SUMMARY
Some organisms, such as adult zebrafish and newborn mice, have the capacity to regenerate heart tissue following injury. Unraveling the mechanisms of heart regeneration is fundamental to understanding why regeneration fails in adult humans. Numerous studies have revealed that nerves are crucial for organ regeneration, thus we aimed to determine whether nerves guide heart regeneration. Here, we show using transgenic zebrafish that inhibition of cardiac innervation leads to reduction of myocyte proliferation following injury. Specifically, pharmacological inhibition of cholinergic nerve function reduces cardiomyocyte proliferation in the injured hearts of both zebrafish and neonatal mice. Direct mechanical denervation impairs heart regeneration in neonatal mice, which was rescued by the administration of Neuregulin 1 (Nrg1) and Nerve growth factor (Ngf) recombinant proteins. Transcriptional analysis of mechanically denervated hearts revealed a blunted inflammatory and immune response following injury. These findings demonstrate that nerve function is required for both zebrafish and mouse heart regeneration.
Graphical abstract
INTRODUCTION
A significant cause of heart failure is the inability of the adult mammalian heart to replace lost cells following injury (Laflamme and Murry, 2011). In contrast to adult mammals, lower organisms are capable of regenerating many of their injured organs. For example, zebrafish can regenerate up to 20% of their hearts after surgical amputation primarily through the dedifferentiation and division of pre-existing cardiomyocytes (Jopling et al., 2010; Kikuchi et al., 2010; Poss et al., 2002). Similarly, newts have a remarkable capacity for regenerating a variety of structures, ranging from the heart and ocular lens to a completely functional limb (Laube et al., 2006; Oberpriller and Oberpriller, 1974; Tsonis et al., 2004).
Recent evidence indicates that mice can regenerate their hearts immediately after birth, though this capacity is lost within the first week of life (Porrello et al., 2011b; Porrello et al., 2013). These data suggest that the mammalian myocardium retains regenerative potential similar to lower organisms, and understanding the regulatory processes that guide this phenomenon could reveal novel approaches to reactivate this dormant regenerative capacity in adults.
For over a century, it has been reported that nerves play a crucial role in guiding regenerative processes in multiple species. Seminal work in salamander limb regeneration has revealed the reliance on nerves in regeneration (Todd, 1823); if the nerve is severed at the base of the limb prior to or shortly after amputation, the limb will fail to regenerate (Singer, 1952). Studies on the regeneration of a variety of structures in the newt and other organisms have revealed that cholinergic neurons often play a crucial role in the reformation of the severed limb (Drachman, 1964; Singer et al., 1960). More importantly, nerves guide the regeneration of multiple organs in diverse species, suggesting some evolutionarily conserved nerve functions in regeneration (Kumar and Brockes, 2012).
Although cholinergic nerves are critical for multiple regenerative processes, their role in heart regeneration has never been studied. Thus, to determine whether nerve activity is essential for heart regeneration, we used the zebrafish and neonatal mouse models of heart regeneration to address this question. Here, we show that reduction of heart innervation in a transgenic zebrafish model diminishes cardiomyocyte cell cycle activity and inhibits heart regeneration following injury. Specifically, disruption of cholinergic signaling by pharmacological approaches leads to a reduction in the cardiomyocyte cell cycle activity of both zebrafish and neonatal mice following injury. We also demonstrate that mechanical ablation of the vagus nerve in the neonatal mouse has a similar effect in inhibiting heart regeneration following apical resection and myocardial infarction. Treatment of mechanically denervated mice with nerve-factors rescued the reduced regenerative capacity following denervation. Finally, transcriptional profiling of the neonatal mouse heart following mechanical denervation revealed patterns for disruption of inflammatory gene expression, which were normally activated during heart regeneration. These factors might contribute to the nerve-dependent regenerative capacity of the heart. These findings reveal that nerve function plays a key role in cardiac regeneration through regulation of cardiomyocyte proliferation.
RESULTS
Innervation Modulates Injury-Induced Cardiomyocyte Proliferation in Zebrafish
The vertebrate heart is innervated during chamber morphogenesis by parasympathetic and sympathetic fibers, which regulate responses of the cardiovascular system to stress. Recent studies indicate that, upon origination from the stellate ganglion, sympathetic nerves are guided along the cardiac surface by neurotrophic signals from developing coronary vascular cells (Nam et al., 2013). We observed extensive innervation of the atrial surface myocardium in adult zebrafish, with less pronounced innervation of the ventricle (Figure 1C, upper panel). After resection of the ventricular apex, new muscle became innervated during regeneration of the new apical wall (Figures 1A and 1B).
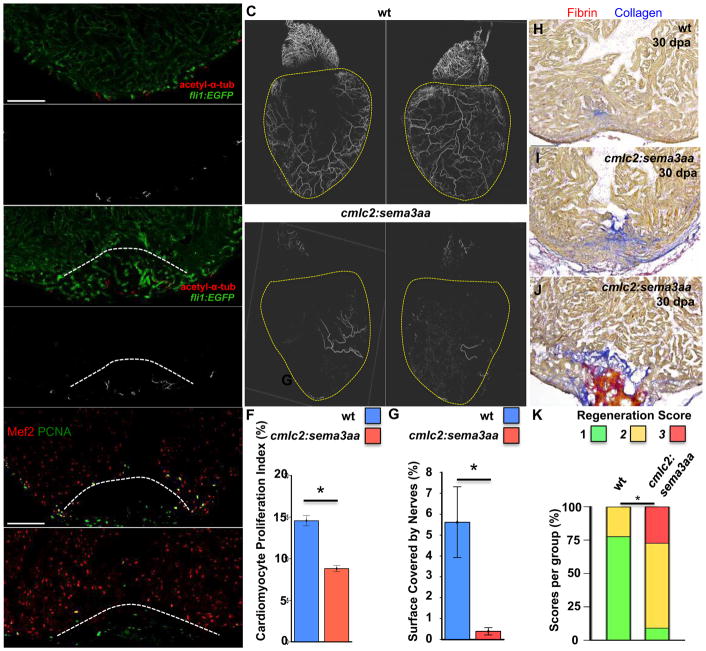
Figure 1. Effects of Cardiac Innervation on Injury-Induced Cardiomyocyte Proliferation.
A and B) Section images of uninjured (A) and regenerated (30 dpa; (B)) ventricular apices visualized for endothelial cells (green) and nerves (red). The approximate regenerated area is indicated by a dashed line. Grayscale images indicating nerves, positive for acetylated alpha-tubulin, are shown in (A′) and (B′). Scale bar represents 100 μm. C) Whole mount images of hearts from wild-type (wt; left) and cmlc2:sema3aa (right) animals, immunostained for acetylated alpha-tubulin to indicate cardiac nerves, which are reduced by sema3aa overexpression. Scale bar represents 100 μm. D and E) Section images of 7 dpa ventricular apices of wild-type (D) or cmlc2:sema3aa (E) animals, stained for Mef2+PCNA+ cells. Wounds are indicated by dashed lines. Scale bar represents 100 μm. F) Quantification of cardiomyocyte proliferation at 7 dpa from hypo- (cmlc2:sema3aa). Wild-type clutchmates (n = 16) were used as controls for cmlc2:sema3aa animals (n = 13). Data are represented as mean ± SEM. *p < 0.05, Mann-Whitney Rank Sum. G) Quantification of surface innervation as measured by acetylated alpha-tubulin staining, in cmlc2:sema3aa (n=6) and Wild-type clutchmate controls (n=5). Data are represented as mean ± SD. *p < 0.05, Mann-Whitney Rank Sum. H–J) Section images of 30 dpa ventricular apices of wild-type (D) or cmlc2:sema3aa (E) animals stained with Acid-Fuschin Orange. Scale bar represents 100 μm. K) Quantification of regeneration between cmlc2:sema3aa and wildtype siblings were compared at 30 dpa. Hearts (H–J) were scored for regeneration, with 1 indicating complete regeneration (H), 2 indicating partial regeneration (I), and 3 indicating a block in regeneration (J). Data represent percent of total heart per score. *p < 0.05, Fisher’s exact.
