Abstract
Oligonucleotide aptamers can specifically bind biomarkers on cancer cells and can be readily chemically modified with different functional molecules for personalized medicine. To target acute myeloid leukemia (AML) cells, we developed a single-strand DNA aptamer specific for the biomarker CD117, which is highly expressed on AML cells. Sequence alignment revealed that the aptamer contained a G-rich core region with a well-conserved functional G-quadruplex structure. Functional assays demonstrated that this synthetic aptamer was able to specifically precipitate CD117 proteins from cell lysates, selectively bound cultured and patient primary AML cells with high affinity (Kd < 5 nM), and was specifically internalized into CD117-expressing cells. For targeted AML treatment, aptamer-drug conjugates were fabricated by chemical synthesis of aptamer (Apt) with methotrexate (MTX), a central drug used in AML chemotherapy regimens. The formed Apt-MTX conjugates specifically inhibited AML cell growth, triggered cell apoptosis, and induced cell cycle arrest in G1 phase. Importantly, Apt-MTX had little effect on CD117-negative cells under the same treatment conditions. Moreover, exposure of patient marrow specimens to Apt-MTX resulted in selective growth inhibition of primary AML cells and had no toxicity to off-target background normal marrow cells within the same specimens. These findings indicate the potential clinical value of Apt-MTX for targeted AML therapy with minimal to no side effects in patients, and also open an avenue to chemical synthesis of new, targeted biotherapeutics.
Keywords: acute myeloid leukemia (AML), CD117 biomarker, aptamer-drug conjugates, methotrexate (MTX), oligonucleotide aptamer, targeted therapy
1. Introduction
Aptamers are a class of molecular ligands composed of synthetic single-stranded oligonucleotides (DNA or RNA) that are able to specifically bind their targets with high affinity [1]. As “chemical antibodies”, oligonucleotide aptamers can be chemically synthesized and easily conjugated with different functional molecules [2–6]. Due to these unique chemical and biological properties, synthetic aptamers have been widely used in biomedical applications to specifically target biomarkers [6–11] for cancer cell detection and targeted therapy [5, 11, 12]. Personalized medicine employing targeted therapeutic approaches not only enhances therapy efficacy, but also reduces adverse side effects in cancer patients. Recent studies have shown that antibody-drug conjugates are a promising technology for targeted cancer therapy [13–16]. However, production of humanized monoclonal antibodies and subsequent drug conjugation are costly and time and labor consuming. In contrast, aptamers can be easily chemically synthesized and, due to their smaller size, exhibit more efficient tissue penetration, faster binding capacity to tumor cells, and no immunogenicity in vivo [17, 18]. These advanced chemical and biological features indicate a potential use for synthetic aptamers in clinical applications.
The current treatment regimen for acute myeloid leukemia (AML) is remission-induction chemotherapy, followed by either consolidation chemotherapy or allogeneic stem cell transplantation [19–22]. As most patients diagnosed with AML are in their sixth or seventh decade of life, many are not candidates for standard induction chemotherapy because of the severe adverse side effects, such as profound myelosuppression, life-threatening infections, and cardiotoxicity [23–26]. Therefore, a personalized medicine specific for AML with fewer side effects is urgently needed [27–31]. CD117 is a transmembrane receptor that is highly expressed on leukemia cells in 95% of patients with relapsed AML [32]. In addition, CD117-expressing AML patients have a worse survival prognosis than CD117-negative patients, and high levels of CD117 expression correlate with low rates of complete remission [33–37]. These data indicate that the CD117 receptor may be a potential therapeutic biomarker for the treatment of AML.
To develop new therapeutics targeting AML, we identified a CD117-specific ssDNA aptamer sequence through a hybrid selection approach. Subsequently, an aptamer-drug conjugate was formulated by synthesizing the aptamer sequence with methotrexate (MTX), a drug used to treat AML. Functional analysis with clinical specimens demonstrated that the aptamer-methotrexate (Apt-MTX) conjugates specifically killed patient primary AML cells and had no toxicity to the off-target background marrow cells of the same specimens.
2. Materials and methods
2.1 Reagents and cell lines
CD117-expressing HEL cells (a leukemia cell line from ATCC, Manassas, VA) were used to select single stranded (ss) DNA aptamers. CD117 recombinant protein with a polyhistidine (his) tag at the C-terminus (Sino Biological Inc. Beijing, China) was used to further enrich ssDNA sequences specific for the CD117. The CD117-negative cell lines included: the B lymphoma cell line CA46 (ATCC, Manassas, VA); the breast cancer cell line 468 (kindly provided by Dr. Haifa Shen, Houston Methodist Hospital); and the prostate cancer cell line LNCaP (ATCC, Manassas, VA). All suspension cells were cultured with RPMI1640 medium (Fisher Scientific, Pittsburgh, PA) with 10% FBS (Atlanta Biologicals, Lawrenceville, GA). All adhesion cell lines were cultured in DMEM (Atlanta Biologicals, Lawrenceville, GA) with 10% FBS.
2.2 Hybrid systematic evolution of ligands by exponential enrichment (SELEX)
For hybrid SELEX (Fig 1A), the ssDNA library consisted of a central, continuous stretch of 35 randomized sequences flanked by PCR primer sequences (5′-GAGGCATACCAGCTTATTCAA-35N-ATAGTAAGTGCAATCTGCGAA-3′). Cy3-labeled 5′ primer (5′-Cy3-GAGGCATACCAGCTTATTCAA-3′) and biotinylated 3′ primer (5′-biotin-TTCGCAGATTGCACTTACTAT-3′) were used in the initial PCR (Fig. 1B). The ssDNA library and primers were purchased from Integrated DNA Technologies (Coralville, IA). The synthesized ssDNA library was purified by reversed-phase ion pairing high-performance liquid chromatography (HPLC, Integrated DNA Technologies).
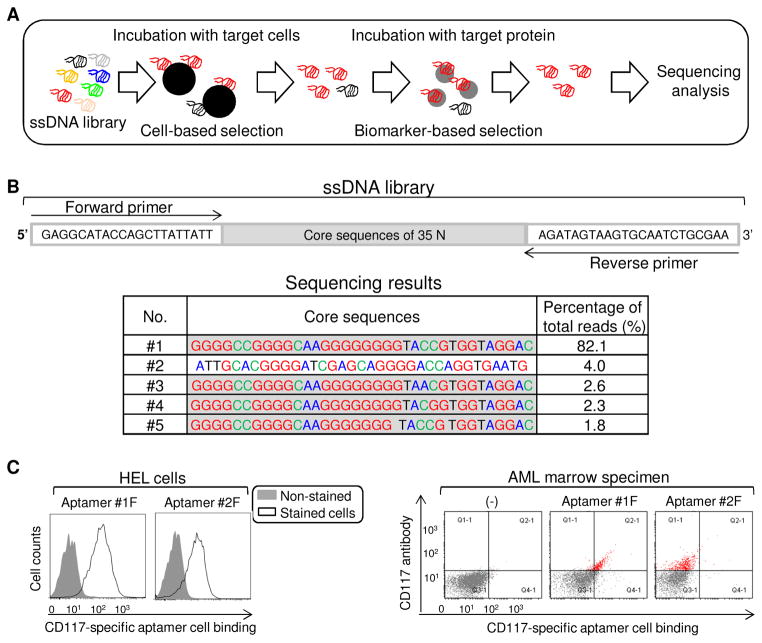
Fig. 1.
