Abstract
We have designed and fabricated a miniature microscope from off-the-shelf components and webcam, with built-in fluorescence capability for biomedical applications. The mini-microscope was able to detect both biochemical parameters such as cell/tissue viability (e.g. Live/Dead assay), and biophysical properties of the microenvironment such as oxygen levels in microfabricated tissues based on an oxygen-sensitive fluorescent dye. This mini-microscope has adjustable magnifications from 8-60X, achieves a resolution as high as <2 μm, and possesses a long working distance of 4.5 mm (at a magnification of 8X). The mini-microscope was able to chronologically monitor cell migration and analyze beating of microfluidic liver and cardiac bioreactors in real time, respectively. The mini-microscope system is cheap, and its modularity allows convenient integration with a wide variety of pre-existing platforms including but not limited to, cell culture plates, microfluidic devices, and organs-on-a-chip systems. Therefore, we envision its widespread applications in cell biology, tissue engineering, biosensing, microfluidics, and organs-on-chips, which can potentially replace conventional bench-top microscopy where long-term in situ and large-scale imaging/analysis is required.
Introduction
Optical microscopy has demonstrated a pivotal role in biology and medicine since the 16th century. For the past few centuries tremendous amount of advancements have been made in enhancing the resolution of the images, improving the penetration depth, and strengthening the instrumentation. For example, various modalities based on interference such as phase contrast microscopy and differential interference contrast microscopy were developed to inspect cellular structures at better contrast.1, 2 The invention of fluorescence microscopy in the 1910s further expedited the entire field at a faster-than-ever pace by introducing the capability to probe specific molecules of interest instead of non-specific observations.3 Several of the more recent advances were due to instrumentation, specifically super-resolution microscopy such as stimulated emission depletion (STED) microscopy,4, 5 as well as volumetric imaging modalities such as confocal and two-photon microscopy.6, 7
Although these techniques provide the high-end capacities in probing biomedical problems, often times they cannot satisfy the need of high-throughput observations due to their bulky volumes. Current modalities available in the laboratory are difficult to mount inside an incubator, forcing one to transfer biological samples between the incubator and the microscope. Frequent disturbance to the cells may adversely affect their behavior and can be detrimental to certain cell types such as cardiomyocytes, which are highly sensitive to external perturbation and temperature alterations. Recent advances in device-based biological systems have further challenged the existing microscopy techniques where long-term, in situ, and simultaneous monitoring of parallel systems is required instead of extended resolution and volume.8-10 As an example, the microfluidic organs-on-a-chip platform builds upon interconnected human organ models, which maximally recapitulate the biology of their in vivo counterparts and the physiology of the circulation system.11-16 Due to its strong similarity with the human body, it has been widely used as a platform to screen potential new pharmaceutical compounds, personalized medicine, and antidotes against biological/chemical weapons.17-23 In order to achieve this ambitious aim, a variety of biosensors need to be embedded into the platform to monitor in real-time the responses of individual organ models and their microenvironment, such as those based on optical methods. The measurement of these parameters in complex microfluidic systems however, would be impractical using conventional bench-top microscopy.
Several compact image sensors have thus become available based on the lens-free technology.24-27 For example, using the lens-free imaging system the beating behavior of neonatal cardiomyocytes upon treatment with drugs doxorubicin and isoprenaline was analyzed in real time.28 Recent advances in the field have further led to the development of an ultra-portable foldable origami microscope29 as well as several mobile phone-based strategies that can image DNA strands,30 fluorescent nanoparticles, and virus.31 Despite being attractive solutions, these miniature imagers are not optimized for long-term monitoring of device-based biological systems in situ.
Based on our prior work on a prototype bright-field mini-microscope,28, 32 here we have further developed a system with built-in fluorescence capability that can be universally mounted at the bottom of any microfluidic bioreactors and devices. With a high resolution of <2 μm and adjustable magnifications of 8-60X, the mini-microscope could monitor in real time cellular behaviors including cell motility and beating analysis of liver- and heart-on-chips, respectively. The fluorescence capability further enabled in situ observation of hepatocyte viability stained with calcein/ethidium homodimer-1 (EthD-1) prior to and upon treatment with acetaminophen. Finally, we demonstrated the possibility to use the fluorescence mini-microscope as an oxygen sensor for intra-organoid measurement of oxygen levels. This microscope is miniaturized at a low-cost, allowing for simultaneous integration of multiple units for in situ, high-throughput imaging and analysis.
Experimental Section
Designing and construction of the mini-microscope
A commercially available, off-the-shelf Logitech C160m USB web camera was dissembled to remove the plastic casing and expose the complementary metal-oxide semiconductor (CMOS) sensor. The base structure of the mini-microscope was fabricated from a 3-mm poly(methyl methacrylate) (PMMA) sheet by laser cutting (VLS 2.30 Desktop Laser System, Universal Laser Systems). Four rectangular PMMA frames were cut out, with circular holes near the edges for the assembling screws/bolts. The bottom PMMA frames held the CMOS unit and lens while the top frames held the sample to be imaged (e.g. a microfluidic bioreactor) and the light source. Four sets of screws/bolts were mounted at the edge between the base and the sample holder for convenient focus adjustment.