To examine the potential involvement of nerves in the regenerative process, we designed transgenic tools to limit the extent of cardiac innervation. We created a transgenic zebrafish line with myocardial overexpression of semaphorin3aa, a known inhibitor of cardiac innervation (Ieda et al., 2007) (Tg(cmlc2:sema3aa)pd106). As expected, hearts from these animals showed clearly reduced innervation on the ventricular surface (Figure 1C, lower panel), but they appeared otherwise grossly normal. Quantification of surface innervation revealed a 90% reduction in cardiac innervation in cmlc2:sema3aa hearts as compared with controls (Figure 1G, n = 5, 6). Following ventricular resection, cmlc2:sema3aa hearts displayed a 40% reduction in cardiomyocyte proliferation at 7 days post amputation (dpa), assessed by Mef2/PCNA staining (n = 13, 16; Figures 1D–1F). When assessed at 30 dpa, a timepoint at which a contiguous wall of heart muscle is typically observed, cmlc2:sema3aa animals had a significantly lower regeneration score than wild-type siblings (Figure 1H–K, n = 9, 11). Thus, hypo-innervated zebrafish hearts display a diminished regenerative response after cardiac injury.
Pharmacological Inhibition of Cholinergic Transmission Reduces Cardiomyocyte Proliferation in Zebrafish and Neonatal Mice
We next designed an experiment to delineate whether cholinergic or adrenergic nerves were responsible for the nerve-dependent heart regeneration phenotypes. We amputated approximately 20% of the ventricle of zebrafish hearts and then exposed zebrafish to the non-selective muscarinic receptor antagonist atropine (50μM) to inhibit cholinergic transmission, or the beta-adrenergic receptor antagonist propranolol (50μM) following apical amputation. We observed many proliferating cardiomyocytes (PCNA/Mef2 double positive) in the regenerate of control zebrafish hearts at 7 dpa (Figure 2A, n = 4). In contrast, resected hearts that were exposed to atropine displayed a marked decrease in the number of proliferating cardiomyocytes (Figure 2B, n = 4). Atropine treatment resulted in a significant reduction in the number of proliferating cardiomyocytes compared to control (Figure 2E, p<0.001). On the other hand, zebrafish treated with the beta-adrenergic antagonist propranolol showed an increase in proliferating cardiomyocytes following amputation similar to controls (Figure 2C, n=4). To further explore the role of cardiac cholinergic transmission in regeneration, we examined the role of the type 2 muscarinic (M2) receptor, which is the most prevalent type of muscarinic receptor in the heart (Dhein et al., 2001). We treated zebrafish with the M2 receptor-specific antagonist methoctramine. Similar to hearts treated with atropine, we found a decrease in the number of proliferating cardiomyocytes at 7 days post-resection (Figure 2D, n=4). Quantification showed a significant reduction in the number of proliferating cardiomyocytes in the methoctramine treated zebrafish (Figure 2E, n=4). Treatment of zebrafish with atropine, propranolol and methoctramine alone without injury was used as an additional control. Pharmacological treatment had no impact on zebrafish well-being or on the heart morphology and baseline myocyte proliferation (Figure S1). These data implicate a specific role for cholinergic nerve activity in the proliferation of cardiomyocytes during regeneration.
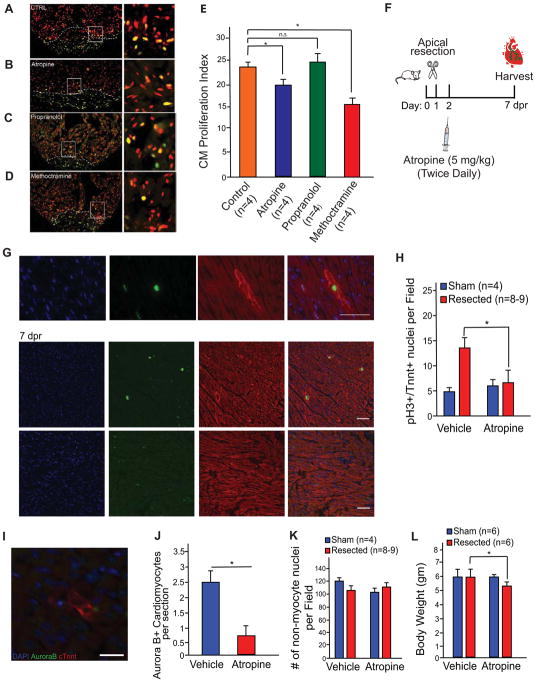
Figure 2. Pharmacological Inhibition of Cholinergic Nerve Function Reduces Cardiomyocyte Proliferation in Zebrafish and Neonatal Mice.
A) Zebrafish hearts were fixed and immunostained for PCNA and Mef2C at 7 days after surgical amputation. Hearts derived from zebrafish treated with water (Control) displayed notable cardiomyocyte proliferation. B) Atropine treated zebrafish exhibited a reduction in proliferating cardiomyocytes. C) PCNA and Mef2c staining at 7 dpa for propranolol treated zebrafish. Control and propranolol treated zebrafish showed equivalent cardiomyocyte proliferation. D) PCNA and Mef2c staining at 7 dpa for methoctramine treated zebrafish showed significant reduction in cardiomyocyte proliferation. Boxed regions in A–D are shown at higher zoom in the left panels. E) Quantification of proliferating cardiomyocytes showing a significant reduction of the number of proliferating cardiomyocytes in atropine and methoctramine treated zebrafish compared to control water and propranolol treated zebrafish. F) Schematic of atropine injection strategy in neonatal mice. G) Mouse hearts were fixed and immunostained for phospho Histone H3 (pH3) and cardiac Troponin T (cTnnt) at 7 dpr in vehicle and atropine treated mice. The upper panel shows a high magnification image of a pH3+ cardiomyocyte. Scale bar, 50 μm. H) Quantification of the number of proliferating cardiomyocytes at 7 days post apical resection showing a significant decrease of proliferating cardiomyocytes in neonatal mice. I) Immunostaining of Aurora B and cTnnt. Scale bar, 50 μm. J) Quantification of the Aurora B+ cardiomyocytes showing a significant reduction in the number of Aurora B+ cardiomyocytes in atropine treated mice. K) Quantification of the number of nuclei in heart sections of vehicle and atropine treated mice showing no significant differences between treatments. L) Body weights of vehicle and atropine treated mice following sham and resection showing a significant reduction in body weights of atropine resected mice but no changes in sham-operated mice. Data presented as mean ± SEM, where p<0.05 was considered statistically significant. See also Figure S1.
We have recently reported extensive analysis of the neonatal mouse regenerative response following apical resection (Bryant et al., 2015). Given the demonstration of nerve function in zebrafish myocyte proliferation, we wanted to determine whether inhibition of cholinergic transmission by atropine halts cardiomyocyte proliferation during neonatal mouse heart regeneration. To test this hypothesis, we resected approximately 15 % of the ventricle of 1-day-old neonatal mice and then injected these mice with atropine (5 mg/kg) or vehicle twice-daily (Figure 2F). We performed immunostaining against the myocyte marker cardiac troponin t (cTnnt) and the mitosis marker phosphorylated histone H3 (pH3) on histological sections at 7 days post resection (dpr), a timepoint when cardiomyocyte proliferation is high following injury. Extensive cardiomyocyte proliferation was detected in vehicle-treated mice, while cardiomyocyte proliferation was markedly reduced in atropine-treated mice (Figure 2G and 2H, n = 6–8, p<0.005). To determine the impact on cytokinesis, we performed immunostaining with an antibody against Aurora B kinase, and we detected a significant reduction in the number of cardiomyocytes undergoing cytokinesis in the atropine treated mice (Figure 2I and J, n=4, p<0.05). There was no significant change in the number of non-myocyte nuclei in both vehicle and atropine treated hearts in both sham-operated and resected mice (Figure 2K). Vehicle-treated and atropine-treated sham-operated mice showed comparable body weights; whereas following resection there was a significant reduction in body weight of atropine treated mice compared to vehicle controls (Figure 2L). Taken together, these results suggest that cholinergic nerve signaling is required for the proliferation of cardiomyocytes during neonatal mammalian cardiac regeneration.