Development of CD117-specific ssDNA aptamers. (A) The process of aptamer selection using a hybrid cell- and protein-based enrichment approach. (B) Sequencing results of selected ssDNA aptamers, including forward primer region (23 nt), 35 nt random core region, and reverse primer region (23 nt). The top five dominant aptamer sequences with percentage of total sequencing reads. (C) Binding ability of aptamers #1 and #2 to cultured CD117-positive HEL cells (left panel) and patient primary AML cells (right panel) assessed by flow cytometry.
The aptamer enrichment process was performed as previously reported [38] and illustrated in Figure 1A. Briefly, CD117-positive HEL cells were used for 10 rounds of positive cell selection. Next, the enriched ssDNA pool underwent protein-based selection by incubation with CD117 recombinant protein. The protein selection process was repeated three times. After a total of 13 rounds of cell- and protein-based selection, binding affinity and specificity of the selected pool was determined by flow cytometry (LSRII, BD Biosciences, San Jose, CA). The final selected pool was sequenced using second-generation sequencing (LC Sciences, Houston, TX).
First, the ssDNA library was incubated with 5–10 × 106 CD117-positive HEL AML cells for 30 min at room temperature (RT). The cells were washed with DPBS buffer (Fisher Scientific) supplemented with 0.1% BSA, 0.1 g/L tRNA, and 5 mM MgCl2, and bound DNA was eluted by heating the mixture to 95°C for 10 min. The eluted sequences were PCR-amplified using the Cy3-labeled or non-fluorescent labeled forward and biotin-labeled reverse primers at the following cycle settings: denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s. The number of PCR cycles was optimized as below for each round of selection and ranged from 10 to 30. PCR products were separated on 2% agarose gels (Bio-Rad, Hercules, CA), and the highest number of cycles that didn’t produce nonspecific bands was used for subsequent amplification steps. Following amplification, ssDNA sequences were separated from the biotinylated antisense ssDNA by alkaline denaturation using 200 mM NaOH, and then purified with streptavidin-coated sepharose beads (Fisher Scientific). The separated ssDNA solution was passed through a desalting Nap-5 column (Fisher Scientific) to remove NaOH, and the purified ssDNA was used for the next round of selection. This cell line-based selection was repeated for total ten times.
Next, the aptamer was further refined through selection with recombinant CD117 protein. Ni-NTA magnetic beads (Qiagen, Valencia, CA) were pre-blocked with 5% bovine serum albumin (BSA, Fisher Scientific), washed with DPBS buffer, incubated with recombinant his-tagged CD117 protein for 30 min, and washed again with DPBS buffer. The CD117 protein-coated beads were incubated with the enriched DNA pool for 30 min, centrifuged for 5 min at 800 rpm, and washed twice. The eluted ssDNA then underwent two additional rounds of the recombinant protein-based selection.
2.3 Cell binding assay
Flow cytometry was used to monitor the enrichment of pools during the selection and to determine the binding affinity and specificity of the synthesized aptamers. The initial Cy3-labeled ssDNA pool or subsequently isolated aptamers were incubated for 30 min at 4°C or room temperature (RT) with 1 × 105 target or control cells at the indicated concentrations. The cells were washed once (500 μL DPBS) and resuspended in 500 μL of DPBS. The fluorescence intensity was determined with a FACScan cytometer (LSRII, BD Biosciences, San Jose, CA) by counting 10,000 events. The Cy3-labeled initial ssDNA library was used as a negative control.
2.4 Thioflavin T staining to determine the G4 structure of the aptamer sequences
To detect the presence of G4 structure, synthetic aptamers were stained with Thioflavin T (Th-T) (Sigma Aldrich, St. Louis, MO) as previously described [39]. Briefly, oligonucleotides and Th-T were mixed at 1 and 0.5 uM final concentrations, respectively, and incubated for 30 min at RT. Fluorescence emission was read at 490 nm after excitation at 425 nm in microplate reader (Biotek, Winooski, VT). Control aptamers that do not have GGG or GGGG repeats (1: CD4-specific DNA aptamer [11], and 2: CD30-specific DNA aptamer [40]) were used as negative controls.
2.5 Cell staining analysis of CD117-specific aptamer
CD117-aptamer binding specificity was further examined using fluorescence microscopy. CD117-negative U937 cells (1 × 105) were incubated with 25 nM carboxyfluorescein diacetate succinimidyl ester (CFSE, Invitrogen, Eugene, OR) in 1 mL of PBS in the dark at RT for 10 min. The labeling reaction was stopped with addition of pre-warmed 100% FBS, and washed with 1 mL 2% FBS/PBS three times and re-cultured in 4 mL fresh RPMI 1640 medium containing 10% FBS for 30 min before use. A cell mixture was prepared by mixing the CFSE-labeled U937 cells (5 × 105) with equal number of fresh HEL cells in RPMI buffer. Cy3-labeled CD117-specific aptamer probe (50 nM) was added to the cell mixture and incubated at room temperature for 30 min. After washing, slides of cell smears were prepared and examined under a fluorescent microscope (Olympus FluoViewTM 1000, Olympus America, Center Valley, PA).
2.6 Formulation and purification of Apt-MTX conjugates
Methotrexate (MTX) was activated by N-hydroxysuccinimide (NHS) ester linkage. Briefly, 15 mg (33 μmol) of MTX was dissolved in 500 μL DMF (dimethylformamide) and 100 μL of NMM (N-methylmorpholine). Next, 11 mg (36 μmol) of TSTU (O-(N-succinimidyl)-1,1,3,3-tetramethyl uranium tetrafluoroborate) was added and stirred for 15 min at RT, then 5 mg (43.5 μmol) of NHS was added and stirred at RT overnight. Activated MTX was then incubated with 5′terminal amino-modified oligonucleotide CD117-specific aptamer. The amino-linked oligonucleotide 5AmMC6-CD117-specific aptamer (3.0 mg, 0.12 μmol) was dissolved in 500 μL of deionized water in a 2 mL Eppendorf centrifuge tube, and 200 μL freshly prepared MTX hydroxysuccinimide ester reaction solution was added. After 1 h reaction, an additional 200 μL of the active ester reaction solution was added (400 μL, 22 μmol total). The reaction solution was mixed overnight at RT, and then purified by HPLC (Varian 920-LC Liquid Chromatograph) on reversed-phase chromatography (PRP-1polymeric reversed Phase HPLC columns, 150X4.1 mm, 10 μm, Hamilton Company, Reno NV). Mobile phase A was 100 mM triethylamine acetate aqueous solution. Mobile phase B was acetonitrile containing 5% of 100 mM triethylamine acetate aqueous solution. The gradient was 0% of B for 5 min; 0% of B to 31% of B 15 min; 31% of B to 85% of B 3 min; 85% of B 3 min; 85% of B to 0% of B 2 min; 0% of B 4 min. at a flow rate of 1 mL min-1. Eluent at 13.5 min was collected. The purification was repeated again to completely remove free MTX and dried by lyophilization to give 1.2 mg of Apt-MTX conjugates as a pale white solid.