To convert the webcam into the mini-microscope, we inverted the lens to achieve magnification, rather than the de-magnifying mechanism adopted by the camera.32 To achieve different magnifications, cylinders of various heights were cut from 2.0-mL Eppendorf tubes to set desired distances between the objective and the CMOS sensor. The tubes were further wrapped in black tape to prevent ambient light from reaching the sensor. Images were acquired with a laptop computer by connecting the camera through the USB port using either the webcam program or custom-written MATLAB (Mathworks) programs.
Quantification of resolution and field-of-view
A positive resolution target (R1DS1P, Thorlabs) was used to evaluate the resolution of the mini-microscope. Bright-field images were captured and the pixel intensities across the patterns on the resolution target were measured using ImageJ (National Institutes of Health) and plotted. Polystyrene microparticles of 16 μm in diameter were also used to qualitatively assess the magnification. Field-of-views were measured by imaging a hemocytometer and calculating the frame size based on the grids.
Digital channel separation with MATLAB
A custom-coded MATLAB script was used to split composite, as-captured images into separate channels of red, green, and blue. Briefly, the obtained image was imported into the MATLAB program and the three co-registering two-dimensional (2D) arrays were extracted from the three-dimensional (3D) matrices that represented images for red, green, and blue channels, respectively. The selected channels were then pseudo-colored to render the colors.
Construction of bioreactors
Resealable microfluidic bioreactors based on polydimethylsiloxane (PDMS, Dow Corning Sylgard 184, Ellsworth) were fabricated by modifying our recently developed protocol. Briefly, the bioreactors had a dual-layer structure with the inlet and organoid culture chamber on the bottom layer while outlet on the top. The two PDMS layers could be separated for organoid seeding, after which closure and sealing were achieved by clamping the pieces using a pair of PMMA surfaces tightened by screw/bolt sets.
HepG2 human hepatocellular carcinoma cells (HB-8065, ATCC) were seeded onto the bottom of the bioreactors at a density of approximately 1000 cells mm−2. Cultures were maintained at a flow rate of 200 μL h−1 in a Dulbecco's Modified Eagle's Medium (DMEM, Life Technologies) supplemented with 10 vol.% fetal bovine serum (FBS, Life Technologies) and 1 vol.% penicillin-streptomycin (P/S, Life Technologies). NIH/3T3 fibroblasts (CRL-1658, ATCC) were seeded onto the bottom of the bioreactors at a density of approximately 200 cells mm−2. Cultures were maintained in the same medium with HepG2 cells at a flow rate of 200 μL h−1. Neonatal rat cardiomyocytes were isolated from Sprague-Dawley rats of 2 days old following our established protocol approved by the Institutional Animal Care and Use Committee.33 The cardiomyocytes were then seeded onto gelatin methacryloyl (GelMA) hydrogel sheets (6 × 6 mm2) containing 1 mg mL−1 carbon nanotubes (CNTs) at a density of 5 × 105 cells.34, 35 The cell-seeded GelMA-CNT mats were first cultured in a petri dish DMEM supplemented with 10 vol.% FBS, 1 vol.% P/S, and 1 vol.% L-glutamine (Life Technologies) for up to 3 days until consistent beating was observed. The constructs were subsequently transferred to the bioreactors and maintained in the same medium at a flow rate of 200 μL h−1.
Live/Dead cell viability assessment
Fluorescence cell viability analysis for liver-on-chips were performed using the Live/Dead kit (Life Technologies) according to manufacturer’s instructions. When necessary, acetaminophen (Sigma-Aldrich) was added into the liver bioreactors at a concentration of 10 mM for 24 h of circulation prior to viability assay.
Oxygen measurements
PDMS microbeads with a uniform size of 30 μm were generated using a microfluidic flow focusing devices. Specifically, we used a three-to-one converging flow-focusing microchannel device to produce PDMS droplets, using uncured PDMS prepolymer (10:1) as the dispersive phase and 5 wt.% sodium dodecyl sulfate (SDS, Sigma-Aldrich) aqueous solution as the continuous phase. Obtained PDMS droplets were then cured at 60 °C before infusion with oxygen-sensitive dye, tris(4,7-diphenyl-1,10-phenanthroline) ruthenium(II) dichloride (ruthenium, Alfa Aesar) and oxygen-insensitive dye, Nile Blue (Sigma-Aldrich). Beads were incubated in a dichloromethane (Sigma-Aldrich) solution of ruthenium (5 mg mL−1), which also served as the swelling agent of PDMS to facilitate the infusion of the dyes into the microbeads. The dichloromethane solution was removed, briefly rinsed with isopropanol containing ruthenium, and extensively washed with distilled water for at least five times to minimize the presence of solvents. The beads were then immersed in an aqueous solution of Nile Blue (1 μg mL−1) for 24 h. The fluorescence of ruthenium was excited at 455 nm while that of Nile blue was excited at 591 nm. In order to improve the sensing resolution of the mini-microscope, a high-pass filter of >610 nm (common emission range of both dyes) was used.
