., Carbone K.M. Comparison of the neurovirulence of a vaccine and a wild-type mumps virus strain in the developing rat brain. J Virol. 1998;72:8037–8042. doi: 10.1128/jvi.72.10.8037-8042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Afzal M.A., Buchanan J., Heath A.B., Minor P.D. Clustering of mumps virus isolates by SH gene sequence only partially reflects geographical origin. Arch Virol. 1997;142:227–238. doi: 10.1007/s007050050073. [DOI] [PubMed] [Google Scholar]
- 33.Malik T.H., Wolbert C., Nerret L., Sauder C., Rubin S. Single amino acid changes in the mumps virus haemagglutinin-neuraminidase and polymerase proteins are associated with neuroattenuation. J Gen Virol. 2009;90:1741–1747. doi: 10.1099/vir.0.009449-0. [DOI] [PubMed] [Google Scholar]
- 34.Committee on the Safety of Medicines . Committee on the Safety of Medicines; 2002–2015. Advice that unlicensed Pavivac single mumps vaccine should not be imported or used.http://webarchive.nationalarchives.gov.uk/20141205150130/http://www.mhra.gov.uk/home/groups/pl-p/documents/websiteresources/con2031109.pdf [Google Scholar]
- 35.Amexis G., Oeth P., Abel K., Ivshina A., Pelloquin F., Cantor C.R. Quantitative mutant analysis of viral quasispecies by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc Natl Acad Sci U S A. 2001;98:12097–12102. doi: 10.1073/pnas.211423298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santak M., Markusic M., Balija M.L., Kopac S.K., Jug R., Orvell C. Accumulation of defective interfering viral particles in only a few passages in Vero cells attenuates mumps virus neurovirulence. Microbes Infect. 2014;17:228–236. doi: 10.1016/j.micinf.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Ivancic-Jelecki J., Santak M., Forcic D. Variability of hemagglutinin-neuraminidase and nucleocapsid protein of vaccine and wild-type mumps virus strains. Infect Genet Evol. 2008;8:603–613. doi: 10.1016/j.meegid.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 38.Tanabayashi K., Takeuchi K., Hishiyama M., Yamada A., Tsurudome M., Ito Y. Nucleotide sequence of the leader and nucleocapsid protein gene of mumps virus and epitope mapping with the in vitro expressed nucleocapsid protein. Virology. 1990;177:124–130. doi: 10.1016/0042-6822(90)90466-5. [DOI] [PubMed] [Google Scholar]
- 39.Rubin S.A., Pletnikov M., Taffs R., Snoy P.J., Kobasa D., Brown E.G. Evaluation of a neonatal rat model for prediction of mumps virus neurovirulence in humans. J Virol. 2000;74:5382–5384. doi: 10.1128/jvi.74.11.5382-5384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauder C.J., Vandenburgh K.M., Iskow R.C., Malik T., Carbone K.M., Rubin S.A. Changes in mumps virus neurovirulence phenotype associated with quasispecies heterogeneity. Virology. 2006;350:48–57. doi: 10.1016/j.virol.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 41.Sauder C.J., Zhang C.X., Ngo L., Werner K., Lemon K., Duprex W.P. Gene-specific contributions to mumps virus neurovirulence and neuroattenuation. J Virol. 2011;85:7059–7069. doi: 10.1128/JVI.00245-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.