2.7 Precipitaion of cellular CD117 protein by synthetic aptamer
HEL and U937 cells (5 × 106) were collected and lysed for 30 min on ice with RIPA lysis and extraction buffer (Thermo Fisher Scientific, Rockford, IL) and centrifuged at 10,000 rpm. The supernatants were incubated with biotin-labeled CD117-specific aptamer (100 nM), control CD4-specific aptamer [11] (100 nM), or beads alone for 30 min at RT. After washing (500 μL DPBS), 100 μL streptavidin beads (Fisher scientific, Rockford, IL) were added and incubation continued for another 30 min on rocker. After DPBS buffer washing, 100 μL 1x loading buffer was added, samples were boiled for 5 min, centrifuged, and supernatants separated by SDS-PAGE. The separated proteins were transferred onto PVDF membranes (Thermo Fisher Scientific, Rockford, IL), and detected by anti-CD117 antibody (Cell Signaling Technology, Danvers, MA).
2.8 Mass spectrometry analysis
The proteins precipitated by the CD117-specific aptamer, CD4-specific aptamer, or beads alone were separated by SDS-PAGE and silver stained (Thermo Fisher Scientific, Rockford, IL). The target protein band was determined by Western blotting, and cutted for mass spectrometry analysis (HMRI Proteomics Programmatic Core, Houston, TX).
2.9 Cell growth inhibition and apoptosis detection
Cell growth inhibition was detected by MTT (MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay post treatment. Briefly, cells (5000) were treated with Apt-MTX or MTX in RPMI 1640 medium for 2 h. After replacement with fresh medium containing 10% serum, cells were cultured for an additional 48 h and then 10 μL of MTT solution (5 mg/mL in PBS, Thermo Fisher Scientific, Rockford, IL) were added to each well. After 4 h of incubation, the culture medium was removed and the formazan crystals in the cells were solubilized with 100 μL DMSO for 15 minutes. The UV absorbance at 570 nm was measured with a microplate reader (Biotek, Winooski, VT). Apoptosis in the treated cells was also assessed using Annexin V-APC staining (BD Biosciences, San Jose, CA) according to the manufacturer’s instructions. The samples were analyzed using a FACS caliber (BD Biosciences, San Jose, CA, USA) flow cytometer equipped with CellQuest software.
2.10 Cell cycle arrest analysis
Cells (1 × 106) treated with Apt-MTX (100 nM final concentration), equal molar free MTX, or aptamer alone, and non-treated cells were collected by centrifugation at 1000 rpm for 5 min, washed with cold PBS buffer without Mg2+, and fixed in 70% ethanol at −20°C for at least 2 h. Fixed cells were then incubated with propidium iodide (PI)/Triton X-100 staining solution: 0.1% Triton X-100, 20 μg/mL PI, 0.2 mg/mL DNase-free RNAse A (Sigma Aldrich, St. Louis, MO) for 30 min at RT. After washing with cold PBS buffer, cells were suspended in 500 μL cold PBS buffer and analyzed using a FACS caliber (BD Biosciences, San Jose, CA, USA) flow cytometer equipped with CellQuest software.
2.11 Cell growth inhibition assays
Under normal culture condition, similar proliferation rates of HEL and U937 cells were confirmed by MTT assay. For treatment, cells were exposed to Apt-MTX (100 nM final concentration), equal molar free MTX, or aptamer alone was added into a cell mixture that contained CD117-expressing HEL cells and CD117-negative U937 cells at an initial ratio of 1:1 (total 1×105 cells). After 2 h of treatment, the mixed cells were seeded in fresh RPMI 1640 medium containing 10% serum and cultured for 48 h. Cells were then harvested and stained by PE-conjugated anti-CD15 antibody (BD Biosciences, San Jose, CA). As CD15 is expressed in U937 but not in HEL cells, the mixed cells were separated by flow cytometry based on their CD15 expression, and each cell population was counted separately. All experiments were performed three times with similar results.
3. Results
3.1 Development of CD117-specific and AML cell-targeting ssDNA aptamer sequences
The ssDNA library was synthesized as described in Materials and Methods and a hybrid SELEX was performed (Fig. 1A and 1B). After 10 rounds of cell-based selection with cultured HEL cells and 3 rounds of recombinant CD117 protein-based enrichment, the resultant ssDNA pool was sequenced. Second generation sequencing identified one dominant sequence (aptamer #1), that accounted for 82% of the 500,000 total reads. An additional four aptamers were also identified (#2 to #5), each of which accounted for >1% of total reads (Fig. 1B). Interestingly, aptamers #1, 3, 4, and 5 have a common conserved core sequence of 5′-GGGGCCGGGGCAAGGGGGGGGTACCGTGGTAGGAC-3′ while aptamer #2 was significantly different (Fig. 1B). To test cell binding capacity, aptamers #1 and #2 were synthesized with Cy3 fluorescence reporter and incubated with HEL cells. Flow cytometry analysis revealed that both aptamers #1 and #2 were able to bind to CD117-expressing HEL cells (left panel of Fig. 1C). However, double-staining of patient marrow specimens revealed that aptamer #1 specifically targeted primary AML cells that were highlighted by anti-CD117 antibody, but aptamer #2 did not react to primary AML cells (right panel of Fig. 1C).