Intra-organoid oxygen levels were measured by encapsulating the oxygen-sensing PDMS beads within a liver organoid made from HepG2 cells inside a GelMA hydrogel. A total of 2 μg of 5 wt.% GelMA prepolymer in phosphate-buffered saline (PBS, Life Technologies) containing 2 × 106 cells mL−1 and 2 × 104 beads mL−1 was printed at the center of a bioreactor and UV-crosslinked for 10 s. Another layer of GelMA containing only the beads but no cells was further crosslinked on top to cover the inner organoid. The culture was maintained for up to 24 h in a closed loop driven by a peristaltic pump at a flow rate of 200 μL h−1.
Results and Discussions
Design and characterization of a mini-microscope
Tracking cellular processes and tissue responses over time requires the use of specialized and expensive microscopes with a temperature, humidity, and CO2-controlled chamber. Laboratories often have limited access to one, which makes the analysis of multiple samples in real-time challenging and time-consuming. To fulfil these needs, we have developed a fluorescence mini-microscope (Figure 1, a and b) with out-of-the-shelf components and encased it within a specially designed PMMA frame for easy integration with existing tissue culture devices such as microfluidic bioreactors. The mini-microscope has a small form factor (4.2 × 5.5 cm2) and is highly portable (65 g). It has a series of applications comparable to a bench-top fluorescence microscope such as live cell imaging, long-term tracking of cellular processes, fluorescence analysis, and biosensing. The reduced cost of each microscope (<$10, see supplementary Table 1 for detailed components and price), together with readily available off-the-shelf parts, enables researchers to manufacture several units in a laboratory setting with minimum investment. This is an ideal solution to economically track multiple samples at once. Additionally, using the mini-microscope directly inside the incubator avoids extra steps that can contribute to cell damage, including changes in temperature, pH, and increased chance of microbial/fungal contaminations. Furthermore, with the emergence of cell-based lab-on-a-chip applications, we highlight the nature of our modular plug-and-play mini-microscope, which is capable of connecting to different setups via screws and bolts.
Fig. 1.

a) Schematic showing design of the mini-microscope integrated with a bioreactor. b) Photograph showing the actual device; the inset shows the device under LED illumination. c, d) Fabrication of the imaging unit of the mini-microscope: a webcam (Logitech C-160) is first disassembled to obtain the CMOS chip, after which the lens is detached, flipped, and then re-attached to the base to achieve magnification.
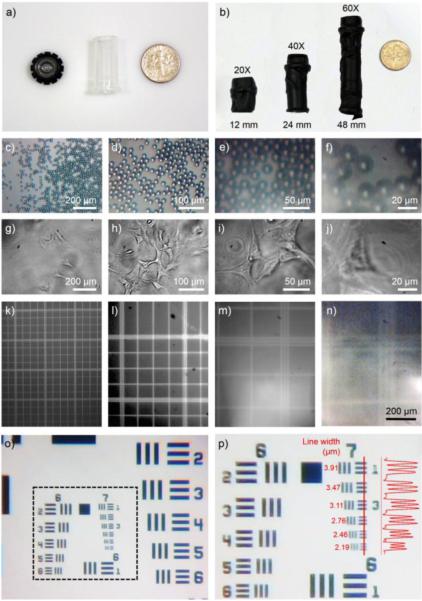
The body of the mini-microscope was constructed by inverting the webcam lens to magnify the object (Figure 1, c and d).32 Interestingly, it was found that, by varying the distance between the lens and the CMOS sensor using a spacer constructed from an Eppendorf tube, we could obtain a continuous gradient of magnifications. Using this approach we were able to equip the mini-microscope with 8X, 20X, 40X, and 60X magnifications at lens-to-sensor distance of 5 mm (no spacer), 12 mm, 24 mm, and 48 mm, respectively (Figure 2, a and b). As shown in Figure 2, c-f and g-j, polystyrene microbeads of 16 μm and NIH/3T3 fibroblasts could be clearly imaged under all these four magnifications. We then imaged a hemocytometer and determined the field-of-views of the mini-microscope at different magnifications to be 0.901 mm2, 0.200 mm2, 0.045 mm2, and 0.014 mm2, respectively (Figure 2, k-n).
Fig. 2.
a, b) Photographs showing the assembly of the objective with different magnifications by varying the height of the tubing. c-f and g-j) Mini-microscopic images of 16-μm polystyrene particles and NIH/3T3 fibroblasts at four different magnifications of 8X, 20X, 40X, and 60X. k-n) Full frame images of a hemocytometer captured from the mini-microscope at different magnifications where the field-of-views were calculated to be 1060 μm × 850 μm = 0.901 mm2, 500 μm × 400 μm = 0.200 mm2, 235 μm × 190 μm = 0.045 mm2, and 130 μm × 105 μm = 0.014 mm2, respectively. o, p) Mini-microscopic images of a resolution target showing Groups 6 and 7 targets where the microscope could easily resolve lines/spacing as small as 2.19 μm; the inset shows the intensity profile across the red line in the Group 7 targets.