Mechanical Cholinergic Denervation in Neonatal Mice Reduces Cardiomyocyte Proliferation and Impairs Heart Regeneration
Although pharmacological agents represent a powerful tool to interfere with nerve function in the heart, secondary effects may arise due to globally antagonizing cholinergic nerve activity and transmission. In order to exclude non-cardiac effects due to systemic atropine administration, we developed a surgical procedure in 1 day-old neonatal mice in which we mechanically ablate the left vagus nerve, which directly innervates the heart (Figure 3A). In order to confirm that we specifically dissect out the nerve, the resected nerve portion was immunostained with the pan neuronal marker beta tubulin III (Tubb3); the resected nerve showed positive nerve staining (Figure 3B). Unilateral left vagotomy, but not right vagotomy, has been shown to upregulate M2 receptor expression as a compensatory response in the left atrium and left ventricle following denervation (Chen et al., 2008). To examine whether neonatal left vagotomy caused a similar effect on the expression of M2 receptors, we performed qPCR on the left ventricle at 7 days following vagotomy in neonatal mice. We detected the anticipated upregulation of M2 receptor gene expression in the left ventricular apex of vagotomized neonatal mice (Figure 3C, n=4 per group, p<0.01). These data confirm that left vagotomy in neonatal mice exhibits a similar response to left vagotomy in adult animals, and thus represents an approach for mechanical denervation.
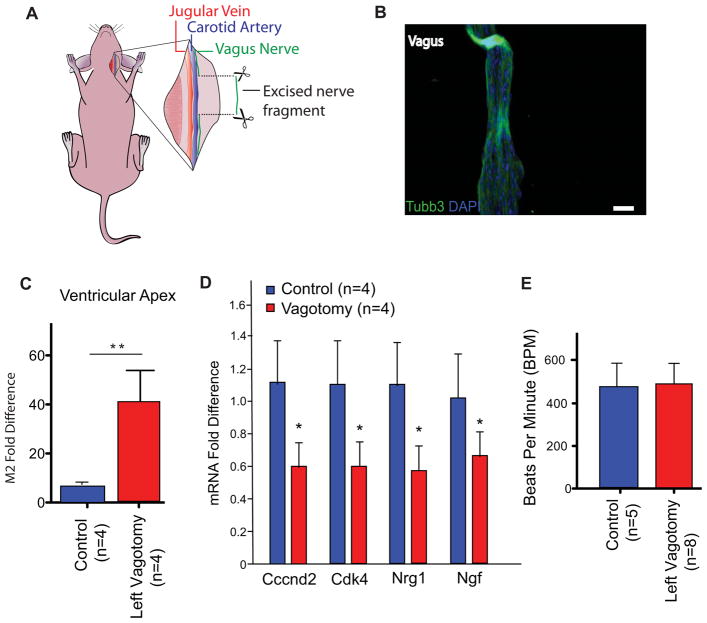
Figure 3. Left Vagotomy as a Model for Mechanical Denervation in the Neonatal Mouse.
A) Schematic depiction of the left vagotomy surgery in neonatal mice. B) Immunohistochemistry of the neuronal marker Tubb3 of the resected vagus nerve, nuclei stained with DAPI. Scale bar, 50 μm. C) qPCR gene expression analysis of the M2 receptor levels in sham operated and vagotomized neonatal mice, showing an upregulation of M2 receptor expression following vagotomy. D) qPCR expression profile of cell cycle and nerve secreted factors show significant downregulation in vagotomy compared to unoperated animals (n=4, p<0.05). E) Heart rate measurements at 7 days following left vagotomy showing similar heart rates in both control and vagotomized mice.
To determine whether mechanical cholinergic denervation alone can impact the levels of cardiomyocyte cell cycle regulators, we performed gene expression analysis of multiple positive regulators of cardiomyocyte cell cycle as Cyclin D2 and Cdk4 by qPCR (Pasumarthi and Field, 2002). Similarly, we studied growth factors as Nrg1 and Ngf that have been previously shown to enhance cardiomyocyte proliferation (Bersell et al., 2009; Lam et al., 2012). Interestingly, positive regulators of cardiomyocyte cell cycle such as Cyclin D2 and Cdk4 were significantly downregulated following vagotomy (Figure 3D). In addition, Nrg1 and Ngf were significantly downregulated in the vagotomized neonatal mice (Figure 3D). These data indicate that neonatal mouse vagotomy directly regulates the cardiomyocyte cell cycle state at the molecular level. To examine whether left vagotomy has an impact on heart rate following vagotomy, we measured the heart rates of mice at 7 days post vagotomy. We found no significant differences in the heart rates of control and vagotomized mice at resting conditions (Figure 3E).
We performed a double surgical procedure where neonates undergo vagotomy paired with apical resection to assess whether vagotomy compromises neonatal heart regeneration. Interestingly, the neonatal mice that underwent apical resection paired with vagotomy of the heart showed lower levels of proliferating cardiomyocytes, similar to both sham operated and vagotomized mice (Figure S2A). Quantification of the pH3 positive cardiomyocytes showed a significant decrease in the number of proliferating cardiomyocytes in resected plus vagotomized mice compared to resection alone (Figure S2B, n=5, p<0.05).
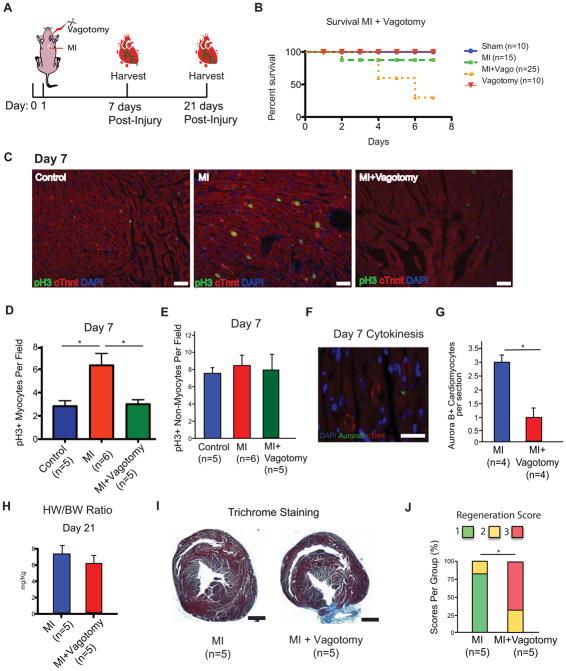
Similar to regeneration following apical resection, neonatal mice have the capacity to regenerate their hearts following myocardial infarction (MI), a major cause of heart failure in humans (Mahmoud et al., 2014; Porrello et al., 2013). We performed both MI + vagotomy on 1 day-old mice (Figure 4A). Interestingly, the mice that underwent the double surgery had a higher mortality rate compared to either vagotomy or MI alone. Kaplan-Meier survival analysis showed that combined vagotomy and MI significantly increased mortality (Figure 4B). At day 7 following MI + vagotomy, immunohistochemistry of pH3 and cTnnt showed reduction of proliferating cardiomyocytes in the MI + vagotomy mice compared to MI alone (Figure 4C and D, n=4–6, p<0.05). No significant changes were observed in the number of proliferating non-myocytes (Figure 4E). In addition, there was a significant reduction in the number of cardiomyocytes undergoing cytokinesis as detected by aurora b staining (Figure 4F and G, n=4, p<0.05). There was a trend toward a decrease in the heart weight to body weight (HW/BW) ratio of MI + vagotomy mice compared to MI only, but this trend was not statistically significant (Figure 4H).