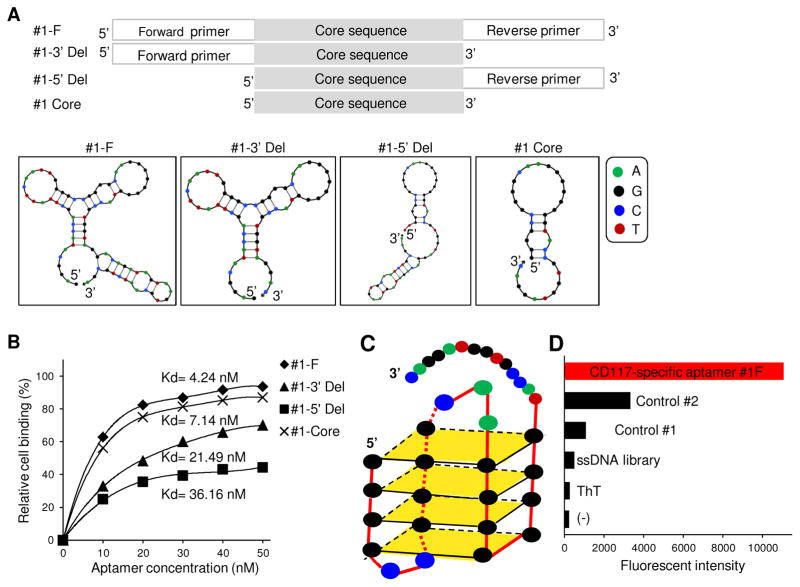
To determine the minimal functional sequence of aptamer #1, three truncated forms were synthesized: #1-3′ Del lacking the 3′ end primer region; #1-5′ Del lacking the 5′ end primer region; and #1-Core lacking both primer regions (Fig. 2A). Aptamer #1 and its truncated forms were synthesized and their 2-D structures predicted (Fig. 2A). To exclude aptamer internalization effect, cell binding assays were performed at 4°C. Figure 2B showed that aptamer #1 had the highest binding ability to HEL cells with a Kd= 4.24 nM, and truncated forms showed decreased cell binding capacity with Kds ranging from 7.14 nM to 36.16 nM. Similar cell bindings of synthetic aptamers were also observed at RT (Data not showed). These findings indicated that the full-length sequence was required for high-affinity cell binding of the aptamer. Moreover, sequence alignment analysis revealed a complete G-quadruplex structure within the core sequence (Fig. 2C), the function of which was also confirmed by the ThT staining assay (Fig. 2D) [39]. Therefore, full-length aptamer #1 (#1-F) was used for the remainder of the studies.
Fig. 2.
Characterization of aptamer sequences. (A) Aptamer #1-F (full length), and aptamers #1-3′ Del, #1-5′ Del (with 3′ or 5′ primer region deletion, respectively), and aptamer #1-35 Core (with only the central core sequence) were synthesized. The 2-dimentional structures of aptamers predicted by IDT software. (B) Cell-binding affinity of synthetic aptamers to cultured HEL cells at 4°C. Aptamer #1-F had the highest binding affinity with Kd =4.24 nM. (C) Predicted 3-dimentional G-quadruplex structure of aptamer core sequence. (D) ThT staining assay confirmed the presence of functional G-quadruplex in aptamer #1-F.
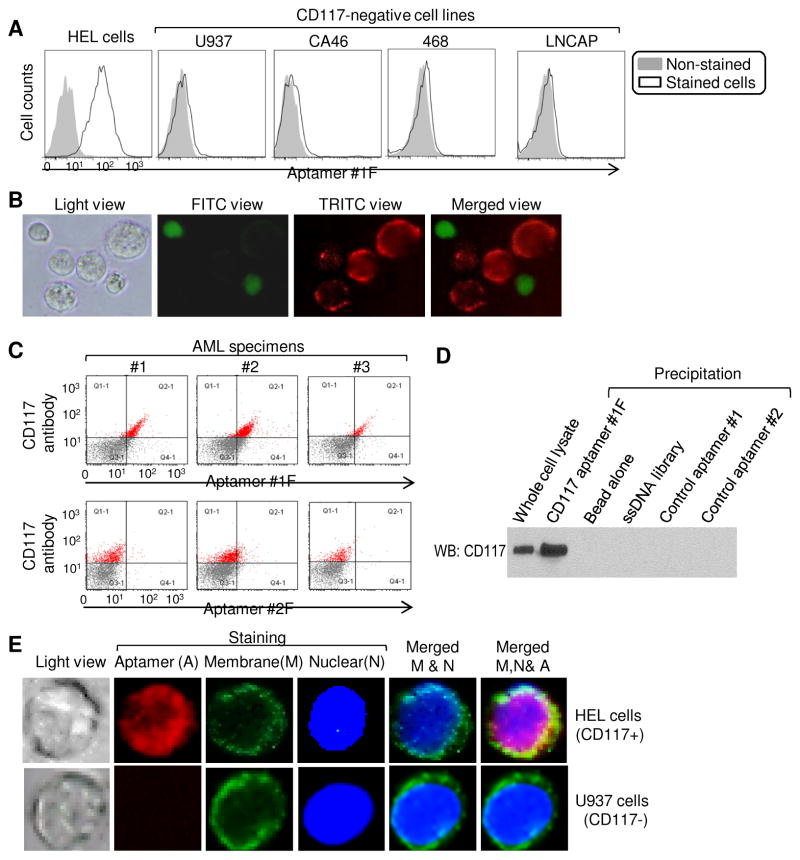
To validate cell binding specificity, multiple CD117-negative cell lines (B-lymphoma CA46, histiocytic lymphoma U937, breast cancer 468, and prostate cancer LNCAP) were incubated with Cy3-labeled aptamer and cell binding specificity was confirmed by flow cytometry (Fig. 3A). In addition, a mixture of HEL cells and U937 cells that were pre-stained with fluorescent dye CFSE was made and treated with aptamer probe. Fluorescent microscopy revealed the aptamer selectively highlighted HEL cells with red fluorescence and did not bind to off-target U937 cells (green fluorescence) in the cell mixture (Fig. 3B). Moreover, three marrow specimens of AML patients were treated with aptamers and FITC-conjugated anti-CD117 antibody simultaneously. Flow cytometry analysis indicated that the aptamer specifically targeted the same populations of AML cells that were detected by anti-CD117 antibody in each marrow specimen (Fig. 3C). Aptamer #2 was used as a negative control, and while it was able to bind cultured HEL cells, did not react to primary AML cells. Moreover, to identify whether the aptamer recognizes cellular CD117 proteins, protein precipitation from HEL cell lysates was performed with aptamer #1F. The bound cellular proteins were separated by gel electrophoresis, and probed with anti-CD117 antibody. Western blotting demonstrated that the aptamer specifically precipitated cellular CD117 proteins and thus, raised the same band as observed in whole cell lysates (Fig. 3D). In contrast, no CD117 proteins were detected in control reactions with ssDNA library or non-relevant aptamer sequences. The gel bands of aptamer-precipitated cellular proteins were also collected and sequenced by mass spectrometry. The presence of CD117 proteins in aptamer-precipitated cellular products was confirmed with a protein ID: P10721, which was not detected in control reactions. These findings indicate that the developed aptamer is AML cell-targeted and CD117-biomarker specific.
Fig. 3.
Specific binding of synthetic aptamer #1-F to HEL cells and cellular CD117 proteins. (A) Cultured HEL cells and CD117-negative control cells were stained with aptamer #1-F and cell binding was detected by flow cytometry. (B) A cell mixture of HEL and pre-stained U937 cells were treated with aptamer #1-F and examined by fluorescence microscopy. Aptamer selectively highlighted HEL cells (red fluorescence), but did not react to CD117-negative U937 cells that were pre-stained (green fluorescence). (C) Marrow cells of AML patients were simultaneously stained with both aptamer probe and FITC-labeled anti-CD117 antibody. Aptamer #1-F targeted AML cells that detected by antibody, but aptamer #2 did not. (D) HEL cell lysates were precipitated using synthetic aptamers or ssDNA library, separated on SDS-PAGE, and proteins were detected by Western blotting with anti-CD117 antibody. Whole HEL cell lysate was used as a positive control. (E) To detect intracellular delivery, cell membrane and nuclei were pre-stained in green and blue, respectively. Cells were then treated with aptamer #1-F and examined under confocal microscope. Aptamer signals (red fluorescence) were detected within HEL cells, but not seen within U937 cells.