In order to suit the majority of the applications, a lower magnification of 8X was chosen for subsequent characterizations. We determined the mini-microscope resolution by imaging a photographic resolution target. As shown in Figure 2, o and p, the microscope was able to resolve lines as closely spaced as 2.19 μm with clear peak separation. Such high resolution is sufficient for most laboratory applications on cell-based measurements. The design of the mini-microscope included four sets of screws/bolts for convenient focus adjustment (Figure 1, a and b). The working distance was determined to be 4.5 mm by imaging targets at pre-set distances from the objective (supplementary Figure S1). Inherited from the webcam specifications, the mini-microscope had a maximal resolution of 1280 × 1024 pixels (1.31 megapixels) and a frame rate of 30 frames per second (fps).
Filter-free color separation and fluorescence capability
One critical functionality for the use of a small and portable mini-microscope is its fluorescence capability. Tagging cells, subcellular compartments, and extracellular matrix (ECM) molecules with fluorescence markers allows extended monitoring with increased sensitivity and precision. Moreover, various cellular functional assays involve the usage of fluorescence tags, which highlights the importance of constructing a fluorescence-capable microscope. A color CMOS sensor is typically constituted by an array of interlacing red, green, and blue (RGB) sensing units (i.e. pixels, Figure 3a), composing a final RGB image when mixed together. A key aspect in using fluorescence is filtering the excitation light from the emission. While conventional methods are all based on the use of filters, we opted for a filter-free approach by digitally separating the red, green, and blue components of an obtained raw image, therefore distinguishing the contributions of the source from the emitter. A custom-coded MATLAB program was used to split the acquired images into separate R/G/B components. In order to demonstrate our concept, we used red, green, and blue LEDs as the emission sources and digitally separated the obtained images into three channels (Figure 3, b-d) and analyzed the emission intensities (Figure 3, e-g). As expected, each LED had a narrow wavelength of emission within its own range and gave a clear high-intensity signal in each respective RGB channel with minimal interference in other channels. To evaluate the fluorescence capability of the mini-microscope on imaging biological samples, we cultured HepG2 cells at the bottom of a liver bioreactor and stained them with calcein AM (1 μM for 20 min). A bright-field image captured under a broadband LED revealed the morphology of the cells (Figure 3h). We then used a blue LED (490 nm) to excite calcein for fluorescence detection. Without channel separation, it is noticed that both the excitation light (blue) and the emission light (green) were mixed together (Figure 3i), rendering the observation of the fluorescence signals highly inefficient. In comparison, when we digitally split the acquired image into distinct R/G/B components (Figure 3, j-l), we could clearly retrieve the blue excitation component in the blue channel (Figure 3k) and the green emission from calcein AM in the green channel (Figure 3l), with slight residual emission in the red channel (Figure 3j). A final pseudo-colored image (Figure 3m) with minimal interference from other channels was then obtained, showing the fluorescently labelled cells in the same way as viewed on a bench-top fluorescence microscope.
Fig. 3.
a) Schematic showing RGB configuration of the CMOS sensor of the mini-microscope. b-d) RGB and R/G/B images showing the filter-free separation of R, G, B colors by taking advantage of the digital channel unmixing using a custom-coded MATLAB program. e-g) Quantification data showing the accuracy and sensitivity of the digital channel separation approach. h) Bright-field image showing HepG2 cells grown in a liver-on-a-chip device. i) The raw image obtained from the mini-microscope showing the same HepG2 cells stained with calcein and illuminated with a monochromatic LED at 490 nm. j-l) Images from R/G/B channels after separation, respectively. m) Pseudo-colored and contrast enhanced image of the G channel showing the fluorescence from the stained cells with minimal interference from the illuminating B channel.
Dual-channel fluorescence imaging with mini-microscope
We next examined the capability of the fluorescent mini-microscope to simultaneously detect green and red fluorescence signals. We co-infused polystyrene beads labelled with fluorescein isothiocyanate (FITC) and rhodamine into a microfluidic channel. The mixture of the beads was then subjected to imaging under both the mini-microscope and a bench-top microscope (Nikon Eclipse Ti-S). It was clear that the mini-microscope could obtain images of the beads with green and red fluorescence, clearly differentiating the two types from a mixture (Figure 4, a-c, left panel). Significantly, the images captured using the mini-microscope were comparable with those from the bench-top microscope (Figure 4, a-c, left versus right panels), revealing the strong fluorescence capacity of the mini-microscope.
Fig. 4.
a-c) Dual-channel fluorescence imaging with the mini-microscope and the bench-top microscope (Nikon Eclipse) for (a) green fluorescent and (b) red fluorescent beads; superimposed images of the two channels are indicated in (c). d, e) Mini-microscopic images of live/dead staining results showing HepG2 cells non-treated or treated with 10 mM acetaminophen for 24 h in the liver bioreactors. f) Bench-top microscope (Zeiss Axio Observer D1) image showing the same cell colony as in (e), indicating the accuracy in the detection of the live/dead cells using the mini-microscope.