Figure 4. Mechanical Denervation Reduces Cardiomyocyte Proliferation and Heart Regeneration in the Neonatal Mouse.
A) Schematic of neonatal vagotomy and myocardial infarction strategy in neonatal mice. B) Kaplan-Meier survival curve of MI and MI + Vagotomy mice. A significant reduction in survival of MI + Vagotomy mice was observed as assessed by Kaplan Meier analysis. C) Immunostaining of pH3 and cTnnt showing high levels of proliferating myocytes in MI hearts at 7 dpr, while a marked decrease in the levels of proliferating cardiomyocytes in MI + Vagotomy hearts. Scale bar, 50 μm. D) Quantification of the number of proliferating cardiomyocytes showing a significant decrease of proliferating cardiomyocytes following vagotomy and myocardial infarction. E) Quantification of the number of proliferating non-myocytes showing no significant changes between groups. F) Immunostaining of Aurora B and cTnnt. G) Quantification of the number of Aurora B cardiomyocytes showing a significant reduction in the number of Aurora B+ cardiomyocytes following MI+Vagotomy. H) Heart weight to body weight (HW/BW) ratio showing slightly reduced but not significant difference in the MI + Vagotomy heart. I) Trichrome staining of MI + Vagotomy and MI hearts at 21 dpr, showing incomplete regeneration and persistence of a fibrotic scar in vagotomized mice. Scale bar, 1 mm. J) Quantification of regeneration in MI and MI + Vagotomy mice. 1 indicates complete regeneration, 2 indicates partial regeneration, and 3 indicates a block in regeneration. Data represent total heart per score, 2×3 contingency analysis, p<0.05. Data presented as mean ± SEM, where p<0.05 was considered statistically significant. See also Figure S2.
To further establish the effects of mechanical denervation on neonatal heart regeneration, we performed Trichrome staining 21 days post injury to assess regeneration and fibrosis in both MI + Vagotomy mice compared to either MI or sham operated mice. The MI + vagotomy mice demonstrated little to no regrowth below the ligated plane of the heart with the presence of a fibrotic scar (Figure 4I). In contrast, animals that had undergone MI alone demonstrated complete heart regeneration with little to no scar formation (Figure 4I). Quantification of regeneration showed a significantly reduced regeneration score following MI + vagotomy (Figure 4J, n=5). To determine whether levels of cardiac innervation are changed beyond the regenerative window, we performed immunostaining of cardiac nerves with the neuronal marker Neurofilament in both P1 and P21 hearts and detected similar patterns of innervation (Figure S3). Collectively, our results show that mechanical ablation of nerves can inhibit the neonatal mouse cardiac regenerative response, which highlights the intimate involvement of nerves during heart regeneration.
Rescue of Mechanically Denervated hearts by Neuregulin1 (Nrg1) and Nerve Growth Factor (Ngf)
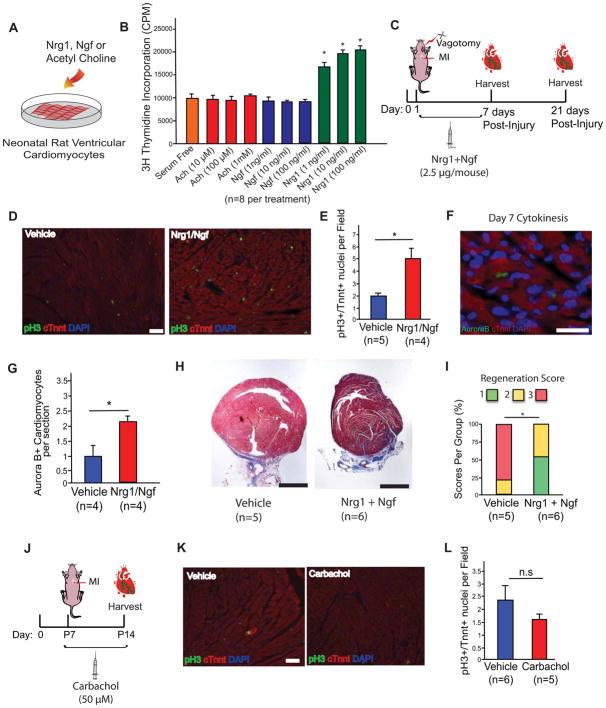
To test whether acetylcholine (Ach) as well as the growth factors Nrg1 and Ngf can stimulate cardiomyocyte proliferation, we treated cultured neonatal rat ventricular cardiomyocytes with these factors for 48 hours and measured DNA synthesis by 3H-thymidine incorporation (Figure 5A). Nrg1 was able to increase cardiomyocyte DNA synthesis, but not acetylcholine or Ngf (Figure 5B). These data suggest that acetylcholine cannot directly stimulate cardiomyocyte proliferation at the M2 muscarinic receptor level, while Nrg1 can directly induce myocyte DNA synthesis as has been previously established (Bersell et al., 2009). Although Ngf did not cause a significant induction of myocyte DNA synthesis, it has been shown to enhance recovery post injury suggesting that Ngf may enhance cardiac repair indirectly (Lam et al., 2012; Meloni et al., 2010). This suggests that Ngf may enhance cardiac repair indirectly.
Figure 5. Rescue of Mechanically Denervated Mice by Nrg1 and Ngf.
A) Schematic of neonatal rat cardiomyocyte treatment with acetylcholine (Ach), Nrg1 and Ngf. B) 3H thymidine incorporation showing increased DNA synthesis in cardiomyocytes treated with Nrg1, but not Acetylcholine or Ngf. C) Schematic of injections of recombinant Nrg1 and Ngf in neonatal mice following MI + Vagotomy. D) Immunostaining of pH3 and cTnnt showing high levels of proliferating myocytes in treated mice at 7 days post injury compared to controls. Scale bar, 50 μm. E) Quantification of the number of proliferating myocytes following Nrg1 and Ngf injection showing higher number of proliferating myocytes compared to controls. F) Immunostaining of Aurora B and cTnnt. Scale bar, 50 μm. G) Quantification of the number of Aurora B cardiomyocytes showing a significant increase in the number of Aurora B+ cardiomyocytes in Nrg1/Ngf treated mice compared to controls. H) Trichrome staining at 21 days post injury, showing reduced scare in Nrg1 and Ngf treated mice. Scale bar, 1 mm. I) Quantification of regeneration in vehicle and Nrg1 + Ngf treated mice showing a significant higher regeneration scores in Nrg1 + Ngf treated mice. 1 indicates complete regeneration, 2 indicates partial regeneration, and 3 indicates a block in regeneration. Data represent total heart per score, 2×3 contingency analysis, p<0.05. J) Schematic of carbachol injections following MI at P7. K) Immunostaining of pH3 and cTnnt to detect mitotic cardiomyocytes. Scale bar, 50 μm. L) Quantification of mitotic cardiomyocytes showing no significant increase in myocyte cell cycle activity in carbachol treated mice. Data presented as mean ± SEM, where p<0.05 was considered statistically significant.