Finally, to examine intracellular delivery potential, cell membrane and nuclei were pre-stained with different fluorescence and cells were treated with aptamers at RT. Merging of confocal microscopic images demonstrated intracellular signal of aptamers in HEL cells through specific surface CD117 mediated internalization. In contrast, no aptamer signal was observed in control U937 cells that do not express CD117 (Fig. 3E).
3.2 Formulation of aptamer-drug conjugates that specifically target AML cells
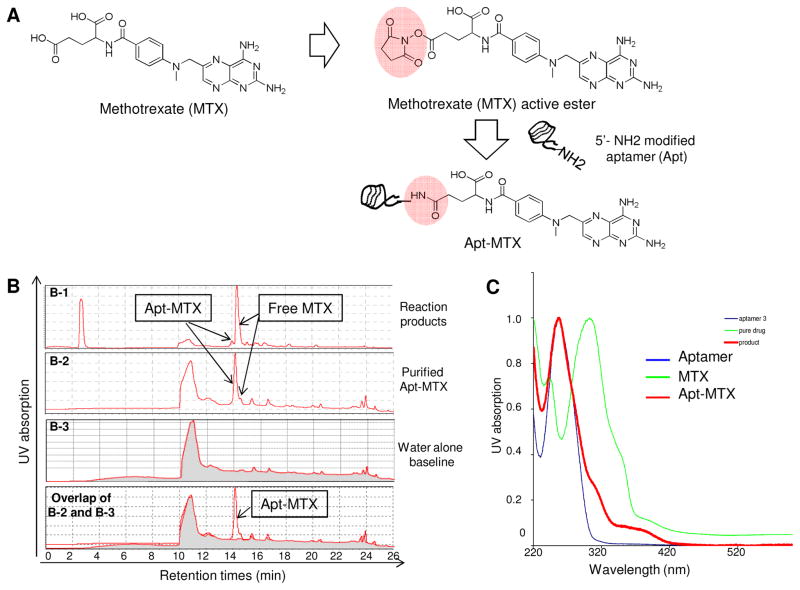
For targeted AML therapy, CD117-specific aptamer #1F was directly conjugated to the chemotherapeutic drug methotrexate (MTX) to form Apt-MTX conjugates. First, MTX molecules were added with an active N-hydroxysuccinimide (NHS) group and then reacted with a synthetic aptamer sequence containing a functional amino-group (Fig. 4A). Initial reaction products were analyzed by HPLC and the formed Apt-MTX (13.7%) was detected with large amount of free MTX (panel B-1 of Fig. 4B). After twice purification, final Apt-MTX products became highly purified and contained less than 3% free MTX (B-2 of Fig. 4B). In addition, the presence of MTX in Apt-MTX was also confirmed by UV detection (Fig. 4C).
Fig. 4.
Synthesis of aptamer-methotrexate (Apt-MTX) conjugate. (A) MTX with a functional NHS ester group was directly conjugated to aptamer #1-F at 5′ through an NH2 group to produce Apt-MTX. (B) Characterization of synthesized Apt-MTX products by HPLC. B-1: initial products; B-2: final Apt-MTX products after twice purification containing highly purified Apt-MTX with <3% free MTX; B-3: baseline of water alone; Overlapping of B-2 and B-3 confirmed purity of final products. (C) UV spectroscopy analysis confirmed the presentation of MTX in Apt-MTX products.
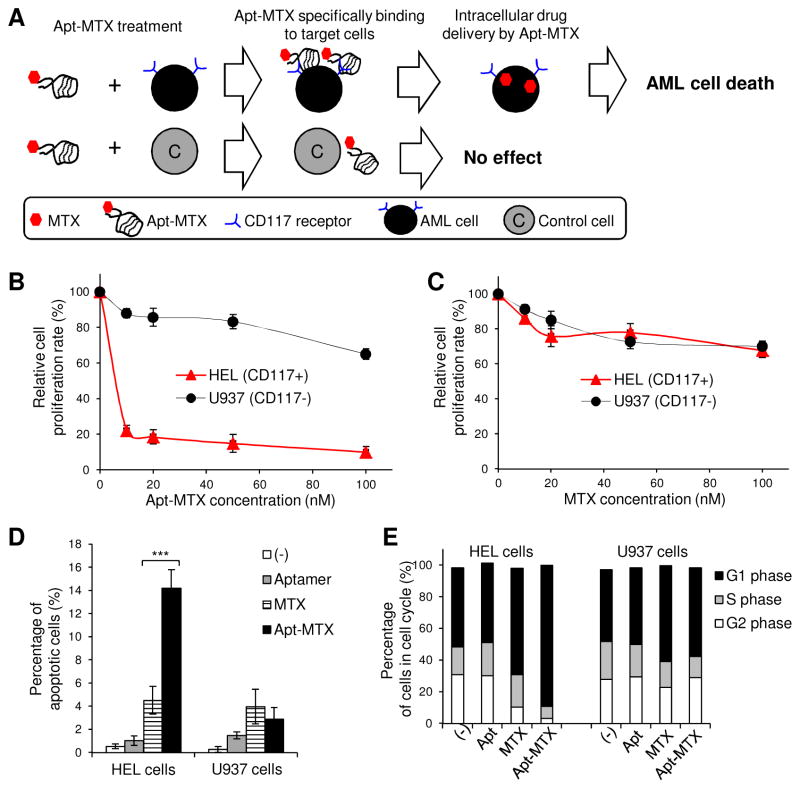
As we already had demonstrated that the aptamer is internalized into target cells (Fig. 3E), it was expected that Apt-MTX would deliver the drug into AML cells with subsequent therapeutic effects (Fig. 5A). To evaluate cellular effects, HEL and U937 cells were treated with Apt-MTX for 2 h to allow targeting and intracellular delivery, and then cultured in fresh medium for 48 hr. Cell proliferation assays showed that Apt-MTX treatment resulted in 80% growth inhibition of HEL cells at 10 nM final concentration of MTX payload with relatively limited effects on U937 control cells (Fig. 5B); however, both cell types had similar sensitivity to free MTX (Fig. 5C). Additionally, the treated cells were also stained with Annexin V and apoptotic cells were quantified by flow cytometry. The Apt-MTX treatment containing 100 nM MTX payloads significantly induced HEL cell apoptosis, but had no toxicity on control U937 cells (Fig. 5D). In addition, cell cycle analysis demonstrated that Apt-MTX treatment induced a G1 phase arrest of more than 90% HEL cells, and showed minimal effect on CD117-negative U937 cells (Fig. 5E).