Assessment of drug toxicity of liver-on-a-chip with fluorescence mini-microscope
To further assess the usability of the fluorescence mini-microscope to track cellular events we performed a cytotoxicity Live/Dead assay. HepG2 cells in a liver bioreactor were treated with 10 mM acetaminophen for 24 h to induce cell death and subsequently stained with a Live/Dead kit. For fluorescence analysis, images were acquired using a blue and a green LED as excitation sources, for emissions in the green (live cells) and red (dead cells) channels, respectively. Filter-free color separation was performed as described above to separate the excitation and emission images. Without any drug administration, we could clearly image cells in green (live cells) with essentially no dead cells present (Figure 4d). At 24 h post administration of acetaminophen, on the other hand, massive cell death was observed in red (Figure 4e) with the mini-microscope. The images were compared with those obtained using a bench-top microscope (Zeiss Axio Observer D1), which exhibited similar results (Figure 4f). We proved with this experiment that our fluorescence mini-microscope could be used to assess cell viability in situ to screen cytotoxic drugs in an organs-on-a-chip platform.
Dynamic examination of liver- and heart-on-chips
The dynamic behaviors of the tissues/organoids in an organs-a-chip platform is a major parameter in assessing their viability and functionality. For example, the attachment, proliferation, and migration of cells in a bioreactor is highly dependent on the local microenvironment, including the supply of nutrients/oxygen, the fluid shear stress, and the administration of pharmaceutical agents.36-38 This is particularly true for heart-on-chip applications, where beating of the cardiac tissues indicates not only the viability but more importantly, also the functionality of the organoids.39, 40 While the beating rate stays constant for the cardiomyocytes under normal conditions, it is easily diminished when an external stimulus is introduced into the system, such as mechanical disturbance or change in temperature. The sensitive nature of these cells further emphasizes the necessity of integrating a miniaturized microscope that can be directly fitted underneath the bioreactor for long-term monitoring of undisturbed responses of the organoids. The need is enhanced by the dependence of beating rate of cardiomyocytes on drug treatment,40, 41 providing a simple but robust way to determine drug toxicity.
We first demonstrated the capability of our mini-microscope to follow the dynamic processes of simplified heart- and liver-on-a-chip models. HepG2 cells were seeded onto the bottom of the microfluidic bioreactor at a density of approximately 1000 cells mm−2. The culture was maintained at a flow rate of 200 μL h−1. Mini-microscope was fitted at the bottom of the hepatic bioreactor for continuous observation. As shown in Figure 5, a-d, HepG2 cells started to aggregate from time zero, and when compact structures of cell aggregates were formed they began to proliferate, well matching the reported motility and growth pattern of the cells.42, 43 The dynamic process of the cell migration is evidenced in supplementary Movie S1. We then cultured NIH/3T3 fibroblasts in another bioreactor and monitored their migration. Again, the mini-microscope demonstrated a very high resolution and could efficiently follow the motility of individual cells over a period of 2.5 h tested (supplementary Movie S2).
Fig. 5.
a-d) Time lapse mini-microscopic images showing the migration of HepG2 cells at the bottom of the bioreactor over a course of 72 h. e) Schematic and photograph showing the preparation of a glass substrate coated with GelMA-CNTs for cardiomyocytes culture. f) Mini-microscopic image showing rat neonatal cardiomyocytes seeded onto the substrate and embedded in the chamber of the bioreactor at Day 5. g) Real-time measurement of cardiomyocytes beating in situ, using custom-coded MATLAB program with the bioreactor-integrated mini-microscope. h) Measurement of cardiomyocytes beating using a bench-top microscope after the cardiac bioreactor was removed from the incubator for 30 min.
The heart is a vital organ in the body that functions as a biological pump to circulate the blood among all organs in the body. Heart-on-a-chip provides a viable platform for not only studying the biology but also screening cardiotoxic pharmaceutical compounds.39, 40, 44 Here we built a cardiac bioreactor to evaluate the capability of the mini-microscope to analyze the beating of the heart-on-a-chip (Figure 5e). A mixture of GelMA prepolymer and CNT was sandwiched between two glass surfaces spaced at 1 mm and UV-cured. Cardiomyocytes were then seeded on top of the substrate to form a confluent cell sheet mimicking the myocardium. In this case, CNTs were incorporated into the GelMA matrix in order to enhance the intercellular communication between the cardiomyocytes, promoting their functionality.34, 45 Figure 5f shows a bright-field image of the cardiomyocytes in the bioreactor, revealing the morphology of the cardiac tissue. The mini-microscope was able to continuously record the beating of the cardiac tissue (supplementary Movie S3), which could further be analyzed in real-time using custom-written MATLAB program based on an intensity or pixel-shift method (Figure 5g). Beating of the cardiomyocytes in the heart-on-a-chip platform was monitored for up to 3 days without any disturbance and the cells maintained stable beating over the entire course of observation. In contrast, when using the conventional bench-top microscope where the cardiac bioreactor was required to stay exposed to a decreased temperature for an extended period of time and/or under mechanical disturbance, the cardiac tissues demonstrated a slowed or irregular beating rate (Figure 5h), greatly interfering with experimental observations of cardiac behaviors when potentially altered beating patterns are expected upon drug treatment.