To determine whether inhibition of regeneration by mechanical denervation could be partly rescued through Nrg1 and Ngf, we injected these recombinant proteins in neonatal mice post MI + Vagotomy (Figure 5C). At 7 days post injury, there was a significant increase in the number of pH3 positive cardiomyocytes in the treated mice compared to controls (Figure 5D and E, n=4–5, p<0.05). A significant increase in the number of cardiomyocytes undergoing cytokinesis as determined by Aurora B staining was detected as well (Figure 5F and G, n=4, p<0.05). At 21 days post injury, there was reduced scar formation in the Nrg1 and Ngf treated mice (Figure 5H, n=6). Quantification of regeneration showed a significantly higher regeneration score in the Nrg1 and Ngf treated mice (Figure 5I). These data suggest that nerves stimulate heart regeneration partly through growth factors as indicated by supplying recombinant Nrg1 and Ngf following mechanical denervation.
To examine whether cholinergic stimulation can enhance regeneration in vivo, we performed MI at P7, a time point when myocyte proliferation and regeneration is limited (Porrello et al., 2013). We injected the mice with the cholinergic agonist carbachol for 7 days (Figure 5J). At 7 days post injury, no significant difference in cardiomyocyte proliferation was detected (Figure 5K and L, n=5–6). These data suggest that cholinergic stimulation in vivo does not appear sufficient to induce cardiomyocyte proliferation.
Mechanical Denervation Disrupts Inflammatory Gene Expression Following Apical Resection in the Mouse
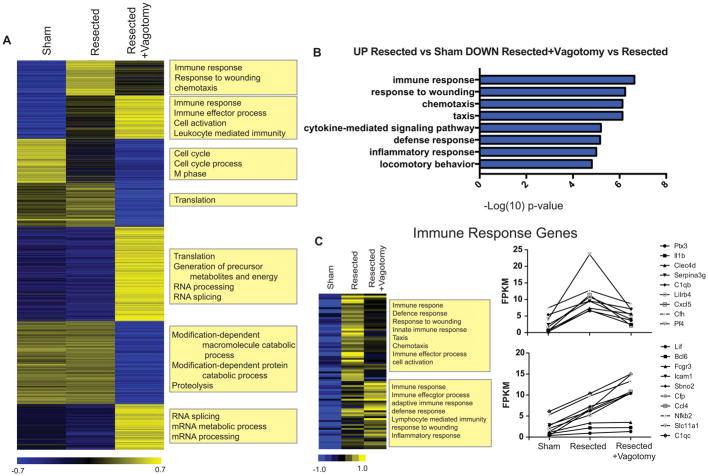
To gain insight into the early factors that might be responsible for nerve-dependent heart regeneration we performed expression analysis on the heart ventricular apex from animals that had undergone sham operation, apical resection, or apical resection and left vagotomy. RNA was extracted from the ventricular apex 24 hours after surgery and RNAsequencing was performed. As previously reported, apical resection stimulates a strong immune response 24 hours post surgery (Aurora et al., 2014; O’Meara et al., 2014). The most striking difference in gene expression between resected and resected plus vagotomy was the blunted expression of inflammatory genes activated upon resection (Figure 6A). Genes that were significantly upregulated in resected versus sham hearts and also significantly downregulated in resected+vagotomy versus resected hearts fell into gene ontology biological processes such as immune response, response to wounding, and chemotaxis (Figure 6B). Specifically, genes involved in the innate immune response and chemotaxis (IL1b, Cxcl5, and Pf4) were downregulated to almost baseline levels (Figure 6C). On the other hand, expression of several immune genes involved in lymphocyte mediated immunity and adaptive immune response (Icam-1, Slc11a1, Bcl6 and Nfkb2) was not blunted in response to vagotomy. A large subset of genes showed similar expression patterns in sham and resected hearts but were upregulated (Translation, RNA processing) or downregulated (Proteolysis, catabolic processes) after vagotomy. These genes are not likely related to the nerve dependent regeneration phenotype since they were not modulated upon resection alone. To determine whether similar transcriptional changes that regulate cholinergic-mediated cardiac regeneration are conserved across species, we performed a microarray on atropine treated zebrafish at 24 hours following resection. Interestingly, there was a significant downregulation of immune response genes in the atropine treated zebrafish compared to control (Figure S4). These data suggest that immune function plays an important role in both zebrafish and heart regeneration, and in both models the immune response is blunted by cholinergic inhibition. Collectively, our results reveal a requirement for nerves in guiding cardiac regeneration in both zebrafish as well as neonatal mice by regulating cardiomyocyte proliferation (Figure 7).
Figure 6. Immune Response Genes are Differentially Expressed Following Mechanical Denervation.
A) K-means clustering of all genes differentially expressed across hearts from animals that had undergone sham operation, apical resection, or apical resection and left vagotomy surgeries. B) Gene ontology biological processes of genes significantly up regulated in resected versus sham hearts and also significantly downregulated in resected+vagotomy versus resected hearts. C) K-means clustering and FPKM plots of Immune response genes significantly up regulated in resected versus sham hearts.
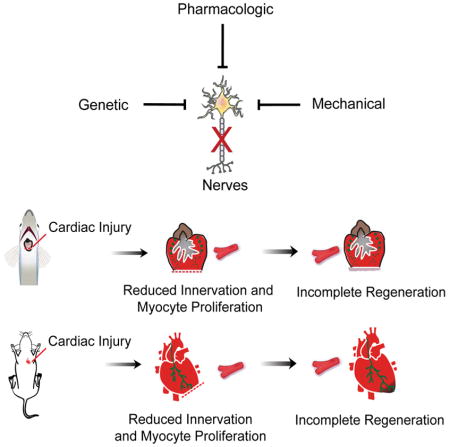
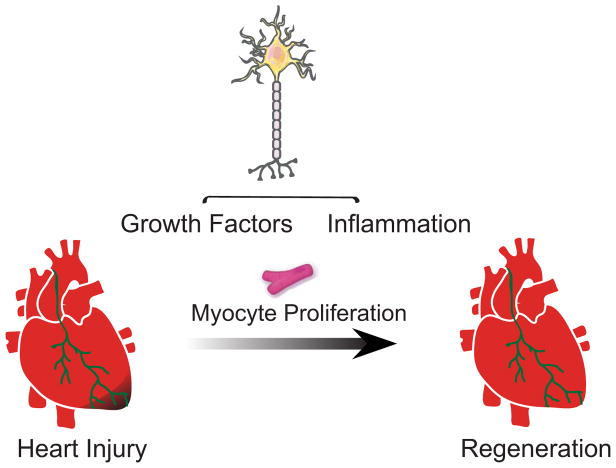
Figure 7. Proposed Model of Nerve Dependence of Heart Regeneration.
A schematic showing the proposed role that nerves play on cardiomyocyte proliferation and the impact of pharmacological, genetic and mechanical denervation on heart regeneration.
DISCUSSION
Multiple species throughout the animal kingdom mediate their regenerative response through nerve activity; we sought to explore whether similar mechanisms are evolutionarily conserved in heart regeneration (Kumar and Brockes, 2012). Our data demonstrate that nerves are required for cardiomyocyte proliferation and heart regeneration of both zebrafish and neonatal mice. Hypo-innervation of the adult zebrafish heart using transgenic zebrafish reduces myocyte proliferation and heart regeneration. Pharmacological inhibition of cholinergic nerve function reduces cardiomyocyte cell cycle activity in both zebrafish and neonatal mice. In addition, we demonstrate that mechanical denervation of the left vagus nerve in neonatal mice reduces cell cycle gene expression, cardiomyocyte proliferation and prevents heart regeneration following both apical resection and myocardial infarction suggesting a critical role for cholinergic nerve activity in heart regeneration. Administration of recombinant proteins of Nrg1 and Ngf increases cardiomyocyte proliferation and enhances regeneration following denervation. RNAseq analysis of mechanically denervated hearts demonstrates a significant impact of denervation on inflammatory pathways gene expression. A similar effect was observed in zebrafish following pharmacological inhibition of cholinergic nerve function.