Fig. 5.
High therapeutic potential of Apt-MTX to treat AML cells. (A) A schematic of Apt-MTX effect on target and off-target cells. (B) Apt-MTX treatment specifically induced 80% inhibition of HEL cell growth at 10 nM final concentration, but had little effect on CD117-negative U937 cells. (C) Both HEL and U937 cells showed similar low sensitivity to free MTX. (D) Apt-MTX specifically stimulated HEL cell apoptosis and had minimal effect on U937 cells. (E) Apt-MTX treatment specifically triggered G1 phase arrest of HEL cells, but not U937 cells.
3.3 Apt-MTX treatment selectively inhibits growth of primary AML cells derived from patient marrow specimens
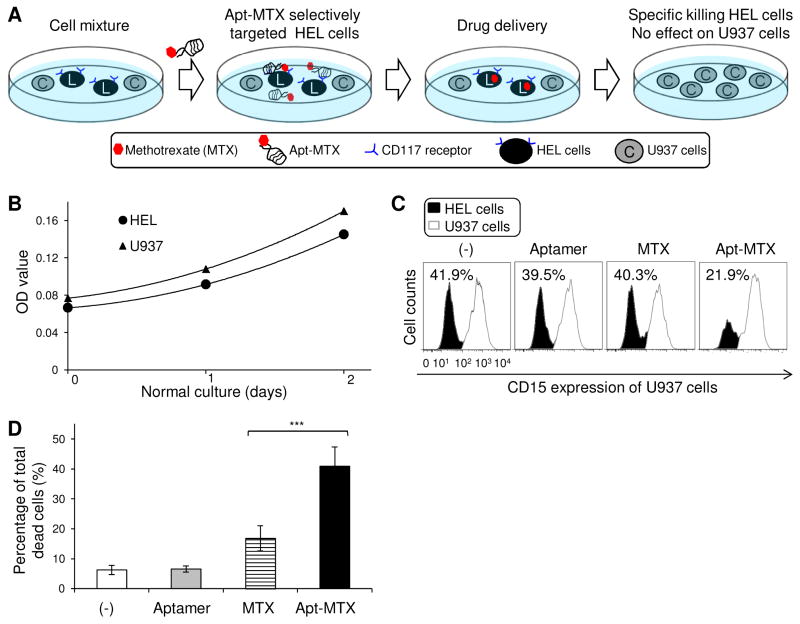
To examine specificity, a cell mixture of HEL and U937 cells (1:1 ratio) was prepared since both cells showed similar sensitivity to free MTX (Fig. 5B) and similar growth rates under normal culture condition (Fig. 6B). The cell mixture was treated with Apt-MTX (final 100nM concentration), equal molar amount of free MTX, or aptamers alone as described above and cultured for 48 hr. The cells were harvested and stained with anti-CD15 antibody to distinguish U937 cells (CD15+) from HEL cells (CD15−). Flow cytometry analysis showed that Apt-MTX selectively inhibited proliferation in HEL cells but not U937 cells, although free MTX or aptamers alone showed similar effects on both cell types (Fig. 6C). In addition, cell mixtures were also stained with propidium iodide (PI) and dead cells under each treatment condition were quantified. The Apt-MTX triggered death of >40% total cells vs. 17% by free MTX treatment (Fig. 6D). Notably, in combination with the findings shown in Figure 6D, the detected dead cells by Apt-MTX treatment were mainly derived from HEL cells, while free MTX induced equal death of both HEL and off-target U937 cells.
Fig. 6.
High specificity of Apt-MTX treatment to AML cells with minimal toxicity on off-target cells. (A) A schematic of the mixed-cell MTX treatment study. (B) HEL and U937 cells had very similar growth rates under normal culture condition. (C) Cell mixtures of HEL and U937 cells (1:1 ratio) were exposed to Apt-MTX, an equal dose of free MTX, aptamers alone, or non-treatment (−). Resultant changes of each cell population (%) post-treatment in the same cell mixtures were quantified by flow cytometry analysis based on CD15-expresion of U937 cells and CD15-negativity of HEL cells. (D) Total dead cells (%) of cell mixtures under individual treatments.
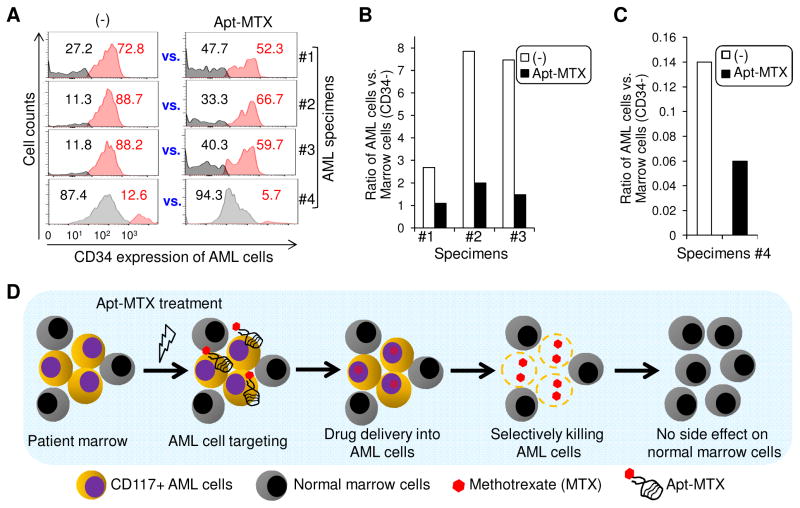
To test the potential clinical utility, primary marrow specimens from AML patients were collected. Each specimen was divided into two parts and exposed to Apt-MTX or non-treatment as a control (−) for two days, respectively. Notably, primary AML cells can be distinguished from background normal marrow cells in the same specimens by high-level expression of CD117 and CD34. Since the presence of Apt-MTX may have unexpected blocking effect on cell binding of anti-CD117 antibody, marrow specimens were stained with antibody against CD34 to identify AML cells. AML cells and background normal marrow cells within each specimen were then quantified by flow cytometry (%) based on cellular CD34 expression. In comparison of Apt-MTX treatment vs. each corresponding non-treatment control (−), Apt-MTX selectively killed patient AML cells and had no effect on growth of background marrow cells (Fig. 7A). The Apt-MTX treatment resulted in significant changes in the ratios of AML cells vs. background normal marrow cells within each patient specimen (Fig. 7B and 7C). These findings demonstrated that Apt-MTX was able to selectively target and specifically inhibit growth of patient AML cells and more importantly, have little to no toxicity on background normal marrow cells (Fig. 7D).
Fig. 7.