Fluorescence mini-microscope-based oxygen sensor
Besides imaging cellular structures, measurement of the physical microenvironment of the platform is equally important in an organs-on-a-chip platform.46 These physical parameters, including but not limited to pH value, oxygen level, temperature, and osmotic pressure, all contribute to maintaining the homeostasis of the system and normal functionality of the embedded organoids. Among these parameters, the oxygen level is critical as it strongly depends on organ type and tissue architecture.13, 47 Detection of oxygen levels in a microfluidic device and particularly oxygen consumption within the organoids however, is challenging. Conventional approaches such as those based on electrochemistry,48 are not suitable due to strong interference caused by proteins and small molecules secreted by cells that lead to rapid biofouling of the sensor surfaces. Optical oxygen sensors are convenient and barely affected by biofouling,49-52 but the methods using bench-top microscopy does not allow in situ measurements based on compact devices. Here again, we proved that our fluorescence mini-microscope could be a versatile tool as an oxygen sensor for measurement of intra-organoid oxygen levels in a liver-on-a-chip platform.
Oxygen-sensitive PDMS microbeads were fabricated using a microfluidic device following our recently developed protocol (Figure 6a, unpublished). The PDMS microbeads had a uniform diameter of 30 μm (Figure 6b), which could be well dispersed inside an organoid for microscopic observation (supplementary Figure S2). The microbeads were then doped with two fluorescent dyes, the oxygen-sensitive ruthenium dye and the inert, oxygen-irresponsive Nile Blue. While the fluorescence intensity of ruthenium is inversely proportional to the oxygen concentration (i.e. its fluorescence is quenched by oxygen molecules), that of Nile Blue is independent on oxygen and would serve as the internal control. In this case, a single piece of high-pass filter of >610 nm (common emission for both dyes) was inserted underneath the bioreactor to improve the sensitivity of the imaging compared to the filter-free approach. In addition, two programmed excitation LEDs were fitted on the top of the bioreactor: 455 nm for exiting ruthenium, and 591 nm for excitation of Nile Blue. Calibration curves were obtained using the setup in the absence of organoids (Figure 6c). It was clear from both the calibration curves and the mini-microscopic image that, the fluorescence intensities of ruthenium followed an exponential decay from 7-21% oxygen (Figure 6, d-i), whereas those of Nile Blue remained constant over the range (supplementary Figure S3). The almost instantaneous responsiveness of the ruthenium fluorescence intensity of the microbeads towards oxygen was further exemplified in supplementary Movies S4 and S5 where oxygen levels were tuned from 21-0% and 0-21%, respectively.
Fig. 6.

a) Microfluidic device to fabricate uniform PDMS microspheres. b) PDMS microbeads fabricated by the microfluidic device and infiltration of ruthenium dye to produce oxygen-sensitive beads. c) Calibration curve showing fluorescence intensity versus oxygen concentration. d-i) Fluorescence images obtained from the mini-microscope showing beads at different oxygen concentrations. Ruthenium channel was used for imaging. j) Schematic diagram showing the setup for real-time sensing of oxygen levels in a liver bioreactor. k, l) Bright-field and fluorescence images showing the cells and the oxygen sensing beads at 24 h post seeding.
We then embedded the oxygen-sensitive PDMS microbeads inside a liver organoid prepared by UV-crosslinking a GelMA spheroid encapsulating a mixture of HepG2 cells and the beads, at the bottom of a liver bioreactor; another layer of GelMA containing only the beads then encased the core (Figure 6j). The system was maintained in a perfusion culture at a flow rate of 200 μL h−1 for 24 h. Individual cells were also clearly discernible in the image captured by the mini-microscope under bright-field (Figure 6k). Mini-microscope images were further taken at both ruthenium channel and Nile Blue channel, and oxygen intensities of beads at different locations were recorded (Figure 6l) to calculate their respective oxygen levels based on the calibration curves. As expected, in the core of the liver organoid the oxygen level dropped significantly to only <10%, with a trend of less oxygen towards the core and more along the periphery. By comparison, in the outer layer of the hydrogel where no cells were present, the oxygen levels were much higher ranging from 12-16%. Such capability of the mini-microscope combined with oxygen-sensing beads have provided a unique opportunity for convenient investigation of the intra-organoid oxygen consumption in an organ-on-a-chip platform, particularly useful for complex organs where multiple cells types are involved. Due to the compact size of the mini-microscope and its easy integration with microfluidic platforms, many other applications in biosensing are also potentially feasible, such as inclusion of multiple modules to monitor oxygen levels up- and downstream of each bioreactor.
Conclusions
We have designed and fabricated a portable miniature microscope from off-the-shelf components and webcam, which had built-in multi-color fluorescence capability to follow cell motility, analyze cell/tissue viability, and as optical sensors to measure biophysical properties of the microenvironment such as oxygen levels inside organoids based on an oxygen-sensitive fluorescent dye. The mini-microscope has adjustable magnifications of 8-60X, a high resolution of <2 μm, and a long working distance of 4.5 mm (at 8X). The cost of the mini-microscope barely exceeds $9, and the modular design allows ready integration with a wide variety of pre-existing platforms including for example, petri dishes, cell culture plates, and microfluidic bioreactors. We believe that our fluorescence mini-microscope can likely replace the conventional bench-top microscopes in many biomedical applications where long-term in situ and high-throughput imaging capacity is required for investigations such as drug screening in organs-on-a-chip systems.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge funding by the Defense Threat Reduction Agency (DTRA) under Space and Naval Warfare Systems Center Pacific (SSC PACIFIC) Contract No. N66001-13-C-2027. The authors also acknowledge funding from the Office of Naval Research Young National Investigator Award, the National Institutes of Health (EB012597, AR057837, DE021468, HL099073, R56AI105024), and the Presidential Early Career Award for Scientists and Engineers (PECASE). The publication of this material does not constitute approval by the government of the findings or conclusions herein. J.R. acknowledges the support from the Portuguese Foundation for Science and Technology (SFRH/BD/51679/2011).