This study sheds light on a requirement for nerves in regulating cardiomyocyte proliferation and heart regeneration in lower vertebrates and mammals. Nerves appear to be essential for the homeostasis of the adult human heart as well, due to re-innervation of the adult human heart post transplantation (Cornelissen et al., 2012; Gallego-Page et al., 2004). Recent studies have uncovered multiple regulators of mammalian cardiac regeneration (Aurora et al., 2014; Chen et al., 2013; Eulalio et al., 2012; Heallen et al., 2013; Mahmoud et al., 2013; Porrello et al., 2011a; Puente et al., 2014; Xin et al., 2013), yet the differences and/or similarities of evolutionary conserved regenerative mechanisms between lower organisms and mammals is not yet clear. We demonstrate that the impact of nerves seems to affect both the expression of growth factors as well as the inflammatory response post injury. Owing to the established role of semaphorin signaling in vascular patterning (Epstein et al., 2015), it is not clear whether a vascular mechanism is involved in the regenerative response as well. Interestingly, modulating expression of growth factors as Ngf by cholinergic signaling has been reported following cholinergic denervation in the brain (da Penha Berzaghi et al., 1993; Lapchak et al., 1993), suggesting interplay between cholinergic nerves and Nrg1 and Ngf activity. Recent reports identified Nrg1 as an inducer of myocyte proliferation in zebrafish, as well as in mice within the first week of birth, through the Nrg1 co-receptor Erbb2 (D’Uva et al., 2015; Gemberling et al., 2015; Polizzotti et al., 2015). Our rescue experiments with Nrg1 and Ngf indicate that administration these factors may partially rescue the effect of denervation, but whether these factors specifically mediate the nerve effect is unclear. Attempts at identifying nerve-derived factors during regeneration have been pursued, but their exact role in different species is not completely understood (Kumar and Brockes, 2012; Kumar et al., 2007). It remains to be determined whether nerves directly release these mitogens, or regulate their expression. In addition, the impact of cholinergic signaling on inflammation has been previously studied (Martelli et al., 2014). The role of the immune system has been shown to mediate a proper regenerative response following injury as well (Aurora et al., 2014; Godwin et al., 2013; Lavine et al., 2014) perhaps suggesting interplay between cholinergic nerves and immune response in facilitating heart regeneration. Collectively, our study identifies a dual role of nerves in heart regeneration in both lower vertebrates and mammals by regulating mitogen and inflammatory gene expression following injury.
EXPERIMENTAL PROCEDURES
Experimental Animals
All experiments were conducted in accordance with the Guide for the Use and Care of Laboratory Animals and approved by the Harvard Medical School Standing Committee on Animals. Timed-Pregnant ICR/CD1 mice used to deliver pups were obtained from Charles River Laboratories.
Zebrafish Amputations
Adult zebrafish from 3–5 months were used for apical amputations as described previously (Poss et al., 2002).
Neonatal Mouse Apical Resection and Myocardial Infarction
Neonatal mouse apical resections and myocardial infarctions were performed on 1-day-old and 7-day-old pups as described previously (Mahmoud et al., 2014).
Neonatal Mouse Vagotomy
Neonatal mice were anesthetized on ice. The neck area was cleaned three times alternately using alcohol and betadine in preparation for surgery. A lateral incision of about 5 mm was made approximately 2–3 millimeters from the clavicle on the left center side of the throat. The lobes of the salivary glands were gently separated and the fascia and fatty tissue were separated until the carotid jugular bundle was visualized. The carotid artery and jugular vein were gently isolated until the vagus nerve was visible. The vagus nerve was firmly grasped and pulled towards the head of the mouse until the vagus broke. A segment of about 3–6 mm is sufficient to ensure complete separation with little chance of reattachment. Mice that had resection of a smaller part of the vagus were excluded from the analysis.
Administration of Pharmacological Agents
For zebrafish experiments, atropine was mixed with tank water and changed daily. Atropine or propranolol was added at a concentration of 50 μM. Methoctramine was added at a concentration of 50 nM.
For neonatal mice, atropine was dissolved in phosphate buffered saline (PBS) and administered to neonatal mice following apical resection at P1 by intraperitoneal injections twice daily at a concentration of 5mg/kg. Carbachol was injected at a concentration of 50 μM. PBS administration was used as vehicle.
Generation of Transgenic Lines
cmlc2:sema3aa
sema3aa cDNA was amplified with the primers 5′-ACAATGGATTACCTTGTTGG-3′ and 5′-ACATTACACGCTGCGTGGTGG-3′ and subcloned behind the 5.1 kb cmlc2 (myl7 – Zebrafish Information Network) promoter (Rottbauer et al., 2002). The cassette was co-injected into one-cell stage wild-type embryos with I-SceI. Two founders were isolated and propagated. The full name of this transgenic line is Tg(cmlc2:sema3aa)pd106.
Histological Analysis
Primary and secondary antibody staining was performed as described (Kikuchi et al., 2011). Whole mount nerve staining was performed as follows. Heart were fixed in 4% PFA for 1 hour at room temperature with atria removed. Heart were then washed 4 X 5 minutes with PBS that contained 0.3% tween-20 (PBT). Hearts were placed in blocking media (0.3% PBT, 2% horse serum, 1% DMSO, and 10% calf-serum (Heat inactivated)) for 2 hours room at 4°C on rotator. Block media was removed and replaced with antibody staining solution (0.3% PBT, 1% DMSO, 1:100 acetylated- alpha-tubulin, and 10% calf-serum (Heat inactivated) overnight at 4°C. Hearts were then placed in mounting media between coverslips for imaging.
Acid Fuchsin-Orange G staining was performed on 10 μm sections as described (Poss et al., 2002). Mef2/PCNA staining on sections from 7 dpa ventricles was performed and imaged as described (Kikuchi et al., 2011). Mef2+ and Mef2+/PCNA+ cells were counted manually. The three largest injuries from each heart were averaged to compute a proliferative index for each animal. Primary antibodies used in this study: anti-PCNA (mouse; sigma) at 1:250, anti-Mef2 (rabbit; Santa Cruz Biotechnology) at 1:75, and anti-acetylated alpha-tubulin (mouse; Sigma) at 1:100. Secondary antibodies used in this study: Alexa Fluor 594 goat anti-mouse IgG (H+L) for anti-Mef2, anti-PCNA, and anti-acetylated alpha-tubulin; and Alexa Fluor 488 goat anti-rabbit IgG (H+L) for anti-Mef2 and anti-PCNA. Secondary antibodies (Invitrogen) were all used at 1:200.
Mouse hearts were fixed in 4% paraformaldehyde and paraffin embedded. Paraffin sections were deparaffinized in xylene and rehydrated by graded alcohols, followed by antigen retrieval with antigen retrieval citrate solution (DAKO) in boiling water for 20 minutes. Slides were cooled at room temperature for 30 minutes; sections were blocked with 10% goat serum, and incubated with pH3 (Millipore, 1:200), Aurora B (Sigma, 1:100) cardiac Tnnt (Abcam, 1:200) and Tubb3 (Abcam, 1:500) antibodies overnight at 4°C. The following day, sections were washed with PBS and incubated with corresponding secondary antibodies conjugated to Alexa Fluor 488 and Alexa Fluor 568 (Invitrogen).