Apt-MTX targeted therapy of AML patient marrow specimens. (A) Primary marrow specimens were collected from AML patients #1 to #4. Each specimen was divided into two parts and exposed to Apt-MTX or non-treatment (−) for two days, respectively. Residual CD34+ AML cells and CD34- background normal marrow cells in each specimen were quantified by flow cytometry (%) and compared between Apt-MTX vs. non-treatment control (−). (B) and (C) Ratios of AML cells to background normal marrow cells with or without Apt-MTX treatment were calculated and showed in graphic. (D) A schematic summary of Apt-MTX medicated targeted therapy of AML patient marrow specimens.
4. Discussion
MTX is a vital component of current chemotherapy regimens for AML. However, the current recommended doses can also cause nephrotoxicity and acute renal failure [41–43]. To reduce toxic MTX side effects yet still remain within its therapeutic range, a new personalized medicine is urgently needed. To this end, a novel Apt-MTX was formulated by chemical conjugation of our CD117-specific aptamer directly to MTX. It is expected that through aptamer guidance, systemic administration of Apt-MTX will specifically target and deliver MTX to AML cells, leading to accumulation of high MTX concentration that reach therapeutic threshold exclusively within the targeted cells.. More importantly, AML cell-targeted delivery will allow us to systemically administer Apt-MTX at a sub-toxic dose and thus, eliminate any adverse toxic effects on off-target normal cells/tissues in patients as illustrated in Fig. 7D. Using primary clinical specimens, we confirmed that Apt-MTX selectively inhibited the growth of primary AML cells and had no effect on normal marrow cells of patients (Fig. 7A), clearly indicating its potential for targeted therapy. Although a single Apt-MTX treatment with a sub-toxic dose did not completely eliminate primary AML cells in patient samples, the therapeutic efficacy could be improved by repeated treatments, and/or in combination with other drugs, which is the standard of care for AML. Moreover, as chemical antibodies, oligonucleotide aptamers can be easily modified and conjugated with multiple therapeutic agents through physical and/or chemical conjugation reactions [44, 45] to achieve a synergistic therapeutic effect.
With many unique chemical and biological properties, synthetic aptamers are highly suitable for in vivo use as a component of targeted therapeutics. This Apt-MTX demonstrates a new approach for targeted AML therapy and also provides a universal platform for developing new targeted therapeutics to different cancers by simply replacing the aptamer sequence specific for the biomarker and therapeutic agents suitable to kill cancer cells of interest.
Acknowledgments
This project was supported in part by NIH grants R01CA151955 and R33CA173382 to Y.Z.
Footnotes
Conflict interest
The authors declare no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Banerjee J, Nilsen-Hamilton M. Aptamers: multifunctional molecules for biomedical research. Journal of molecular medicine. 2013;91(12):1333–42. doi: 10.1007/s00109-013-1085-2. [DOI] [PubMed] [Google Scholar]
- 2.Kanwar JR, Shankaranarayanan JS, Gurudevan S, Kanwar RK. Aptamer-based therapeutics of the past, present and future: from the perspective of eye-related diseases. Drug discovery today. 2014 doi: 10.1016/j.drudis.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Xing H, Hwang K, Li J, Torabi SF, Lu Y. DNA Aptamer Technology for Personalized Medicine. Current opinion in chemical engineering. 2014;4:79–87. doi: 10.1016/j.coche.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J, Rossi JJ. Cell-type-specific, Aptamer-functionalized Agents for Targeted Disease Therapy. Molecular therapy Nucleic acids. 2014;3:e169. doi: 10.1038/mtna.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong B, Zu Y. Detecting circulating tumor cells: current challenges and new trends. Theranostics. 2013;3(6):377–94. doi: 10.7150/thno.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Zhao Q, Qiu L. Smart ligand: aptamer-mediated targeted delivery of chemotherapeutic drugs and siRNA for cancer therapy. Journal of controlled release: official journal of the Controlled Release Society. 2013;171(2):152–62. doi: 10.1016/j.jconrel.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Barbas AS, Mi J, Clary BM, White RR. Aptamer applications for targeted cancer therapy. Future oncology. 2010;6(7):1117–26. doi: 10.2217/fon.10.67. [DOI] [PubMed] [Google Scholar]
- 8.Lassalle HP, Marchal S, Guillemin F, Reinhard A, Bezdetnaya L. Aptamers as remarkable diagnostic and therapeutic agents in cancer treatment. Current drug metabolism. 2012;13(8):1130–44. doi: 10.2174/138920012802850038. [DOI] [PubMed] [Google Scholar]
- 9.Pednekar PP, Jadhav KR, Kadam VJ. Aptamer-dendrimer bioconjugate: a nanotool for therapeutics, diagnosis, and imaging. Expert opinion on drug delivery. 2012;9(10):1273–88. doi: 10.1517/17425247.2012.716421. [DOI] [PubMed] [Google Scholar]
- 10.Radom F, Jurek PM, Mazurek MP, Otlewski J, Jelen F. Aptamers: molecules of great potential. Biotechnology advances. 2013;31(8):1260–74. doi: 10.1016/j.biotechadv.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Zhao N, Pei SN, Parekh P, Salazar E, Zu Y. Blocking interaction of viral gp120 and CD4-expressing T cells by single-stranded DNA aptamers. The international journal of biochemistry & cell biology. 2014;51:10–18. doi: 10.1016/j.biocel.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao N, You J, Zeng Z, Li C, Zu Y. An ultra pH-sensitive and aptamer-equipped nanoscale drug-delivery system for selective killing of tumor cells. Small. 2013;9(20):3477–84. doi: 10.1002/smll.201202694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nature biotechnology. 2008;26(8):925–32. doi: 10.1038/nbt.1480. [DOI] [PubMed] [Google Scholar]
- 14.Adem YT, Schwarz KA, Duenas E, Patapoff TW, Galush WJ, Esue O. Auristatin antibody drug conjugate physical instability and the role of drug payload. Bioconjugate chemistry. 2014;25(4):656–64. doi: 10.1021/bc400439x. [DOI] [PubMed] [Google Scholar]
- 15.Fauvel B, Yasri A. Antibodies directed against receptor tyrosine kinases: Current and future strategies to fight cancer. mAbs. 2014;6(4):838–51. doi: 10.4161/mabs.29089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sievers EL, Senter PD. Antibody-drug conjugates in cancer therapy. Annual review of medicine. 2013;64:15–29. doi: 10.1146/annurev-med-050311-201823. [DOI] [PubMed] [Google Scholar]