Footnotes
Notes
The authors declare no conflict of interest.
Electronic Supplementary Information (ESI) available: Table S1, materials, part numbers, and cost analysis on the construction of fluorescence mini-microscope; Figure S1, procedure of constructing the mini-microscope body from a webcam; Figure S2, measurement of working distance; Figure S3, microscopic images showing the setup of intra-organoid oxygen measurement; Figure S4, fluorescence images obtained from the mini-microscope showing Nile blue fluorescence at different oxygen concentrations; Movie S1, the migration of HepG2 cells cultured at the bottom of the bioreactor over a course of 24 h; Movie S2, the migration of NIH/3T3 fibroblasts cultured at the bottom of a bioreactor over a course of 2.5 h; Move S3, the change in fluorescence intensity from the ruthenium-PDMS microbeads over 21-0% O2 levels captured by the mini-microscope; Move S4, the change in fluorescence intensity from the ruthenium-PDMS microbeads over 0-21% O2 levels captured by the mini-microscope. See DOI: 10.1039/b000000x/
References
- 1.Zernike F. Physica. 1942;9:686–698. [Google Scholar]
- 2.Pluta M. Nomarski's DIC microscopy: a review. 1994 [Google Scholar]
- 3.Lichtman JW, Conchello J-A. Nat. Methods. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 4.Hell SW, Wichmann J. Opt. Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 5.Klar TA, Hell SW. Opt. Lett. 1999;24:954–956. doi: 10.1364/ol.24.000954. [DOI] [PubMed] [Google Scholar]
- 6.Denk W, Strickler JH, Webb WW. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 7.Minsky M. Scanning. 1988;10:128–138. [Google Scholar]
- 8.Figeys D, Pinto D. Anal. Chem. 2000;72:330 A–335 A. doi: 10.1021/ac002800y. [DOI] [PubMed] [Google Scholar]
- 9.Whitesides GM. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 10.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 11.Huh D, Hamilton GA, Ingber DE. Trends Cell Biol. 2011;21:745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moraes C, Mehta G, Lesher-Perez SC, Takayama S. Ann. Biomed. Eng. 2012;40:1211–1227. doi: 10.1007/s10439-011-0455-6. [DOI] [PubMed] [Google Scholar]
- 13.Bhatia SN, Ingber DE. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 14.Moya ML, George SC. Curr. Opin. Chem. Eng. 2014;3:102–111. doi: 10.1016/j.coche.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sei Y, Justus K, LeDuc P, Kim Y. Microfluid. Nanofluid. 2014:1–14. [Google Scholar]
- 16.Wikswo JP. Exp. Biol. Med. 2014;239:1061–1072. doi: 10.1177/1535370214542068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esch M, King T, Shuler M. Annu. Rev. Biomed. Eng. 2011;13:55–72. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]
- 18.Ghaemmaghami AM, Hancock MJ, Harrington H, Kaji H, Khademhosseini A. Drug Discov. Today. 2012;17:173–181. doi: 10.1016/j.drudis.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selimović Š, Dokmeci MR, Khademhosseini A. Curr. Opin. Pharmacol. 2013;13:829–833. doi: 10.1016/j.coph.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Bhise NS, Ribas J, Manoharan V, Zhang YS, Polini A, Massa S, Dokmeci MR, Khademhosseini A. J. Controlled Release. 2014;190:82–93. doi: 10.1016/j.jconrel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebrahimkhani MR, Young CL, Lauffenburger DA, Griffith LG, Borenstein JT. Drug Discov. Today. 2014;19:754–762. doi: 10.1016/j.drudis.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polini A, Prodanov L, Bhise NS, Manoharan V, Dokmeci MR, Khademhosseini A. Expert opinion on drug discovery. 2014;9:335–352. doi: 10.1517/17460441.2014.886562. [DOI] [PubMed] [Google Scholar]
- 23.Zhang YS, Khademhosseini A. Nanomedicine. 2015;10:685–688. doi: 10.2217/nnm.15.18. [DOI] [PubMed] [Google Scholar]
- 24.Greenbaum A, Luo W, Su TW, Gorocs Z, Xue L, Isikman SO, Coskun AF, Mudanyali O, Ozcan A. Nat. Methods. 2012;9:889–895. doi: 10.1038/nmeth.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenbaum A, Zhang Y, Feizi A, Chung PL, Luo W, Kandukuri SR, Ozcan A. Science Transl. Med. 2014;6:267ra175. doi: 10.1126/scitranslmed.3009850. [DOI] [PubMed] [Google Scholar]
- 26.Ozcan A, Demirci U. Lab Chip. 2008;8:98–106. doi: 10.1039/b713695a. [DOI] [PubMed] [Google Scholar]