Trichrome staining was performed on neonatal mouse hearts fixed in 4% paraformaldehyde and paraffin embedded. Sections were deparaffinized, placed in Bouin’s fluid at 60°C for 1 hour, and then rinsed for 5 minutes in deionized water. Sections were then stained with iron hematoxylin for 10 minutes then rinsed in deionized water for 10 minutes. Sections were stained with Biebrich Scarlet-Acid Fuchsin solution for 5–10 minutes then rinsed in deionized water for 30 seconds. Sections were placed in Phosphotungstic-Phosphomolybdic Acid solution for 5 minutes, and then stained in Aniline Blue for 5–10 minutes. Slides were placed in 1% Acetic Acid for 1 minute, rinsed in deionized water for 30 seconds, followed by dehydration in alcohol for two minutes then mounted with a coverslip.
Quantification of Innervation
Whole mount images obtained from cmlc2:sema3aa hearts and controls stained with acetylated-alpha tubulin, were imported into Photoshop and merged using the photomerge function. Subsequent brightness and contrast adjustments were made to allow for proper masking of the nerves. Images were then imported to ImageJ, where thresholding was set to define the area of the nerves. Masks for total area of the heart and nerve area were quantified using ImageJ. Nerves were quantified by calculating the total area of the nerves divided by the total area of the surface of the heart to calculate a percent nerve area. Quantification was performed for the surface of the heart opposite the valve.
Quantification of Regeneration
Regenerates were scored on a scale from 1 to 3 after Acid-Fuschin Orange staining for zebrafish and trichrome staining for mice. “1” indicated complete regeneration (signified by contiguous muscle and little to no scarring), “2” indicated partial regeneration (signified by moderate scarring and partial muscle regeneration), and “3” indicated a block in regeneration (clear gap in ventricular wall, filled by fibrin and scar). Representative images from each animal were scored by 3 individuals blinded to the identities of the animals. The average score for each heart was calculated and rounded to the nearest whole number.
Real-Time PCR
Total RNA was isolated from zebrafish and mouse hearts using Trizol (Invitrogen) and reverse transcription for cDNA synthesis was performed using random hexamer primers with the High-Capacity cDNA Reverse Transcription Kit (Invitrogen). Validated primers from MGH primer bank (http://pga.mgh.harvard.edu/primerbank/) were ordered from Integrated DNA technologies. Real time PCR was performed with SYBR Green (Applied Biosystems) on CFX384 Touch Real-Time PCR detection system (Bio-Rad). GAPDH, Beta-actin, and 18S were used as a loading control to normalize gene expression using the ΔΔCt method.
RNA Sequencing and Analysis
At 24 hours post-procedure, neonatal mouse hearts were isolated and perfused in 1X PBS. Total RNA was extracted from the lower half of the ventricle using Trizol (Invitrogen) according to the manufacturer’s protocol. The Poly(A) mRNA Isolation Module (Wafergen, Cat# 400047) was used to isolate poly-adenylated RNA from 0.2 to 1.0 μg of total RNA. The polyadenylated RNA was fragmented, and the first and second strand synthesis were performed on a Wafergen Apollo 324 System using the IntegenX Directional PrepX mRNA kit (IntegenX). The addition of barcodes and minimal library amplification were done using PCR. Library clustering and paired-end (50 bp read lengths) sequencing were performed on an Illumina HiSeq 2500. Three biological replicates for each group (sham, resection, and resection + vagotomy) were processed and sequenced as described above. Sequencing reads were aligned to the GRCm38 mouse genome using Tophat (Kim et al., 2013). Cufflinks was used to assemble transcripts and to estimate transcript abundances, which are measured in fragments per kilobase per million fragments mapped (FPKM) (Trapnell et al., 2010). A q-value cutoff of 0.05 was used to determine the statistical significance of differentially expressed genes. The heatmaps were generated in Java Treeview (Saldanha, 2004). Gene ontology (GO) categories were determined using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (Huang da et al., 2009a, b). Benjamini-Hochberg-corrected p-values are reported for enriched biological processes. RNAseq data has been uploaded to the gene expression omnibus (GEO accession GSE69855).
Isolation, Culture and Treatment of Neonatal Rat Cardiomyocytes
Rat ventricular cardiomyocytes were isolated from P1 neonatal Sprague Dawley rats as previously described (Liao and Jain, 2007). Cardiomyocytes in serum free media were treated with multiple doses of acetylcholine, Nrg1 (R&D) and Ngf (Sigma) followed by quantification of 3H thymidine incorporation.
3H Thymidine Incorporation
To quantify 3H thymidine incorporation, cardiomyocytes were treated with 3H thymidine 24 hours before collection. Cells were washed with PBS and lysed with 0.1M NaOH + 0.1% SDS. Quantification was performed using the LS 6500 Scintillation Counter (Beckman Coulter).
Statistical Analysis
All data are presented as mean ± SEM. Student’s unpaired t test or one way ANOVA was used for comparisons between two groups unless otherwise noted. A value of P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank the Lee and Poss laboratory members for helpful discussions, J. Burris, N. Blake, S. Davies, and A. Dunlap for zebrafish care, and A. Dickson for quantitative analysis of images. This work was supported by an American Heart Association Postdoctoral fellowship 15POST21870000 (A.I.M.), NIH F32HL117595 Postdoctoral Fellowship (C.C.O.), and an HHMI Gilliam Fellowship (D.M.B.). M.G. was supported by a fellowship from the AHA. L.Z. was supported by a fellowship from the AHA (14POST20380738). W.Y.C. was supported by a fellowship from NIH (F32 HL106987-02). This work was supported by a grant from the NIH (HL127067) to C.E.B. and C.G.B., (HL081674) to K.D.P. and NIH grants (AG040019) and (HL117986) to R.T.L.
Footnotes
AUTHOR CONTRIBUTIONS
A.I.M. and C.C.O. designed and performed the experiments, analyzed the data, made the figures and wrote the paper. M.G., W.C., G.F.E. and K.D.P. generated the cmlc2:sema3aa zebrafish and assessed cardiac innervation and heart regeneration in these animals. L.Z., R.Z., D.M.B., J.B.G. and L.C. performed experiments. C.E.B., C.G.B. and C.A.M. supervised experiments and edited the paper. R.T.L. conceived the study, designed experiments and wrote and edited the paper.