- 17.Zeng Z, Zhang P, Zhao N, Sheehan AM, Tung CH, Chang CC, Zu Y. Using oligonucleotide aptamer probes for immunostaining of formalin-fixed and paraffin-embedded tissues. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2010;23(12):1553–8. doi: 10.1038/modpathol.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shigdar S, Lin J, Yu Y, Pastuovic M, Wei M, Duan W. RNA aptamer against a cancer stem cell marker epithelial cell adhesion molecule. Cancer science. 2011;102(5):991–8. doi: 10.1111/j.1349-7006.2011.01897.x. [DOI] [PubMed] [Google Scholar]
- 19.Mayer RJ, Davis RB, Schiffer CA, Berg DT, Powell BL, Schulman P, Omura GA, Moore JO, McIntyre OR, Frei E., 3rd Intensive postremission chemotherapy in adults with acute myeloid leukemia. Cancer and Leukemia Group B The New England journal of medicine. 1994;331(14):896–903. doi: 10.1056/NEJM199410063311402. [DOI] [PubMed] [Google Scholar]
- 20.Burnett AK. Treatment of acute myeloid leukemia: are we making progress? Hematology/the Education Program of the American Society of Hematology American Society of Hematology Education Program. 2012;2012:1–6. doi: 10.1182/asheducation-2012.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Klimek VM. Recent advances in the management of therapy-related myelodysplastic syndromes and acute myeloid leukemia. Current opinion in hematology. 2013;20(2):137–43. doi: 10.1097/MOH.0b013e32835d82e6. [DOI] [PubMed] [Google Scholar]
- 22.Stone RM. Consolidation chemotherapy for adults with AML in first remission: is there a best choice? Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31(17):2067–9. doi: 10.1200/JCO.2013.48.6886. [DOI] [PubMed] [Google Scholar]
- 23.Martner A, Thoren FB, Aurelius J, Hellstrand K. Immunotherapeutic strategies for relapse control in acute myeloid leukemia. Blood reviews. 2013;27(5):209–16. doi: 10.1016/j.blre.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Peloquin GL, Chen YB, Fathi AT. The evolving landscape in the therapy of acute myeloid leukemia. Protein & cell. 2013;4(10):735–46. doi: 10.1007/s13238-013-3057-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nazha A, Ravandi F. Acute myeloid leukemia in the elderly: do we know who should be treated and how? Leukemia & lymphoma. 2014;55(5):979–87. doi: 10.3109/10428194.2013.828348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrara F. Conventional chemotherapy or hypomethylating agents for older patients with acute myeloid leukaemia? Hematological oncology. 2014;32(1):1–9. doi: 10.1002/hon.2046. [DOI] [PubMed] [Google Scholar]
- 27.Snauwaert S, Vandekerckhove B, Kerre T. Can immunotherapy specifically target acute myeloid leukemic stem cells? Oncoimmunology. 2013;2(2):e22943. doi: 10.4161/onci.22943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sweet K, Lancet JE. Novel therapeutics in acute myeloid leukemia. Current hematologic malignancy reports. 2014;9(2):109–17. doi: 10.1007/s11899-014-0199-0. [DOI] [PubMed] [Google Scholar]
- 29.Li K, Lv XX, Hua F, Lin H, Sun W, Cao WB, Fu XM, Xie J, Yu JJ, Li Z, et al. Targeting acute myeloid leukemia with a proapoptotic peptide conjugated to a Toll-like receptor 2-mediated cell-penetrating peptide. International journal of cancer Journal international du cancer. 2014;134(3):692–702. doi: 10.1002/ijc.28382. [DOI] [PubMed] [Google Scholar]
- 30.Konig H, Levis M. Is targeted therapy feasible in acute myelogenous leukemia? Current hematologic malignancy reports. 2014;9(2):118–27. doi: 10.1007/s11899-014-0198-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter RB. The role of CD33 as therapeutic target in acute myeloid leukemia. Expert opinion on therapeutic targets. 2014;18(7):715–18. doi: 10.1517/14728222.2014.909413. [DOI] [PubMed] [Google Scholar]
- 32.Hans CP, Finn WG, Singleton TP, Schnitzer B, Ross CW. Usefulness of anti-CD117 in the flow cytometric analysis of acute leukemia. American journal of clinical pathology. 2002;117(2):301–5. doi: 10.1309/RWCG-E5T9-GU95-LEWE. [DOI] [PubMed] [Google Scholar]
- 33.Doepfner KT, Boller D, Arcaro A. Targeting receptor tyrosine kinase signaling in acute myeloid leukemia. Critical reviews in oncology/hematology. 2007;63(3):215–30. doi: 10.1016/j.critrevonc.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Stirewalt DL, Meshinchi S. Receptor tyrosine kinase alterations in AML - biology and therapy. Cancer treatment and research. 2010;145:85–108. doi: 10.1007/978-0-387-69259-3_6. [DOI] [PubMed] [Google Scholar]
- 35.Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(5):475–86. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 36.Ashman LK, Griffith R. Therapeutic targeting of c-KIT in cancer. Expert opinion on investigational drugs. 2013;22(1):103–15. doi: 10.1517/13543784.2013.740010. [DOI] [PubMed] [Google Scholar]
- 37.Liang J, Wu YL, Chen BJ, Zhang W, Tanaka Y, Sugiyama H. The C-kit receptor-mediated signal transduction and tumor-related diseases. International journal of biological sciences. 2013;9(5):435–43. doi: 10.7150/ijbs.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Chen Y, Han D, Ocsoy I, Tan W. Aptamers selected by cell-SELEX for application in cancer studies. Bioanalysis. 2010;2(5):907–18. doi: 10.4155/bio.10.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renaud de la Faverie A, Guedin A, Bedrat A, Yatsunyk LA, Mergny JL. Thioflavin T as a fluorescence light-up probe for G4 formation. Nucleic acids research. 2014;42(8):e65. doi: 10.1093/nar/gku111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parekh P, Kamble S, Zhao N, Zeng Z, Portier BP, Zu Y. Immunotherapy of CD30-expressing lymphoma using a highly stable ssDNA aptamer. Biomaterials. 2013;34(35):8909–17. doi: 10.1016/j.biomaterials.2013.07.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ackland SP, Schilsky RL. High-dose methotrexate: a critical reappraisal. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1987;5(12):2017–31. doi: 10.1200/JCO.1987.5.12.2017. [DOI] [PubMed] [Google Scholar]
- 42.Widemann BC, Adamson PC. Understanding and managing methotrexate nephrotoxicity. The oncologist. 2006;11(6):694–703. doi: 10.1634/theoncologist.11-6-694. [DOI] [PubMed] [Google Scholar]
- 43.Buchen S, Ngampolo D, Melton RG, Hasan C, Zoubek A, Henze G, Bode U, Fleischhack G. Carboxypeptidase G2 rescue in patients with methotrexate intoxication and renal failure. British journal of cancer. 2005;92(3):480–87. doi: 10.1038/sj.bjc.6602337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruno JG. A review of therapeutic aptamer conjugates with emphasis on new approaches. Pharmaceuticals. 2013;6(3):340–57. doi: 10.3390/ph6030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Estevez MC, Huang YF, Kang H, O’Donoghue MB, Bamrungsap S, Yan J, Chen X, Tan W. Nanoparticle-aptamer conjugates for cancer cell targeting and detection. Methods in molecular biology. 2010;624:235–48. doi: 10.1007/978-1-60761-609-2_16. [DOI] [PubMed] [Google Scholar]