- 27.Coskun AF, Su T-W, Ozcan A. Lab Chip. 2010;10:824–827. doi: 10.1039/b926561a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SB, Bae H, Cha JM, Moon SJ, Dokmeci MR, Cropek DM, Khademhosseini A. Lab Chip. 2011;11:1801–1807. doi: 10.1039/c1lc20098d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cybulski JS, Clements J, Prakash M. PloS one. 2014;9:e98781. doi: 10.1371/journal.pone.0098781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei Q, Luo W, Chiang S, Kappel T, Mejia C, Tseng D, Chan RY, Yan E, Qi H, Shabbir F, Ozkan H, Feng S, Ozcan A. ACS Nano. 2014;8:12725–12733. doi: 10.1021/nn505821y. [DOI] [PubMed] [Google Scholar]
- 31.Wei Q, Qi H, Luo W, Tseng D, Ki SJ, Wan Z, Gorocs Z, Bentolila LA, Wu TT, Sun R, Ozcan A. ACS Nano. 2013;7:9147–9155. doi: 10.1021/nn4037706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SB, Koo K.-i., Bae H, Dokmeci MR, Hamilton GA, Bahinski A, Kim SM, Ingber DE, Khademhosseini A. Lab Chip. 2012;12:3976–3982. doi: 10.1039/c2lc40345e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khademhosseini A, Eng G, Yeh J, Kucharczyk P, Langer R, Vunjak-Novakovic G, Radisic M. Biomed. Microdevices. 2007;9:149–157. doi: 10.1007/s10544-006-9013-7. [DOI] [PubMed] [Google Scholar]
- 34.Shin SR, Jung SM, Zalabany M, Kim K, Zorlutuna P, Kim S. b., Nikkhah M, Khabiry M, Azize M, Kong J, Wan K.-t., Palacios T, Dokmeci MR, Bae H, Tang X, Khademhosseini A. ACS Nano. 2013;7:2369–2380. doi: 10.1021/nn305559j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A. Biomaterials. 2010;31:5536–5544. doi: 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khetani SR, Bhatia SN. Nat. Biotechnol. 2008;26:120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 37.Lan S-F, Safiejko-Mroczka B, Starly B. Toxicol. In Vitro. 2010;24:1314–1323. doi: 10.1016/j.tiv.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Mao S, Gao D, Liu W, Wei H, Lin J-M. Lab Chip. 2012;12:219–226. doi: 10.1039/c1lc20678h. [DOI] [PubMed] [Google Scholar]
- 39.Grosberg A, Alford PW, McCain ML, Parker KK. Lab Chip. 2011;11:4165–4173. doi: 10.1039/c1lc20557a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal A, Goss JA, Cho A, McCain ML, Parker KK. Lab Chip. 2013;13:3599–3608. doi: 10.1039/c3lc50350j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menna P, Salvatorelli E, Minotti G. Chem. Res. Toxicol. 2008;21:978–989. doi: 10.1021/tx800002r. [DOI] [PubMed] [Google Scholar]
- 42.Manley P, Lelkes PI. J. Biotechnol. 2006;125:416–424. doi: 10.1016/j.jbiotec.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 43.Chang TT, Hughes-Fulford M. Tissue Eng. A. 2008;15:559–567. doi: 10.1089/ten.tea.2007.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang YS, Aleman J, Arneri A, Bersini S, Piraino F, Shin SR, Dokmeci MR, Khademhosseini A. Biomed. Mater. 2015 doi: 10.1088/1748-6041/10/3/034006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin SR, Bae H, Cha JM, Mun JY, Chen Y-C, Tekin H, Shin H, Farshchi S, Dokmeci MR, Tang S. ACS nano. 2011;6:362–372. doi: 10.1021/nn203711s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wikswo JP, Block FE, Cliffel DE, Goodwin CR, Marasco CC, Markov DA, McLean DL, McLean JA, McKenzie JR, Reiserer RS, Samson PC, Schaffer DK, Seale KT, Sherrod SD. Biomedical Engineering, IEEE Transactions on. 2013;60:682–690. doi: 10.1109/TBME.2013.2244891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ. Lab Chip. 2013;13:3496–3511. doi: 10.1039/c3lc50243k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thévenot DR, Toth K, Durst RA, Wilson GS. Biosensors Bioelectron. 2001;16:121–131. doi: 10.1016/s0956-5663(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 49.Chan C-M, Chan M-Y, Zhang M, Lo W, Wong K-Y. Analyst. 1999;124:691–694. [Google Scholar]
- 50.Jiang K, Thomas PC, Forry SP, DeVoe DL, Raghavan SR. Soft Matter. 2012;8:923–926. [Google Scholar]
- 51.Sud D, Mehta G, Mehta K, Linderman J, Takayama S, Mycek M. J. Biomed. Opt. 2006;11:50504–50506. doi: 10.1117/1.2355665. [DOI] [PubMed] [Google Scholar]
- 52.Mehta G, Mehta K, Sud D, Song JW, Bersano-Begey T, Futai N, Heo YS, Mycek M-A, Linderman JJ, Takayama S. Biomed. Microdevices. 2007;9:123–134. doi: 10.1007/s10544-006-9005-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.