Supplemental information includes supplemental materials and methods and two figures and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aurora AB, Porrello ER, Tan W, Mahmoud AI, Hill JA, Bassel-Duby R, Sadek HA, Olson EN. Macrophages are required for neonatal heart regeneration. The Journal of clinical investigation. 2014;124:1382–1392. doi: 10.1172/JCI72181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- Bryant DM, O’Meara CC, Ho NN, Gannon J, Cai L, Lee RT. A systematic analysis of neonatal mouse heart regeneration after apical resection. Journal of molecular and cellular cardiology. 2015;79:315–318. doi: 10.1016/j.yjmcc.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, Hu X, Wang G, Lin Z, Wang S, et al. mir-17–92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circulation research. 2013;112:1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LN, Zang WJ, Yu XJ, Liu J, Li DL, Kong SS, Lu J, Xu XL. Compensatory recovery of vagal control of hemodynamics after unilateral vagotomy. Physiological research/Academia Scientiarum Bohemoslovaca. 2008;57:119–132. doi: 10.33549/physiolres.931095. [DOI] [PubMed] [Google Scholar]
- Cornelissen VA, Vanhaecke J, Aubert AE, Fagard RH. Heart rate variability after heart transplantation: a 10-year longitudinal follow-up study. Journal of cardiology. 2012;59:220–224. doi: 10.1016/j.jjcc.2011.12.002. [DOI] [PubMed] [Google Scholar]
- D’Uva G, Aharonov A, Lauriola M, Kain D, Yahalom-Ronen Y, Carvalho S, Weisinger K, Bassat E, Rajchman D, Yifa O, et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nature cell biology. 2015;17:627–638. doi: 10.1038/ncb3149. [DOI] [PubMed] [Google Scholar]
- da Penha Berzaghi M, Cooper J, Castren E, Zafra F, Sofroniew M, Thoenen H, Lindholm D. Cholinergic regulation of brain-derived neurotrophic factor (BDNF) and nerve growth factor (NGF) but not neurotrophin-3 (NT-3) mRNA levels in the developing rat hippocampus. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1993;13:3818–3826. doi: 10.1523/JNEUROSCI.13-09-03818.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhein S, Van Koppen CJ, Brodde OE. Muscarinic receptors in the mammalian heart. Pharmacological Research. 2001;44:161–182. doi: 10.1006/phrs.2001.0835. [DOI] [PubMed] [Google Scholar]
- Drachman DB. Atrophy of Skeletal Muscle in Chick Embryos Treated with Botulinum Toxin. Science. 1964;145:719–721. doi: 10.1126/science.145.3633.719. [DOI] [PubMed] [Google Scholar]
- Epstein JA, Aghajanian H, Singh MK. Semaphorin signaling in cardiovascular development. Cell metabolism. 2015;21:163–173. doi: 10.1016/j.cmet.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- Gallego-Page JC, Segovia J, Alonso-Pulpon L, Alonso-Rodriguez M, Salas C, Ortiz-Berrocal J. Re-innervation after heart transplantation: a multidisciplinary study. The Journal of heart and lung transplantation: the official publication of the International Society for Heart Transplantation. 2004;23:674–682. doi: 10.1016/j.healun.2003.07.011. [DOI] [PubMed] [Google Scholar]
- Gemberling M, Karra R, Dickson AL, Poss KD. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. eLife. 2015:4. doi: 10.7554/eLife.05871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin JW, Pinto AR, Rosenthal NA. Macrophages are required for adult salamander limb regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9415–9420. doi: 10.1073/pnas.1300290110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL, Martin JF. Hippo signaling impedes adult heart regeneration. Development. 2013;140:4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ieda M, Kanazawa H, Kimura K, Hattori F, Ieda Y, Taniguchi M, Lee JK, Matsumura K, Tomita Y, Miyoshi S, et al. Sema3a maintains normal heart rhythm through sympathetic innervation patterning. Nature medicine. 2007;13:604–612. doi: 10.1038/nm1570. [DOI] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Major RJ, Blum N, Dahn RD, Begemann G, Poss KD. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Developmental cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Brockes JP. Nerve dependence in tissue, organ, and appendage regeneration. Trends in neurosciences. 2012;35:691–699. doi: 10.1016/j.tins.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Kumar A, Godwin JW, Gates PB, Garza-Garcia AA, Brockes JP. Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science. 2007;318:772–777. doi: 10.1126/science.1147710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam NT, Currie PD, Lieschke GJ, Rosenthal NA, Kaye DM. Nerve growth factor stimulates cardiac regeneration via cardiomyocyte proliferation in experimental heart failure. PloS one. 2012;7:e53210. doi: 10.1371/journal.pone.0053210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapchak PA, Araujo DM, Hefti F. Cholinergic regulation of hippocampal brain-derived neurotrophic factor mRNA expression: evidence from lesion and chronic cholinergic drug treatment studies. Neuroscience. 1993;52:575–585. doi: 10.1016/0306-4522(93)90407-7. [DOI] [PubMed] [Google Scholar]
- Laube F, Heister M, Scholz C, Borchardt T, Braun T. Re-programming of newt cardiomyocytes is induced by tissue regeneration. Journal of cell science. 2006;119:4719–4729. doi: 10.1242/jcs.03252. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, Epelman S, Uchida K, Weber KJ, Nichols CG, Schilling JD, Ornitz DM, Randolph GJ, Mann DL. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:16029–16034. doi: 10.1073/pnas.1406508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao R, Jain M. Isolation, culture, and functional analysis of adult mouse cardiomyocytes. Methods in molecular medicine. 2007;139:251–262. doi: 10.1007/978-1-59745-571-8_16. [DOI] [PubMed] [Google Scholar]
- Mahmoud AI, Kocabas F, Muralidhar SA, Kimura W, Koura AS, Thet S, Porrello ER, Sadek HA. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497:249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud AI, Porrello ER, Kimura W, Olson EN, Sadek HA. Surgical models for cardiac regeneration in neonatal mice. Nature protocols. 2014;9:305–311. doi: 10.1038/nprot.2014.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli D, McKinley MJ, McAllen RM. The cholinergic anti-inflammatory pathway: a critical review. Autonomic neuroscience: basic & clinical. 2014;182:65–69. doi: 10.1016/j.autneu.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Meloni M, Caporali A, Graiani G, Lagrasta C, Katare R, Van Linthout S, Spillmann F, Campesi I, Madeddu P, Quaini F, et al. Nerve growth factor promotes cardiac repair following myocardial infarction. Circulation research. 2010;106:1275–1284. doi: 10.1161/CIRCRESAHA.109.210088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J, Onitsuka I, Hatch J, Uchida Y, Ray S, Huang S, Li W, Zang H, Ruiz-Lozano P, Mukouyama YS. Coronary veins determine the pattern of sympathetic innervation in the developing heart. Development (Cambridge, England) 2013;140:1475–1485. doi: 10.1242/dev.087601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Meara C, Wamstad JA, Gladstone R, Fomovsky G, Butty V, Shrikumar A, Gannon J, Boyer L, Lee RT. Transcriptional Reversion of Cardiac Myocyte Fate During Mammalian Cardiac Regeneration. Circulation research. 2014 doi: 10.1161/CIRCRESAHA.116.304269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberpriller JO, Oberpriller JC. Response of the adult newt ventricle to injury. The Journal of experimental zoology. 1974;187:249–253. doi: 10.1002/jez.1401870208. [DOI] [PubMed] [Google Scholar]
- Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circulation research. 2002;90:1044–1054. doi: 10.1161/01.res.0000020201.44772.67. [DOI] [PubMed] [Google Scholar]
- Polizzotti BD, Ganapathy B, Walsh S, Choudhury S, Ammanamanchi N, Bennett DG, Dos Remedios CG, Haubner BJ, Penninger JM, Kuhn B. Neuregulin stimulation of cardiomyocyte regeneration in mice and human myocardium reveals a therapeutic window. Science translational medicine. 2015;7:281ra245. doi: 10.1126/scitranslmed.aaa5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Johnson BA, Aurora AB, Simpson E, Nam YJ, Matkovich SJ, Dorn GW, 2nd, van Rooij E, Olson EN. MiR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circulation research. 2011a;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011b;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Johnson BA, Grinsfelder D, Canseco D, Mammen PP, Rothermel BA, Olson EN, Sadek HA. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Puente BN, Kimura W, Muralidhar SA, Moon J, Amatruda JF, Phelps KL, Grinsfelder D, Rothermel BA, Chen R, Garcia JA, et al. The Oxygen-Rich Postnatal Environment Induces Cardiomyocyte Cell-Cycle Arrest through DNA Damage Response. Cell. 2014;157:565–579. doi: 10.1016/j.cell.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Singer M. The influence of the nerve in regeneration of the amphibian extremity. The Quarterly review of biology. 1952;27:169–200. doi: 10.1086/398873. [DOI] [PubMed] [Google Scholar]
- Singer M, Davis MH, Scheuing MR. The influence of atropine and other neuropharmacological substances on regeneration of the forelimb in the adult urodele, Triturus. J Exp Zool. 1960;143:33–45. [Google Scholar]
- Todd JT. On the process of reproduction of the members of the aquatic salamander. The Quarterly Journal of Science, Literature, and the Arts. 1823;16:84–96. [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsonis PA, Madhavan M, Tancous EE, Del Rio-Tsonis K. A newt’s eye view of lens regeneration. The International journal of developmental biology. 2004;48:975–980. doi: 10.1387/ijdb.041867pt. [DOI] [PubMed] [Google Scholar]
- Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.